-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
GAPDH-A Recruits a Plant Virus Movement Protein to Cortical Virus Replication Complexes to Facilitate Viral Cell-to-Cell Movement
Intercellular movement of plant viruses is the crucial step during systemic viral infections. Red clover necrotic mosaic virus (RCNMV), a bipartite positive-strand RNA plant virus, forms movement protein (MP)-containing punctate structures on the cortical endoplasmic reticulum in infected cells, which are required for efficient intercellular movement of the virus. We provide evidence that these cortical punctate structures constitute the viral replication complex (VRC), which forms during the early stage of virus infection. Furthermore, we show that a host protein of Nicotiana benthamiana, glyceraldehyde 3-phosphate dehydrogenase-A (NbGAPDH-A), possibly intercalates between the cortical VRC and MP. Knockdown of NbGAPDH-A diffused subcellular localization of MP and reduced intercellular movement of the virus. Chloroplastic NbGAPDH-A relocalized to the cortical VRC after infection with the virus. Our results suggest that the cortical VRC serves not only as the replication factory of viral RNA but also as a transportation hub, which transports viral RNA to neighboring uninfected cells via plasmodesmata.
Published in the journal: . PLoS Pathog 10(11): e32767. doi:10.1371/journal.ppat.1004505
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004505Summary
Intercellular movement of plant viruses is the crucial step during systemic viral infections. Red clover necrotic mosaic virus (RCNMV), a bipartite positive-strand RNA plant virus, forms movement protein (MP)-containing punctate structures on the cortical endoplasmic reticulum in infected cells, which are required for efficient intercellular movement of the virus. We provide evidence that these cortical punctate structures constitute the viral replication complex (VRC), which forms during the early stage of virus infection. Furthermore, we show that a host protein of Nicotiana benthamiana, glyceraldehyde 3-phosphate dehydrogenase-A (NbGAPDH-A), possibly intercalates between the cortical VRC and MP. Knockdown of NbGAPDH-A diffused subcellular localization of MP and reduced intercellular movement of the virus. Chloroplastic NbGAPDH-A relocalized to the cortical VRC after infection with the virus. Our results suggest that the cortical VRC serves not only as the replication factory of viral RNA but also as a transportation hub, which transports viral RNA to neighboring uninfected cells via plasmodesmata.
Introduction
Eukaryotic positive-strand RNA viruses replicate their genomes using membrane-bound virus replication complexes (VRC), which contain viral replicase proteins, viral RNA templates, and host-factor proteins [1]–[5]. Viral replicase proteins modify the host intracellular membrane morphology, including swelling, invagination, and the formation of spherules. Thus, VRC formation is accompanied by the remodeling of intracellular membranes [3]. Studies of the subcellular localization of viral proteins using immunoelectron microscopy or confocal laser scanning microscopy (CLSM) have shown that the movement proteins (MPs) of several plant viruses colocalize with the viral replicase protein [6]–[10], thereby suggesting that MPs are also components of VRCs. However, the MPs that localize to VRCs are not likely to be involved in the replication of viral genomic RNA because mutant viruses that do not encode functional MP can accumulate viral genomic RNA similar to that of the wild type virus with functional MP in infected protoplasts [11], [12]. MPs play central roles in the cell-to-cell and systemic movement of plant viruses, and they have been investigated intensively to determine their biochemical characteristics; their subcellular localization, including the cellular pathways that target them to the plasmodesmata (PD), the cytoplasmic channels connecting plant cells; and their interactions with host membranes or proteins [13]–[17].
The early process of VRC formation has been well characterized in Tobacco mosaic virus (TMV), which is the type member of the well-studied genus Tobamovirus [18]. TMV virions that enter host plant cells are partially uncoated, and the exposed 5′ cap structures are assumed to be recruited by unknown host factors to form small particles that contain viral RNA on the cortical endoplasmic reticulum (ER), before moving along the ER and actin filaments. The replication cycles starts after translation of the replicase proteins and the VRC increases in size. The punctate-like TMV VRC forms on the cortical ER and moves along actin filaments [18]–[20] or microtubules [21], [22]. The intracellular movement of VRC on actin filaments requires motor proteins such as myosin, and it is considered to be necessary for the targeting of viral genomic RNA to the PD [23]–[25]. The roles of TMV MP in the VRC are unknown. The “126-bodies”, which comprise the fusion protein of TMV 126-kDa replicase component protein and green fluorescent protein (GFP), can move along actin filaments without MP [20].
Red clover necrotic mosaic virus (RCNMV) is a positive-strand RNA virus with a bipartite genome that belongs to the genus Dianthovirus, in the family Tombusviridae [26]. Genomic RNA1 encodes p27 auxiliary replication protein, p88 RNA-dependent RNA polymerase (RdRp), and coat protein (CP), while RNA2 encodes MP (Figure S1A). p27 and p88 induce the production of an aggregate structure from ER membrane and they form the 480-kDa replication complex, which is a key enzyme complex for virus replication, via interactions with host chaperone proteins such as heat shock protein (HSP) 70 and HSP90, and membrane traffic-associated proteins such as Arf1 and Sar1 [27]–[31]. Using a bimolecular fluorescence complementation (BiFC) assay, these four host factors were shown to interact directly with p27 in the large aggregate adjacent to the nucleus in Nicotiana benthamiana cells [30], [31]. However, when the p27-GFP fusion protein was expressed with p88 and RNA2, it formed small punctate structures on the cortical ER and later formed large aggregates adjacent to the nucleus [32]. These results suggest that RCNMV VRC forms small punctate structures on the cortical ER, which then change their subcellular localization to form a large aggregate adjacent to the nucleus.
RCNMV MP belongs to the 30K superfamily and it is required for viral cell-to-cell and systemic movement [33], [34]. RCNMV is considered to pass through PD in the form of a viral RNA-MP complex because CP is dispensable for viral cell-to-cell movement [34] and MP also has the ability to bind single-stranded nucleic acids [35]. Microinjected RCNMV MP can increase the size-exclusion limit of PD and enable the transport of coinjected viral RNA into neighbor cells [36]. Alanine-scanning mutant analysis was used to determine the functional domains of the MP that bind RNA and that target it to PD, both of which are required for viral cell-to-cell movement [37], [38]. However, the cellular pathway that allows MP and/or MP-viral RNA complexes to target PD is unknown.
Previously, we reported the subcellular localization of the fusion protein of RCNMV MP and GFP (MP-GFP) in N. benthamiana [10]. In addition to PD localization, MP-GFP expressed by a recombinant virus formed punctate structures with p27 on the cortical ER. Transiently expressed MP-GFP also localized to punctate structures on the cortical ER, which was associated with the replication of RNA1, but not with that of RNA2. These results suggest that MP is recruited to the cortical ER by the viral replicase complexes formed with RNA1. To demonstrate the importance of cortical punctate structures containing MP, we conducted a deletion analysis of MP and showed that 70 C-terminus amino acids are required for both cortical punctate structure formation and viral cell-to-cell movement [39]. Based on these results, we hypothesized that the recruitment of MP by the viral replication complex might help MP to acquire viral genomic RNA1 that does not encode MP, thereby leading to the efficient cell-to-cell movement of RNA1.
To further investigate the mechanism that facilitates the movement of RCNMV, we performed tandem affinity purification of MP from virus-infected N. benthamiana leaves and analyzed the co-purified host proteins, by mass spectrometry. One of these host proteins was glyceraldehyde 3-phosphate dehydrogenase subunit A (GAPDH-A). GAPDHs are ubiquitous enzymes involved in glycolysis and gluconeogenesis, and GAPDH-A is a component of the Calvin-Benson cycle of photosynthetic organisms [40]. GAPDH-A and another subunit, GAPDH-B, are both located in the chloroplast in plants and algae [41]. Thus, we isolated the full-length cDNA of N. benthamiana GAPDH-A (NbGAPDH-A) and investigated its involvement in RCNMV multiplication. Our results demonstrate that NbGAPDH-A is involved in virus cell-to-cell movement by influencing MP localization to the VRC. We discuss the possible mechanism that underlies this process.
Results
In the course of this study, we used many recombinant RCNMVs listed in Figure S1. Fluorescent proteins (FPs) or fusion proteins of MP with FP were expressed from the recombinant RNA1, in which the CP gene had been replaced. In those cases RNA2 or MP-frameshifted RNA2 were included in the inocula, because the stem-loop structure in RNA2 is required for the transcription of CP-subgenomic RNA from RNA1 [42].
Cortical punctate structures containing RCNMV MP are the sites of viral RNA replication
Previously, we reported that RCNMV MP colocalized with the viral replicase protein p27 to the punctate structures on the cortical ER in virus-infected N. benthamiana cells during the early stage of infection. Later, most of these cortical punctates disappeared and a large aggregate was formed adjacent to the nucleus in epidermal cells [10], [32]. These aggregates contained newly synthesized viral RNAs and the host-factor proteins essential for replication, and they were shown to be the sites of RCNMV RNA replication [30], [31]. However, no evidence of viral RNA replication in the cortical punctates has been reported. Thus, we detected double-stranded RNA (dsRNA), the replication intermediates of positive-stranded RNA viruses, by immunostaining using the antibody against double-stranded RNA (J2 antibody) in N. benthamiana protoplasts. J2 antibody has been widely used to detect the replication sites of animal and plant RNA viruses, and cellular RNAs such as ribosomal RNA are below the limit of detection [43]–[45]. N. benthamiana protoplasts were inoculated with in vitro transcripts of recombinant RCNMV, which expressed the fusion protein of MP and a red FP, mCherry (MP-mCherry, Figure S1B).
Using CLSM, MP-mCherry and dsRNA were detected as overlapping small punctate signals near the surfaces of protoplasts at 16 h post inoculation (hpi) (Figure 1, left 2 rows of panels). At 24 hpi, most of these small punctates disappeared and large aggregates were detected adjacent to the nucleus, which contained both MP and dsRNA (Figure 1, center 2 rows of panels), thereby confirming the results reported previously [30], [31]. No fluorescent signals for dsRNA were detected in mock-inoculated protoplasts (Figure 1, right panels). These results indicate that both the cortical punctates formed during an early stage of RCNMV infection and the aggregates formed adjacent to the nucleus during the later stage of infection are the sites of RCNMV RNA replication. Subsequently, we refer to the small punctate-like structures that contain the MP in the cortical region as ‘cortical VRC.’
Fig. 1. Cortical punctate structures that contain RCNMV MP are sites of viral RNA replication. 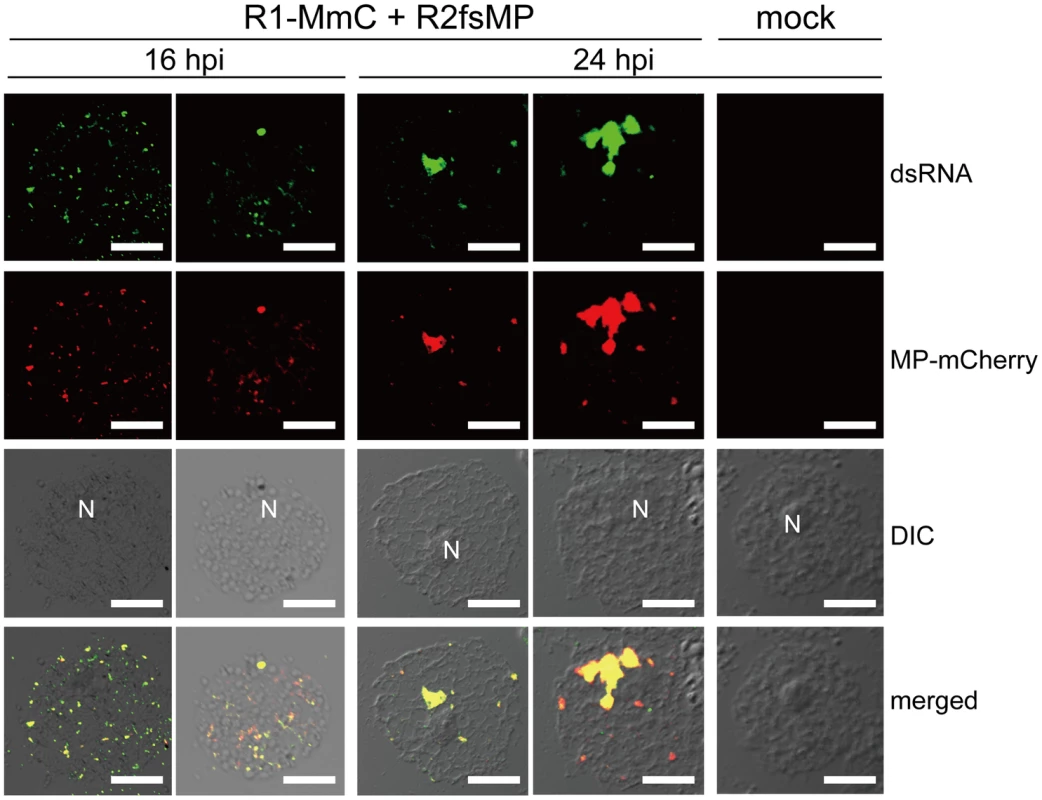
Nicotiana benthamiana protoplasts were inoculated with recombinant RCNMV RNAs that expressed the MP-mCherry fusion protein (Figure S1B) and subjected to immunostaining with anti-dsRNA primary antibody followed by Alexa Fluor 488-conjugated secondary antibody at an early stage of infection (16 hpi, left 2 rows of panels) and at a late stage of infection (24 hpi, center 2 rows of panels). The right-most panels show the results for mock-inoculated protoplasts treated with the same antibodies. Images present confocal projections of five optical sections at 1 µm intervals, which range from the surface to the middle of the protoplasts. DIC: differential interference contrast, N: nucleus. Scale bar = 20 µm. Identification of host plant proteins that interact with RCNMV MP
To identify the host proteins that interact with RCNMV MP, we performed two-step affinity purification of MP fused to a tandem affinity purification tag sequence. The tagged MP was functional because it supported virus cell-to-cell and systemic movement with the same efficiency as the native MP in N. benthamiana plants (Figure S2).
Binary vector plasmid pBICR12/MP-TAP (Figure S1D), and pBICR12 (Figure S1C) as the negative control, were infiltrated via Agrobacterium into N. benthamiana. The tandem affinity purified fraction prepared from pBICR12/MP-TAP-infiltrated leaves contained several silver-stained bands, which were not detected in the negative control (Figure 2A). The clear silver-stained band that represented the MP-FLAG was not detected in the MP-TAP lane for unknown reason. Considering its size (35.6 kDa), the band is probably masked in the broad range of the stained area below the 42 kDa marker. Actually nano-liquid chromatography-tandem mass spectrometry (LC/MS/MS) analysis demonstrated that a piece of wide gel cut out from MP-TAP lane (Figure 2A, red arrow) contained the MP (Table S1). MP-FLAG was also detected by Western blotting analysis in the tandem affinity purified fraction prepared from pBICR12/MP-TAP-infiltrated leaves but not from the negative control leaves (Figure 2B).
Fig. 2. Identification of proteins that were copurified with RCNMV MP. 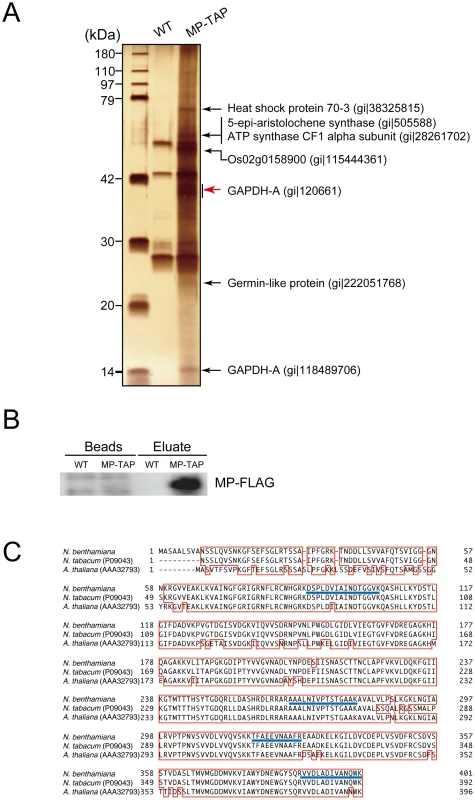
Protein extracts prepared from Agrobacterium-infiltrated leaves that expressed RCNMV RNA1 plus RNA2 (WT, pBICR12, Figure S1C), or RNA1 plus recombinant RNA2 encoding MP tagged with TAP tag sequences (MP-TAP, pBICR12/MP-TAP, Figure S1D), were subjected to two-step affinity purification using an anti-HA antibody followed by an anti-FLAG antibody. (A) One fifth of the affinity-purified fractions eluted from the anti-FLAG beads (Eluate) and the proteins contained with the beads after elution (Beads) were subjected to Western blotting using anti-FLAG antibody. (B) Four fifths of the affinity-purified fractions were subjected to SDS-PAGE and stained using MS-compatible silver staining. The protein bands of interests were excised, subjected to in-gel digestion, and analyzed by tandem mass spectrometry. Proteins with Mascot search scores >50, which were absent from the control protein bands, and proteins with significantly higher scores than the control proteins are indicated on the right-hand sides of the panels. The NCBI accession numbers of the identified proteins are also indicated. (C) The GAPDH-A gene of N. benthamiana was cloned and the deduced amino acid sequence was aligned using GENETYX-Mac ver. 14.0.1 with its closest homolog from N. tabacum (accession number P09043) and that from Arabidopsis thaliana (accession number AAA32793). The blue lines show the peptide sequences detected. These silver-stained bands in the MP-TAP lane, and the similar regions of the gel for the negative control lane were excised and subjected to in-gel trypsin digestions and LC/MS/MS analyses. We identified RCNMV MP and several host proteins from the stained bands, and these proteins were not detected from the negative control gels. Among these, we focused on GAPDH-A. A partial GAPDH-A sequence was amplified by RT-PCR using the total RNA of N. benthamiana, where the primer designs were based on N. tabacum GAPDH-A. The full-length cDNA of GAPDH-A was cloned according to 5′ and 3′ RACE methods, which we refer to as NbGAPDH-A (accession number AB937979). The deduced amino acid sequence of NbGAPDH-A was almost identical to the reported partial GAPDH-A of N. tabacum (96.9% shared identity, except for nine N terminal amino acids) and very similar to that of Arabidopsis thaliana, except for 60 N terminal amino acids (Figure 2C).
VIGS of NbGAPDH-A negatively affects the multiplication of RCNMV
To investigate the possible involvement of NbGAPDH-A in RCNMV multiplication, we downregulated the gene using the Apple latent spherical virus (ALSV) vector [46]. A plasmid that expressed wild type ALSV, or that expressed the recombinant ALSV containing 294 nucleotides of NbGAPDH-A (ALSV/gsGAP vector), was mobilized into Agrobacterium and the bacterium was used to inoculate young N. benthamiana plants. The accumulation level of NbGAPDH-A mRNA in the newly developed leaves was determined 2–3 weeks later by real time RT-PCR. NbGAPDH-A was silenced effectively in ALSV/gsGAP vector-infected plants; the mRNA level of NbGAPDH-A was reduced to 3% of that in the wild type ALSV-infected plants (Figure 3A). This result coincided with that by semi-quantitative RT-PCR in which mRNA level in ALSV/gsGAP vector-infected plants was about 1/32 of that in wild type ALSV-infected plants (Figure S3). No symptoms or growth inhibition were detected in the NbGAPDH-A-silenced plants and wild type ALSV-infected plants (Figure S4, see Discussion). Hereafter, all of the ALSV/gsGAP-infected plants and the protoplasts prepared from those plants were tested by real time or semi-quantitative RT-PCR to confirm the NbGAPDH-A gene was silenced.
Fig. 3. Multiplication of RCNMV is inhibited in NbGAPDH-A-silenced N. benthamiana leaves. 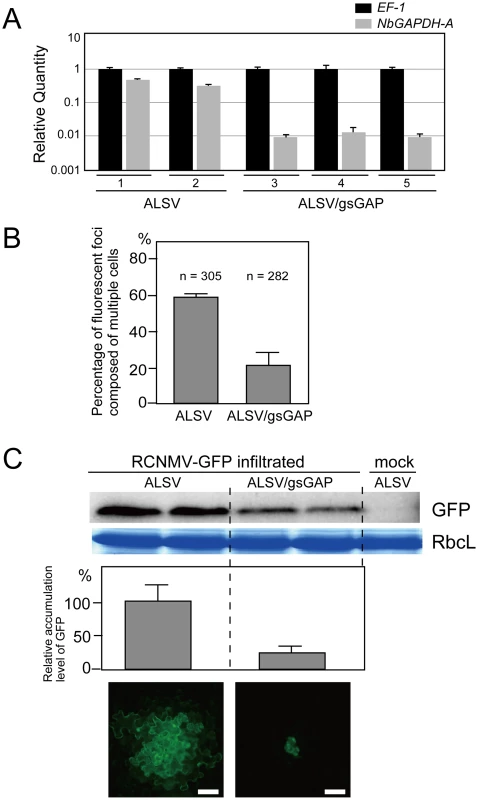
(A) Gene silencing of NbGAPDH-A was induced in N. benthamiana plants inoculated with the ALSV vector, which harbored a 294 bp partial fragment (nucleotides 352–645 from start codon) of NbGAPDH-A (ALSV/gsGAP), via Agrobacterium. The empty ALSV vector (ALSV) was used as a control. Total RNA was prepared from two ALSV-infected and three ALSV/gsGAP-infected plants. NbGAPDH-A mRNA levels were determined by real time PCR using primers specific to NbGAPDH-A (nucleotides 755–954 from start codon). The real time PCR results for the EF-1 mRNA (closed column) were used to adjust the relative accumulation levels of NbGAPDH-A mRNA (gray column). (B) In vitro transcripts of a recombinant RCNMV that expressed GFP from its subgenomic RNA (RCNMV-GFP, Figure S1E) were inoculated mechanically onto ALSV- or ALSV/gsGAP-infected N. benthamiana plants. At 20 hpi, the percentages of fluorescent foci that comprised multiple cells were measured using epifluorescence microscopy. ‘n’ represents the total number of fluorescent foci in 4 inoculated leaves (about 25 square centimeters). (C) An Agrobacterium culture that contained the pBICR1sG2 plasmid, which expressed RCNMV-GFP (Figure S1F), was diluted to OD600 = 0.03 and infiltrated into ALSV- or ALSV/gsGAP-infected N. benthamiana plants. At 35 hpi, protein was extracted from the infiltrated leaves and subjected to Western blotting using anti-GFP antibody. RbcL is a Coomassie brilliant blue-stained gel image, which shows the large subunit of Rubisco proteins. The accumulated levels of GFP from three separate experiments were quantified using the Image Gauge program and plotted in the graph. The lowest two panels show representative epifluorescence microscopy images of the infiltrated leaves at 35 hpi. Scale bar = 50 µm. We then subjected the ALSV - and ALSV/gsGAP-infected plants to challenge via the mechanical inoculation of in vitro transcripts of the recombinant RCNMV containing the GFP gene (RCNMV-GFP; Figure S1E). The percentage of fluorescent foci with multiple cells in the ALSV/gsGAP-infected plants was about 1/3 of that in the ALSV-infected plants at 20 hpi (Figure 3B). The result suggests that RCNMV multiplication was negatively affected by the silencing of NbGAPDH-A.
In order to evaluate the effect of the gene silencing on RCNMV multiplication more objectively, we further performed challenge inoculation with pBICR1sG2 (Figure S1F), which expressed RCNMV-GFP, via Agrobacterium infiltration, and the multiplication level of the recombinant virus was estimated by western blot analysis for GFP. The level of GFP accumulation at 35 hpi in the leaves of ALSV/gsGAP-infected plants was approximately 20% of that in the leaves of the ALSV-infected plants (Figure 3C). The majority of the fluorescent foci were comprised of more than 10 cells in the latter plants, whereas such a wide spread of fluorescence was barely detected in the former plants (Figure 3C, lower panels). At 48 hpi, most of the fluorescent foci in the ALSV/gsGAP-infected plants became larger and the level of GFP accumulation was about 80% of that in the wild type ALSV-infected plants (Figure S5). Thus, RCNMV multiplication was impaired in the NbGAPDH-A-silenced N. benthamiana leaves, at an early stage of infection.
To investigate whether downregulation of the NbGAPDH-A gene could affect the multiplication of viruses other than RCNMV, we inoculated ALSV - and ALSV/gsGAP-infected N. benthamiana plants with a recombinant Tomato mosaic virus (ToMV), where the CP gene was replaced with the GFP gene. The spread of GFP fluorescence was indistinguishable at 40 and 48 hpi by epifluorescence microscopy and the GFP accumulation level was also similar in both plants (Figure S6). These results indicate that the NbGAPDH-A gene is not involved in the multiplication of ToMV.
VIGS of NbGAPDH-A does not affect RCNMV replication
To investigate the effect of NbGAPDH-A silencing on RCNMV accumulation at the single cell level, we infiltrated ALSV-, or ALSV/gsGAP-infected plants with Agrobacterium that contained pBICR12fsMP, which expressed movement-deficient RCNMV RNAs (Fig. S1G) [10]. At 26 and 43 hpi, similar amounts of positive-stranded viral RNAs accumulated (Figure 4A), thereby suggesting that RCNMV multiplied at similar levels in the initially infected cells.
Fig. 4. NbGAPDH-A does not affect the accumulations of RCNMV RNA and MP at the single-cell level. 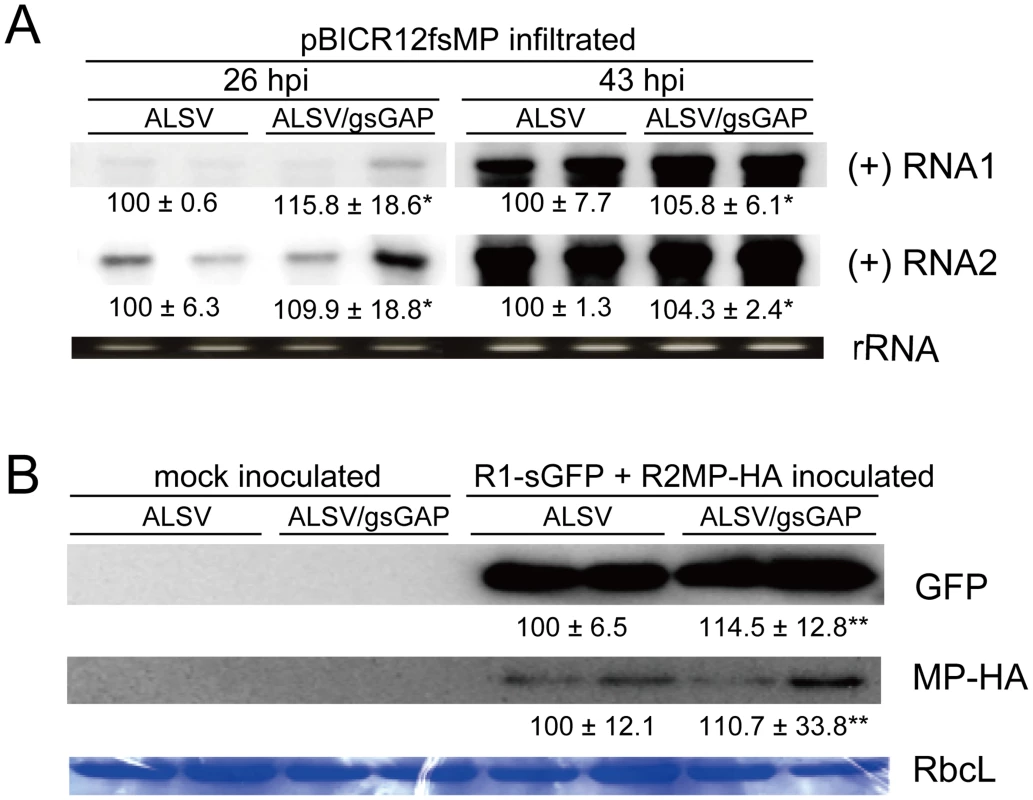
(A) An Agrobacterium culture that contained the pBICR12fsMP plasmid, that expressed movement-deficient RCNMV (Figure S1G), was diluted to OD600 = 0.8 and infiltrated into ALSV- or ALSV/gsGAP-infected N. benthamiana plants (two plants for each). At 26 hpi and 43 hpi, the total RNA was extracted from the infiltrated leaves and subjected to Northern blotting using DIG-labeled riboprobes specific for the plus-strand (+) RNA1 or RNA2 of RCNMV. rRNA is an ethidium bromide-stained agarose gel image of 1 µg total RNA, which was used as the loading control. The numbers below the images represent the relative accumulation levels (means ± SE) of viral RNAs using the Image Gauge program (Fuji Film), which were calculated based on two independent experiments. Asterisk indicates a not significant (P>0.05; Student's t-test) difference compared with the viral RNA accumulation level in the protoplasts from ALSV-infected N. benthamiana. (B) Protoplasts prepared from ALSV- or ALSV/gsGAP-infected N. benthamiana plants (each two plants) were inoculated with a mixture of in vitro transcripts of the recombinant RCNMV RNA1, which expressed GFP from subgenomic RNA; and RNA2, which expressed MP tagged with HA (Figure S1H). Proteins extracted from 2×104 protoplasts were loaded in each lane. GFP and MP-HA were detected using a rabbit polyclonal antibody against GFP and a rat polyclonal antibody against HA, respectively. RbcL is a Coomassie brilliant blue-stained gel image of proteins extracted from 2×104 protoplasts, which shows the large subunit of Rubisco proteins. The numbers below the images represent the relative accumulation levels (means ± SE) of the proteins using the Image Gauge program (Fuji Film), which were calculated from two independent experiments. Double asterisk indicates a not significant (P>0.05; Student's t-test) difference compared with the protein accumulation level in the protoplasts from ALSV-infected N. benthamiana. To further investigate the multiplication levels of RCNMV in single cells, protoplasts were prepared from ALSV - and ALSV/gsGAP-infected plants and inoculated with in vitro transcripts of the recombinant RCNMV, which expressed GFP and MP tagged with HA (MP-HA) (Figure S1H). Similar amounts of GFP accumulated in both protoplasts (Figure 4B, upper panel), thereby indicating that gene silencing of NbGAPDH-A did not affect the accumulation of the recombinant virus at the single cell level. We also analyzed the accumulation of MP-HA. As shown in the middle panel of Figure 4B, the levels of MP-HA were similar with either inoculation, which suggests that NbGAPDH-A is not involved in the translational control or stability of MP. Overall, these results suggest that NbGAPDH-A is unlikely to be involved in the replication of RCNMV RNAs and that it is involved in the cell-to-cell movement of RCNMV via its interaction with MP.
Subcellular localization of NbGAPDH-A
NbGAPDH-A is assumed to localize to the chloroplasts. However, RCNMV replication occurs in association with the ER membrane and no relationship with the chloroplasts has been reported previously. To investigate the possible interaction between NbGAPDH-A and RCNMV proteins in vivo, we examined the subcellular localization of NbGAPDH-A in the absence or presence of RCNMV factors. When NbGAPDH-A tagged with GFP (NbGAPDH-A-GFP) alone was expressed transiently in N. benthamiana leaves via agroinfiltration, the protein localized exclusively to chloroplasts (Figure 5A, left two panels and Figure 5B, panels 1 and 2). The localization pattern of NbGAPDH-A-GFP was not altered by coexpression with RCNMV MP-mCherry. NbGAPDH-A-GFP signals were detected in the chloroplasts and were never detected in PD (Figure 5A, right four panels). This suggests that the transiently expressed MP does not interact with NbGAPDH-A in vivo.
Fig. 5. NbGAPDH-A changes its subcellular localization in association with RCNMV RNA replication, but not with viral proteins. 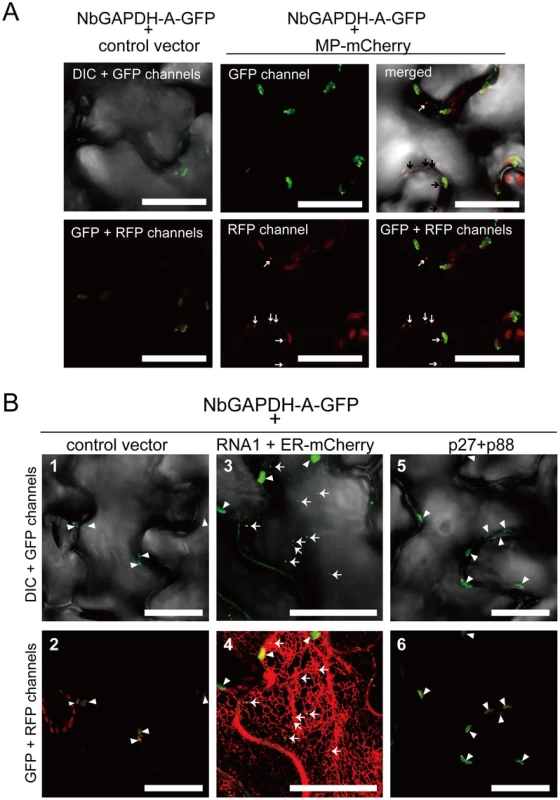
Representative CLSM images of agroinfiltrated N. benthamiana epidermal cells. Each Agrobacterium culture was diluted to OD600 = 0.8 and equal volume of cultures were mixed for infiltration. Confocal microscopy images were taken at 40 h after infiltration. Images present confocal projections composed of 15 optical sections taken at 1 µm intervals, from the surface to the middle of epidermal cells. Chloroplast autofluorescence was detected in all the RFP-channel images by the use of 610IF emission filter. Scale bar = 20 µm. (A) Epidermal cells transiently expressing NbGAPDH-A-GFP (left two panels), and NbGAPDH-A-GFP with MP-mCherry (right four panels). Arrows represent PD-localized MP-mCherry signals. (B) Epidermal cells, which transiently expressed NbGAPDH-A-GFP (panels 1 and 2), or NbGAPDH-A-GFP with RCNMV RNA1 and ER marker (ER-mCherry) (panels 3 and 4), or NbGAPDH-A-GFP with viral replicase component proteins p27 and p88 (panels 5 and 6). Arrowheads represent chloroplast-localized NbGAPDH-A-GFP signals and arrows represent cortical ER-localized NbGAPDH-A-GFP signals. Next to investigate whether the subcellular localization of NbGAPDH-A-GFP could be affected by RCNMV RNA replication, NbGAPDH-A-GFP was coexpressed with RNA1. The GFP signals were detected in punctate structures that formed near the surface regions of epidermal cells, as well as in chloroplasts (Figure 5B, panel 3). These cortical signals colocalized with ER marker signals (Figure 5B, panel 4). Similar cortical punctate signals of NbGAPDH-A-GFP were also detected when it was coexpressed with both RNA1 and RNA2 (Figure S7), but not with the viral replicase component proteins, p27 and p88 (Figure 5B, panels 5 and 6). Lack of the cortical punctate signals of NbGAPDH-A-GFP in the latter leaves does not seem to be due to the low level of viral replicase proteins. p27 accumulated efficiently in the latter leaves (Figure S8). p88 was below the limit of detection in these agroinfiltrated leaves, as described previously [28], [29]. These results suggested the association between the localization of NbGAPDH-A-GFP to punctates on the cortical ER and the replication of RNA1. To examine this association is specific to NbGAPDH-A-GFP, we investigated the localization of free GFP or the GFP with chloroplast-targeting signal peptide in the presence of RNA1. No cortical punctate signals were detected in the leaves expressing these GFP proteins with RNA1 (Figure 6 and Figure S9). These results suggested that NbGAPDH-A was recruited to the cortical punctate structures in association with the replication of RNA1.
Fig. 6. Subcellular localization of free GFP and GFP with chloroplast targeting signal is not affected by coexpression of RCNMV RNA1. 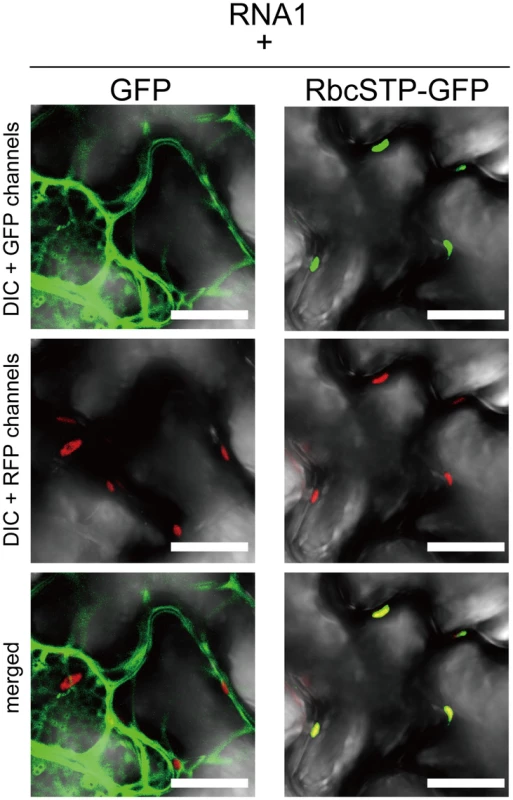
Representative CLSM images of agroinfiltrated N. benthamiana epidermal cells. Epidermal cells transiently expressing RCNMV RNA1 together with GFP (left panels), or with GFP with chloroplast targeting signal peptide of Rubisco small subunit (RbcSTP-GFP, right panels). Scale bar = 20 µm. Conditions for infiltration, plant incubation, and CLSM were similar to those in Figure 5. NbGAPDH-A interacts with RCNMV MP in vivo
The interaction between NbGAPDH-A and RCNMV MP in vivo was confirmed by BiFC assays in N. benthamiana epidermal cells. NbGAPDH-A was fused to the C-terminal half of yellow fluorescent protein (YFP) at the C terminus (NbGAPDH-A-cYFP) and was expressed with Tomato bushy stunt virus (TBSV) silencing suppressor p19 in N. benthamiana via agroinfiltration. Recombinant RCNMV transcripts that expressed the MP fused to the N-terminal half of YFP at the C terminus (MP-nYFP, Figure S1I) was mechanically inoculated at 16 h post infiltration. At 28 hpi with the recombinant virus, fluorescence was observed using CLSM. YFP fluorescence was reconstituted in the presence of NbGAPDH-A-cYFP and the MP-nYFP (Figure 7A, left panel). No YFP fluorescence was detected in control experiments (Figure 7A, center and right panels; Figure S1J). With higher magnification, reconstituted YFP signals were observed as punctate structures in the cortical region (Figure 7B, left panel) and were also detected in the cell wall (Figure 7B, right panel, see Discussion). Reconstituted YFP signals in the cortical punctates were confirmed to overlap with ER marker signals (Figure 7C). These results, together with the localization results of the MP expressed from recombinant virus to the cortical VRC ([10] and Figure 1) show that the reconstituted YFP signals are on the cortical VRC.
Fig. 7. Bimolecular fluorescence complementation analyses of the interaction between RCNMV MP and NbGAPDH-A. 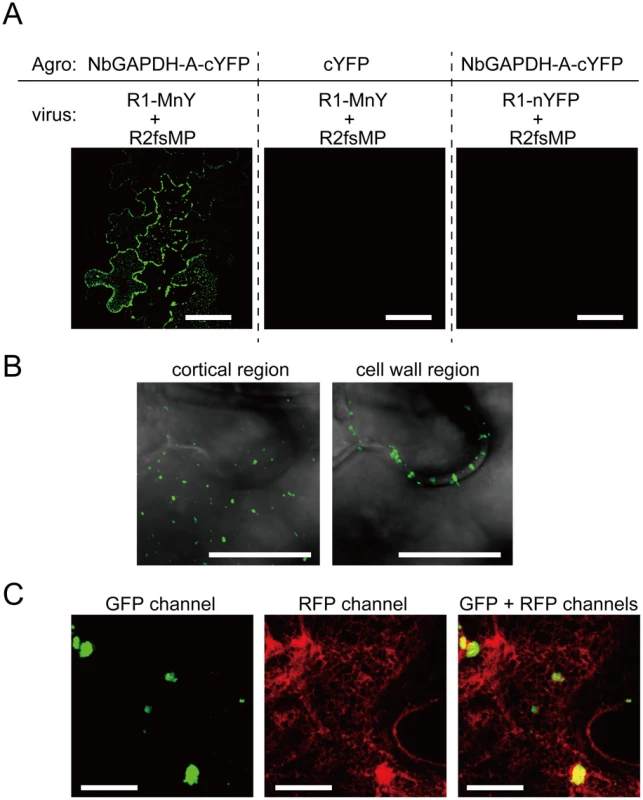
NbGAPDH-A fused to the C-terminal half of YFP at the C-terminus (NbGAPDH-A-cYFP), or C-terminal half of YFP (cYFP) as the negative control, was expressed with TBSV silencing suppressor p19 in N. benthamiana leaves via Agrobacterium infiltration. 18 h after infiltration, in vitro transcripts of the recombinant RCNMV that expressed fusion protein of the MP and N-terminal half of YFP at the C-terminus (R1-MnY+R2fsMP, Figure S1I), or the recombinant virus that expressed N-terminal half of YFP (R1-nYFP+R2fsMP, Figure S1J) as the negative control, was mechanically inoculated. At 28 hpi of the recombinant virus, reconstructed YFP signal was visualized using CLSM. (A) Reconstituted YFP signals were detected as foci composed of 5–10 cells in the leaves that expressed NbGAPDH-A-cYFP and inoculated with R1-MnY+R2fsMP (left panel). No YFP signals were detected in the leaves that expressed unfusedcYFP (center panel), or those inoculated with R1-nYFP+R2fsMP (right panel). Scale bar = 50 µm (B) Large magnification images of the reconstituted YFP in the cortical (left panel) and inner cell wall region (right panel). Images are from optical sections taken at upper part for cortical region and middle part for cell wall region in the same site of the leaf and mergers of DIC and GFP channel. Scale bar = 10 µm (C) Reconstituted YFP signals as cortical punctates (left panel), ER-mCherry signals (center panel) and overlapped (right panel). Most YFP-punctates are larger than those in (B), because this cell is closer to the center of infection and that shows the cell is at a later stage of virus infection. Scale bar = 10 µm. NbGAPDH-A interacts with RCNMV MP and p27 in vitro
Subcellular localization results (Figure 5) and BiFC results (Figure 7) suggest that NbGAPDH-A interacts with both viral replicase protein(s) and MP in association with the replication of viral RNA. To confirm the direct interaction between NbGAPDH-A and RCNMV MP, or NbGAPDH-A and p27, we performed GST pulldown assays in vitro. Bacterially expressed and purified NbGAPDH-A with an N-terminal 6× His tag and C-terminal myc tag (His-GAP-myc) was incubated with N-terminally GST - and C-terminally HA-tagged MP (GST-MP-HA), N-terminally GST-fused p27 (GST-p27), or GST, which were captured on glutathione-bound beads. Immunoblot analyses using an anti-myc antibody demonstrated that His-GAP-myc was pulled down by GST-MP-HA and GST-p27, but not by GST (Figure 8), thereby indicating that His-GAP-myc binds to both MP and p27 in vitro. To rule out the possibility that coprecipitation in the GST-pulldown experiment was mediated by interaction with any unspecific RNA that bound to MP or p27, we included RNaseA to the reaction. Addition of 50 µg/ml of RNaseA did not affect the result (Figure S10), suggesting that NbGAPDH-A interacted with the MP and p27 directly.
Fig. 8. NbGAPDH-A interacts with both RCNMV MP and p27 in vitro. 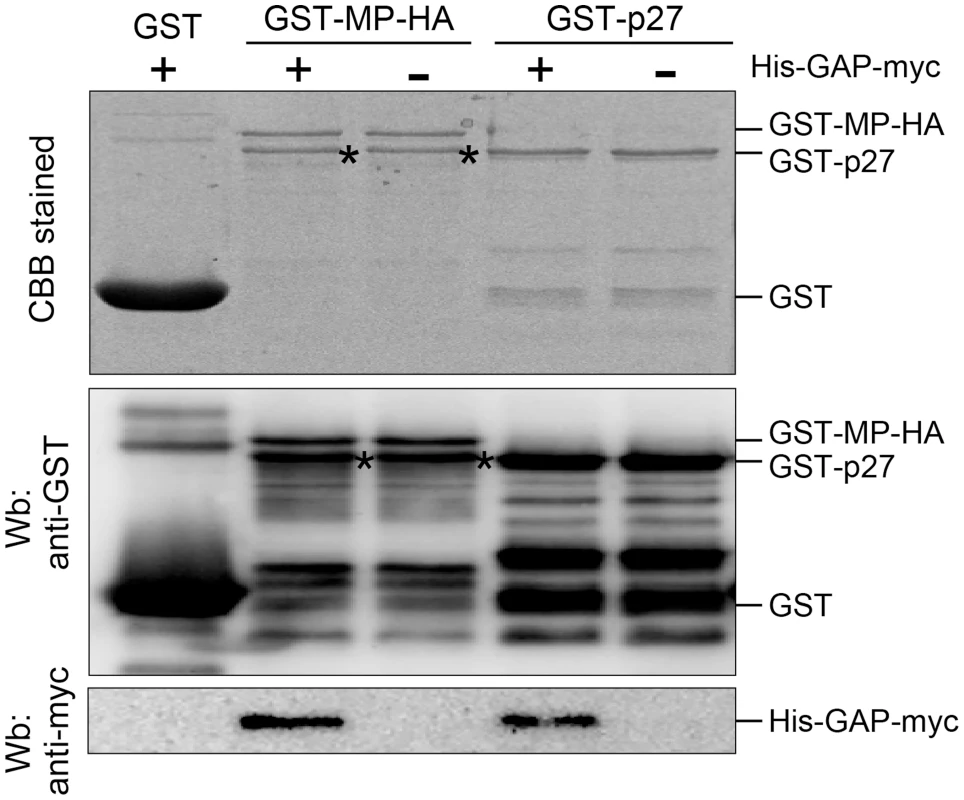
Glutathione resin-bound GST, GST-fused MP (GST-MP-HA) and GST-fused p27 (GST-p27) was incubated with the purified recombinant NbGAPDH-A (His-GAP-myc). After washing, the pulled-down complexes were subjected to SDS-PAGE and analyzed by Western blotting (Wb) using anti-GST and anti-myc antibodies. Same samples were subjected to SDS-PAGE and stained with Coomasie brilliant blue CBB). The bands that appeared in GST-MP-HA lanes (*) with almost similar mobility as GST-p27 are probably the degradation product of GST-MP-HA. Silencing of NbGAPDH-A compromises MP localization to the VRC
To address the possible effects of NbGAPDH-A on the subcellular localization of MP, we investigated whether MP targeting to the PD or to the cortical VRC was affected by the silencing of NbGAPDH-A. Our previous results showed that the transient expression of MP-GFP in N. benthamiana cells resulted in its localization exclusively to the PD, while infection with recombinant RCNMV RNAs that encoded MP-GFP resulted in the formation of cortical VRC and localization to the VRC as well as to the PD [10]. Agroinfiltration of pBICRMsG that expressed MP-GFP fusion protein [10] into ALSV/gsGAP-infected N. benthamiana plants resulted in the same localization to PD that was found in ALSV-infected plants (Figure S11). This showed that NbGAPDH-A had no effect on the intracellular transportation of RCNMV MP to the PD.
Next, we investigated the effects of NbGAPDH-A-silencing on the localization of MP to the cortical VRC. The pBICR1/MsG2fsMP plasmid, which expressed recombinant RCNMV RNAs that encoded MP-GFP (Figure S1K) [10], was agroinfiltrated into ALSV - or ALSV/gsGAP-infected plants. During the early stage of infection at 38 h post infiltration, cortical fluorescent punctates were detected in most of fluorescent mesophyll and epidermal cells in ALSV-infected plants (Figure 9A, left planels), whereas the majority of the fluorescence exhibited a dispersed cytoplasmic pattern in mesophyll cells of ALSV/gsGAP-infected plants (Figure 9A, upper right panels). In epidermal cells, cortical punctates were detected rarely and the PD localization of MP-GFP was detected in ALSV/gsGAP-infected plants (Figure 9A, lower right panel). The ratio of fluorescent cells with cortical punctates was 7.4 times higher in ALSV-infected plants compared with ALSV/gsGAP-infected plants (Figure 9B). At 44 h post infiltration with the recombinant RCNMV, the ratio of fluorescent cells with cortical punctates increased to 41.0% in ALSV/gsGAP-infected plants, although the number of cortical punctates in a single fluorescent cell was lower compared to that in ALSV-infected plants (Figure S12).
Fig. 9. Gene silencing of NbGAPDH-A inhibits the localization of RCNMV MP to the cortical VRCs. 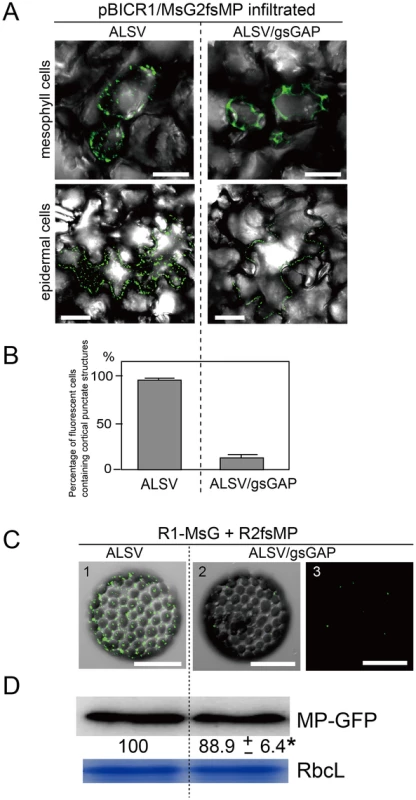
(A) Representative CLSM images of ALSV- or ALSV/gsGAP-infected N. benthamiana mesophyll (upper panels) and epidermal cells (lower panels) infiltrated with Agrobacterium cultures that contained pBICR1/MsG2fsMP, which expressed recombinant RCNMV RNAs encoding MP-GFP (Figure S1K). The images were obtained at 38 h after infiltration. Scale bar = 30 µm. The images include the merged DIC and GFP channels, and they represent confocal projections of 20 optical sections at 1 µm intervals, ranging from the surface to the middle of the cells. (B) The leaf samples in (A) were subjected to epifluorescence microscopy and the percentage of fluorescent cells containing cortical punctates was determined. The data shown are the totals from three replicate assays. (C) Protoplasts prepared from ALSV- or ALSV/gsGAP-infected N. benthamiana plants were inoculated with a mixture of in vitro transcripts of pUCR1-MsG and pRNA2fsMP (Figure S1L) [10]. Representative CLSM images of the protoplasts at 12 hpi. Scale bar = 30 µm. The images include the merged DIC and GFP channels (panels 1 and 2), or GFP channel (panel 3). They represent confocal projections of 15 optical sections at 2 µm intervals, ranging from the surface to the center of the cells. (D) The protoplast samples in (C) were subjected to Western blotting. Proteins extracted from 2×104 protoplasts were loaded in each lane. MP-GFP was detected using a rabbit polyclonal antibody against GFP. The numbers below the image represent the relative accumulation levels (means ± SE) of the proteins obtained using the Image Gauge program (Fuji Film), which were calculated from two independent experiments. Asterisk indicates a not significant (P>0.05; Student's t-test) difference compared with the accumulation of proteins in the protoplasts from ALSV-infected N. benthamiana. RbcL is a Coomassie brilliant blue-stained gel image, which shows the large subunit of Rubisco proteins. The negative effect of NbGAPDH-A-silencing on the localization of MP to the cortical VRC was confirmed using protoplasts. At 12 h post infection with the transcripts of pUCR1-MsG and pRNA2fsMP (Figure S1L) [10], cortical fluorescent punctates with MP-GFP were detected in the protoplasts prepared from ALSV-infected plants, whereas they were barely detectable in the protoplasts prepared from ALSV/gsGAP-infected plants (Figure 9C). Probably MP-GFP molecules that were diffused in the cytoplasm could not be detected by CLSM. Despite the reduced fluorescence, MP-GFP accumulated at similar levels in both protoplasts (Figure 9D), thereby showing that NbGAPDH-A silencing did not affect the expression, or stability of MP-GFP. These results suggest that NbGAPDH-A is involved in the recruitment of RCNMV MP to the cortical VRC, or that it may stabilize the interaction between MP and VRC.
Finally, these protoplasts were subjected to immunofluorescent staining of dsRNA for the detection of VRC. In the protoplasts prepared from ALSV/gsGAP-infected plants, MP-GFP was rarely detected. However, cortical punctate-like structures of dsRNA were detected in these cells, as well as in the protoplasts prepared from ALSV-infected plants (Figure 10). The accumulation level of p27 protein was also similar in both protoplasts (Figure S13). These results suggest that NbGAPDH-A does not affect the formation of cortical VRC and that it is associated with the recruitment of RCNMV MP to the cortical VRC.
Fig. 10. Cortical VRC formation in the NbGAPDH-A-silenced N. benthamiana protoplasts. 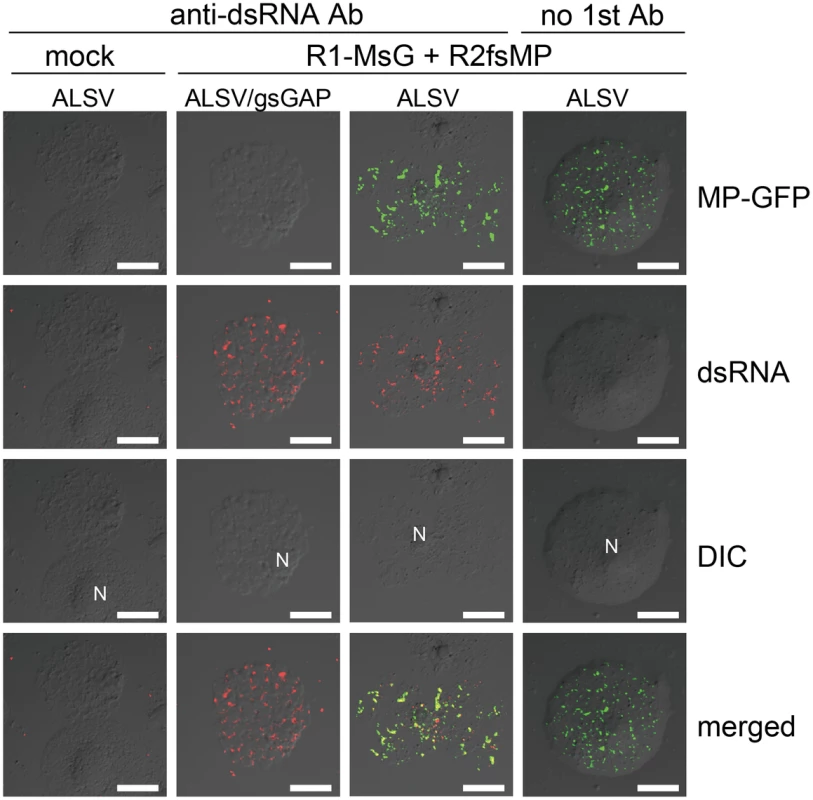
Protoplasts prepared from ALSV- or ALSV/gsGAP-infected N. benthamiana plants were inoculated with recombinant RCNMV RNAs that expressed the MP-GFP fusion protein (Figure S1L) and subjected to immunostaining with anti-dsRNA primary antibody followed by Alexa Fluor 594-conjugated secondary antibody at an early stage of infection (16 hpi). The left-most panels show the results for mock-inoculated protoplasts treated with the same antibodies. The right-most panels show the results for protoplasts prepared from ALSV-infected plants treated with only secondary antibody. Images present confocal projections of five optical sections at 1 µm intervals, which range from the surface to the middle of the protoplasts. N: nucleus. Scale bar = 20 µm. Discussion
Replication and movement processes are assumed to be linked to facilitate successful infection by plant viruses. In addition to the temporal regulation of MP expression [47]–[49], spatial regulation is required to allow MPs to encounter the viral genomes. Thus, localized MP synthesis at the VRC, or specific MP recruitment to the VRC, would facilitate efficient and specific virus cell-to-cell movement [50]. In the present study, we showed that MP-containing cortical punctate structures formed during the early stage of RCNMV infection and large aggregates assembled adjacent to the nucleus during the late stage of infection in N. benthamiana cells, and both were sites of viral RNA replication (Figure 1). These results suggest that RCNMV VRC changes its location from cortical to perinuclear ER-containing structures, while the VRC also increases in size, as the infection stage proceeds.
A division of roles between the VRCs formed during the early and late stages of infection has recently been proposed for Potato virus X (PVX). The early VRCs of PVX are formed in a membranous structure called the ‘cap’ at the orifice of PD, and triple gene block (TGB)-type MPs that accumulate at the cap and PD pore play roles in trafficking the replicated viral genomic RNA via the PD [51]. Furthermore, the X-body formed during the late stage of infection compartmentalizes the TGB1 protein and prevents it from having roles in translational activation, which could lead to the destabilization of PVX virions, while the VRCs that surround the TGB1 core maximize the replication of the viral RNA and the production of virions [52]. The cap structure at the PD orifice and MP compartmentalization in the X-body were not detected in RCNMV-infected cells [10], but it is likely that the two types of RCNMV VRCs have distinct roles. Given that RCNMV cell-to-cell movement occurs before the large aggregate-type VRCs form in virus-infected N. benthamiana epidermal cells [10], [39, unpublished results], it is probable that only the cortical VRC contributes to virus cell-to-cell movement whereas the large aggregate-type VRC (X-body) might maximize the production of progeny virions.
Our previous studies showed that the host proteins that contribute to the replication of RCNMV RNAs colocalized with p27 in the perinuclear large aggregates [30], [31], rather than the cortical VRC. It is possible that the modes of VRC formation differ between the cortical VRC and the perinuclear large aggregates. Further studies using specific antibodies against the host factors associated with the VRC are required to answer this question.
We identified NbGAPDH-A as an interacting partner for RCNMV MP (Figure 2). Although VIGS of NbGAPDH-A using ALSV vector that contained 294 bases of the gene fragment reduced the accumulation of the mRNA to 3% of that in the empty ALSV infected plants (Figure 3A), the silencing had no effect on plant growth (Figure S4). This result contradicted a previous report where transgenic tobacco (Nicotiana tabacum) plants with silenced GAPDH-A exhibited severe growth inhibition compared with the wild type plants [53]. Over 1,000 bases of the GAPDH-A coding region had been introduced into these transgenic plants to express an antisense RNA that was complementary to GAPDH-A mRNA. The induction of gene silencing using such a long sequence might have affected the expression levels of unidentified GAPDH orthologs, which could have led to growth inhibition. Alternatively, N. tabacum GAPDH-A might have a greater impact on growth than that of N. benthamiana, or transgenic plants in which the gene was silenced had a greater effect on the phenotype than VIGS. Our preliminary results showed that induction of VIGS of NbGAPDH-A by the other widely-used VIGS vector based on Tobacco rattle virus (TRV) caused the same symptoms as those by the empty TRV vector (Figure S14). This supports that VIGS of NbGAPDH-A in N. benthamiana does not cause severe symptoms.
We showed that NbGAPDH-A is a host protein that is involved in the cell-to-cell movement of RCNMV (Figures 3 and 4). In addition to chloroplast localization, the NbGAPDH-A-GFP fusion protein also localized to cortical VRCs. The localization to VRC was associated with viral RNA replication, not the replicase component proteins alone (Figure 5B). Gene silencing of NbGAPDH-A inhibited the targeting of RCNMV MP to cortical VRCs (Figure 9), but it did not affect the targeting of MP-GFP to the PD (Figure S11), or the stability of MP (Figures 4 and 9). Based on the overall results obtained in the present study, we propose that NbGAPDH-A is contained in the VRC without influencing viral RNA replication and is an interstitial agent between RCNMV MP and the VRC. NbGAPDH-A probably plays a role in recruiting MP to the VRC, or stabilizing the interaction between MP and VRC. BiFC assays confirmed the in vivo interaction between NbGAPDH-A and the MP (Figure 7). BiFC assays also showed that the interaction occurred not only in the cortical VRC, but also in the large aggregates and in the cell wall (Figure 7B). The distribution pattern of the reconstituted YFP signal was quite similar to that of the MP-GFP expressed from the recombinant virus [10]. The signals of the reconstituted YFP observed in the cell wall could be due to the NbGAPDH-A-YFP-MP complexes that had been transported to PD by the function of MP. However, significance of the observed colocalization of NbGAPDH-A and MP in the cell wall is ambiguous. This is because the reconstitution of YFP is irreversible [54], and because NbGAPDH-A-GFP did not localize in the cell wall when coexpressed with RCNMV RNA1 and RNA2 (Figure S7). Further study is needed to elucidate the role, if any, of NbGAPDH-A in the cell wall in virus infection.
GST pulldown assays confirmed that NbGAPDH-A interacted with both p27 and MP in vitro (Figure 8). This suggests that NbGAPDH-A may be a bridge between MP and p27 that is a constituent of the VRC. In vivo, however, relocalization of NbGAPDH-A-GFP to the cortical VRC and interaction of NbGAPDH-A-cYFP and MP-nYFP occurred only in association with the viral RNA replication (Figures 5B and 7). By contrast, coexpression of NbGAPDH-A-GFP and p27 fused with DsRed-monomer in N. benthamiana cells using agroinfiltration resulted in different localization patterns, with the former in chloroplasts and the latter in the ER-containing large aggregate ([32] and Figure S15). Furthermore, coexpression with MP-mCherry did not affect the localization of NbGAPDH-A-GFP to the chloroplasts (Figure 5A). These results suggest that unidentified factors might be involved in the in vivo interaction between NbGAPDH-A and viral proteins. Three possibilities can be considered, 1) Enhancement of the local concentration of viral proteins: RNA1 replicates autonomously, and the replication coupled with the translation of replicase proteins p27 and p88, followed by the formation of 480 kDa replication complex [30], [55], [56]. This replication cycle might increase the local concentration of p27 in or near the VRC on the cortical ER membrane to higher levels than transiently expressed p27. Such a process might improve the probability that p27 and NbGAPDH-A will encounter in the cortical VRC. In association with this assumption, transiently expressed p27-GFP alone forms a large aggregate ([32] and Figure S15). Formation of such aggregates might sequester p27 and prevent the interaction with NbGAPDH-A. 2) Involvement of unknown host proteins associated with the VRC: The formation of the 480 kDa replication complex of RCNMV requires not only p27 and p88 but also viral RNAs in host cells [30]. The 480 kDa replication complex contains many host proteins that have not been identified yet. It is possible that such unknown proteins are involved in the recruitment of NbGAPDH-A to the VRC or in the stabilization of the interaction in vivo. 3) Involvement of viral RNA: Cytoplasmic GAPDH (GAPDH-C) has been reported to interact with the cis-acting elements of many RNA viruses, some of which affect the multiplication of viruses ([57], [58] and references therein). Although chloroplastic and cytoplasmic GAPDHs are assumed to have evolved from different lineages [59], their amino acid sequence identity is as high as ca 45% in Arabidopsis thaliana (NCBI Gene ID: 819567 and 822277) and Zea mays (NCBI Gene ID: 542367 and 542368). It is possible that NbGAPDH-A also has an RNA-binding ability and that it is recruited to VRCs by interacting with RCNMV RNAs in vivo, as shown in Hepatitis delta virus-infected cultured cells [60]. These three possibilities may not be mutually exclusive. Further studies on the molecular mechanisms of the VRC formation are awaited.
In addition to glycolysis, cytoplasmic GAPDH-C has a variety of functions, such as membrane fusion and vesicular transport (reviewed in [61], [62]). In contrast, only the classical functions associated with carbon fixation have been reported previously for GAPDH-A. Thus, further experimental evidence is required to explain the involvement of GAPDH-A in the intracellular transport mechanism of RCNMV RNA. Alternatively, RCNMV might have evolved to utilize highly expressed and ubiquitous GAPDH-A by diverting it from its natural functions. Several plant RNA viruses use host metabolic enzymes and housekeeping proteins in ways that are unrelated to their original functions [63]. Several chloroplast-localizing proteins have recently been shown to regulate virus multiplication. Among these, chloroplastic phosphoglycerate kinase (PGK) was isolated from RNA-dependent RNA polymerase (RdRp) fraction prepared from Bamboo mosaic virus-infected N. benthamiana. PGK positively regulates multiplication of the virus through the interaction with the 3′ untranslated region of the viral genomic RNA and transportation to the chloroplasts where the viral RNA replication occurs [64], [65]. ATP synthase-γ subunit (AtpC) and Rubisco activase (RCA) were also isolated from the RdRp fraction prepared from the TMV-infected N. tabacum. AtpC and RCA negatively regulate the movement and accumulation of the virus, respectively [66]. Interestingly, gene silencing of these genes led to the increased number and the smaller size of VRC. These results are in contrast to our results that VIGS of NbGAPDH-A did not affect the number and the size of the cortical VRC in RCNMV infected cells and that it interfered with the recruitment of the MP to the cortical VRC (Figures 9 and 10). Plant RNA viruses might have evolved to utilize abundant chloroplast-localizing proteins as the positive or negative regulators through the interaction with viral proteins.
Materials and Methods
Plasmids given the prefix “pBIC” were used for Agrobacterium infiltration, “pUC”, “pRC” and “pR” were used for in vitro transcription, “pCold” was used for protein expression in Escherichia coli. pUCR1 [67] and pRC2|G [68] are full-length cDNA clones of RNA1 and RNA2 of an RCNMV Australian strain, respectively (Figure S1A). The plasmids described previously used in this study include pUCR1-MsG (Figure S1L) [39], pBICR12fsMP (Figure S1G) [10], pBICRMsG [10], pBICR1/MsG2fsMP (Figure S1K) [10], pRNA2fsMP (Figure S1B) [10], pBICP35 [39], pBICp27-iFTH [28], pColdGST (where GST is glutathione S-transferase) [29], pColdGSTp27 [29], pBICHA:cYFP [30], pBICp27 [67], pBICp88 [67], and pBICp19 [67].
E. coli DH5α was used for the construction of all plasmids. All PCR reactions were performed using a high fidelity proofreading KOD Plus-Ver.2 polymerase (Toyobo), and all the PCR-amplified regions were verified by sequencing. The primers used in this study are listed in Table S2.
Construction of plasmids expressing recombinant virus RNAs
mCherry gene was amplified from pmCherry-N1 (Clontech) using primers 1 and 2. The amplified PCR product was digested with ClaI/MluI and inserted into the same sites of pUCR1-MsG [39], producing pUCR1-MmC (Figure S1B) that expresses MP-mCherry fusion protein from the subgenomic RNA.
AscI/SacI fragment of pUC118RA1(AscI) [10] that contains the expression cassette of RCNMV RNA1 and AscI/SmaI fragment of pUC118RA2(AscI) [10] that contains the expression cassette of RCNMV RNA2 was inserted into SacI/SmaI site of pBIC18 [67] binary vector, producing pBICR12 (Figure S1C) that expresses full-genome of RCNMV.
pBICp27-iFTH is a binary vector plasmid expressing p27 tagged with FLAG-TEV protease recognition peptide-HA [28]. The TEV protease recognition peptide was replaced by 3C protease recognition peptide by recombinant PCR to produce pBICp27TEP (Mine and Okuno, unpublished). The tag sequence for tandem affinity purification was amplified from pBICp27TEP using primers 3 and 4. The cauliflower mosaic virus 35S promoter and 5′ half of RNA2 was amplified from pBICR12 using primers 5 and 6. A DNA fragment containing the 3′ half of RNA2 and the 35S terminator sequence was amplified from pBICR12 using primers 7 and 8. These three fragments were mixed and used as the template for recombinant PCR using primers 6 and 8. The generated PCR product was digested with AscI/SmaI and inserted into the same sites of pBICR12, producing pBICR12/MP-TAP (Figure S1D).
R1-MP:GFP plasmid [38] was digested with ClaI and MP coding sequence was removed. The larger fragment was self-ligated, producing pR1-sGFP (Figure S1E).
pR1-sGFP was digested with BglII/MluI and the 0.8 kb fragment containing GFP gene was inserted into the same sites of pUC118RA1(AscI), producing pUC118RA1sGC(AscI). pUC118RA1sGC(AscI) was digested with AscI/SacI and the 4.4 kb fragment was inserted to the same sites of pBICR12, producing pBICR1sG2 (Figure S1F).
A DNA fragment containing HA and the 3′ non-coding region of RNA2 was amplified from pRC|2G using primers 9 and 10. A DNA fragment containing T7 promoter and the 5′ half of RNA2 and HA was amplified from pRC2|G using primers 11 and 12. These fragments were mixed and used as the template for recombinant PCR using primers 10 and 12. The generated PCR product was digested with EcoRI/SmaI and inserted into the same site of pUC119, producing pUCR2MP-HA (Figure S1H).
EcoRI/HindIII fragment of pBE2113 [69] that contains a 35S promoter-∧ sequence-nos terminator cassette was inserted to the same sites of pUC19 (Takara Bio Inc.) producing pUC2113. The XbaI site downstream of ∧ sequence in pUC2113 was digested and filled in with T4-polymerase, and the linker sequence containing SacI site was ligated, producing pUC2113(SacI). EcoRV fragment of piL:G3 (0.4 kb) [70] containing the 35S promoter and the 5′ sequence of Tomato mosaic virus (ToMV) was inserted to the same site of pUC2113(SacI), producing pUC:ToMVrec. EcoRI/HindIII fragment of pBE2113 was replaced by the EcoRI/HindIII fragment of pUC:ToMVrec, producing pBE:ToMVrec. The KpnI/MluI fragment of piL:G3 containing ToMV MP and GFP sequence was inserted to the same sites of pTLW3 [71], producing pTLWdCP-GFP. The ribozyme sequence of Tobacco ringspot virus satellite RNA was PCR-amplified from pUCBR1R plasmid [72] and introduced to MluI site downstream of ToMV 3′ noncoding sequence in pTLWdCP-GFP, producing pTLWdCP-GFP-rib. Finally, StuI/SacI fragment of pTLWdCP-GFP-rib, containing most of the recombinant ToMV and the ribozyme sequences was inserted into the same sites of pBE:ToMVrec, producing pToMVdCP-GFP.
Plant growth conditions
N. benthamiana plants were grown on commercial soil (Tsuchi-Taro, Sumirin-Nosan-Kogyo Co. Ltd.) at 25±2°C and 16 hours illumination per day.
Protoplast preparation and viral RNA inoculation using polyethylene glycol
N. benthamiana protoplasts were prepared according to Li et al. (2013) [73] and Navas-Castillo et al. (1997) [74] with minor modifications. Briefly, young expanded leaves from 5 week-old plants were cut into 1-mm strips with a razor blade and digested in 15 ml of enzyme solution (1% cellulase RS [Yakult Pharmaceutical Ind. Co. Ltd.], 0.5% macerozyme R-10 [Yakult], 0.5 M mannitol, 10 mM CaCl2, 5 mM MES, pH 5.7) within a petri dish at 25°C in the dark with gentle shaking (40 rpm) for 4 to 5 h. After being filtered through 4 layers of cheesecloth, protoplasts were precipitated by centrifugation at 80× g for 2 min and were suspended with 10 ml of MMC solution (0.5 M mannitol, 10 mM CaCl2, 5 mM MES, pH 5.7). Concentration of cells were counted using hemacytometer. Protoplasts (1×105 cells in 100 µl) were mixed with 5 µg of viral RNAs and 200 µl of PEG solution (1 g of PEG4000 [Sigma-Aldrich #81240], 125 µl of sterile distilled water, 1.25 ml of 0.8 M mannitol, 250 µl of 1 M Ca(NO3)2) and mixed completely by gently tapping the tube. Then 2 ml of MMC solution was added and mixed. After 15 min of incubation on ice, protoplasts were precipitated by centrifugation at 80× g for 2 min, resuspended in 4 ml of MMC solution and precipitated by centrifugation again. Protoplasts were resuspended in 0.5 ml of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7) and incubated.
Preparation of protoplasts from NbGAPDH-A-silenced or ALSV-infected N benthamiana plants was essentially described above, except that the plants were 6–8 weeks old.
Immunofluorescent labeling
Fixation of N. benthamiana protoplasts and immunolabeling procedure were as described by Liu et al. (2005) [20]. For the detection of double-stranded RNA, formaldehyde-fixed protoplasts were incubated with mouse monoclonal antibody J2 (diluted 1∶200; Scicons) for 16 h in a moisturized chamber at 4°C. The samples were washed three times and then incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (diluted 1∶200; Invitrogen) for 2 h at room temperature. After washing three times, the samples were subjected to CLSM.
Affinity purification of RCNMV MP-containing fraction
N. benthamiana plants and Agrobacterium tumefaciens GV3101 (pMP90) were used for infiltration experiments as described previously [67]. A. tumefaciens transformed by pBICR12/MP-TAP, or negative control pBICR12 was used for expression of MP-HA from viral context. 1.67 g of Agrobacterium-infiltrated leaves at 48 h post infiltration were ground in liquid nitrogen and homogenized in 5 ml of extraction buffer A (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5% glycerol, 0.5% Triton X-100, 1 tablet of Complete Mini protease inhibitor cocktail [EDTA-free, Roche Diagnostics]/10 ml), followed by centrifugation at 21,000× g for 10 min at 4°C to remove cell debris. The supernatant (4.0 ml) was divided into 5 tubes (800 µl each), and each incubated with 20 µl of Anti-HA Affinity Matrix (Roche #11815016001) for 4 h at 4°C with gentle rotation. The resin was washed three times with 1 ml of washing buffer 1 (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5% glycerol, 0.1% Triton X-100), and equilibrated with 3C buffer (washing buffer 1 containing 1 mM DTT). Then the resin was incubated with 20 units of PreScission protease (GE Healthcare) in 1 ml of 3C buffer for 16 h at 4°C with gentle rotation. The resins were centrifuged at 500× g for 1 min, and the supernatant (1 ml) was immunoprecipitated again with 50 µl of ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich #A2220) for 4 h at 4°C with gentle rotation. The gel was then washed three times with washing buffer 1. The bound proteins were eluted by 125 µl of elution solution (washing buffer 1 containing 150 ng/µl FLAG peptide [Sigma-Aldrich #F3290]) for 30 min at 4°C with gentle rotation. This elution process was repeated once again, and the total of 250 µl was precipitated with trichloroacetic acid.
The affinity-purified preparation and its control preparation were subjected to SDS-PAGE, and the several bands that were not detected in the negative control lane (Figure 2A) were cut out and subjected to liquid chromatography-tandem mass spectrometry analysis, as described previously [28].
Cloning of NbGAPDH genes
RNA extraction from N. benthamiana leaves was performed using PureLink Plant RNA Reagent (Invitrogen) and treated with DNase (RQ1 RNase-free DNase; Promega). Reverse transcription was carried out using PrimeScript RT reagent Kit (Takara) using oligo-dT.
Based on the GAPDH-A sequence of N. tabacum (gi|120661), primers 13 and 14 were designed. An 1176-bp nearly full-length cDNA fragment of GAPDH-A gene was amplified from cDNA derived from N. benthamiana RNA using primers 13 and 14. The 5′ and 3′ sequences of NbGAPDH-A were amplified by SMARTer RACE cDNA amplification Kit (Clontech) using the gene-specific primers 15 and 16, respectively, and cloned into pGEM-T Easy (Promega). From each of 8 clones the 5′ and 3′ ends of NbGAPDH-A gene were determined. Nucleotide sequence data of NbGAPDH-A gene is available in the DDBJ/EMBL/Genebank databases under accession number AB937979.
Based on the GAPDH-B sequence of N. tabacum (gi|120665), primers 17 and 18 were designed. A 1270-bp partial fragment of GAPDH-B cDNA was amplified from cDNA derived from N. benthamiana RNA using primers 17 and 18, and cloned into pGEM-T Easy. Partial sequence of NbGAPDH-B gene was determined.
Construction of plasmids that express NbGAPDH-A-derivatives
Full-length cDNA of NbGAPDH-A was amplified from cDNA derived from N. benthamiana RNA using primers 19 and 20. The generated PCR product was then cloned into the BamHI site of pBICP35, producing pBICNbGA-myc.
Full-length cDNA of NbGAPDH-A was amplified from pBICNbGA-myc using primers 14 and 19. The generated PCR product was digested with BamHI/ClaI and cloned into the same sites of pUB/RMsG [39], producing pUBNbGA-sG. BamHI/HindIII fragment of pUBNbGA-sG, containing NbGAPDH-A-GFP and 35S terminator, was cloned into the same sites of pBICP35, producing pBICNbGA-sG. This was used for transient expression of NbGAPDH-A-GFP fusion protein by agroinfiltration.
Construction of plasmids that express fluorescent protein-tagged markers
Chloroplast targeting sequence of RbcS was amplified from cDNA derived from A. thaliana RNA using primers 21 and 22. sGFP sequence was amplified from pUBsGFP [39] using primers 23 and 24. Recombinant PCR fragment was amplified using primers 21 and 24. The generated PCR product was then cloned into the BamHI and KpnI sites of pUBP35 [67], producing pUBTPRbcS-sGFP. HindIII/SalI fragment of pUBTPRbcS-sGFP was cloned into the same sites of pBICP35, producing pBICRbcSTP-sGFP.
MP-mCherry sequence was amplified from pUCR1-MmC using primers 25 and 26. The generated PCR product was digested with BamHI/EcoRI and cloned into the same sites of pBICP35, producing pBICRMmC.
pBICRbcSTP-sGFP and pBICRMmC was introduced into A. tumefaciens and used for the transient expression of RbcSTP-GFP and MP-mCherry, respectively.
Virus-induced gene silencing
Construction of ALSV vector pBICAL1, pBICAL2 and pBICAL2gsPDS was described previously [75]. Partial fragment of NbGAPDH-A (294 nucleotides) was amplified from cDNA derived from N. benthamiana RNA using primers 27 and 28. The fragment was digested with BamHI/XhoI and inserted into the same sites of pBICAL2, producing pBICAL2gsNbGAP-A.
The plasmids containing the ALSV expression cassette were introduced into A. tumefaciens GV3101 (pMP90). Similar amount of fresh colonies of Agrobacterium containing pBICAL1 and each of pBICAL2, pBICAL2gsPDS, or pBICAL2gsNbGAP-A were collected using sterile toothpicks and suspended in 0.2 ml of Agro Incubation Buffer (10 mM MgCl2, 10 mM MES-KOH, pH 5.7, 0.15 mM Acetosyringone) at OD600 of 2.0–3.0, and were incubated at 20°C for more than 3 h in the dark. Sterile toothpick was soaked in the Agrobacterium suspension and stuck 4 times into 1st, 2nd and 3rd true leaves of 17–21 days old N. benthamiana plants. Three to 4 days later, toothpick-inoculation was repeated to a newly developed leaf. After inoculation, the plants were incubated in a moist chamber at 22°C overnight and transferred to a plant growth room at 25°C. Two to 3 weeks later, silencing of PDS or NbGAPDH-A was induced in the non-inoculated upper leaves.
Quantitative and semi-quantitative RT-PCR analysis
Total RNA extracted from N. benthamina leaves or protoplasts were subjected to reverse transcription using PrimeScript RT reagent Kit (Takara) using oligo-dT according to manufacturer's protocol. Real-time PCR was carried out using SYBR Premix Ex Taq (RR420A, Takara) using primers 29 and 30 for EF1 and primers 16 and 31 for NbGAPDH-A. Quantitative analysis of each mRNA was performed using a Thermal cycler Dice Real Time System TP800 (Takara). Semi-quantitative RT-PCR was performed using the same cDNA and primers and amplified by Ex Taq polymerase (Takara).
Western and northern blot analyses
Protein extraction and western blot analyses were performed as described previously [76]. Total RNA extraction from N. benthamiana leaves or protoplasts and northern blot analysis were performed as described previously [76]. Probes used for detection of positive-strand RCNMV RNA1 and RNA2 were as described previously [49]. The signals were detected with a luminescent image analyzer (LAS 1000 plus, Fuji Film Co. Ltd.) and the signal intensities were quantified using the Image Gauge program version 3.1 (Fuji Film).
Microscopy
The spread of GFP fluorescence was observed using an Olympus BX53 fluorescence microscope equipped with an Olympus DP72 camera using the imaging program Olympus cellSens.
Subcellular localizations of proteins tagged with FPs and dsRNA that was detected with fluorescent antibodies were observed using an Olympus FluoView FV500 confocal microscope. Both a Nikon 60× Plan Apo oil immersion objective lens (numerical aperture 1.4) and a Nikon 40× UPlan Apo oil immersion objective lens (numerical aperture 1.0) were used. The sets of dichroic mirror, beam splitter, and emission filter used were DM488/543, SDM560, and BA505-525 for GFP, and DM488/543/633, SDM630, and BA560-600 for mCherry For the detection of mCherry signal and chloroplast autofluorescence simultaneously, emission filter BA610IF was used. In experiments for detecting dual localization, scanning was performed in sequential mode to minimize signal bleed-through. All images shown are from optical sections taken at 1 or 2 µm intervals and were processed using Adobe Photoshop CS6 software.
Construction of protein-expression vectors in E. coli
BamHI/EcoRI fragment of pBICRMP-HA [39] that contains MP-HA gene was inserted into pCold-1 (Takara), producing pCold-MP-HA. EcoRI/KpnI fragment of pCold-MP-HA that contains MP-HA gene was inserted into the same sites of pColdGST, producing pColdGST/MP-HA.
pBICNbGAP-myc was digested with BamHI and the smaller fragment containing NbGAPDH-A-myc was cloned into the same site of pCold-1 in the correct orientation, producing pColdNbGAP-myc.
GST pulldown assay
E. coli BL21(DE3) strain was transformed with plasmids containing the prefix pCold and used for the expression of GST and GST-fused viral proteins and NbGAPDH-A tagged with a myc. All the conditions and procedures are described previously [31].
Construction of BiFC vectors
The sequence of the C-terminal half of YFP was amplified from pBICHA:cYFP [30] using primers 32 and 33 [30]. The amplified DNA was digested with StuI and cloned into StuI-digested pBICAsc2 [30], producing pBICHA-cYFPAsc2. Full-length cDNA of NbGAPDH-A was amplified from pUBNbGA-sG using primers 34 and 35. The generated PCR product was digested with BamHI and cloned into pBICHA-cYFPAsc2, producing pBICGAP-HA-cYFP.
myc-nYFP sequence was amplified from pBICMP-myc-nYFP using primers 36 and 37. The amplified PCR products were digested with ClaI/MluI and cloned into the same sites of pUCR1-MsG, producing pUCR1-MnY (Figure S1I).
p88-myc-nYFP sequence was amplified from pUCR1-MnY using primers 37 and 38. p88 sequence was amplified using primers 39 and 40 The recombinant PCR products were generated from mixture of these products using primers 37 and 39, and were digested with MluI/XhoI and cloned into the same sites of pUCR1-MsG, producing pUCR1-nYFP (Figure S1J).
BiFC assay
Twenty-five to 28 days old N. benthamiana plants were used for BiFC assays. pBICGAP-HA-cYFP, or control pBICHA-cYFPAsc2 plasmid, together with pBICp19 that expresses TBSV silencing suppressor protein p19 was infiltrated via A. tumefaciens GV3101 (pMP90) as described above. The plants were incubated in a moist chamber at 22°C for 18 h. Then in vitro transcripts (1 µg/µl) of the recombinant RCNMV that expresses MP-nYFP (Figure S1I), or negative control virus that expresses nYFP (Figure S1J) were mechanically inoculated onto the leaves. The plants were incubated in a moist chamber at 17°C for 27–30 h and were subjected to CLSM.
Accession number
NbGAPDH-A was registered through DDBJ and accession number AB937979 was given on May 27 2014.
Supporting Information
Zdroje
1. den BoonJA, AhlquistP (2010) Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64 : 241–256.
2. den BoonJA, DiazA, AhlquistP (2010) Cytoplasmic viral replication complexes. Cell Host Microbe 8 : 77–85.
3. LalibertéJF, SanfaçonH (2010) Cellular remodeling during plant virus infection. Annu Rev Phytopathol 48 : 69–91.
4. MineA, OkunoT (2012) Composition of plant virus RNA replicase complexes. Curr Opin Virol 2 : 669–675.
5. HyodoK, OkunoT (2014) Host factors used by positive-strand RNA plant viruses for genome replication. J Gen Plant Pathol 80 : 123–135.
6. HeinleinM, PadgettHS, GensJS, PickardBG, CasperSJ, et al. (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10 : 1107–1120.
7. SzécsiJ, DingXS, LimCO, BendahmaneM, ChoMJ, et al. (1999) Development of tobacco mosaic virus infection sites in Nicotiana benthamiana. Mol Plant Microbe Interact 12 : 143–152.
8. DohiK, MoriM, FurusawaI, MiseK, OkunoT (2001) Brome mosaic virus replicase proteins localizes with the movement protein at infection-specific cytoplasmic inclusions in ifected barley leaf cells. Arch Virol 146 : 1607–1615.
9. BamunusingheD, HemenwayCL, NelsonRS, SanderfootAA, YeCM, et al. (2009) Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 393 : 272–285.
10. KaidoM, TsunoY, MiseK, OkunoT (2009) Endoplasmic reticulum targeting of the Red clover necrotic mosaic virus movement protein is associated with the replication of viral RNA1 but not that of RNA2. Virology 395 : 232–242.
11. MeshiT, WatanabeY, SaitoT, SugimotoA, MaedaT, et al. (1987) Function of the 30 kd protein of tobacco mosaic virus: involvement in cell-to-cell movement and dispensability for replication. EMBO J 6 : 2557–2563.
12. TatsutaM, MizumotoH, KaidoM, MiseK, OkunoT (2005) The Red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J Virol 79 : 978–986.
13. WaigmannE, UekiS, TrutnyevaV, CitovskyV (2004) The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Curr Rev Plant Sci 23 : 195–250.
14. LucasWJ (2006) Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344 : 169–184.
15. HarriesPA, SchoelzJE, NelsonRS (2010) Intracellular transport of viruses and their components: Utilizing the cytoskeleton and membrane highways. Mol Plant Microbe Interact 23 : 1381–1393.
16. NiehlA, HeinleinM (2011) Cellular pathways for viral transport through plasmodesmata. Protoplasma 248 : 75–99.
17. UekiS, CitovskyV (2011) To gate or not to gate: Regulatory mechanisms for intercellular protein transport and virus movement in plants. Mol Plant 4 : 782–793.
18. ChristensenN, TilsnerJ, BellK, HammannP, PartonR, et al. (2009) The 5′ cap of Tobacco mosaic virus (TMV) is required for virion attachment to the actin/endoplasmic reticulum network during early infection. Traffic 10 : 536–551.
19. LiuC, NelsonRS (2013) The cell biology of Tobacco mosaic virus replication and movement. Front Plant Sci 4 : 12.
20. LiuJ, BlancaflorEB, NelsonRS (2005) The Tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone or within the complex aligns with and traffics along microfilaments. Plant Physiol 138 : 1853–1865.
21. BoykoV, HuQ, SeemanpillaiM, AshbyJ, HeinleinM (2007) Validation of microtubule-associated Tobacco mosaic virus RNA movement and involvement of microtubule-aligned particle trafficking. Plant J 51 : 589–603.
22. SambadeA, BrandnerK, HofmannC, SeemanpillaiM, MuttererJ, et al. (2008) Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata. Traffic 9 : 2073–2088.
23. KawakamiS, WatanabeY, BeachyRN (2004) Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc Natl Acad Sci U S A 101 : 6291–6296.
24. HarriesPA, ParkJW, SasakiN, BallardKD, MauleAJ, et al. (2009) Differing requirements for actin and myosin by plant viruses for sustained intercellular movement. Proc Natl Acad Sci U S A 106 : 17594–17599.
25. SchoelzJE, HarriesPA, NelsonRS (2011) Intracellular transport of plant viruses: Finding the door out of the cell. Mol Plant 4 : 813–831.
26. OkunoT, HirukiC (2013) Molecular biology and epidemiology of dianthoviruses. Adv Virus Res 87 : 36–71.
27. TurnerKA, SitTL, CallawayAS, AllenNS, LommelSA (2004) Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320 : 276–290.
28. MineA, TakedaA, TaniguchiT, TaniguchiH, KaidoM, et al. (2010) Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of Red clover necrotic mosaic virus. J Virol 84 : 6070–6081.
29. MineA, HyodoK, TakedaA, KaidoM, MiseK, et al. (2010) Interactions between p27 and p88 replicase proteins of Red clover necrotic mosaic virus play an essential role in viral RNA replication and suppression of RNA silencing via the 480-kDa viral replicase complex assembly. Virology 407 : 213–224.
30. MineA, HyodoK, TajimaY, KusumanegaraK, TaniguchiT, et al. (2012) Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J Virol 86 : 12091–12104.
31. HyodoK, MineA, TaniguchiT, KaidoM, MiseK, et al. (2013) ADP ribosylation factor 1 plays an essential role in the replication of a plant RNA virus. J Virol 87 : 163–176.
32. KusumanegaraK, MineA, HyodoK, KaidoM, MiseK, et al. (2012) Identification of domains in p27 auxiliary replicase protein essential for its association with the endoplasmic reticulum membranes in Red clover necrotic mosaic virus. Virology 433 : 131–141.
33. LommelSA, Weston-FinaM, XiongZ, LomonossoffGP (1988) The nucleotide sequence and gene organization of red clover necrotic mosaic virus RNA-2. Nucleic Acids Res 16 : 8587–8602.
34. XiongZ, KimKH, Giesman-CookmeyerD, LommelSA (1993) The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology 192 : 27–32.
35. OsmanTAM, HayesRJ, BuckKW (1992) Cooperative binding of the red clover necrotic mosaic virus movement protein to single-stranded nucleic acids. J Gen Virol 73 : 223–227.
36. FujiwaraT, Giesman-CookmeyerD, DingB, LommelSA, LucasWJ (1993) Cell-to-cell trafficking of macromolecules through plasmodesmata potentiated by the red clover necrotic mosaic virus movement protein. Plant Cell 5 : 1783–1794.
37. Giesman-CookmyerD, LommelSA (1993) Alanine scanning mutagenesis of a plant virus movement protein identifies three functional domains. Plant Cell 5 : 973–982.
38. TremblayD, VaewhongsAA, TurnerKA, SitTL, LommelSA (2005) Cell wall localization of Red clover necrotic mosaic virus movement protein is required for cell-to-cell movement. Virology 333 : 10–21.
39. KaidoM, FunatsuN, TsunoY, MiseK, OkunoT (2011) Viral cell-to-cell movement requires formation of cortical punctate structures containing Red clover necrotic mosaic virus movement protein. Virology 413 : 205–215.
40. MicheletL, ZaffagniniM, MorisseS, SparlaF, Pérez-PérezME, et al. (2013) Redox regulation of the Calvin-Benson cycle: something old, something new. Front Plant Sci 4 : 470.
41. FiggeRM, SchubertM, BrinkmannH, CerffM (1999) Glyceraldehyde-3-phosphate dehydrogenase gene diversity in eubacteria and eukaryotes: evidence for intra - and inter-kingdom gene transfer. Mol Biol Evol 16 : 429–440.
42. SitTL, VaewhongsAA, LommelSA (1998) RNA-mediated trans-activation of transcription from a viral RNA. Science 281 : 829–832.
43. WeberF, WagnerV, RasmussenSB, HartmannR, PaludanSR (2006) Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80 : 5059–5064.
44. MiorinL, Romero-BreyI, MaiuriP, HoppeS, Krijnse-LockerJ, et al. (2013) Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J Virol 87 : 6469–6481.
45. ChoiSH, SeoJ-K, KwonS-J, RaoALN (2012) Helper virus-independent transcription and multimerization of a satellite RNA associated with Cucumber mosaic virus. J Virol 86 : 4823–4832.
46. IgarashiA, YamagataK, SugaiT, TakahashiY, SugawaraE, et al. (2009) Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 386 : 407–416.
47. JoshiS, PleijCWA, HaenniAL, ChapevilleF, BoschL (1983) Properties of the tobacco mosaic virus intermediate length RNA-2 and its translation. Virology 127 : 100–111.
48. WatanabeY, EmoriY, OoshimaI, MeshiT, OhnoT, et al. (1984) Synthesis of TMV-specific RNAs and proteins at the early stage of infection in tobacco protoplast: transient expression of the 30K protein and its mRNA. Virology 133 : 18–24.
49. MizumotoH, IwakawaH, KaidoM, MiseK, OkunoT (2006) Cap-independent translation mechanism of Red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J Virol 80 : 3781–3791.
50. TilsnerJ, OparkaKJ (2012) Missing links? – The connection between replication and movement of plant RNA viruses. Curr Opin Virol 2 : 705–711.
51. TilsnerJ, LinnikO, LouveauxM, RobertsIM, ChapmanSN, et al. (2013) Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 201 : 981–995.
52. TilsnerJ, LinnikO, WrightKM, BellK, RobertsAG, et al. (2012) The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol 158 : 1359–1370.
53. PriceGD, EvansJR, von CaemmererS, YuJ-W, BadgerMR (1995) Specific reduction of chloroplast glyceraldehyde-3-phosphate dehydrogenase activity by antisense RNA reduces CO2 assimilation via a reduction in ribulose bisphosphate regeneration in transgenic tobacco plants. Planta 195 : 369–378.
54. MaglieryTJ, WilsonCGM, PanW, MishlerD, GhoshI, et al. (2005) Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc 127 : 146–157.
55. OkamotoK, NaganoH, IwakawaH, MizumotoH, TakedaA, et al. (2008) cis-Preferential requirement of a −1 frameshift product p88 for the replication of Red clover necrotic mosaic virus RNA1. Virology 375 : 205–212.
56. IwakawaH-O, MineA, HyodoK, AnM, KaidoM, et al. (2011) Template recognition mechanism by replicase proteins differ between bipartite positive-strand genomic RNAs of a plant virus. J Virol 85 : 497–509.
57. WangRY-L, NagyPD (2008) Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host Microbe 3 : 178–187.
58. PrasanthKR, HuangY-W, LiouM-R, WangRY-L, HuC-C, et al. (2011) Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of Bamboo mosaic virus and its associated satellite RNA. J Virol 85 : 8829–8840.
59. ShihM-C, LazarG, GoodmanHM (1986) Evidence in favor of the symbiotic origin of chloroplasts: Primary structure and evolution of tobacco glyceraldehyde-3-phophate dehydrogenase. Cell 47 : 73–80.
60. LinS-S, ChangSC, WangY-H, SunC-Y, ChangM-F (2000) Specific interaction between the Hepatitis delta virus RNA and glyceraldehyde 3-phophate dehydrogenase: An enhancement on ribozyme catalysis. Virology 271 : 46–57.
61. SiroverMA (1999) New insight into an old protein: The functional diversity of mammalian glyceraldehyde-3 - phosphate dehydrogenase. Biochim Biophys Acta 1432 : 159–184.
62. SiroverMA (2005) New nuclear functions of the glycolytic protein, glyceraldehyde-3 - phosphate dehydrogenase, in mammalian cells. J Cell Biochem 95 : 45–52.
63. HuangY-W, HuC-C, LinN-S, HsuY-H (2012) Unusual roles of host metabolic enzymes and housekeeping proteins in plant virus replication. Curr Opin Virol 2 : 676–682.
64. LinJ-W, DingM-P, HsuY-H, TsaiC-H (2007) Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucl Acids Res 35 : 424–432.
65. ChengS-F, HuangY-P, ChenL-H, HsuY-H, TsaiC-H (2013) Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol 163 : 1598–1608.
66. BhatS, FolimonovaSF, ColeAB, BallardKD, LeiZ, et al. (2013) Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement. Plant Physiol 161 : 134–147.
67. TakedaA, TsukudaM, MizumotoH, OkamotoK, KaidoM, et al. (2005) A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J 24 : 3147–3157.
68. XiongZG, LommelSA (1991) Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology 182 : 388–392.
69. MitsuharaI, UgakiM, HirochikaH, OhshimaM, MurakamiT, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37 : 49–59.
70. TamaiA, MeshiT (2001) Tobamoviral movement protein transiently expressed in a single epidermal cell functions beyond multiple plasmodesmata and spreads multicellularly in an infection-coupled manner. Mol Plant Microbe Interact 14 : 126–134.
71. HamamotoH, SugiyamaY, NakagawaN, HashidaE, MatsunagaY, et al. (1993) A new tobacco mosaic virus vector and its use for the systemic production of angiotensin-I-converting enzyme inhibitor in transgenic tobacco and tomato. Nature Biotech 11 : 930–932.
72. KaidoM, MoriM, MiseK, OkunoT, FurusawaI (1997) Auto-cleavable ribozyme sequence attached to brome mosaic virus cDNAs enhances accumulation of viral RNAs transcribed in vivo from the cDNAs. Ann Phytopathol Soc Jpn 63 : 95–98.
73. LiJ-F, ChungHS, NiuY, BushJ, McCormackM, et al. (2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25 : 1507–1522.
74. Navas-CastilloJ, Albiach-MartiMR, GowdaS, HilfME, GarnseySM, et al. (1997) Kinetics of accumulation of citrus tristeza virus RNAs. Virology 228 : 92–97.
75. KawaiT, GonoiA, NittaM, KaidoM, YamagishiN, et al. (2014) Virus-induced gene silencing in Apricot (Prunus armeniaca L.) and Japanese apricot (P. mume Siebold & Zucc.) with the Apple latent spherical virus vector system. J Jpn Soc Hort Sci 83 : 23–31.
76. MizumotoH, TatsutaM, KaidoM, MiseK, OkunoT (2003) Cap-independent translational enhancement by the 3′ untranslated region of Red clover necrotic mosaic virus RNA1. J Virol 77 : 12113–12121.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Acidification Activates Motility and Egress by Enhancing Protein Secretion and Cytolytic ActivityČlánek Plasticity between MyoC- and MyoA-Glideosomes: An Example of Functional Compensation in InvasionČlánek Coronavirus Cell Entry Occurs through the Endo-/Lysosomal Pathway in a Proteolysis-Dependent MannerČlánek NK Cell Activation in Human Hantavirus Infection Explained by Virus-Induced IL-15/IL15Rα Expression
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Peculiarities of Prion Diseases
- Inhibitors of Peptidyl Proline Isomerases As Antivirals in Hepatitis C and Other Viruses
- War and Infectious Diseases: Challenges of the Syrian Civil War
- Microbial Contamination in Next Generation Sequencing: Implications for Sequence-Based Analysis of Clinical Samples
- Acidification Activates Motility and Egress by Enhancing Protein Secretion and Cytolytic Activity
- Co-dependence of HTLV-1 p12 and p8 Functions in Virus Persistence
- Shed GP of Ebola Virus Triggers Immune Activation and Increased Vascular Permeability
- Plasticity between MyoC- and MyoA-Glideosomes: An Example of Functional Compensation in Invasion
- The Type III Translocon Is Required for Biofilm Formation at the Epithelial Barrier
- Retromer Regulates HIV-1 Envelope Glycoprotein Trafficking and Incorporation into Virions
- IFI16 Restricts HSV-1 Replication by Accumulating on the HSV-1 Genome, Repressing HSV-1 Gene Expression, and Directly or Indirectly Modulating Histone Modifications
- Coronavirus Cell Entry Occurs through the Endo-/Lysosomal Pathway in a Proteolysis-Dependent Manner
- Silencing by H-NS Potentiated the Evolution of
- Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions
- GAPDH-A Recruits a Plant Virus Movement Protein to Cortical Virus Replication Complexes to Facilitate Viral Cell-to-Cell Movement
- Genomic Insights into the Fungal Pathogens of the Genus : Obligate Biotrophs of Humans and Other Mammals
- Unravelling Human Trypanotolerance: IL8 is Associated with Infection Control whereas IL10 and TNFα Are Associated with Subsequent Disease Development
- The Skin Microbiome: A Focus on Pathogens and Their Association with Skin Disease
- Human Cytomegalovirus Vaccine Based on the Envelope gH/gL Pentamer Complex
- IL-37 Inhibits Inflammasome Activation and Disease Severity in Murine Aspergillosis
- Host-Specific Parvovirus Evolution in Nature Is Recapitulated by Adaptation to Different Carnivore Species
- Activation of HIV Transcription with Short-Course Vorinostat in HIV-Infected Patients on Suppressive Antiretroviral Therapy
- PUL21a-Cyclin A2 Interaction is Required to Protect Human Cytomegalovirus-Infected Cells from the Deleterious Consequences of Mitotic Entry
- Programmed Ribosomal Frameshift Alters Expression of West Nile Virus Genes and Facilitates Virus Replication in Birds and Mosquitoes
- Aminoterminal Amphipathic α-Helix AH1 of Hepatitis C Virus Nonstructural Protein 4B Possesses a Dual Role in RNA Replication and Virus Production
- NK Cell Activation in Human Hantavirus Infection Explained by Virus-Induced IL-15/IL15Rα Expression
- Structure and Specificity of the Bacterial Cysteine Methyltransferase Effector NleE Suggests a Novel Substrate in Human DNA Repair Pathway
- Genetics, Receptor Binding Property, and Transmissibility in Mammals of Naturally Isolated H9N2 Avian Influenza Viruses
- A Gatekeeper Chaperone Complex Directs Translocator Secretion during Type Three Secretion
- A Conserved Peptide Pattern from a Widespread Microbial Virulence Factor Triggers Pattern-Induced Immunity in
- Succinate Dehydrogenase is the Regulator of Respiration in
- The Plasmodesmal Protein PDLP1 Localises to Haustoria-Associated Membranes during Downy Mildew Infection and Regulates Callose Deposition
- Dysregulated B Cell Expression of RANKL and OPG Correlates with Loss of Bone Mineral Density in HIV Infection
- Restriction of Genetic Diversity during Infection of the Vector Midgut
- The Epithelial αvβ3-Integrin Boosts the MYD88-Dependent TLR2 Signaling in Response to Viral and Bacterial Components
- The Relationship between Host Lifespan and Pathogen Reservoir Potential: An Analysis in the System
- Multiple Roles of the Cytoskeleton in Bacterial Autophagy
- The Evolution and Genetics of Virus Host Shifts
- ChIP-seq and In Vivo Transcriptome Analyses of the SREBP SrbA Reveals a New Regulator of the Fungal Hypoxia Response and Virulence
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Coronavirus Cell Entry Occurs through the Endo-/Lysosomal Pathway in a Proteolysis-Dependent Manner
- Peculiarities of Prion Diseases
- Host-Specific Parvovirus Evolution in Nature Is Recapitulated by Adaptation to Different Carnivore Species
- War and Infectious Diseases: Challenges of the Syrian Civil War
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



