-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
Accurate segregation of homologous chromosomes of different parental origin (homologs) during the first division of meiosis (meiosis I) requires inter-homolog crossovers (COs). These are produced at the end of meiosis I prophase, when recombination intermediates that contain Holliday junctions (joint molecules, JMs) are resolved, predominantly as COs. JM resolution during the mitotic cell cycle is less well understood, mainly due to low levels of inter-homolog JMs. To compare JM resolution during meiosis and the mitotic cell cycle, we used a unique feature of Saccharomyces cerevisiae, return to growth (RTG), where cells undergoing meiosis can be returned to the mitotic cell cycle by a nutritional shift. By performing RTG with ndt80 mutants, which arrest in meiosis I prophase with high levels of interhomolog JMs, we could readily monitor JM resolution during the first cell division of RTG genetically and, for the first time, at the molecular level. In contrast to meiosis, where most JMs resolve as COs, most JMs were resolved during the first 1.5–2 hr after RTG without producing COs. Subsequent resolution of the remaining JMs produced COs, and this CO production required the Mus81/Mms4 structure-selective endonuclease. RTG in sgs1-ΔC795 mutants, which lack the helicase and Holliday junction-binding domains of this BLM homolog, led to a substantial delay in JM resolution; and subsequent JM resolution produced both COs and NCOs. Based on these findings, we suggest that most JMs are resolved during the mitotic cell cycle by dissolution, an Sgs1 helicase-dependent process that produces only NCOs. JMs that escape dissolution are mostly resolved by Mus81/Mms4-dependent cleavage that produces both COs and NCOs in a relatively unbiased manner. Thus, in contrast to meiosis, where JM resolution is heavily biased towards COs, JM resolution during RTG minimizes CO formation, thus maintaining genome integrity and minimizing loss of heterozygosity.
Published in the journal: . PLoS Genet 7(5): e32767. doi:10.1371/journal.pgen.1002083
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002083Summary
Accurate segregation of homologous chromosomes of different parental origin (homologs) during the first division of meiosis (meiosis I) requires inter-homolog crossovers (COs). These are produced at the end of meiosis I prophase, when recombination intermediates that contain Holliday junctions (joint molecules, JMs) are resolved, predominantly as COs. JM resolution during the mitotic cell cycle is less well understood, mainly due to low levels of inter-homolog JMs. To compare JM resolution during meiosis and the mitotic cell cycle, we used a unique feature of Saccharomyces cerevisiae, return to growth (RTG), where cells undergoing meiosis can be returned to the mitotic cell cycle by a nutritional shift. By performing RTG with ndt80 mutants, which arrest in meiosis I prophase with high levels of interhomolog JMs, we could readily monitor JM resolution during the first cell division of RTG genetically and, for the first time, at the molecular level. In contrast to meiosis, where most JMs resolve as COs, most JMs were resolved during the first 1.5–2 hr after RTG without producing COs. Subsequent resolution of the remaining JMs produced COs, and this CO production required the Mus81/Mms4 structure-selective endonuclease. RTG in sgs1-ΔC795 mutants, which lack the helicase and Holliday junction-binding domains of this BLM homolog, led to a substantial delay in JM resolution; and subsequent JM resolution produced both COs and NCOs. Based on these findings, we suggest that most JMs are resolved during the mitotic cell cycle by dissolution, an Sgs1 helicase-dependent process that produces only NCOs. JMs that escape dissolution are mostly resolved by Mus81/Mms4-dependent cleavage that produces both COs and NCOs in a relatively unbiased manner. Thus, in contrast to meiosis, where JM resolution is heavily biased towards COs, JM resolution during RTG minimizes CO formation, thus maintaining genome integrity and minimizing loss of heterozygosity.
Introduction
Recombination has a major role during meiosis, as it is necessary for accurate homolog segregation at the first meiotic division [1]. Meiotic recombination is initiated by DNA double strand breaks (DSBs) that are formed by the Spo11 nuclease [2], [3]. Single stranded DNA, produced at break ends by 5′ to 3′ resection [4], then interacts with complementary sequences on the homolog or on the sister chromatid [5], [6]. Some interhomolog recombination events produce a noncrossover (NCO), in which both interacting chromosomes retain parental flanking sequence configurations, whereas other events produce a reciprocal exchange of flanking sequences, or crossover (CO). COs, in combination with sister chromatid cohesion, form the inter-homolog linkage that is required for proper homolog segregation [1]. In Saccharomyces cerevisiae, COs comprise about one half of all interhomolog recombination events [7]. Meiotic COs are produced by the resolution of joint molecule (JM) intermediates [8]–[10], most of which contain two Holliday junctions [11], here called double Holliday junction JMs (dHJ-JMs).
In most organisms, including S. cerevisiae, meiotic DSB formation and recombination are also necessary for progressive colocalization and alignment of homologs during prophase. This process culminates at pachytene, where homologs are joined at sites of recombination and linked tightly along their entire length by a meiosis-specific tripartite protein structure called the synaptonemal complex (SC; [12]).
Although genome-wide programmed DSB formation is central to normal meiosis, it does not usually occur during the mitotic cell cycle. During the budding yeast mitotic cell cycle, most breaks are repaired by recombination between sister chromatids [13]–[15], and the inter-homolog homologous recombination (HR) events that do occur during the mitotic cell cycle produce COs less frequently than in meiosis [13], [16].
The lower yield of COs during mitotic recombination, as compared to meiotic recombination, can be explained in two ways. First, fewer dHJ-JMs are produced per DSB repair event during mitosis than during meiosis [15], and it is possible that most mitotic DSB repair does not involve dHJ-JM formation. Second, it is possible that JMs are produced at significant levels during mitotic HR, but are resolved differently than are JMs produced during meiosis. In S. cerevisiae, most meiotic JMs are resolved as COs [8]–[10] in a process that most likely involves endonuclease cleavage of Holliday junctions, and that is triggered by Cdc5, the budding yeast polo-like kinase homolog [17], [10]. Much less is known about JM resolution during the mitotic cell cycle, since the products of intersister recombination cannot be distinguished from the precursor molecules.
Several structure-selective nucleases have been suggested as having a role in JM resolution by Holliday junction cleavage [18]. The most extensively studied of these is a structure-selective heterodimeric endonuclease, hereafter called the Mus81 complex, that contains the conserved Mus81 nuclease in complex with a second protein, called Mms4 in S. cerevisiae and Drosophila, and Eme1 in fission yeast, mammals and plants [19]–[21]. Meiotic progression defects are evident in S. pombe and S. cerevisiae mutants lacking the Mus81 complex, but the nature of these defects differs in the two organisms. In S. pombe, mutants lacking the Mus81 complex show a strong CO defect and accumulate unresolved JMs [19],[22]–[24], while in S. cerevisiae, mus81 or mms4 mutants show only a minor CO loss and resolve the vast majority of JMs [25]–[29]. Thus, in budding yeast, most meiotic JMs must be resolved by other, yet unidentified endonucleases. It also is not clear whether or not the Mus81 complex resolves JMs that form during the mitotic cell cycle. A recent study of I-Sce1 endonuclease-promoted mitotic recombination in S. cerevisiae suggested redundant roles for the Mus81 complex and for the Yen1 endonuclease in interhomolog CO formation [30], but it remains to be established that these crossovers are produced by dHJ-JM resolution.
dHJ-JMs can also be resolved by an endonuclease-independent process, called dissolution, that uses a RecQ-family helicase and a type 1 topoisomerase to disassemble JMs and to produce only NCOs [31]–[34]. Dissolution has been demonstrated in biochemical studies of the human BLM helicase combined with the TOPOIIIα/BLAP75 heterodimer, and of the corresponding budding yeast proteins Sgs1 and Top3/Rmi1 [35], [33], [36]. Dissolution has not yet been directly demonstrated in vivo, but is consistent with observations that loss of BLM or Sgs1 helicase activity is accompanied by a substantial increase in mitotic sister chromatid exchange [37]–[39], and that sgs1 mutants show increased JM accumulation and CO formation during mitotic DSB repair [16], [15]. During meiosis, sgs1 single mutants show only a slight increase in COs, but produce “abnormal” JMs involving 3 or 4 chromatids at elevated levels [40], [41]. In addition, the CO and JM formation defects of mutants lacking SC components are partially suppressed by sgs1 mutation [40], [42], [41]. These findings are consistent with the suggestion that the Sgs1/BLM helicase prevents COs by reducing JM levels. However, because this helicase also has the potential to disassemble early strand invasion intermediates that are precursors to JMs [43], [44], it remains to be determined if Sgs1/BLM act primarily to prevent JM formation, or to disassemble JMs once they form.
Finally, JMs that form during the G1 phase of the mitotic cell cycle can, in theory, also be resolved passively by chromosome replication [45], producing a CO if the original JM contains an odd number of HJs and an NCO if the original JM contains an even number of HJs.
In the current study, we present experiments aimed at examining how JMs are resolved during the S. cerevisiae mitotic cell cycle. Although several groups have detected JMs in S. cerevisiae undergoing vegetative growth [46], [47], [15], definitive study of their resolution has been precluded by their relatively low levels and by the fact that most form between sister chromatids. However, interhomolog JMs can be recovered at high levels during meiosis, especially in cells that lack Ndt80, a transcription factor required for expression of many mid - and late-meiosis proteins, including the Cdc5 polo-like kinase which is required for meiotic JM resolution [48], [17]. ndt80 mutant cells arrest at the pachytene stage of meiosis, with duplicated but unseparated spindle pole bodies [49], with homologs tightly paired by SC [49], and, most important to this study, with a high level of unresolved JMs [8]. To examine resolution of these JMs in a cellular environment that mimics the mitotic cell cycle, we used a singular property of S. cerevisiae, called return to growth (RTG). When cells in meiosis I prophase are shifted to rich medium, they rapidly exit meiosis, adopt a G1-like transcription pattern, and ultimately resume the mitotic cell cycle [50]–[58].
We report here the first molecular characterization of JM resolution during RTG. We show here that, unlike in meiosis, most JMs are resolved after RTG in a manner that does not produce COs. Examination of JM resolution in sgs1 and in mus81 mutants suggest that, during RTG of wild-type cells, the majority of JMs are resolved by Sgs1-mediated dissolution, with a minor fraction of JMs being resolved by Mus81 complex-dependent cleavage to produce both CO and NCO products.
Results
To determine how JMs are resolved after RTG, we used ndt80Δ mutant cells, which arrest at pachytene with fully-formed SC and high levels of JMs [49], [8]. In general, RTG experiments involved incubating ndt80Δ cells in nutrient-poor sporulation medium (1% potassium acetate) for 7 hr to allow cells to initiate meiosis and arrest at pachytene, and then shifting cells to nutrient-rich growth medium (YPD) to induce RTG. We confirmed that ndt80Δ cells retain viability after RTG [49]; virtually all cells produced colonies when a culture incubated 7 hours in sporulation medium was plated on YPD agar plates (colonies/visible cells = 1.0+/−0.1; strain MJL3164—see Table S1). To examine the timing and efficiency of RTG in greater detail, we monitored progression of the first cell cycle after RTG (Figure 1). Budded cells were first observed 2 hr after RTG, and half of the cells had produced a bud by 2.5 hr. Nuclear division occurred about 1 hr after bud emergence, with half of the cells having undergone nuclear division by 3.5 hr after RTG. By 4 hr after RTG, virtually all cells had undergone nuclear division, consistent with the high viability seen in plating experiments.
Fig. 1. Cell cycle progression and SC breakdown after RTG. 
a. Representative images of ndt80 cells (MJL3430) at various stages of RTG, visualized by differential interference contrast (DIC) or by DAPI-staining to detect nuclei (DNA). Note that the daughter cell is elongated as compared to the round mother cell. Scale bar—4µm. b. Time of bud emergence and nuclear division after RTG using SPO11 ndt80Δ (MJL3164, top) or spo11-Y135F ndt80Δ (MJL2807, bottom); the latter do not form SC or JMs. Circles – unbudded cells; squares – cells with a bud and one nucleus; triangles – cells that are undergoing or have finished nuclear division. Values for MJL3164 are from 4 independent determinations. c. SC breakdown upon RTG. Nuclei (MJL3163) were surface-spread and probed with anti-Zip1 antisera. Representative images of nuclei classified as full SC (long, continuous Zip1 lines), partial SC (discontinuous or dotty Zip1) and no SC (no Zip1 chromosomal staining) are shown together with DNA staining. Extrachromosomal Zip1 aggregates (polycomplex) were also detected as a bright-staining body. Scale bar—4 µm. d. Time of SC breakdown after RTG (MJL3163). At least 150 nuclei were scored for each time point. Circles – nuclei with full SC; squares – nuclei with partial SC; triangles – nuclei with no SC. Values are from a single experiment. Cells of the SK1 strain background used here complete a mitotic cell cycle every 80 minutes while growing in YPD (M. L., unpublished data), whereas in the current experiments, the first cell division did not occur until at least 2.5 hr after the shift from sporulation to YPD growth medium (Figure 1b). This difference might be explained if nuclear division during RTG was delayed by the presence of unresolved interhomolog connections that were formed during meiosis. To test this suggestion, we examined RTG in spo11 mutant cells (strain MJL2807), which do not initiate recombination or produce SC [59], [60]. Bud emergence and nuclear divisions occurred at times similar to those seen in SPO11 cells (Figure 1b), indicating that the extended gap phase seen upon RTG is not caused by a need to resolve recombination-dependent meiotic chromosome structures.
The SC rapidly breaks down after RTG
ndt80Δ cells arrest with chromosomes that are fully paired by SC [49]. It was previously shown that the SC formed in NDT80 cells breaks down rapidly after RTG [56]. We confirmed this observation in ndt80Δ strains by staining surface-spread nuclei for Zip1, a central component of the SC [61]. Most cells lose full-length linear SC within 15 minutes of transfer to YPD, and less than 30% of cells contained even residual (dotty) Zip1-containing structures 1.5 hr after RTG, before bud emergence and well before nuclear division (Figure 1c, 1d).
Sister chromatids segregate during the nuclear division after RTG
The first nuclear division of meiosis involves segregation of homologs (reductional division), whereas during mitotis, sister chromatids separate from each other (equational division). To determine if the first nuclear division after RTG is reductional or equational, we used a TRP1/trp1 heterozygous strain. TRP1 is tightly linked to the centromere of chromosome IV (<0.5cM; [62]), so chromosome segregation in the first division after RTG can be determined by examining TRP1 allele segregation (Figure 2a). If the first division is reductional, one daughter cell will inherit both copies of the TRP1 allele, whereas the other will inherit both copies of the trp1 allele, resulting in a sectored Trp+/Trp− colony. If the first division is equational, both daughter cells will inherit one TRP1 and one trp1 allele, resulting in a uniform Trp+ colony. A TRP1/trp1 ndt80Δ/ndt80Δ diploid (strain MJL3163) was induced to undergo meiosis for 7 hr, returned to growth by plating on YPD, and the resulting colonies were replica plated onto medium lacking tryptophan. Only one colony in 2767 was sectored, and the rest were uniformly Trp+ (Figure 2b). Thus, the first nuclear division after RTG involves a mitosis-like equational chromosome segregation.
Fig. 2. The first cell division after RTG involves equational chromosome segregation without replication. 
a. Outcome of different types of chromosome segregation after RTG. One homolog is shown as solid line and the other as dashed line. Black and diagonal hatched boxes indicate dominant TRP1 and recessive trp1 alleles, respectively. Reductional chromosome segregation (left) separates homologs, producing a sectored colony with TRP1/TRP1 and trp1/trp1 cells. Equational chromosome segregation (right) separates sister chromatids, producing homogenous TRP1/trp1 colonies. b. Meiotic cells (MJL3163) were plated on YPD, inducing RTG, and 2767 colonies were replica-plated to medium lacking tryptophan. The single Trp+/Trp− colony observed is shown. c. Expected outcomes if DNA replication occurs or does not occur before the first nuclear division after RTG. A strain hemizygous for a CEN5-GFP array (black rectangles, see text for details) is illustrated. After 7 hr in meiosis, each cell includes two copies of CEN5-GFP (middle). Replication followed by equational chromosome segregation (left) results in two copies of CEN5-GFP in each cell. Equational chromosome segregation without prior replication (right) leaves a single copy of CEN5-GFP in each cell. d. Upper panel—post-mitotic cells with a hemizygous CEN5-GFP array (MJL3312), from a sample taken 3.5 hr after RTG. All 282 post-mitotic G1 cells examined had a single GFP spot. Lower panel—control cells with a homozygous CEN5-GFP array (MJL3313) growing vegetatively in YPD. An unbudded cell in G1 is shown. 28/104 G1 cells had two GFP dots. Left—Nuclei detected by DNA/DAPI fluorescence; right—GFP fluorescence. Cells do not replicate DNA before the first nuclear division after RTG
Because DNA replication can resolve JMs, it was important to determine whether or not cells undergo replication before the first division after RTG. During the mitotic cell cycle, bud emergence is closely followed by initiation of DNA replication [63]. We asked if bud emergence after RTG was also associated with DNA replication. ndt80Δ cells arrest after meiotic DNA replication, and thus have a 4C DNA content. Therefore, DNA re-replication before the first division after RTG will result in tetraploid daughter cells. On the other hand, if DNA re-replication does not occur after RTG, diploid daughter cells will be produced. To determine whether DNA re-replication occurs after RTG, we monitored the copy number of chromosome V, using a centromere-linked array of tet operator (tetO) repeats that bind a constitutively-expressed tet repressor-green fluorescent protein fusion [64], [65], referred to here as CEN5-GFP. To check the efficiency of detection of individual CEN5-GFP signals, diploids that were hemizygous (strain MJL3312) or homozygous (strain MJL3313) for CEN5-GFP were grown to log phase, and the number of GFP dots per nucleus was scored in unbudded cells (G1-phase of the cell cycle). As expected, unbudded cells with a hemizygous CEN5-GFP showed one dot per nucleus (133/133). In contrast, 28/104 unbudded cells homozygous for CEN5-GFP showed two dots in their nuclei (Figure 2d), indicating that two copies of CEN5-GFP are detected with about 25% efficiency. The reduced efficiency of detection of two GFP spots is most likely a result of the limited separation of centromeres during interphase in yeast, due to the close attachment of centromeres to the spindle pole body [66].
Using this assay, we determined the number of GFP dots in unbudded cells produced from the first or second division after RTG of a diploid with a hemizygous CEN5-GFP (strain MJL3312). Re-replication followed by an equational division would result in each daughter cell inheriting two copies of CEN5-GFP, and two GFP dots will be observed in the nucleus (Figure 2c). However, if no re-replication occurs, each daughter cell will inherit one copy of CEN5-GFP, resulting in one GFP dot in the nucleus. All cells examined (282/282) showed only one dot in each nucleus. Thus, cells do not undergo DNA replication before the first nuclear division after RTG.
To confirm the conclusion that cells do not undergo DNA replication before the first nuclear division after RTG, we monitored the copy number of the loosely centromere linked MAT locus. Re-replication, followed by an equational division, would result in most daughter cells being MATa/MATa/MATα/MATα tetraploids. However, if no re-replication occurs, most daughter cells will be MATa/MATα diploids. Sporulation of MATa/MATa/MATα/MATα tetraploid cells would frequently produce MATa/MATα nonmating diploid spores. On the other hand, sporulation of MATa/MATα diploid cells will only produce haploid spores with a single MATa or MATα allele (Figure S1).
To sporulate cells that are phenotypically Ndt80−, we used a strain (strain MJL3430, pGPD1-GAL4-ER pGAL1-NDT80; [67], [68], [10]) where NDT80 is normally not expressed, but where NDT80 expression can be induced by the addition of estradiol (ED). Seven independent segregants from RTG performed without NDT80 expression (without ED) were induced to undergo a second meiosis with NDT80 expression (with ED), and tetrads produced by these strains were dissected. All spores from 4 spore-viable tetrads (at least 10 tetrads per primary segregant; n = 400) were either MATa or MATα maters, and none were MATa/MATα nonmaters, confirming the conclusion that re-replication does not occur before the first nuclear division after RTG.
Genetic evidence that COs are infrequently produced after RTG
Since unresolved JMs are expected to interfere with chromosome segregation at mitosis, the observation that most ndt80 mutant cells retain viability after RTG ([49]; see above) suggests that meiotic JMs must be resolved before the first cell division after RTG. During meiosis, JMs are mainly resolved to produce COs [8]–[10]. To ask if JMs are resolved similarly after RTG, we monitored segregation of the recessive cycloheximide–resistance allele, cyh2-z, in a cyh2-z/CYH2 heterozygous diploid. In wild-type meiosis, 66% of cells undergo second division segregation for cyh2-z, resulting from crossing over between the CYH2 locus and the centromere of chromosome VII (CEN7; see Materials and Methods). If JMs are similarly resolved as COs during RTG, 66% of cells are expected to have a CO between CYH2 and CEN7. Assuming random sister chromatid segregation at the first division after RTG, as it is in mitosis [69], half of the cells with a CO between CEN7 and CYH2 will produce cycloheximide-resistant cyh2-z/cyh2-z daughter cells (33% of total colonies; Figure 3a).
Fig. 3. Few COs are produced after RTG. 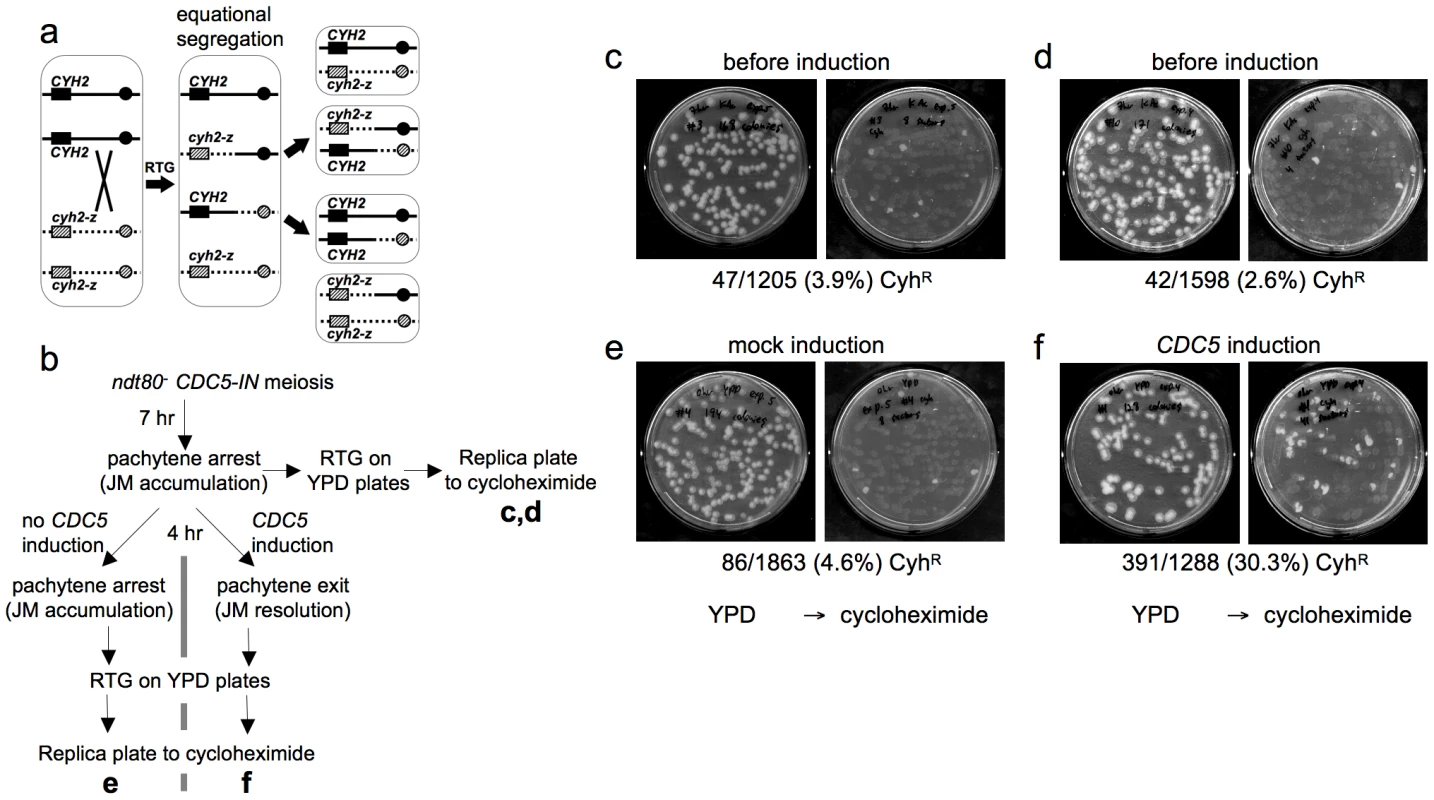
a. CO detection after RTG. Chromosome VII homologs are shown as solid and dashed lines. Black and grey boxes indicate dominant CYH2 and recessive cyh2-z cycloheximide sensitive and resistant alleles, respectively. If a CO occurs between CYH2 and the centromere, equational chromosome segregation produces either a colony that is uniformly CYH2/cyh2-z (cycloheximide-sensitive), or a colony with a CYH2/CYH2 (cycloheximide-sensitive) sector and a cyh2-z/cyh2-z (cycloheximide-resistant) sector. b. Experimental design. ndt80Δ CDC5-IN (MJL3267) cells are incubated in sporulation medium for 7 hr to uniform pachytene arrest, and aliquots are plated on YPD for RTG (c and d). The culture is then incubated for an additional 4 hr without CDC5 induction and plated on YPD (e), or the culture is incubated for 4hr in the presence of estradiol to induce CDC5 expression before plating on YPD (f). Colonies on YPD are replica-plated to YPD + cycloheximide to detect cyh2-z/cyh2-z recombinants. c, d. Control aliquots plated directly on YPD before replica-plating to YPD + cycloheximide. e. Pachytene-arrested cells were incubated for 4 hr without CDC5 induction before plating on YPD. f. Pachytene-arrested cells were incubated for 4 hr with estradiol to induce CDC5 expression before plating on YPD. Note the marked increase in the frequency of cycloheximide-resistant segregants. To directly compare JM resolution after RTG and during meiosis, we used an ndt80Δ/ndt80Δ CYH2/cyh2-z strain that contains an estrogen-inducible CDC5 gene (ndt80Δ pGPD1-GAL4-ER pGAL1-CDC5; strain MJL3267), to allow conditional JM resolution [10]. In the absence of inducer (-ED), cells accumulate in pachytene with unresolved JMs. ED addition induces CDC5 expression, and cells exit from pachytene and resolve JMs to produce COs, but do not progress further through meiosis [10]. Thus, if CDC5 is expressed before RTG, JMs will be resolved and COs will be produced at a level similar to that seen in meiosis. Thus, 33% of colonies are expected to be cycloheximide resistant (Figure 3a). Cells were induced to undergo meiosis for 7 hr, and then aliquots were plated on YPD to undergo RTG (Figure 3b). The remainder of the culture was incubated for another 4 hr in sporulation medium, either with ED to induce pachytene exit, or in the absence of ED as a control, and aliquots were plated on YPD. Colonies on YPD were replica plated onto YPD with cycloheximide to score for sectored colonies produced by crossovers. Only a small fraction of the RTG colonies from samples taken before mock or CDC5 induction contained cycloheximide-resistant sectors (3.9% and 2.6%, respectively, Figure 3c, 3d), and cells plated after a 4 hr incubation without ED also produced few cycloheximide-resistant sectors (4.6%, Figure 3e). In contrast, when CDC5 was expressed and JMs resolved as COs, 30% of colonies contained cycloheximide-resistant sectors (Figure 3f). The relatively low frequencies of colonies with cycloheximide-resistant sectors in all samples that underwent RTG without CDC5 induction indicates that the majority of JMs are not resolved as COs after RTG.
Molecular evidence that most JMs are not resolved as COs after RTG
Reduced CO formation after RTG was confirmed by molecular analysis. To allow direct comparison between events that occur during meiosis and during RTG, we used a recombination-reporter strain, described below, that also contained the estrogen-inducible NDT80 allele described above (strain MJL3430) that confers reversible pachytene arrest [68]. Pachytene-arrested cells can be transferred to YPD without estradiol addition to undergo RTG in the absence of NDT80 expression. Alternatively, they can be kept in sporulation medium, and by adding ED to induce NDT80 expression, be made to complete meiosis (Figure 4a, 4b). Meiotic NDT80 expression resulted in meiotic divisions (Figure 4d), spore formation (data not shown), and the rapid expression of CDC5, a known target of Ndt80 [70]. Cdc5 was detected one hr after addition of ED to meiotic cultures, whereas Cdc5 was not present in RTG cultures until 2–2.5 hr after the shift to YPD, about 30 min before nuclear division (Figure 4c, 4e). The mitotic cyclin Clb2, which is not produced during meiosis [71], was observed only in the RTG culture, at about the same time as Cdc5 (Figure 4c).
Fig. 4. JM resolution and recombinant product formation during meiosis and RTG. 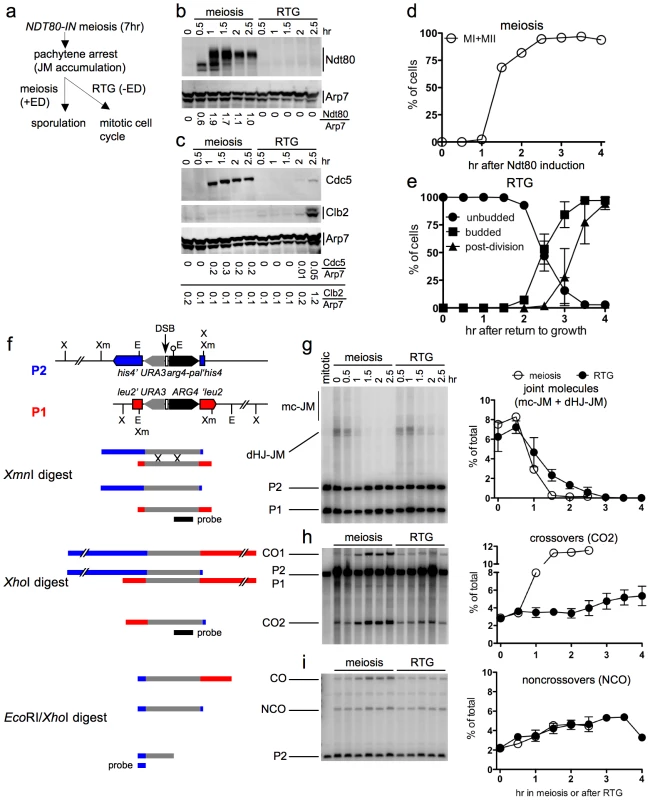
a. Experimental design. Cells with an estrogen-inducible NDT80 allele (MJL3430) are incubated in sporulation medium for 7 hr to uniform pachytene arrest. Estradiol (ED) is added to half of the culture to induce NDT80 expression and the completion of meiosis, while the other half is transferred to YPD to undergo RTG in the absence of NDT80 expression. b. Western blot showing Ndt80 production after addition of ED (meiosis) or after RTG. Arp7 is used as a loading control. Relative Ndt80 levels (arbitrary units) are shown below each lane. c. Western blot showing production of Ndt80-regulated polo-like kinase, Cdc5, and of the G2/M cyclin, Clb2, which is not expressed during meiosis. Arp7 is used as loading control. Relative protein levels (arbitrary units) are shown below each lane. d. Meiotic progression after NDT80 induction by ED addition. The percentage of cells completing meiosis I in a single experiment was determined by DAPI staining and counting the fraction of cells with more than one nucleus (MI + MII). Values are from a single experiment. e. Cell cycle progression after RTG. Cell cycle events were scored as in Figure 1. Values are from three independent experiments. f. Recombination reporter system used to detect recombination intermediates and products [7]. A 3.5 Kb insert with the URA3 (grey) and ARG4 (black) genes is inserted at LEU2 (red) on one chromosome III homolog and at HIS4 (blue), 16.7 Kb away, on the other. 65 nt of yeast telomere sequences (open box), inserted between URA3 and ARG4, create a strong meiotic DSB site (vertical arrow). A short palindrome containing an EcoRI site (lollipop) ∼0.6 kb from the DSB site, creates the arg4-pal allele in the insert at his4. Arrows denote the direction of transcription. Restrictions sites: Xm—XmnI; X—XhoI; E—EcoRI. An XmnI digest probed with ARG4 sequences (black bar) detects dHJ-JMs. A XhoI digest probed with the same sequences detects CO products. An EcoRI/XhoI double digest, probed with HIS4 sequences (blue bar) detects NCO events where the arg4-pal allele is converted to ARG4 (full conversion shown), as well as a subset of COs (CO). It should be noted that a subset of NCOs are detected by this assay. Based on tetrad data from similar strains [7], we estimate that about 1/6 of total NCOs are detected. g–i. DNA was prepared from NDT80-IN cells (MJL3430) that were either induced to complete meiosis by ED addition or shifted to YPD to undergo RTG, as illustrated in a. Samples were analyzed for JMs, COs and NCOs as illustrated in f. Values for meiosis are from a single experiment; values for RTG are from three independent experiments (for JMs and COs) and two independent experiments for NCOs. g. JM intermediates. Left: blots of XmnI digests probed with ARG4 sequences. In addition to dHJ-JMs, JMs containing 3 or 4 chromatids (multichromatid, mc-JMs) were detected at low levels. Right: frequencies of all JMs, plotted as a percent of total lane signal. h. COs. Left: blots of XhoI digests probed with ARG4 sequences. Right: CO product 2 (CO2) plotted as a percent of total lane signal. i. Noncrossover recombinants. Left: blots of XhoI/EcoRI digests probed with HIS4 sequences. Right: NCOs, plotted as a percent of total lane signal. Recombination intermediate resolution and recombinant product formation were monitored at the molecular level, using a recombination reporter system [7] (Figure 4f). JM resolution initiated at similar times in both ED-induced meiotic and RTG cultures (Figure 4g). However, the two cultures differed markedly in terms of CO production. JM resolution in the meiotic culture was accompanied by a marked increase in crossovers in the same time interval, and was complete by 1.5 hr after Ndt80 induction (Figure 4h). In contrast, no increase in COs was seen in the first 2 hr after RTG, during which JMs decreased by five-fold. After two hr, a time that corresponded to the time of bud emergence (Figure 4e), resolution of the remaining JMs was accompanied by a modest increase in COs (Figure 4h). NCO products were produced in meiotic and in RTG cultures at similar levels (Figure 4i). Similar results were observed in RTG experiments using ndt80Δ cells lacking the inducible NDT80 system (strain MJL3164; Figure S2).
The data presented here support the conclusion from genetic experiments described above, that most JMs are resolved after RTG without producing COs. The CO increase seen after 2 hr indicates that surviving JMs can be resolved as COs during the later stages of RTG.
Efficient JM resolution without CO production after RTG in the absence of Mus81
The Mus81 complex plays a major role in JM resolution during meiosis in S. pombe and a less prominent role in meiotic JM metabolism in S. cerevisiae [19], [26], [20], [27], [22], [24], [72], [28]. To determine if the Mus81 complex resolves JMs after RTG, ndt80Δ mus81Δ cells (strain MJL3389) were induced to undergo meiosis for 7 hr and then transferred to YPD. Bud emergence and nuclear division occurred at times similar to those seen in ndt80Δ MUS81 cells (Figure 5a, compare to Figure 1b). JMs were resolved completely after RTG (Figure 5b). A modest net increase in noncrossovers was seen (Figure 5d), similar to that seen in MUS81 cells (see Figure 4i). Unlike in wild-type, where JM resolution after two hr was accompanied by an increase in COs, no significant CO increase was observed after RTG in mus81Δ mutants (Figure 5c). These data indicate that the Mus81 complex is not required for JM resolution after RTG, but it may play an important role in the limited JM resolution as COs that occurs at later stages.
Fig. 5. Efficient JM resolution without CO production after RTG in the absence of Mus81. 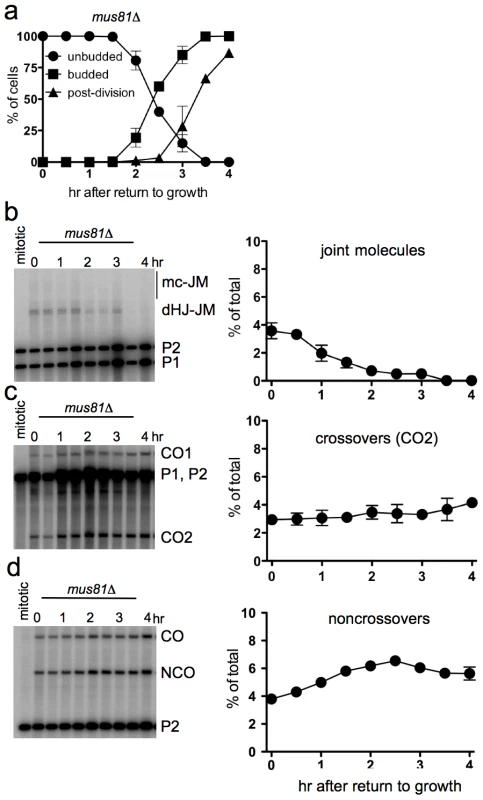
a. Cell cycle progression of ndt80Δ mus81Δ cells (MJL3389) after RTG. Cell cycle events were scored as in Figure 1. b. JM intermediates. Left: blot of XmnI digests probed with ARG4 sequences as in Figure 4. Right: total JMs plotted as a percentage of total lane signal. c. COs. Left: blot of XhoI digests probed with ARG4 sequences, as in Figure 4. Right: CO product 2 (CO2), plotted as a percentage of total lane signal. d. NCOs. Left: blots of XhoI/EcoRI digests probed with HIS4 sequences, as in Figure 4. Right: NCO products plotted as a percentage of total lane signal. Delayed JM resolution after RTG in the absence of Sgs1 helicase activity
The BLM and Sgs1 helicases, in combination with topoisomerase III and Rmi1/BLAP45, resolve dHJs in vitro as NCOs [33], [36]. To ask if Sgs1 has a similar role in JM resolution after RTG, we used an sgs1 mutant allele (strain MJL3388; sgs1-ΔC795) that expresses only the first 652 amino acids of the protein [73], and which lacks both the helicase domain and a region (the HRDC domain) which in BLM interacts with Holliday junctions [74]. Although bud emergence occurred at a similar time after RTG in sgs1-ΔC795 and in SGS1 cells, nuclear division was 1.5–2 hr later in sgs1-ΔC795 than in SGS1 (Figure 6a, compare to Figure 1b). A recombination-null ndt80Δ sgs1-ΔC795 spo11 triple mutant (strain MJL3428), which does not produce JMs, underwent nuclear division without this delay (Figure 6a), suggesting that the delay in nuclear division seen in sgs1-ΔC795 might result from a delay in JM resolution.
Fig. 6. Delayed JM resolution and increased CO formation after RTG in the absence of the Sgs1 helicase. 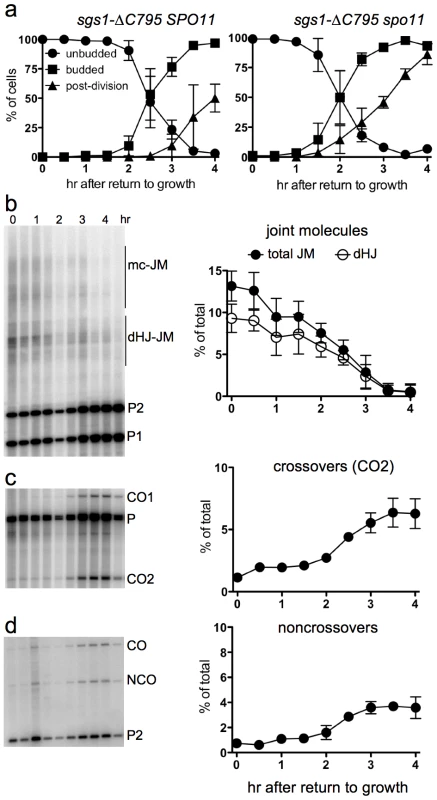
a. Delayed nuclear division during RTG of in the absence of Sgs1 helicase activity is due to meiotic recombination. Panels show cell cycle progression of ndt80Δ sgs1-ΔC795 cells that are meiotic recombination competent (SPO11, left; MJL3388) or recombination null (spo11, right; MJL3428). b. Joint molecule intermediates. Left: blots of XmnI digests probed with ARG4 sequences. Right: frequencies of total JMs (multichromatid JMs, mcJMs plus dHJ-JMs, filled circles) and of dHJ intermediates (dHJ; empty circles) plotted as a percentage of total lane signal. c. Crossovers. Left: blots of XhoI digests probed with ARG4 sequences. Right: CO product 2 (CO2) are plotted as a percentage of total lane signal. d. Noncrossovers. Left: blots of XhoI/EcoRI digests probed with HIS4 sequences. Right: NCOs, plotted as a percentage of total lane signal. To ask if JM resolution is delayed in ndt80Δ sgs1-ΔC795 cells, we monitored JMs and recombination products, using the molecular assay system described above. As was previously described [41], ndt80Δ sgs1-ΔC795 cells accumulate high levels of intersister JMs, and JMs with more than two chromatids (multi-chromatid JMs; mcJMs), in addition to the dHJ-JMs that accumulate in ndt80Δ SGS1 cells (Figure 6b). Resolution of all JM species was delayed by about 1 hr in sgs1-ΔC795 as compared to SGS1. While the vast majority of JMs resolved in SGS1 by about 2.5 hr after RTG (Figure 4g), more than half of total JMs remained unresolved in sgs1-ΔC795 at the same time, although all JMs resolved by 4 hr (Figure 6b). Thus, loss of the Sgs1 helicase results in a substantial delay in JM resolution after RTG.
Delayed JM resolution after RTG in sgs1-ΔC795 was accompanied by altered recombinant product formation. COs increased only slightly in the first 1.5 hr after RTG (Figure 6c), but there was also only a slight increase in NCOs during the same period (Figure 6d). After 1.5 hr, JM resolution was accompanied by an increase in both COs and NCOs (Figure 6c, 6d). Thus, in both SGS1 and in sgs1-ΔC795, few COs are produced during the first 1.5–2 hr after RTG, with substantially greater CO formation at later times. However, unlike in SGS1, where most NCOs appear in the first 1.5–2 hr after RTG, NCO production in sgs1-ΔC795 is delayed until the time that COs also appear.
Discussion
Most JM intermediates formed during budding yeast meiosis are produced by interhomolog recombination and are resolved as COs, and the majority of meiotic COs derive from interhomolog JMs [8], [9], [17], [10]. In contrast, interhomolog JMs and COs are less prominent during the mitotic cell cycle. Most JMs produced during mitotic DSB repair involve sister chromatids [15], and only a minor fraction (typically 5–10%) of mitotic recombination involves crossing-over, as would be expected if interhomolog JMs are rarely resolved as COs during the mitotic cell cycle [16], [75]. Testing this suggestion has, to date, been limited by the very low levels of interhomolog JMs produced in vegetatively-growing cells, even when initiating DSBs occur at levels similar to those seen in meiosis [15].
In this paper, we used RTG as an alternate approach to the study of JM resolution during the mitotic cell cycle. Although aspects of RTG have been examined in many studies [50]–[58], interpretation has been complicated by the relatively poor synchrony of yeast meiotic cultures. Thus, RTG samples from normal meiotic cultures can contain cells with unrepaired DSBs, cells with repaired DSBs but unresolved recombination intermediates, and cells where intermediates already have been resolved. To avoid complications inherent in the analysis of such a complex mixture, we performed RTG using meiotic cultures of ndt80 mutant cells, which arrest at a single stage of meiosis (pachytene), with chromosomes fully paired by synaptonemal complex and with high levels of interhomolog JMs. This has provided insight into features of the mitosis-like cell cycle that immediately follows exit from meiosis, and into mechanisms of the recombination intermediate resolution.
Return to growth involves a mitosis-like division without an intervening S-phase
When transferred from sporulation to growth medium, yeast cells degrade most meiotic transcripts within 20 min, and return to a pattern of gene expression that roughly resembles the G1 phase of the mitotic cell cycle [57]. Despite this rapid change in transcription patterns, cells spend an extended lag period (1.5 to 3 hours, equivalent to one or two normal mitotic cell cycles) before they undergo bud emergence, the first outward sign of resumed growth (Figure 1). Although cells disassemble synaptonemal complex and resolve meiotic recombination intermediates during this period ([56], this work), a similar lag before bud emergence is seen in spo11 mutants (this work), and also if SC disassembly and JM resolution occur before RTG, by virtue of Cdc5 induction in ndt80Δ CDC5-IN cells (Y.D. and M.L., unpublished observations). It is therefore likely that this extended gap phase represents the time needed for metabolic adjustment to the shift from acetate to glucose, and from nitrogen-depleted to nitrogen-rich medium, rather than the time needed to disassemble meiosis-specific chromosome and DNA structures.
During the mitotic cell cycle, bud emergence is accompanied by the initiation of chromosome replication [63], but this is not the case during RTG. We used two different approaches to confirm that bud emergence occurs without DNA replication after RTG [53]. This could be the consequence of a failure to express completely the ensemble of proteins necessary for DNA replication. While some replication protein-encoding genes are transcribed after RTG ([57], Lea Jessop and M. L., unpublished observations), transcripts of DBF4 and CDC7, which encode a kinase critical for replication origin firing, are rapidly reduced upon RTG [57]. Re-replication may also be blocked if cyclin-dependent kinase remains at post-S phase levels throughout RTG, which would prevent origin re-licensing [76]–[78].
We also find that the first nuclear division after RTG involves an equational division, unlike the reductional division that occurs during meiosis I. Reductional division at meiosis I requires the loading, at kinetochores, of the meiosis-specific protein complex monopolin, which promotes co-orientation of sister kinetochores towards a single spindle pole [79], [80]. Monopolin contains a meiosis-specific protein, Mam1, and two nucleolar proteins, Csm1 and Lrs4, whose kinetochore localization requires Cdc5 activity [79], . Meiotic CDC5 transcription requires NDT80, and MAM1 transcripts are reduced in ndt80 mutants [70] and rapidly decline upon RTG [57]. In addition, monopolin loading at kinetochores requires active Cdc7/Dbf4 kinase [82], which is most likely not produced after RTG [57]. Therefore, it is unlikely that monopolin is loaded at kinetochores during RTG of ndt80Δ cells, and thus it is not surprising that the first nuclear division after RTG is equational.
Recombination intermediate resolution during RTG is biased against crossovers
Most of the Holliday junction-containing JMs that accumulate during meiosis in ndt80 mutants are resolved as COs upon restoration of either NDT80 or CDC5 gene expression ([10], this work). In contrast, our genetic and molecular analyses show that most of the JMs that form during wild-type meiosis are resolved without crossover formation during RTG. This indicates that mechanisms of JM resolution that operate during RTG differ from those that operate during meiosis.
There are three general mechanisms for dHJ-JM resolution: endonuclease cleavage; helicase/topoisomerase-mediated dissolution; and replication (Figure 7a–7c). Of these, replication and dissolution produce only NCO products, while endonuclease cleavage can, in principle, produce either COs or NCOs, depending upon the orientation of the two cleavage reactions. Since most dHJ-JMs resolve as COs during meiosis, meiotic resolution must involve endonuclease cleavage, and this cleavage must be constrained so that the two Holliday junctions are usually cut in opposite directions (see Figure 7a).
Fig. 7. Modes of dHJ-JM resolution and summary of data. 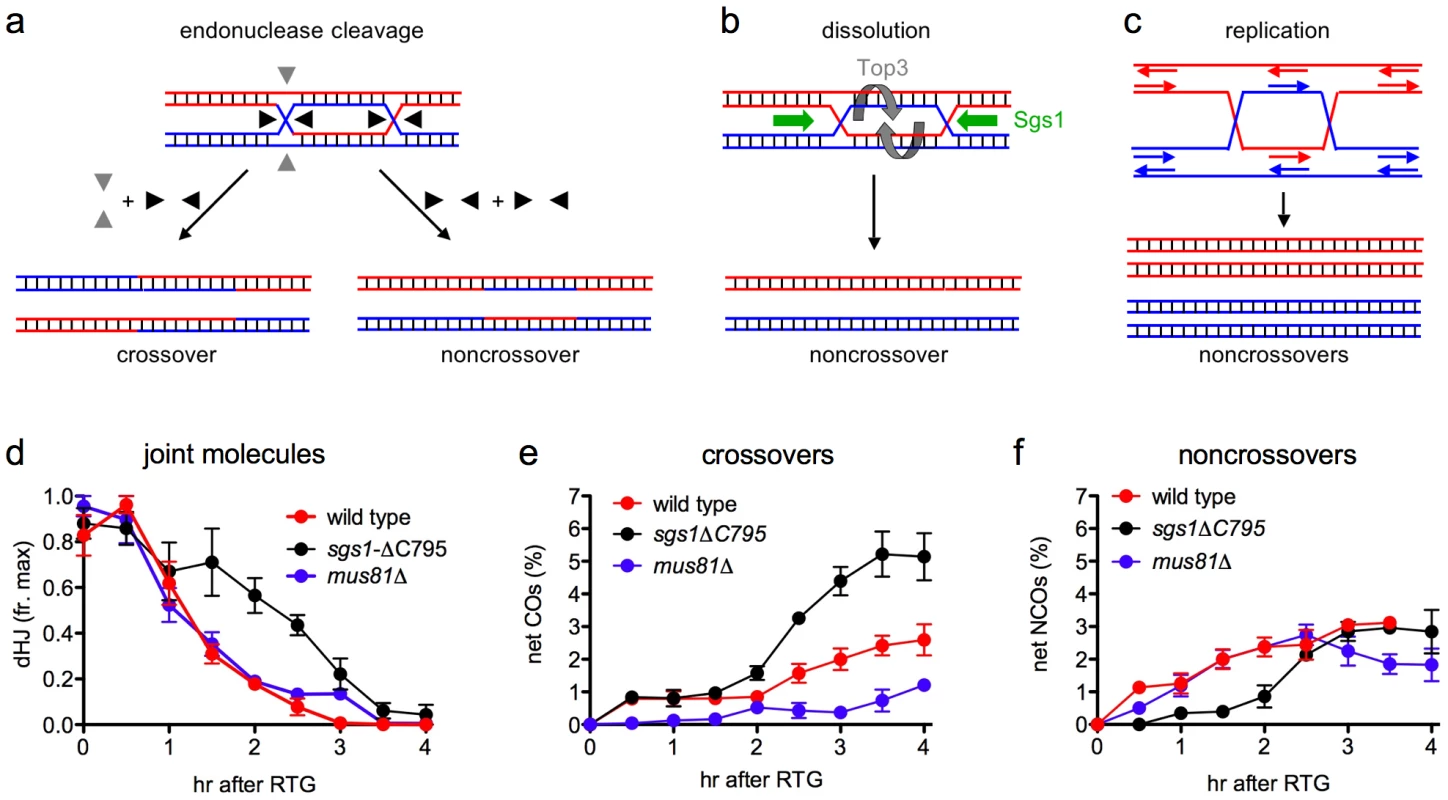
a. Resolution by junction cleavage [83]. Cleavage of both Holliday junctions in the same orientation (black arrows) yields noncrossovers; cleavage of the two junctions in orthogonal orientations (black and grey arrows) yields crossovers. For simplicity, only one of the two patterns for each type of cleavage is shown. b. Resolution by dissolution [31], [32]. Helicase-driven convergent junction branch migration, coupled with topoisomerase-removal of superhelical stress, produces only noncrossovers. c. Resolution by replication produces only noncrossovers. d. Summary of JM resolution during RTG. Maximum JM levels in each individual experiment (3 for wild-type, 2 for sgs1-ΔC795 and mus81Δ) were set to 1. For sgs1-ΔC795, 2-chromatid JM values were used, although similar results are obtained with total JMs (2-chromatid + multichromatid). Plotted values represent averages; error bars indicate standard error of the mean. e. Net CO production during RTG. CO levels at 0 hr (the time of RTG) were subtracted from each time-point value and plotted as in d. f. Net NCO production during RTG. NCO levels at 0 hr (the time of RTG) were subtracted from each time-point value and plotted as in d. In contrast, JM resolution during RTG appears to occur in two phases with different outcomes (Figure 7d–7f). In wild-type cells, about 80% of JMs disappear during the first 1.5–2 hr after RTG. Few COs are produced during this period, and NCOs increase to near-final levels. The greatest net increase in COs occurs at 2 hr and later (Figure 7e), when the remaining 20% of JMs are resolved (Figure 7d). Thus, RTG appears contain an initial period (hereafter called early RTG) that precedes bud emergence, during which SC breaks down (Figure 1c) and the majority of JMs resolve without CO formation (Figure 7d, 7e). During the second period (hereafter called late RTG), between bud emergence and nuclear division, JM resolution is accompanied by CO formation.
Sgs1-dependent dissolution as a mechanism for JM resolution during early RTG
JM resolution without CO formation, which predominates during early RTG, could occur by endonucleolytic cleavage that is constrained to produce only NCOs, by dissolution, or by replication (Figure 7a–7c). Resolution by replication is unlikely, since all available evidence indicates that the first cell division after RTG occurs without prior replication (this work, [53]). Both JM resolution and NCO formation are significantly reduced during early RTG in sgs1-ΔC795 mutant cells (Figure 7d, 7f), which lack both the helicase and Holliday junction-binding domains of this RecQ helicase [73], [74]. The most parsimonious interpretation of these data is that, in wild-type cells, JM resolution during early RTG occurs primarily by dissolution, catalyzed by Sgs1 and Top3/Rmi1, as has been observed in vitro [36]. However, it is formally possible that other activities are responsible for the initial phase of JM resolution in wild-type, and that, unlike in wild-type, the majority JMs that form during sgs1-ΔC795 meiosis have structures that are refractory to resolution by these hypothetical activities.
During budding yeast meiosis, the Sgs1 helicase acts with Mus81/Mms4 to prevent the accumulation of abnormal recombination intermediates [28], [29]. Normal JM intermediates are protected from Sgs1 by components of the synaptonemal complex, and sgs1-ΔC795 partially suppresses the JM deficit observed in mutants lacking SC components [40], [42], [41]. These and other observations have been interpreted as indicating that Sgs1 acts primarily to prevent JM formation during meiosis. Our current data indicate that, in addition to preventing JM formation, Sgs1 can also dissolve JMs in vivo, but is prevented from doing so during meiosis by the SC. This suggestion is also supported by the finding that most JMs are resolved without CO production upon Cdc5-independent SC breakdown in pachytene-arrested meiotic cells (Anuradha Sourirajan, Arnaud de Muyt and M. L., unpublished observations).
JM resolution by endonucleolytic cleavage during late RTG
While JM resolution during early RTG is rarely accompanied by CO production, JMs that survive this initial phase appear to be resolved frequently as COs. This is seen in wild-type, but is most evident in sgs1-ΔC795 mutant cells, where an increase in the rate of JM resolution during late RTG is accompanied by a marked increase in both CO and NCO recombinants (Figure 7e, 7f). Because COs can only be produced by endonuclease-mediated JM cleavage, this suggests that a Holliday junction resolvase is activated 1.5–2 hr after RTG, a time that is also marked by bud emergence. We do not know the regulatory change that is responsible for this change in modes of JM resolution, but it is worth noting that both Cdc5 and the G2/M phase cyclin, Clb2, are first produced at this time (Figure 4c).
During meiosis, the Cdc5 kinase triggers JM resolution as COs [10], suggesting an obligate cleavage of JM Holliday junctions in opposite directions (Figure 7a). In contrast, JM resolution during late RTG of sgs1-ΔC795 mutants produces both COs and NCOs (Figure 7e, 7f), as would be expected for the mixed parallel and opposite cleavage patterns contained in the original DSBR model ([83], see Figure 7a). This apparent difference in resolution mechanisms may reflect the chromosome environment in which intermediates reside. While JM resolution during late RTG occurs in the absence of detectable SC, crossover-designated meiotic JMs are thought to reside in SC-associated structures, called late recombination nodules, that contain the Holliday junction-binding proteins Msh4/Msh5 and associated Mlh1, Mlh3 and Exo1 proteins [84]–[86]. In mlh1, mlh3, and exo1 mutants, meiotic JM levels are normal but crossover formation is reduced roughly two-fold [87], [88], consistent with the suggestion that the Mlh1/Mlh3/Exo1 components of late recombination nodules direct nuclease-mediated meiotic JM resolution towards a crossover-only outcome. In the absence of such specialized chromosome structures, nuclease-mediated JM resolution may be more evenly divided between COs and NCOs, in both mitotic and meiotic cells.
A role for Mu81/Mms4 in JM resolution during RTG?
Although the nuclease(s) responsible for dHJ resolution during either meiosis or during RTG remain to be determined, it is worth noting that CO formation during RTG is even more reduced in mus81Δ mutants than in wild-type (Figure 7e), and the increase in COs seen during late RTG in wild-type and in sgs1-ΔC795 is not seen in mus81Δ mutants. In many organisms, including S. cerevisiae, the Mus81 nuclease complex is dispensable for most meiotic COs [26], [89]–[91], and the majority of meiotic JMs resolve in a timely manner in S. cerevisiae mus81 or mms4 mutants [27], [28]. In addition, it has been reported that intact Holliday junctions are a relatively poor substrate for the Mus81/Mms4 nuclease, while junctions with one nicked strand are resolved efficiently [92], [22], [93]. On the other hand, MUS81 is required for timely disappearance of X-shaped DNA molecules that form in methyl methanesulfonate-treated rmi1-ts cells [94]. This would suggest a role for Mus81/Mms4 in resolving these JMs, whose structure remains to be determined.
Our data suggest that Mus81/Mms4 has a role in resolving the JMs that survive until late RTG, but it does not appear to be active during early RTG. It is possible that either Mus81/Mms4 or a junction nicking activity that converts HJs into a Mus81/Mms4 substrate are absent during early RTG. Alternatively, the Mus81 complex may be modified during late RTG so that it resolves intact Holliday junctions unassisted. The latter suggestion, if correct, might explain the failure to observe robust Holliday junction resolution activity in most biochemical studies [95].
Concluding remarks
In this work, we have shown that Holliday junction-containing recombination intermediates, formed during meiosis, are resolved during RTG in a manner that substantially reduces CO production. To the extent that recombination is regulated similarly during RTG and during the mitotic cell cycle, and to the extent that similar recombination intermediates are present, this finding can help explain the relatively low yield of COs during mitotic recombination. In particular, our findings reinforce the identification of the BLM family of RecQ helicases as playing an important role in suppressing CO recombination during the mitotic cell cycle [38]. Our findings also suggest that the Mus81 complex is the primary nuclease responsible for mitotic CO recombination [30]. Our finding, that these two enzymes act during different phases of the period before the first cell dvision after RTG, raises the intriguing possibility that the mitotic cell cycle may be similarly partitioned. It is attractive to suggest that helicase-mediated dissolution predominates during most of the mitotic cell cycle, with endonuclease-mediated JM cleavage being activated at the end. This would minimize the potential for CO-mediated loss of heterozygosity and chromosome entanglement, while preserving the ability to resolve JMs that escape dissolution before the initiation of mitosis.
In applying conclusions regarding JM resolution during RTG to the mitotic cell cycle, it should be kept in mind that these processes are not identical. For example, RTG involves the disassembly of chromosome structures that are not present during the mitotic cell cycle, as well as S-phase bypass, and both of these differences have the potential to affect modes of JM resolution. It will be of considerable interest to examine, during RTG, patterns of expression and modification of proteins involved in recombination, repair, and cell cycle progression during meiosis and the mitotic cell cycle.
Materials and Methods
Yeast strains and media
Strains are listed in Table S1 and are SK1 derivatives [96]. The URA3-ARG4 recombination interval has been described [7]; cyh2-z is a spontaneous cycloheximide resistance mutation (CyhR); spo11-Y135F [97] was a gift from S. Keeney; mus81Δ and sgs1-ΔC795 have been described [42], [28]. Strains with estrogen-inducible CDC5 and NDT80 alleles (pGPD1-GAL4-ER pGAL1-CDC5 and pGPD1-GAL4-ER pGAL1-NDT80, respectively) have been described [10]. Strains were constructed by genetic crosses, or by transformation. Media formulae were as described [98], [99].
Liquid sporulation and return to growth
Sporulation was as described [99] using 400 ml cultures in a 2.8 liter baffled Fernbach flask (BellCo Glass) with a cell density of 2x 107 cells per ml at the beginning of sporulation. For RTG experiments, cells were induced to undergo meiosis for 7 hr, harvested by centrifugation, resuspended in an equal volume of liquid YPD (prewarmed to 30°C) and aerated with vigorous shaking at 30°C in conditions similar to those used for sporulation. For plating experiments, samples were sonicated twice for 5 seconds at baseline power (Microson XL 2005), diluted appropriately and then plated on YPD plates. To determine colony-forming units, samples were counted in a hematocytometer and the concentration of cells was determined; cells with unseparated buds were counted as a single entity. For Ndt80 or Cdc5 induction, β-estradiol (ED; Sigma; 5 mM stock in ethanol) was added to a final concentration of 1 µM. For no Cdc5-indcuation control experiments, the same amount of ethanol (without ED) was added. For RTG after Cdc5 induction during meiosis, cells were washed twice with sporulation medium lacking ED at 30°C before resuspension in YPD.
Unless stated otherwise, all data presented are the average of two independent experiments; error bars in plots indicate standard error.
Cytology
To score bud emergence and nuclear division, 1 ml of a culture was mixed with 1 ml of ethanol and stored at 4°C. Just before examination, 1 µl of 1 mg/ml 4′,6-diamidino-2-phenylindole (DAPI) was added and samples were left for 5 min at room temperature, washed once with an equal volume of water and resuspended in 0.5 ml water. Cell morphology was scored using phase contrast or differential interference contrast microscopy and nuclear morphology by DAPI epifluorescence microscopy, using a Zeiss Axioplan 2 epifluorescence microscope and a QICAM camera. Images were acquired using QCapture 3.1.1 and processed with Adobe Photoshop CS3.
GFP chromosome dot visualization was done using cells fixed in 3.7% formaldehyde as described [65]. Vectashield with DAPI (Vector Laboratories) was used to simultaneously stain DNA. Cells were counted as having two GFP dots if two separated GFP dots could be clearly visualized. Sample fluorescence was visualized using a Zeiss Axioplan 2 epifluorescence microscope and a Micromax 1300 CCD camera. Images were acquired using IPlab 3.7 and processed with Adobe Photoshop CS3.
Nuclear spreads were performed and stained as described [100] using cells from 5 ml of culture. Zip1 was detected using anti-Zip1 rabbit polyclonal sera (a gift from G.S. Roeder, 1∶100 dilution) as the primary antibody and Alexafluor 488 conjugated goat anti-rabbit IgG (Molecular Probes #A11034) at 1∶100 as the secondary antibody. To visualize DNA, 40 µl of Vectashield with DAPI (Vector Laboratories) was added. Sample fluorescence was visualized using a Zeiss Axioplan 2 epifluorescence microscope and a Micromax 1300 CCD camera. Images were acquired using IPlab 3.7 and processed with Adobe Photoshop CS3.
Calculation of cumulative curves for bud emergence and nuclear division
During RTG, cells lose synchrony and continue to further cell cycles, complicating calculation of a cumulative cell division curve. We assumed that bud emergence and nuclear division occur with the same relative timing in the first and second cell division after RTG. To distinguish between daughter and mother cells, we took advantage of the fact that after RTG, ndt80Δ cells produce an elongated bud that can be easily distinguished from the round mother cell (Figure 1a). The fraction of cells that had not yet budded (unbudded cells) was calculated according to the equation: unbudded cells = (X1-Y1)/Z1 where X1 = unbudded round cells (i.e. cells before the first mitotic division), Y1 = unbudded elongated cells (i.e. products of the first mitotic division) and Z1 = total cells counted. At late times, due to continuous division of the cells, the number of cells that have already undergone the first mitotic division (Y1) can exceed the number of cells that have not undergone a mitotic division (X1). In such a case, (X1-Y1) was set to zero.
The fraction of cells that had undergone the first nuclear division (post-division) was calculated according to the equation: post-division = X2/Y2 where X2 = round cells that were undergoing mitosis (detected as budded with a nucleus stretched between the mother and daughter cells) plus all elongated cells with a nucleus (i.e. cells that have already completed the first mitotic division) and Y2 = all round cells. At late times, due to continuous cell division, X2 may be greater than Y2. In such a case, the fraction of post-division cells was set to one.
DNA extraction and digestion
DNA preparation and analysis on Southern blots were as described [101], [8]. XhoI and XmnI digests were probed with ARG4 coding sequences (+165 to +1413). XhoI/EcoRI double digests were probed with HIS4 coding sequences (+538 to +718).
Protein analysis
Protein was prepared from 4 ml of sporulating culture by TCA precipitation [102]. 5 µl samples of each extract were displayed on 7.5% polyacrylamide Tris-Glycine pre-cast gels (Bio-Rad) and electroblotted to a PVDF membrane (Invitrogen), using an iBlot Dry Blotting System (Invitrogen) as recommended by the manufacturer. Blots were washed for at least one hr on an orbital shaker at room temperature in blocking buffer, 0.2% I-block (Tropix) in PBST (0.15 M NaCl, 0.053 M Na2HPO4, 0.008 M KH2PO4, 0.05% v/v Tween-20, pH 7.4). Primary antibody, diluted in blocking buffer, was added to the blot and incubated on an orbital shaker at room temperature for at least one hr. Blots were washed four times for 15 min with blocking buffer, incubated with secondary antibody for one hr with shaking at room temperature, and wash steps were repeated. Signal was developed using the chemiluminescent CDP-star substrate (Applied Biosystems), detected using a Fuji LAS3000 CCD camera, and quantified using ImageGauge V4.22 software (Fuji). Blots were stripped with OneMinute Western Blot Stripping Buffer (GM Biosciences) and reprobed for Arp7 as a loading control. Primary antisera were as follows: Arp7 – goat polyclonal (Santa Cruz Biotechnology, Inc; Sc-8961), 1∶500; influenza hemagglutinin (HA) – mouse monoclonal (5 µg/µl; Roche Applied Science; 12CA5), 1∶10,000; Cdc5 – goat polyclonal (Santa Cruz Biotechnology, Inc; Sc-6733), 1∶500; Ndt80 – rabbit polyclonal (a gift from K. Benjamin), 1∶10,000; Clb2 – rabbit polyclonal (Santa Cruz Biotechnology, Inc; Sc-9071), 1∶500. Secondary antibodies were alkaline phosphatase conjugates of goat-anti-mouse (Sigma, A3562), goat-anti-rabbit (Sigma, A3687) and rabbit-anti-goat (Sigma, A4187), all used at 1∶10,000.
Measuring crossovers between CYH2 and the centromere
To measure the frequency of recombination between the CYH2 locus and the centromere of chromosome VII, we measured second division segregation pattern of the TRP1 and CYH2 alleles in dissected tetrads from strain MJL3548 (CYH2/cyh2-z TRP1/trp1), using TRP1 as a centromere-linked marker [62]. Of 72 tetrads with 4 viable spores, 12 tetrads were parental ditypes, 12 were non-parental ditypes and 47 were tetratypes. One tetrad had gene conversion of cyh2-z and was not counted. Thus, as expected for a locus far removed from its centromere, the vast majority of cells undergo at least one crossover between CYH2 and CEN7, and about two thirds of cells produce spores with a crossover between the CYH2 locus and its centromere.
Supporting Information
Zdroje
1. PetronczkiMSiomosMFNasmythK 2003 Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell 112 423 440
2. BergeratAde MassyBGadelleDVaroutasPCNicolasA 1997 An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386 414 417
3. KeeneySGirouxCNKlecknerN 1997 Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 375 384
4. SunHTrecoDSzostakJW 1991 Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64 1155 1161
5. GoldfarbTLichtenM 2010 Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol 8 e1000520 doi:10.1371/journal.pbio.1000520
6. LaoJPHunterN 2010 Trying to avoid your sister. PLoS Biol 8 e1000519 doi:10.1371/journal.pbio.1000519
7. JessopLAllersTLichtenM 2005 Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics 169 1353 1367
8. AllersTLichtenM 2001 Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106 47 57
9. HunterNKlecknerN 2001 The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106 59 70
10. SourirajanALichtenM 2008 Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev 22 2627 2632
11. SchwachaAKlecknerN 1995 Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83 783 791
12. ZicklerD 2006 From early homologue recognition to synaptonemal complex formation. Chromosoma 115 158 174
13. LichtenMHaberJE 1989 Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics 123 261 268
14. KadykLCHartwellLH 1992 Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132 387 402
15. BzymekMThayerNHOhSDKlecknerNHunterN 2010 Double Holliday junctions are intermediates of DNA break repair. Nature
16. IraGMalkovaALiberiGFoianiMHaberJE 2003 Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115 401 411
17. ClyneRKKatisVLJessopLBenjaminKRHerskowitzI 2003 Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol 5 480 485
18. Jinks-RobertsonS 2010 Seeking resolution: budding yeast enzymes finally make the cut. Mol Cell 40 858 859
19. BoddyMNGaillardPHMcDonaldWHShanahanPYatesJR 2001 Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107 537 548
20. KaliramanVMullenJRFrickeWMBastin-ShanowerSABrillSJ 2001 Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev 15 2730 2740
21. CicciaAConstantinouAWestSC 2003 Identification and characterization of the human Mus81-Eme1 endonuclease. J Biol Chem 278 25172 25178
22. OsmanFDixonJDoeCLWhitbyMC 2003 Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell 12 761 774
23. SmithGRBoddyMNShanahanPRussellP 2003 Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics 165 2289 2293
24. CromieGAHyppaRWTaylorAFZakharyevichKHunterN 2006 Single Holliday junctions are intermediates of meiotic recombination. Cell 127 1167 1178
25. InterthalHHeyerWD 2000 MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV - and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet 263 812 827
26. de los SantosTLoidlJLarkinBHollingsworthNM 2001 A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159 1511 1525
27. de los SantosTHunterNLeeCLarkinBLoidlJ 2003 The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164 81 94
28. JessopLLichtenM 2008 Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell 31 313 323
29. OhSDLaoJPTaylorAFSmithGRHunterN 2008 RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell 31 324 336
30. HoCKMazonGLamAFSymingtonLS 2010 Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell 40 988 1000
31. NasmythKA 1982 Molecular genetics of yeast mating type. Annu Rev Genet 16 439 500
32. GilbertsonLAStahlFW 1996 A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics 144 27 41
33. WuLHicksonID 2003 The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426 870 874
34. PlankJLWuJHsiehTS 2006 Topoisomerase IIIα and Bloom's helicase can resolve a mobile double Holliday junction substrate through convergent branch migration. Proc Natl Acad Sci U S A 103 11118 11123
35. WuLHicksonID 2002 The Bloom's syndrome helicase stimulates the activity of human topoisomerase IIIα. Nucleic Acids Res 30 4823 4829
36. CejkaPPlankJLBachratiCZHicksonIDKowalczykowskiSC 2010 Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol 17 1377 1382
37. ChagantiRSSchonbergSGermanJ 1974 A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci U S A 71 4508 4512
38. HicksonID 2003 RecQ helicases: caretakers of the genome. Nat Rev Cancer 3 169 178
39. MankouriHWHicksonID 2004 Understanding the roles of RecQ helicases in the maintenance of genome integrity and suppression of tumorigenesis. Biochem Soc Trans 32 957 958
40. RockmillBFungJCBrandaSSRoederGS 2003 The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr Biol 13 1954 1962
41. OhSDLaoJPHwangPYTaylorAFSmithGR 2007 BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130 259 272
42. JessopLRockmillBRoederGSLichtenM 2006 Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of Sgs1. PLoS Genet 2 e155 doi:10.1371/journal.pgen.0020155
43. van BrabantAJYeTSanzMGermanIJEllisNA 2000 Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39 14617 14625
44. BachratiCZBortsRHHicksonID 2006 Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res 34 2269 2279
45. EspositoMS 1978 Evidence that spontaneous mitotic recombination occurs at the two-strand stage. Proc Natl Acad Sci U S A 75 4436 4440
46. ZouHRothsteinR 1997 Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90 87 96
47. LiberiGMaffiolettiGLuccaCChioloIBaryshnikovaA 2005 Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 19 339 350
48. ChuSHerskowitzI 1998 Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell 1 685 696
49. XuLAjimuraMPadmoreRKleinCKlecknerN 1995 NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol 15 6572 6581
50. GanesanATHolterHRobertsC 1958 Some observations on sporulation in Saccharomyces. C R Trav Lab Carlsberg Chim 31 1 6
51. ShermanFRomanH 1963 Evidence for two types of allelic recombination in yeast. Genetics 48 255 261
52. SimchenGPinonRSaltsY 1972 Sporulation in Saccharomyces cerevisiae: premeiotic DNA synthesis, readiness and commitment. Exp Cell Res 75 207 218
53. EspositoREEspositoMS 1974 Genetic recombination and commitment to meiosis in Saccharomyces. Proc Natl Acad Sci U S A 71 3172 3176
54. HonigbergSMConicellaCEspositioRE 1992 Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics 130 703 716
55. HonigbergSMEspositoRE 1994 Reversal of cell determination in yeast meiosis: postcommitment arrest allows return to mitotic growth. Proc Natl Acad Sci U S A 91 6559 6563
56. ZenvirthDLoidlJKleinSArbelAShemeshR 1997 Switching yeast from meiosis to mitosis: double-strand break repair, recombination and synaptonemal complex. Genes Cells 2 487 498
57. FriedlanderGJoseph-StraussDCarmiMZenvirthDSimchenG 2006 Modulation of the transcription regulatory program in yeast cells committed to sporulation. Genome Biol 7 R20
58. SimchenG 2009 Commitment to meiosis: what determines the mode of division in budding yeast? Bioessays 31 169 177
59. MoensPBMowatMEspositoMSEspositoRE 1977 Meiosis in a temperature-sensitive DNA-synthesis mutant and in an apomictic yeast strain (Saccharomyces cerevisiae). Philos Trans R Soc Lond B Biol Sci 277 351 358
60. KlapholzSWaddellCSEspositoRE 1985 The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110 187 216
61. SymMEngebrechtJARoederGS 1993 ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72 365 378
62. MortimerRKHawthorneDC 1966 Genetic mapping in Saccharomyces. Genetics 53 165 173
63. WilliamsonDH 1965 The timing of deoxyribonucleic acid synthesis in the cell cycle of Saccharomyces cerevisiae. J Cell Biol 25 517 528
64. MichaelisCCioskRNasmythK 1997 Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 35 45
65. LeeBHKiburzBMAmonA 2004 Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr Biol 14 2168 2182
66. JinQTrelles-StickenEScherthanHLoidlJ 1998 Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol 141 21 29
67. BenjaminKRZhangCShokatKMHerskowitzI 2003 Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev 17 1524 1539
68. CarlileTMAmonA 2008 Meiosis I is established through division-specific translational control of a cyclin. Cell 133 280 291
69. ChuaPJinks-RobertsonS 1991 Segregation of recombinant chromatids following mitotic crossing over in yeast. Genetics 129 359 369
70. ChuSDeRisiJEisenMMulhollandJBotsteinD 1998 The transcriptional program of sporulation in budding yeast. Science 282 699 705
71. GrandinNReedSI 1993 Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol 13 2113 2125
72. GaskellLJOsmanFGilbertRJWhitbyMC 2007 Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? EMBO J 26 1891 1901
73. MullenJRKaliramanVBrillSJ 2000 Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154 1101 1114
74. WuLChanKLRalfCBernsteinDAGarciaPL 2005 The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J 24 2679 2687
75. PrakashRSatoryDDrayEPapushaASchellerJ 2009 Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev 23 67 79
76. DahmannCDiffleyJFNasmythKA 1995 S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol 5 1257 1269
77. NguyenVQCoCLiJJ 2001 Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411 1068 1073
78. SawarynskiKENajorNAKepselACBrushGS 2009 Sic1-induced DNA rereplication during meiosis. Proc Natl Acad Sci U S A 106 232 237
79. TothARabitschKPGalovaMSchleifferABuonomoSB 2000 Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell 103 1155 1168
80. RabitschKPPetronczkiMJaverzatJPGenierSChwallaB 2003 Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4 535 548
81. LeeBHAmonA 2003 Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 300 482 486
82. MatosJLippJJBogdanovaAGuillotSOkazE 2008 Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135 662 678
83. SzostakJWOrr-WeaverTLRothsteinRJStahlFW 1983 The double-strand-break repair model for recombination. Cell 33 25 35
84. LipkinSMMoensPBWangVLenziMShanmugarajahD 2002 Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet 31 385 390
85. MarconEMoensP 2003 MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165 2283 2287
86. HoffmannERBortsRH 2004 Meiotic recombination intermediates and mismatch repair proteins. Cytogenet Genome Res 107 232 248
87. WangTFKlecknerNHunterN 1999 Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci U S A 96 13914 13919
88. ZakharyevichKMaYTangSHwangPYBoiteuxS 2010 Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol Cell 40 1001 1015
89. TrowbridgeKMcKimKBrillSJSekelskyJ 2007 Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176 1993 2001
90. HigginsJDBucklingEFFranklinFCJonesGH 2008 Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54 152 162
91. HollowayJKBoothJEdelmannWMcGowanCHCohenPE 2008 MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4 e1000186 doi:10.1371/journal.pgen.1000186
92. GaillardPHNoguchiEShanahanPRussellP 2003 The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell 12 747 759
93. EhmsenKTHeyerWD 2008 Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res 36 2182 2195
94. AshtonTMMankouriHWHeidenblutAMcHughPJHicksonID 2011 Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol Cell Biol in press
95. HollingsworthNMBrillSJ 2004 The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev 18 117 125
96. KaneSMRothR 1974 Carbohydrate metabolism during ascospore development in yeast. J Bacteriol 118 8 14
97. ChaRSWeinerBMKeeneySDekkerJKlecknerN 2000 Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev 14 493 503
98. GuthrieCFinkGR 1991 Guide to yeast genetics and molecular biology. San Diego Academic Press
99. GoyonCLichtenM 1993 Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol 13 373 382
100. BishopDK 1994 RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79 1081 1092
101. AllersTLichtenM 2000 A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res 28 e6
102. FoianiMMariniFGambaDLucchiniGPlevaniP 1994 The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol Cell Biol 14 923 933
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Structural and Functional Differences in the Long Non-Coding RNA in Mouse and Human
- Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue
- A −436C>A Polymorphism in the Human Gene Promoter Associated with Severe Childhood Malaria
- A Decline in p38 MAPK Signaling Underlies Immunosenescence in
- The Operon Balances the Requirements for Vegetative Stability and Conjugative Transfer of Plasmid R388
- Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in
- Ixr1 Is Required for the Expression of the Ribonucleotide Reductase Rnr1 and Maintenance of dNTP Pools
- Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
- A Deficiency of Ceramide Biosynthesis Causes Cerebellar Purkinje Cell Neurodegeneration and Lipofuscin Accumulation
- A Latent Pro-Survival Function for the Mir-290-295 Cluster in Mouse Embryonic Stem Cells
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time
- Prion Formation and Polyglutamine Aggregation Are Controlled by Two Classes of Genes
- Integrated Genome-Scale Prediction of Detrimental Mutations in Transcription Networks
- Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation
- A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
- A Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
- A Missense Mutation in Causes a Major QTL Effect on Ear Size in Pigs
- A Flurry of Folding Problems: An Interview with Susan Lindquist
- Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
- A Nervous Origin for Fish Stripes
- The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
- The Telomerase Subunit Est3 Binds Telomeres in a Cell Cycle– and Est1–Dependent Manner and Interacts Directly with Est1
- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



