-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Structural and Functional Differences in the Long Non-Coding RNA
in Mouse and Human
Long non-coding RNAs regulate various biological processes such as dosage
compensation, imprinting, and chromatin organization. HOTAIR, a paradigm of this
new class of RNAs, is localized within the human HOXC gene
cluster and was shown, in human cells, to regulate HOXD genes
in trans via the recruitment of Polycomb Repressive Complex
2 (PRC2), followed by the trimethylation of lysine 27 of histone H3. We looked
for the presence of Hotair in mice to assess whether this
in trans mechanism was conserved, in particular at the
developmental stages, when Hoxd genes must be tightly
regulated. We show that the cognate mouse Hotair is poorly
conserved in sequence; and its absence, along with the deletion of the
HoxC cluster, has surprisingly little effect in
vivo, neither on the expression pattern or transcription
efficiency, nor on the amount of K27me3 coverage of different
Hoxd target genes. We conclude that Hotair
may have rapidly evolved within mammals and acquired a functional importance in
humans that is not easily revealed in mice. Alternatively, redundant or
compensatory mechanisms may mask its function when studied under physiological
conditions.
Published in the journal: . PLoS Genet 7(5): e32767. doi:10.1371/journal.pgen.1002071
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002071Summary
Long non-coding RNAs regulate various biological processes such as dosage
compensation, imprinting, and chromatin organization. HOTAIR, a paradigm of this
new class of RNAs, is localized within the human HOXC gene
cluster and was shown, in human cells, to regulate HOXD genes
in trans via the recruitment of Polycomb Repressive Complex
2 (PRC2), followed by the trimethylation of lysine 27 of histone H3. We looked
for the presence of Hotair in mice to assess whether this
in trans mechanism was conserved, in particular at the
developmental stages, when Hoxd genes must be tightly
regulated. We show that the cognate mouse Hotair is poorly
conserved in sequence; and its absence, along with the deletion of the
HoxC cluster, has surprisingly little effect in
vivo, neither on the expression pattern or transcription
efficiency, nor on the amount of K27me3 coverage of different
Hoxd target genes. We conclude that Hotair
may have rapidly evolved within mammals and acquired a functional importance in
humans that is not easily revealed in mice. Alternatively, redundant or
compensatory mechanisms may mask its function when studied under physiological
conditions.Introduction
Genomes contain a large number of RNAs, which do not encode any protein [1]–[5]. While some of these non-coding RNAs such as XIST, TSIX and AIR associate with epigenetic modifying complexes [6]–[11], the functions of others remain poorly understood. Many of the predicted long non coding RNAs (lincRNAs) are thought to be spliced and polyadenylated, thus resembling protein coding RNAs [12]–[15] and have been proposed to impact on gene regulation [16], [17].
Recent studies have shown that distinct lincRNAs are involved in diverse biological processes such as dosage compensation, imprinting or cancer metastasis [10], [18]–[20]. More specifically, they may function at the interface between DNA and its epigenetic regulation by targeting remodeling complexes to their target sites [21]. HOTAIR, one such lincRNA located within the human HOXC cluster, regulates HOXD cluster genes in trans via the recruitment of PRC2, a silencing complex responsible for the deposition of trimethyl groups on lysine 27 of histone H3 (H3K27me3) [10]. Knock-down of HOTAIR in human fibroblasts induced gain of expression of different members of the HOX family, associated with a loss of K27me3 decorating part of the HOXD locus in these cells [10].
In addition, HOTAIR has been shown to co-immunoprecipitate with members of the PRC2 complex such as SUZ12 and EZH2, but not with the putative PRC1 member YY1, suggesting a primary role in the initiation of silencing, rather than in its maintenance [6], [10], [21]. Subsequent studies have suggested that distinct sub-domains of HOTAIR are essential for the binding of either EZH2, or of LSD1 and that HOTAIR functions as a bridge to bring both complexes together. In the absence of these two binding domains, the epigenetic functionalities of this lincRNA are indeed completely abrogated [21].
Altogether, these results indicate that human HOTAIR is an important regulator of the HOX epigenetic landscape in skin fibroblasts. Given both the importance of this lincRNA in adult tissues and the critical dynamics of H3K27 trimethylation for the early control of Hoxd gene activation [22], we investigated its role in developing mouse embryos. Here, we describe the mouse Hotair cognate lincRNA and show that its complete depletion in vivo has no severe effect upon Hoxd gene activation, neither during early trunk development, nor in the course of limb morphogenesis, two sites where HOTAIR was seen expressed.
Results
The mouse Hotair lincRNA
We first looked for the presence of Hotair in the mouse genome. Because the human RNA locates between HOXC12 and HOXC11, i.e. within a region of very high micro-synteny amongst all vertebrates, we performed a pair-wise sequence alignment with the cognate mouse DNA segment, using the rVISTA software [23]. Alignment of the entire mouse Hoxc11 to Hoxc12 region with the human genome revealed various domains of strong sequence homology (Figure 1A). Expectedly, the Hoxc11 and Hoxc12 exons are highly conserved, with more than 95% homology between the mouse and human sequences. However, the intergenic region between Hoxc11 and Hoxc12 showed more variability, with some peaks of conservation, but also segments close to random variability, as previously described [15], [24].
Sequence alignment revealed that the human HOTAIR lincRNA most likely has a mouse ortholog RNA, referred to as AC160979. This EST (mHotair from now onwards) is indeed located at the expected micro-syntenic position and exhibits partial homology with human HOTAIR. mHotair derives from the Vega Protein Coding Annotation and corresponds to the UCSC gene based on RefSeq AK035706 transcript. However, and even though mHotair and HOTAIR are clearly cognate transcripts, several important differences were scored. First, while the RefSeq annotation of HOTAIR indicates six exons, mHotair derives from two exons only. The second half of the first exon of mHotair seems to match exon 4 of HOTAIR, whereas the second exon clearly matches exon 6 of HOTAIR (Figure 1A). Blasts of the first three human exons against the mouse Hoxc11 to Hoxc12 intergenic region did not give any significant homology.
Secondly, the level of sequence similarity between different exons is highly variable. The first exon of mHotair, which is 234 base pairs long, shows significant conservation (>80% over >100 bp) with the human sequence. However, the second exon, which is 1770 bases long, is poorly related to the human sequence and shows conservation higher than 75% only in a sub-domain of ca. 400 bp. Altogether, this large exon, which contains the LSD1 binding region of HOTAIR, is rather poorly conserved in its mouse counterpart, ranging from 50 to 70% homology. In addition, human HOTAIR contains several binding sites for the SET domain containing PRC2 component EZH2, responsible for the histone H3 methyltransferase activity (HMTase) of this enzyme, which are absent from mHotair. Although it is unclear as to whether the primary nucleotide sequence or the tertiary RNA structure is involved in binding EZH2, it nevertheless suggests that the function of this RNA in mice is not identical to that described for its human cognate. Transcriptome analyses by deep sequencing confirmed that mHotair was most likely encoded by two exons only, instead of six in humans (see below).
Expression of mHotair
Hox genes are clustered at four different genomic loci (HoxA, B, C and D) and are crucial in organizing the metazoan body plans. They encode transcription factors, which work in various combinations to allocate morphogenetic identities to groups of cells. To properly coordinate their transcription, these contiguous genes are activated following a collinear regulatory strategy, whereby genes positioned at the 3′ end of the cluster are activated earlier in time and more anteriorly, whereas more 5′ located genes are activated later in time and more posteriorly [25]. This sequential activation in time and space thus follows the physical positions of genes along their respective clusters. This property, which may in part depend upon chromatin modifications [22] also applies either to transgenes, when introduced into the gene clusters, or for non coding intergenic transcripts, regardless of their sense of transcription. These non-coding transcripts associated with Hox genes were proposed to regulate the collinear opening and maintenance of the epigenetic status of the cluster [5].
We looked at the expression of mHotair by whole mount in situ hybridization (WISH) on developing mouse embryos at embryonic day 11.5, 12.5 and 13.5, and compared with the expression of Hoxc11, the gene located immediately 3′ from the mHotair promoter. The mHotair probe was selected within the region showing the highest conservation with the human ortholog (Figure 1A), i.e. the middle half of the second exon, such as to compare as accurately as possible with previously published data where the distantly related human HOTAIR sequence was used as a probe for WISH on mouse embryos [10]. Experiments using sense and antisense probes confirmed that mHotair is solely transcribed from the opposing canonical Hox DNA strand, as is human HOTAIR.
As expected from its position within the ‘posterior’ part of the HoxC cluster, mHotair expression was scored in posterior and distal sites. It was readily detected in E11.5 embryos with marked staining in the posterior part of the hindlimbs, in the genital bud and in the tail. At E12.5, the expression pattern was mainly restricted to the posterior aspect of the intermediate part of the hindlimbs, as well as to the genital bud, whereas it became barely detectable at E13.5. In parallel experiments, Hoxc11 transcripts showed a comparable distribution, yet with stronger signals at all three stages (Figure 1C and 1D), in agreement with previously published data. Given the strong similarities of expression patterns between mHotair and its closest 3′ neighbor Hoxc11, we concluded that mHotair is expectedly regulated in coordination with other posterior Hoxc genes. mHotair expression, however, was quite distinct from that reported in similar staged mouse embryos when using a human HOTAIR probe [10].
Regulation of Hoxd genes in trans
Human HOTAIR was shown to act in trans by tethering Polycomb Repressive Complex 2 (PRC2) to a subset of its targets, amongst which the HOXD locus [10], [21]. HOTAIR thus acts as a scaffold for the repression of a number of genes in this region via the recruitment of these silencing proteins, with a particular impact on the expression levels of human HOXD8, HOXD9, HOXD10, HOXD11 and HOXD13, while having no impact neither on the HOXA, nor on the HOXB and HOXC clusters [10]. To investigate whether this mechanism was conserved throughout mammals, we looked at the expression of these potential target genes in the absence of mHotair. We used a full deletion of the HoxC cluster whereby all Hoxc genes and intergenic transcripts are missing (Figure 2A) [26]. We isolated HoxCDel/Del embryos at embryonic day 13.5 (E13.5), derived from a cross between heterozygous animals, and dissected them into four distinct pieces; the forebody, hindbody, forelimbs and hindlimbs. We performed quantitative RT-PCR analyses on these various samples using wild type and heterozygous littermates as controls for homozygous mutant samples.
As expected, mHotair was detected neither in HoxCDel/Del mutant embryos, nor in forebody samples of all three genotypes, which we used as negative controls. In the three other samples, mHotair transcripts were scored, though at very low levels. However, no difference was noted in the expression levels of the presumptive mHotair targets Hoxd8, Hoxd9, Hoxd10, Hoxd11 or Hoxd13 (Figure 2B). The expression level of Hoxd12 remained unchanged too, as well as those of Evx2 and Lunapark, two neighboring genes largely co-regulated with Hoxd genes [27].
A change in the expression of different Hoxd genes could nevertheless remain unnoticed, should a spatial shift in their transcript patterns occur, rather than variations in their RNA steady state levels. We thus performed in situ hybridization on mutant animals to reveal the distribution of Hoxd10 transcripts, which was reported as the main HOXD target for a HOTAIR-mediated de-repression in human cells. At all three stages examined (E11.5, E12.5, E13.5), Hoxd10 transcripts showed wild type patterns in mutant animals (Figure 2C and 2D). Taken together, these observations indicate that mHotair has little or no detectable regulatory effect in trans over Hoxd cluster genes in mice, at least in these conditions.
Tri-methylation of H3K27 at the HoxD locus
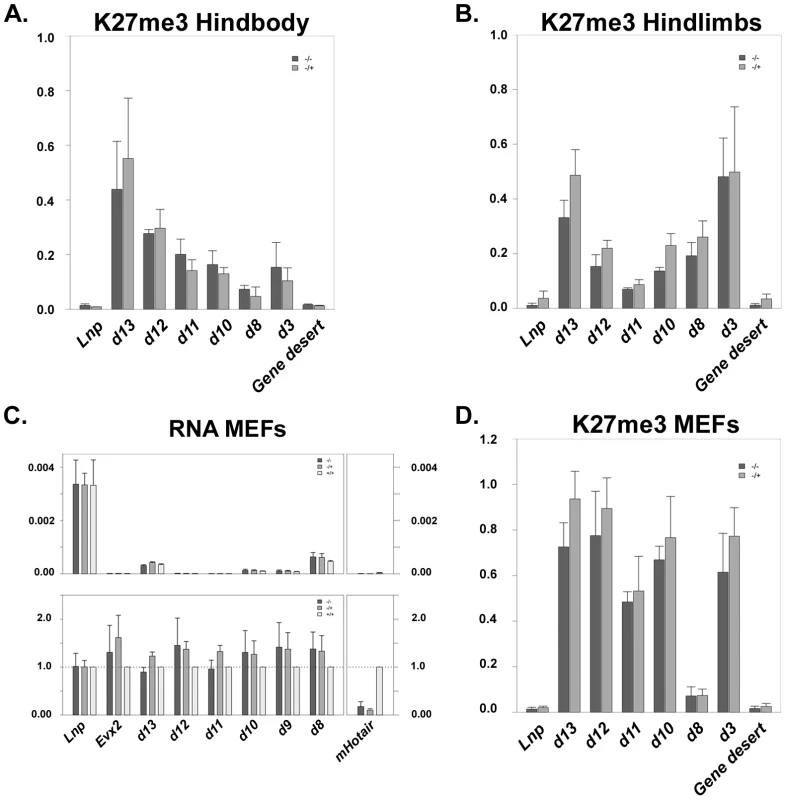
HOTAIR was reported to regulate several HOXD genes by tethering PcG proteins (the PRC2 complex) to the posterior HOXD cluster [10], [21]. Knock-down of HOTAIR in human fibroblasts indeed showed a decreased trimethylation of lysine 27 on histone H3, in particular at the HOXD locus, with the strongest effect observed over the region between HOXD3 and HOXD8. Since a loss of H3K27me3 may not necessarily be translated into a detectable increase in Hoxd gene transcription in mouse embryos, we investigated the chromatin status of the HoxD locus in mutant animals. We used chromatin immunoprecipitation (ChIP) on E13.5 embryos, a stage at which mHotair is transcribed (see below), followed by quantitative RT-PCR to quantify the enrichment of H3K27me3 over the gene cluster. Here again, the parallel loss of both HoxC and mHotair alleles did not significantly alter the amount of K27me3 covering this presumptive target locus (Figure 3A and 3B). From this set of experiments, we concluded that although human HOTAIR might be essential for the recruitment of PRC2 and subsequent tri-methylation of H3K27 in cultured fibroblast, its role in the regulation of mouse Hoxd genes in embryo seems to be minor, if any, at least at this developmental stage.
Function of mouse Hotair in MEFs
As the reported effects of human HOTAIR were not observed in the absence of the mouse counterpart in vivo, we derived mouse embryonic fibroblast (MEFs) from E13.5 embryos, either heterozygous or homozygous mutant for the HoxC cluster, to try and better match the conditions wherein HOTAIR's functions had been originally elucidated. We quantified both the amount of transcription of different Hoxd genes and the enrichment of H3K27me3 at this locus. Results obtained with MEFs heterozygous for the deletion of the HoxC cluster were indistinguishable from those obtained from MEFs lacking both copies of HoxC and mHotair. Analyses of both lines of MEFs gave similar amounts of Hoxd gene transcripts and no significant variations was scored in the enrichments of H3K27me3 marks, indicating that the presence of mHotair is not critical for the regulation of Hoxd genes in this context (Figure 3C and 3D).
Comparative transcriptome analyses with and without Hotair in vivo
To assess the global impact of mHotair on the gene regulation, we looked at the transcriptomes of those tissues where mHotair was clearly transcribed at E13.5 in our whole mount in situ hybridization, namely the hindbody, the hindlimbs and the genital bud. Embryonic tissues were micro-dissected and total messenger RNA isolated from both control and HoxC mutant animals and sequenced using an Illumina Genome Analyzer. Nearly 15 million high quality single reads were mapped on the mouse mm9 genome, using Tophat [28] and visualized using the integrative genome viewer [29]. In this way, we could confirm that, as annotated in RefSeq, mHotair is a two-exons transcript initiating from the opposite strand of the canonical HoxC genes, at least in this context. No additional 5′ located exons were used, unlike in human.
We compared mutant and wild type transcript profiles genome wide and observed significant changes. These modifications, which may reflect direct or indirect targets either of mHotair, or of Hoxc gene products, were either up - or down regulated and broadly distributed over all gene ontology categories. Hox genes were included, along with housekeeping genes and genes from unrelated structures and functions (Figure 4A and 4B). We looked at the HoxD cluster and the strongest variation in steady-state level of transcripts was observed for Hoxd8, Hoxd9, Hoxd10 and Hoxd11, as previously reported for HOTAIR in human cells, though the amplitudes were significantly lower (Figure 4A). While these results appeared at first to somehow correlate with the reported effect of human HOTAIR on this gene cluster, Rinn et al. [10] observed a substantial increase in expression of these genes by down-regulating HOTAIR by a factor of two thirds, whereas we detected a maximum of three-fold difference in the complete absence of this lincRNA.
To assess whether these differences could be partly explained by the relatively low expression of mHotair at this particular stage (E13.5) or a dilution effect, we isolated RNA from the same set of tissues, i.e. hindbody, hindlimbs and genital bud, from E11.5 embryos and quantified the RNAs by reverse transcription PCR. Differences in absolute expression levels of the different Hox genes analyzed were comparable to those obtained in our RNA-seq experiment at E13.5, suggesting that the observed effects of mHotair and HoxC deletions on gene regulation are reproducible, at least between these two developmental stages (Figure 4D).
The discrepancies between our results and those reported previously may reflect a dilution effect due to only few cells expressing mHotair in our samples. However, we also observed a slight up-regulation of Hoxd1, Hoxd3 and Hoxd4 and, surprisingly, our mutants exhibited no change in Hoxd13 transcripts (Figure 4B and 4D), neither in downstream-located non coding RNAs, a region significantly up-regulated in previous work. Also, we observed a similar de-repression of Hox genes belonging to other clusters, with Hoxa7 and Hoxb9 showing comparable up-regulations (two fold, Figure 4B), unlike previously reported. Of note, a substantial increase of transcripts matching the second exon of Hoxc4, i.e. the most 3′ part remaining after the deletion of the HoxC gene cluster. This unexpected burst likely reflects the presence of ‘posterior-acting’ regulation, which are now re-routed towards this sequence, in the absence of the intervening HoxC cluster, as describe in similar contexts [30]. Taken together, while these observations support a general, though rather moderate, effect of removing the HoxC gene cluster, including mHotair, in the posterior part of the developing embryo, transcriptome analyses confirmed the difficulty to attribute to mHotair the same regulatory capacities during embryonic development, than those associated to its human counterpart in cultured fibroblasts.
Even though the structure of mHotair showed substantial differences with its human ortholog, we looked for additional evidence of a potential role as a molecular scaffold to bridge PcG proteins to their target sites. We assessed whether or not the group of genes that displayed a clear transcriptional de-repression in HoxC mutant animals was enriched in genes known to recruit PRC2 in ES cells, i.e. in conditions where Hox clusters are covered by H3K27me3. We applied a stringent cut-off with a significance window of 1 kb and obtained 263 genes up-regulated in the mutant sample, whereas 105 genes were down-regulated. We looked at which fraction of these genes represented known PcG targets, as defined by binding to SUZ12 [31]. Of the 263 genes defined as up-regulated in the HoxC null mice, only 35 (13%) had been determined as being bound by SUZ12 in ES cells (Figure 4C). Likewise, out of a total of 105 genes down-regulated, only 16 were bound by SUZ12 (15%), a figure that was down to 8.6% after Hoxc genes were removed from the list (since they are deleted in the mutant) (Figure 4C).
Discussion
The importance of long non-coding RNAs (lincRNAs) for gene regulation has been recently emphasized in many different contexts. One of the paradigms of this novel class of transcripts is the human HOTAIR RNA, which is encoded from within the HOXC gene cluster and acts in trans to regulate HOXD target genes via the recruitment of PRC2 and further tri-methylation of H3K27 [10]. Interestingly, the mouse counterpart shows little sequence conservation with HOTAIR. While such lincRNAs are known to be moderately conserved in sequence between different species, sequence alignment between the mouse and human HoxC clusters reveals that the DNA fragments included in both HOTAIR and mHotair are amongst the less conserved within the Hoxc12 to Hoxc11 DNA interval, as if they would correspond to the less constrained sequences in terms of evolution. Yet some intron-exon borders are conserved, as well as the direction of transcription, which suggests that the mouse HoxC cluster does contain a genuine cognate HOTAIR RNA.
Interestingly, the first three exons of HOTAIR seem to be absent from mHotair, which appears to contain two exons only, a first exon related to the fourth exon of HOTAIR, followed by a larger exon 2, related to the large sixth exon of HOTAIR. Even though an increase in the number of sequence reads may reveal the presence of either additional, poorly spliced 5′ located exons or alternative start sites, mHotair is thus quite distinct in structure from its human cognate. Such a divergence may underlie important differences in function since the first three exons of HOTAIR (absent from mHotair) contain binding sites for EZH2. Likewise, the LSD1 binding sequences, localized at the 3′ extremity of human HOTAIR, is part of the least conserved DNA sequence within mHotair exon 2 (below 70% conservation). Altogether, based on DNA sequence analyses, it is difficult to reconcile the structure of mHotair with the potential function previously attributed to HOTAIR, even though binding of both EZH2 and LSD1 proteins may mostly rely on tri-dimensional structures rather than upon specific RNA sequences.
This conclusion was re-enforced by the expression analyses during mouse development, which revealed patterns different from those previously reported when a human probe was used to assess the presence of mouse transcripts [10]. As expected, mHotair is expressed very much like the neighboring Hoxc11 gene, i.e. in parts of the proximal hindlimbs, in the posterior part of the body and in the emerging presumptive external genital organs. We think that this discrepancy in expression patterns can be explained by the very low sequence conservation between the human RNA antisense probe and the mouse target RNA. Coordinated expression of RNA or transgenes introduced within Hox gene clusters has been reported in several instances [32] and illustrates the strong global regulation that controls these groups of genes. Non-Hox promoters located in - or introduced into - these loci tend to adopt the shared expression specificities and thus behave like their nearest neighbors.
The genetic ablation of mHotair, under physiological condition, confirmed the apparent difference between the functions of this lincRNA in mice and humans. Firstly, Hoxd genes expression remained moderately affected in most tissues analyzed, as assessed by quantitative PCR, in situ hybridization and RNA-seq, in particular in those tissues of the developing body where steady-state levels of mHotair were the highest. Secondly, the group of genes that was either up - or down-regulated in the absence of mHotair, as scored by transcriptome analyses, did not particularly overlap with known PcG targets as described in ES cells, nor was it enriched in any of the GO terms. Thirdly, no significant difference was scored in the amount of H3K27me3 decorating the HoxD locus, neither by using embryonic tissues, nor when assessing MEFs derived from homozygous null fetuses. This latter point may reflect the fact that mouse Hotair lacks most of the cognate human 5′ RNA fragment, which was shown to be necessary for the binding of EZH2 [21]. Although we cannot exclude that the deletion of mHotair may have induced a subtle effect upon Hox gene expression, these genes would need to be affected much more severely for a phenotypic outcome to be observed, as animals heterozygous for a deletion of the entire HoxD cluster are virtually of wild type phenotype [33]. Therefore, only a robust impact of mHotair on Hoxd genes regulation would make this lincRNA a candidate regulator of these developmental genes in mice, at least at the time when critical changes in chromatin status are observed [22].
How can we explain this unexpected difference in the functional importance of cognate non-coding RNAs in two mammalian species where both the structure and function of Hox genes appear to be highly conserved? First, our mutant configuration not only lacks mHotair, but also all Hoxc genes as well as the potential mouse ortholog of FRIGIDAIR, another lincRNA located within the HoxC cluster [3] and whose deletion could counterbalance the effect of removing mHotair. However, our transcriptome dataset indicates that mFrigidair, if present in the mouse genome, is not transcribed at detectable level in our posterior body sample, unlike mHotair, which makes this possibility unlikely. Also, there is no evidence supporting a strong effect of HOXC proteins over Hoxd genes regulation. If any, this effect would need to exactly compensate for a potential effect of mHotair such that the situation in the mutant samples would look like wild type.
Secondly, the function of mHotair could be restricted to a limited number of cells within the expression domains of Hox genes, in which case our selection of a rather large piece of tissue would reduce the sensitivity of our functional assays via a dilution effect, which would not occur in cultured fibroblasts. While this is a serious possibility, it would imply that only a small subset of Hox positive cells would be ‘exposed’ to mHotair, questioning its general importance in the recruitment of PRC2 during development. Alternatively, human HOTAIR may be required for HOXD gene regulation at later stages and in different contexts, rather than in the early recruitment of PRC2 over the HOXD cluster. As for all other posterior Hox genes, Hoxc11 and Hoxc12 expression is restricted towards the posterior part of the developing body in early mouse embryos. It is nonetheless conceivable that mHotair be transcribed subsequently, in a tissue or organ where it may have a functional importance, such as in foreskin fibroblasts where its function was originally described. This would imply that the recruitment of PRC2 and subsequent tri-methylation of H3K27 over Hoxd cluster genes would be achieved by different mechanisms in different contexts or, at least, by using various components to recruit PRC2.
Another possibility is that mHotair and HOTAIR may have importantly diverged and no longer share any functional similarity. Non-coding RNAs are generally rather poorly conserved in sequences amongst different species and this possibility may not be overtly surprising. The fact that RNA sequences present in HOTAIR and associated with the binding of either EZH2 or LSD1 do not seem to be present in mHotair supports this view. However, this would be difficult to reconcile with HOTAIR being a key player in the regulation of HOX genes in human, since this gene family has been the paradigm of the structural and functional conservation of genetic circuitries in vertebrates, not talking about mammals.
Alternatively, mHotair may have a genuine function in organizing the chromatin landscape over Hox genes, but its deletion in vivo could activate redundant or compensatory pathways still allowing proper PcG-mediated silencing to occur, a mechanism absent from cultured human fibroblasts. Silencing of Hox genes during early development must be tightly achieved, to prevent precocious activation leading to mis-identification of structures. Yet this repression will have to be easily reversed subsequently, in the many different contexts where these genes will be activated. Whether or not this epigenetic versatility would be best implemented by redundant silencing mechanisms or by a preponderant strategy relying upon PRC2 dependent tri-methylation of H3K27 is difficult to evaluate. In both cases, mHotair may be recruited to the HoxD cluster to help this silencing to be established, in those regions where it is expressed. However, our results argue against this mechanism being a fundamental process in Hox gene silencing, in particular as these gene clusters are tightly covered by PcG proteins and decorated by tri-methylated H3K27 in all embryonic contexts analyzed so far where these genes must be repressed, i.e. mostly in tissues where mHotair transcripts were below our detection level.
Materials and Methods
Ethics statement
All experiments involving living animals were authorized by - and carried out following - the swiss legal framework.
Mutant mice
Mice carrying a deletion of the HoxC gene cluster were published previously [26]. They were purchased from the RIKEN BioResource Center (BRC), in Japan. Heterozygous mice were crossed to obtain wild type, heterozygous and homozygous mutant embryos. Genotyping was performed on individual yolk sacs with the following primers:
-
WTforward: CGCTCTGGGAGTGGTCTTCAGAAG;
-
WTreverse: GTGCTACGATCTGTTATGTATGTG;
-
delCforward: GATGGAGTTTCCCCACACTGAGTG;
-
delCreverse: CGTGAGGAAGAGTTCTTGCAGCTC.
Sequence comparison
Sequences alignments between the mouse and human HoxC loci were performed using the pairwise Lagan analysis from the Vista website [23].
In situ hybridization
Mid-day of vaginal plug was considered as E0.5. Embryos were dissected in PBS and fixed overnight at 4° in 4% PFA. Whole mount in situ hybridization was performed according to standard protocols. The decreasing signal intensity observed for the oldest processed embryos is partially due to the somewhat lower permeability of the probe, along with tissue differentiation. Mutant, heterozygous and wild type animals were processed simultaneously to ensure identical conditions. The Hoxd10 probe was as previously described [34]. The murine Hotair and Hoxc11 probes were PCR-subcloned into pGEM-T Easy vector (Promega), sequence verified, linearized and in vitro transcribed with either SalI-T7 (antisense) or NcoI-SP6 (sense), using the DIG RNA Labeling Mix (Roche).
-
mHotair forward: GAGCCAGAGCTGAAGGTATG
-
mHotair reverse: AAGACACGCACGGAGAAAGG
-
Hoxc11 forward: CCCCGCACCCGCAAGAAGC
-
Hoxc11 reverse: GTCCAGTTTTCCACCCGCGG
Chromatin immunoprecipitation
Chromatin immunoprecipitation followed by quantitative reverse transcription was performed as previously described [35]. Briefly, cells or tissues were fixed for 15 minutes in 1% formaldehyde, washed three times in cold PBS and stored at −80° before being processed using polyclonal anti-H3K27me3 antibody (Millipore, 17-622).
Cell culture
Mouse embryonic fibroblasts were derived from heterozygous crosses of E13.5 embryos using standard protocols. Cells were cultured in MEF culture conditions in DMEM supplemented with 10% FBS. Isolated lines were first genotyped using tissues from the embryos and subsequently confirmed with DNA extraction procedures. Passage No 4 MEFs were used for further experiments.
Expression analysis
The posterior parts of embryos including the hindlimbs, the genital bud and the developing tail at day 11.5 and the forebody, hindbody, forelimbs and hindlimbs at day 13.5, were dissected and stored in RNAlater (Qiagen) until genotyped. Cells or tissues were first disrupted and homogenized using a Polytron (kinematic) before RNA was extracted using the RNeasy Microkit (Qiagen, 74034), followed by qRT-PCR with SYBR Green. Mean values derive from two (MEFs) or four (tissues) biological replicates, processed in triplicates and normalized to a housekeeping gene (Rps9).
RNA–seq and downstream analysis
The most posterior parts of fetuses at day E13.5 were dissected, including the hindlimbs, the genital bud and the developing tail, and total RNA was extracted as for expression analysis. Wild type and mutant samples were deep sequenced using the Illumina Genome Analyzer. Reads were mapped onto the mouse mm9 genome using Tophat and visualized with the integrative genome viewer (mean value of 25 bp windows). Mis-regulated genes were identified using a 200 bp binning approach across the genome. Significance was measure by the presence of probes showing a difference between wt and mutant profiles greater than 6 over at least 5 probes (1 kb).
Zdroje
1. BirneyEStamatoyannopoulosJADuttaAGuigoRGingerasTR
2007
Identification and analysis of functional elements in 1%
of the human genome by the ENCODE pilot project.
Nature
447
799
816
2. ChengJKapranovPDrenkowJDikeSBrubakerS
2005
Transcriptional maps of 10 human chromosomes at 5-nucleotide
resolution.
Science
308
1149
1154
3. GuttmanMAmitIGarberMFrenchCLinMF
2009
Chromatin signature reveals over a thousand highly conserved
large non-coding RNAs in mammals.
Nature
458
223
227
4. MattickJS
2009
The genetic signatures of noncoding RNAs.
PLoS Genet
5
e1000459
doi:10.1371/journal.pgen.1000459
5. SessaLBreilingALavorgnaGSilvestriLCasariG
2007
Noncoding RNA synthesis and loss of Polycomb group repression
accompanies the colinear activation of the human HOXA
cluster.
RNA
13
223
239
6. KanekoSLiGSonJXuCFMargueronR
2010
Phosphorylation of the PRC2 component Ezh2 is cell
cycle-regulated and up-regulates its binding to ncRNA.
Genes Dev
24
2615
2620
7. NaganoTMitchellJASanzLAPaulerFMFerguson-SmithAC
2008
The Air noncoding RNA epigenetically silences transcription by
targeting G9a to chromatin.
Science
322
1717
1720
8. PandeyRRMondalTMohammadFEnrothSRedrupL
2008
Kcnq1ot1 antisense noncoding RNA mediates lineage-specific
transcriptional silencing through chromatin-level
regulation.
Mol Cell
32
232
246
9. PlathKFangJMlynarczyk-EvansSKCaoRWorringerKA
2003
Role of histone H3 lysine 27 methylation in X
inactivation.
Science
300
131
135
10. RinnJLKerteszMWangJKSquazzoSLXuX
2007
Functional demarcation of active and silent chromatin domains in
human HOX loci by noncoding RNAs.
Cell
129
1311
1323
11. ZhaoJSunBKErwinJASongJJLeeJT
2008
Polycomb proteins targeted by a short repeat RNA to the mouse X
chromosome.
Science
322
750
756
12. RinnJLEuskirchenGBertonePMartoneRLuscombeNM
2003
The transcriptional activity of human Chromosome
22.
Genes Dev
17
529
540
13. BertonePStolcVRoyceTERozowskyJSUrbanAE
2004
Global identification of human transcribed sequences with genome
tiling arrays.
Science
306
2242
2246
14. KapranovPDrenkowJChengJLongJHeltG
2005
Examples of the complex architecture of the human transcriptome
revealed by RACE and high-density tiling arrays.
Genome Res
15
987
997
15. CarninciPKasukawaTKatayamaSGoughJFrithMC
2005
The transcriptional landscape of the mammalian
genome.
Science
309
1559
1563
16. BernsteinEAllisCD
2005
RNA meets chromatin.
Genes Dev
19
1635
1655
17. HuttenhoferASchattnerPPolacekN
2005
Non-coding RNAs: hope or hype?
Trends Genet
21
289
297
18. GuptaRAShahNWangKCKimJHorlingsHM
2010
Long non-coding RNA HOTAIR reprograms chromatin state to promote
cancer metastasis.
Nature
464
1071
1076
19. KhalilAMGuttmanMHuarteMGarberMRajA
2009
Many human large intergenic noncoding RNAs associate with
chromatin-modifying complexes and affect gene expression.
Proc Natl Acad Sci U S A
106
11667
11672
20. PontingCPOliverPLReikW
2009
Evolution and functions of long noncoding RNAs.
Cell
136
629
641
21. TsaiMCManorOWanYMosammaparastNWangJK
2010
Long noncoding RNA as modular scaffold of histone modification
complexes.
Science
329
689
693
22. SoshnikovaNDubouleD
2009
Epigenetic regulation of vertebrate Hox genes: a dynamic
equilibrium.
Epigenetics
4
537
540
23. LootsGGOvcharenkoI
2004
rVISTA 2.0: evolutionary analysis of transcription factor binding
sites.
Nucleic Acids Res
32
W217
221
24. EngstromPGSuzukiHNinomiyaNAkalinASessaL
2006
Complex Loci in human and mouse genomes.
PLoS Genet
2
e47
doi:10.1371/journal.pgen.0020047
25. KmitaMDubouleD
2003
Organizing axes in time and space; 25 years of colinear
tinkering.
Science
301
331
333
26. SuemoriHNoguchiS
2000
Hox C cluster genes are dispensable for overall body plan of
mouse embryonic development.
Dev Biol
220
333
342
27. SpitzFGonzalezFDubouleD
2003
A global control region defines a chromosomal regulatory
landscape containing the HoxD cluster.
Cell
113
405
417
28. TrapnellCPachterLSalzbergSL
2009
TopHat: discovering splice junctions with
RNA-Seq.
Bioinformatics
25
1105
1111
29. RobinsonJTThorvaldsdottirHWincklerWGuttmanMLanderES
2011
Integrative genomics viewer.
Nat Biotechnol
29
24
26
30. KmitaMTarchiniBZakanyJLoganMTabinCJ
2005
Early developmental arrest of mammalian limbs lacking HoxA/HoxD
gene function.
Nature
435
1113
1116
31. CreyghtonMPMarkoulakiSLevineSSHannaJLodatoMA
2008
H2AZ is enriched at polycomb complex target genes in ES cells and
is necessary for lineage commitment.
Cell
135
649
661
32. HeraultYBeckersJGerardMDubouleD
1999
Hox gene expression in limbs: colinearity by opposite regulatory
controls.
Dev Biol
208
157
165
33. SpitzFHerkenneCMorrisMADubouleD
2005
Inversion-induced disruption of the Hoxd cluster leads to the
partition of regulatory landscapes.
Nat Genet
37
889
893
34. GerardMChenJYGronemeyerHChambonPDubouleD
1996
In vivo targeted mutagenesis of a regulatory element required for
positioning the Hoxd-11 and Hoxd-10 expression boundaries.
Genes Dev
10
2326
2334
35. LeeTIJohnstoneSEYoungRA
2006
Chromatin immunoprecipitation and microarray-based analysis of
protein location.
Nat Protoc
1
729
748
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 5- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Intrauterinní inseminace a její úspěšnost
- Růst a vývoj dětí narozených pomocí IVF
- Pánevní endometrióza spojená s volnou tekutinou v peritoneální dutině snižuje úspěšnost otěhotnění po intrauterinní inseminaci
-
Všechny články tohoto čísla
- Structural and Functional Differences in the Long Non-Coding RNA in Mouse and Human
- Identification, Replication, and Functional Fine-Mapping of Expression Quantitative Trait Loci in Primary Human Liver Tissue
- A −436C>A Polymorphism in the Human Gene Promoter Associated with Severe Childhood Malaria
- A Decline in p38 MAPK Signaling Underlies Immunosenescence in
- The Operon Balances the Requirements for Vegetative Stability and Conjugative Transfer of Plasmid R388
- Novel and Conserved Protein Macoilin Is Required for Diverse Neuronal Functions in
- Ixr1 Is Required for the Expression of the Ribonucleotide Reductase Rnr1 and Maintenance of dNTP Pools
- Genome of Strain SmR1, a Specialized Diazotrophic Endophyte of Tropical Grasses
- A Deficiency of Ceramide Biosynthesis Causes Cerebellar Purkinje Cell Neurodegeneration and Lipofuscin Accumulation
- A Latent Pro-Survival Function for the Mir-290-295 Cluster in Mouse Embryonic Stem Cells
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- DNA Methylation Dynamics in Human Induced Pluripotent Stem Cells over Time
- Prion Formation and Polyglutamine Aggregation Are Controlled by Two Classes of Genes
- Integrated Genome-Scale Prediction of Detrimental Mutations in Transcription Networks
- Post-Embryonic Nerve-Associated Precursors to Adult Pigment Cells: Genetic Requirements and Dynamics of Morphogenesis and Differentiation
- A Novel Mouse Synaptonemal Complex Protein Is Essential for Loading of Central Element Proteins, Recombination, and Fertility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
- A Genetic and Structural Study of Genome Rearrangements Mediated by High Copy Repeat Ty1 Elements
- A Missense Mutation in Causes a Major QTL Effect on Ear Size in Pigs
- A Flurry of Folding Problems: An Interview with Susan Lindquist
- Meiotic Recombination Intermediates Are Resolved with Minimal Crossover Formation during Return-to-Growth, an Analogue of the Mitotic Cell Cycle
- A Nervous Origin for Fish Stripes
- The ISWI Chromatin Remodeler Organizes the hsrω ncRNA–Containing Omega Speckle Nuclear Compartments
- The Telomerase Subunit Est3 Binds Telomeres in a Cell Cycle– and Est1–Dependent Manner and Interacts Directly with Est1
- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Characterizing Genetic Risk at Known Prostate Cancer Susceptibility Loci in African Americans
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Nodal-Dependent Mesendoderm Specification Requires the Combinatorial Activities of FoxH1 and Eomesodermin
- SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs
- Association of Genetic Variants in Complement Factor H and Factor H-Related Genes with Systemic Lupus Erythematosus Susceptibility
- STAT Is an Essential Activator of the Zygotic Genome in the Early Embryo
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání