-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Nutrient Limitation Governs Metabolism and Niche Adaptation in the Human Nose
Colonization of the human nose by Staphylococcus aureus in one-third of the population represents a major risk factor for invasive infections. The basis for adaptation of S. aureus to this specific habitat and reasons for the human predisposition to become colonized have remained largely unknown. Human nasal secretions were analyzed by metabolomics and found to contain potential nutrients in rather low amounts. No significant differences were found between S. aureus carriers and non-carriers, indicating that carriage is not associated with individual differences in nutrient supply. A synthetic nasal medium (SNM3) was composed based on the metabolomics data that permits consistent growth of S. aureus isolates. Key genes were expressed in SNM3 in a similar way as in the human nose, indicating that SNM3 represents a suitable surrogate environment for in vitro simulation studies. While the majority of S. aureus strains grew well in SNM3, most of the tested coagulase-negative staphylococci (CoNS) had major problems to multiply in SNM3 supporting the notion that CoNS are less well adapted to the nose and colonize preferentially the human skin. Global gene expression analysis revealed that, during growth in SNM3, S. aureus depends heavily on de novo synthesis of methionine. Accordingly, the methionine-biosynthesis enzyme cysteine-γ-synthase (MetI) was indispensable for growth in SNM3, and the MetI inhibitor DL-propargylglycine inhibited S. aureus growth in SNM3 but not in the presence of methionine. Of note, metI was strongly up-regulated by S. aureus in human noses, and metI mutants were strongly abrogated in their capacity to colonize the noses of cotton rats. These findings indicate that the methionine biosynthetic pathway may include promising antimicrobial targets that have previously remained unrecognized. Hence, exploring the environmental conditions facultative pathogens are exposed to during colonization can be useful for understanding niche adaptation and identifying targets for new antimicrobial strategies.
Published in the journal: . PLoS Pathog 10(1): e32767. doi:10.1371/journal.ppat.1003862
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003862Summary
Colonization of the human nose by Staphylococcus aureus in one-third of the population represents a major risk factor for invasive infections. The basis for adaptation of S. aureus to this specific habitat and reasons for the human predisposition to become colonized have remained largely unknown. Human nasal secretions were analyzed by metabolomics and found to contain potential nutrients in rather low amounts. No significant differences were found between S. aureus carriers and non-carriers, indicating that carriage is not associated with individual differences in nutrient supply. A synthetic nasal medium (SNM3) was composed based on the metabolomics data that permits consistent growth of S. aureus isolates. Key genes were expressed in SNM3 in a similar way as in the human nose, indicating that SNM3 represents a suitable surrogate environment for in vitro simulation studies. While the majority of S. aureus strains grew well in SNM3, most of the tested coagulase-negative staphylococci (CoNS) had major problems to multiply in SNM3 supporting the notion that CoNS are less well adapted to the nose and colonize preferentially the human skin. Global gene expression analysis revealed that, during growth in SNM3, S. aureus depends heavily on de novo synthesis of methionine. Accordingly, the methionine-biosynthesis enzyme cysteine-γ-synthase (MetI) was indispensable for growth in SNM3, and the MetI inhibitor DL-propargylglycine inhibited S. aureus growth in SNM3 but not in the presence of methionine. Of note, metI was strongly up-regulated by S. aureus in human noses, and metI mutants were strongly abrogated in their capacity to colonize the noses of cotton rats. These findings indicate that the methionine biosynthetic pathway may include promising antimicrobial targets that have previously remained unrecognized. Hence, exploring the environmental conditions facultative pathogens are exposed to during colonization can be useful for understanding niche adaptation and identifying targets for new antimicrobial strategies.
Introduction
Staphylococcus aureus is a major cause of human invasive infections ranging from superficial skin and soft tissue infections to severe disseminated diseases such as sepsis and endocarditis [1]. S. aureus is also a human commensal and part of the microbiota in healthy individuals, which facilitates its access to sterile tissues via open wounds and catheter entry sites. S. aureus can be isolated from various human body surfaces such as the pharynx, axillae and perineum but its main ecological niche and reservoir is known for long to be the human nose [2]–[4]. In contrast, coagulase-negative staphylococci (CoNS), such as Staphylococcus epidermidis, have a much lower virulence potential and use different areas of the skin as their major habitats [5]. The basis of staphylococcal host and niche-specificity has remained unknown.
Analysis of nasal carriage over long time periods has identified three types of S. aureus carriers [6]. About 20% of the human population can be regarded as non-carriers, who are never or only in very rare instances colonized with low bacterial numbers. In contrast, intermittent carriers show alternating periods of non-carrier status and colonisation by various S. aureus strains. The number of bacteria per isolation can be highly variable. The third group of roughly 20% persistent carriers is characterised by the presence of S. aureus in nearly all nasal swabs, usually at high bacterial numbers and with one specific strain per person over time. Recently, it has been suggested to distinguish only between carriers and non-carriers because of similar S. aureus nasal elimination kinetics and anti-staphylococcal antibody profiles in intermittent - and non-carriers [7].
Recent studies have shown that being an S. aureus carrier bears a higher risk of invasive S. aureus infections, predominantly by the carriers' own strain [8], but a lower risk of infection-associated mortality compared to being a non-carrier [9]. The reasons for the underlying predisposition, which may involve individual differences in epithelial ligands for bacterial adhesins, local host defense, or availability of nutrients in the nose, have remained unclear. While some polymorphisms in immunity-related genes are weakly associated with the carrier status, the human predisposition appears to have multifactorial reasons [10]. On the bacterial side several factors required for nasal colonization have been identified. Wall teichoic acid (WTA) polymers at the staphylococcal surface [11], [12] and cell-wall anchored proteins, such as ClfB [13], [14] and IsdA [15], have been found to be required for nasal colonization and adhesion to nasal epithelial cells. While the epithelial receptor for WTA is still unknown, ClfB and IsdA bind to cytokeratin 10 and loricrin, major components of human squamous epithelial cells, and IsdA also binds to the matrix protein involucrin [15], [16]. Recently, up-regulation of WTA-biosynthetic genes tagO and tarK and of clfB and isdA during nasal colonisation has been shown in nasal swab samples from human volunteers [17] and in the cotton rat model of S. aureus nasal colonisation [18], underscoring the importance of these factors in nasal colonization. S. aureus encounters iron-limiting conditions in the nose because isdA expression is strictly dependent on iron limitation [19], and haemoglobin has recently been shown to promote S. aureus nasal colonization [20].
The human body can be regarded as a chemostat, where the nutrients required for bacterial growth are replenished over time and allow growth of bacteria within human microenvironments [21]. While several S. aureus nasal adhesion factors have been studied in the past, nothing is known about the growth conditions in nasal fluid such as the availability of carbon and nitrogen sources and the metabolic activities of S. aureus in its nasal habitat. Knowledge of metabolite availabilities and utilisation patterns could direct the identification of important metabolic enzymes that are essential during infection or colonization by S. aureus and could serve as targets for new antibiotics. Along this line enzymes from the folic acid biosynthetic pathway or the isoleucyl-tRNA synthetase are valuable targets for widely used anti-staphylococcal antibiotics such as cotrimoxazole or mupirocin, respectively. Mupirocin is frequently used to eliminate S. aureus from the noses of high-risk patients [22], but the increasing resistance to mupirocin [23] and almost all antibiotics used to prevent or treat staphylococcal infections puts urgency to the development of new antimicrobial compounds. For this purpose the most critical and ‘drugable’ metabolic pathways of S. aureus need to be identified.
In this study we elucidated the abundance of potential nutrients for S. aureus in the human nose by metabolomics analysis of nasal secretions. We found a high diversity but low concentrations of metabolites and no significant differences in the composition of secretions from S. aureus carriers and non-carriers. A synthetic nasal medium (SNM3) was composed based on the metabolomics data. S. aureus growth in SNM3 led to similar gene expression patterns as during in vivo colonization. While S. aureus isolates grew steadily in SNM3, CoNS did not, indicating that S. aureus is particularly well adapted to life in the human nose. Analysis of global gene expression in SNM3 revealed that the methionine-biosynthetic pathway may be a critical target for new anti-colonization drugs. In support of this notion an inhibitor of methionine biosynthesis had antimicrobial activity against S. aureus in SNM3 but not in complex media. The inhibitor's staphylococcal target gene was strongly up-regulated during human nasal colonization, and deletion of the target gene led to reduced colonisation ability of the respective mutant in the cotton rat colonisation model. Thus, deciphering the in vivo metabolism of pathogens represents a valuable strategy for defining new antimicrobial targets.
Results
Human nasal secretions have similar composition in S. aureus carriers and non-carriers
The abundance of potential nutrients in nasal secretions has never been described. In order to explore the metabolic lifestyle of S. aureus during nasal colonization, the amounts of small organic compounds in secretions from eight volunteers who were not carriers of S. aureus, were analyzed by metabolomics (Fig. 1A, Table 1). Similar amounts of amino acids and organic acids were found in the micromolar range in the eight samples. Urea was by far the most abundant organic substance at concentrations of 2.5–7.5 mM. Glucose exhibited the highest concentrations among carbohydrates with large variation between 35 µM and ca. 1 mM, while only very low amounts of other mono - or disaccharides were detected. Most of the proteinogenic amino acids and ornithine were present at average concentrations between 50 and 150 µM, while tryptophan and cysteine were detected only at very low concentrations around 10 µM. Some amino acids were not found (methionine, glutamine, tyrosine, isoleucine, asparagine, and aspartate). Whereas the carboxylic acids fumarate, malate and citrate were detected at about 5–25 µM, pyruvate and succinate were usually present at much higher concentrations (Table 1). No lactate and only trace amounts of several other substances, including fatty acids, cholesterol and pyrimidines, were found (Fig. 1B).
Fig. 1. Metabolite composition of nasal secretions in S. aureus carriers and non-carriers. 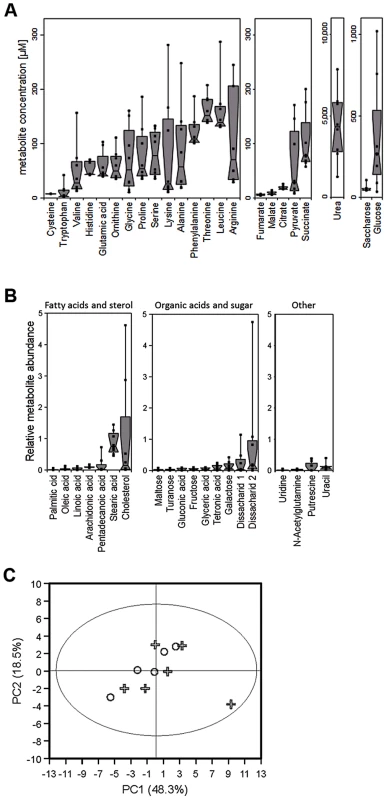
(A, B) Metabolite quantification by GC-MS from eight sample donors, micromolar concentrations are given for compounds with standard dilutions measured (A), for other substances relative abundances are given against internal standards (B). Upper and lower box limits and the horizontal lines within the boxes represent 25 and 75% percentiles and the medians, respectively. The whiskers of the plots indicate minimum and maximum range, additionally showing the data points. (C) Differences in metabolite profiles in nasal secretions from six carriers (crosses) and seven non-carriers (circles) analyzed by principal component analysis of all metabolite levels. Principal component 1 vs. 2 is shown, explaining in total 67.9% of the PCA model. Metabolite values were log-transformed and scaled by unit variance. Tab. 1. Concentration ranges of metabolites and inorganic ions in human nasal secretions and amounts of these compounds in the chemically defined medium SNM. 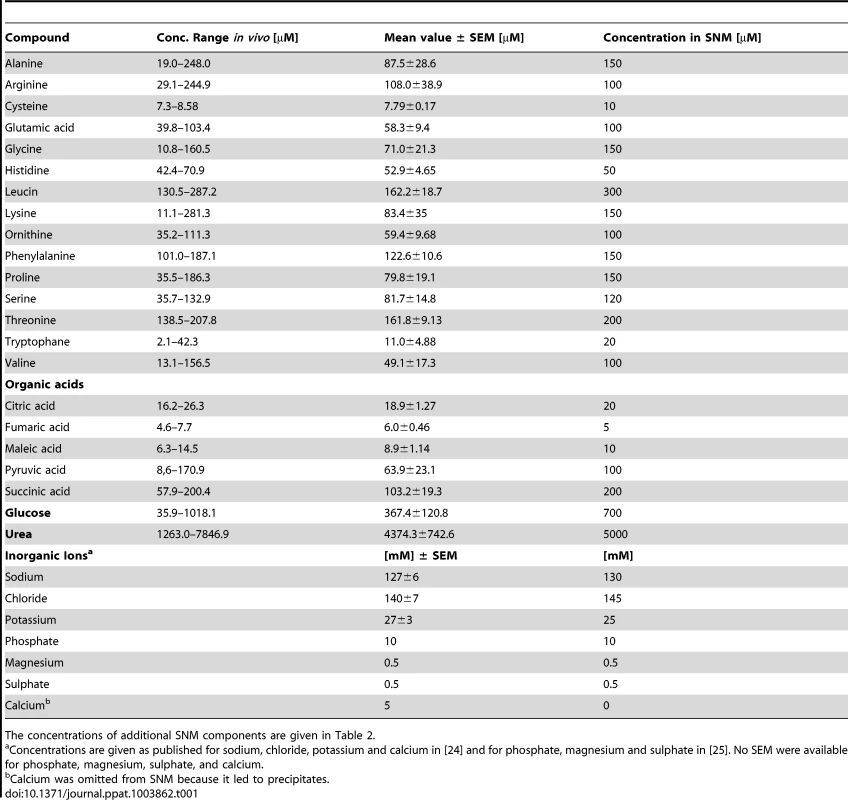
The concentrations of additional SNM components are given in Table 2. To confirm the observed results and to investigate potential differences between S. aureus carriers and non-carriers, the metabolite composition in nasal secretions from six S. aureus non-carriers were compared with those from seven S. aureus carriers. The metabolite patterns of carriers exhibited a similar degree of variation as in non-carriers but no significant difference between the two groups of donors for any of the detected compounds (Fig. 1C). Thus, the human S. aureus carrier status is not associated with a notable difference in nasal nutrient supply, and the metabolic activities of S. aureus do not seem to have a major impact on the overall metabolite concentrations in the nose.
Synthetic nasal medium (SNM) permits steady growth of S. aureus
The average nutrient concentrations found in human nasal secretions were used to compose a synthetic medium for simulating S. aureus growth in the nose (Table 1). Amino acids and glucose were added to SNM at the upper limit of the detected concentration ranges found in the samples but not at higher amounts than twice the mean concentration values. The amounts of inorganic ions in nasal secretions have previously been described [24], and these values served as a basis for the salt content of SNM. The synthetic medium was buffered with 10 mM phosphate buffer (pH 7.2), which corresponds to the previously described nasal phosphate content [25]. The concentrations of essential cofactors such as vitamins and trace elements in nasal secretions were below detection limits. Since earlier studies had shown that S. aureus requires minimal amounts of these compounds [26], highly diluted standard vitamin and trace element solutions were included in SNM. Moreover, because recent gene expression data have shown that S. aureus encounters iron-limited conditions in the human nose [17], iron was omitted from the trace element solution, and SNM was supplemented with 200 µM of the iron-complexing agent 2, 2′-bipyridin, which has been shown to confer iron limitation in S. aureus [27]. The concentrations of inorganic salts, 2, 2′-bipyridine, trace elements, and cofactors in SNM are listed in Table 2.
Tab. 2. Inorganic salt, 2, 2′-bipyridine, trace element, and cofactor concentrations in SNM. 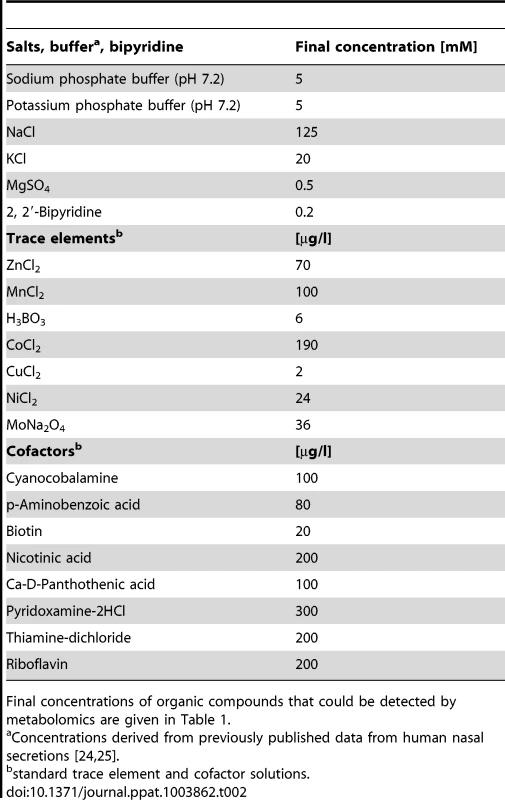
Final concentrations of organic compounds that could be detected by metabolomics are given in Table 1. The community-associated methicillin-resistant S. aureus (CA-MRSA) clinical isolate USA300 LAC and the laboratory strain Newman were used to evaluate if S. aureus is able to grow in SMN. Despite the very low amount of 238 mg amino acids per liter, both strains showed reproducible but moderate growth in SNM. This is in agreement with the rather low bacterial numbers found in swabs from the human nose [28]. In contrast, much higher bacterial densities have been found in microbiomes which are in contact with ingested food e.g. in the human mouth or gut [29], [30]. When we increased the concentration of amino acids, organic acids and glucose in SNM, the maximal bacterial densities also increased until they reached a plateau at approximately 20-fold nutrient concentration for S. aureus USA300 (Fig. 2A). Because epithelial secretions are continuously produced and removed, nutrient concentrations should remain more or less constant at the surface of nasal tissues, while they are continuously decreasing in a test tube culture. In agreement with this notion S. aureus Newman reached 2.4-fold higher bacterial numbers when grown in SNM in a continuous flow system compared to static SNM (Supplementary Figure S1). Because several of the subsequent experiments involved growth on SNM agar plates or in multiple parallel cultures, which could not be performed in a continuous flow system, we used SNM with threefold increased amino acid, organic acid and glucose concentration (SNM3) in subsequent experiments.
Fig. 2. Capacities of S. aureus and CoNS strains to grow in synthetic nasal medium (SNM). 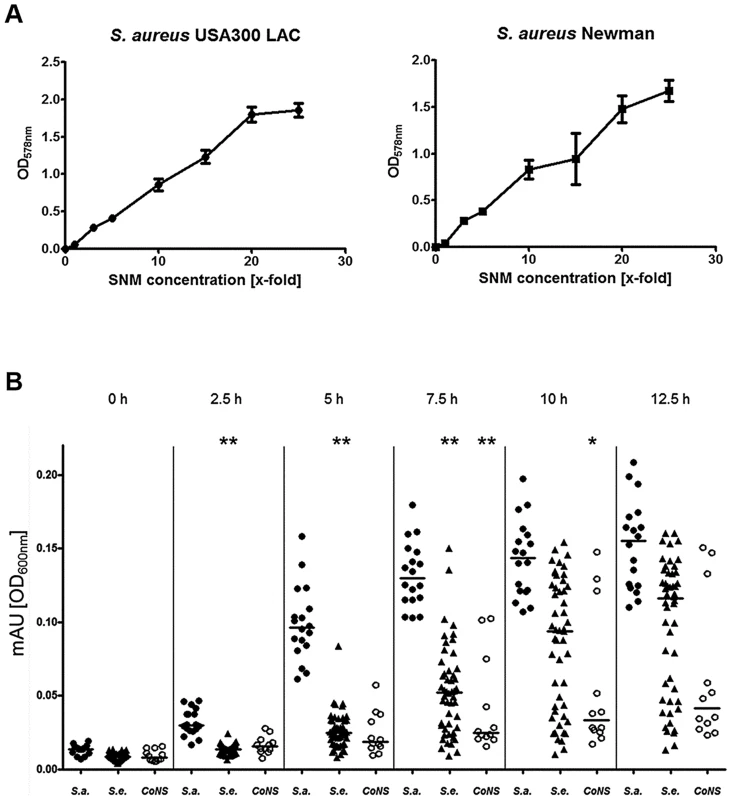
(A), growth of S. aureus USA300 LAC and S. aureus Newman in SNM with increasing (x-fold) concentrations of amino acids, organic acids and glucose after 90 h growth. The mean values and SEM for three independent cultures of each concentration is shown. (B) Growth of nasal isolates of S. aureus (filled circles), S. epidermidis (triangles) and other coagulase-negative staphylococci (CoNS) (open circles) in liquid SNM3 after the indicated incubation times. The means at each time point are shown as horizontal lines for each group. Statistically significant differences vs. S. aureus calculated by the ANOVA Kruskal-Wallis test with Post-test Dunns and 95% confidence intervals are indicated: *, p≤0.05; **, p≤0.01. Cultures were vigorously shaken at 37°C in A and B. S. aureus is better adapted to growth in SNM3 than CoNS
In order to validate the capacity of SNM3 to simulate living conditions for staphylococci in the human nose, 87 different staphylococcal strains from the anterior nares of 37 human volunteers were isolated, and growth of the corresponding 18 S. aureus, 57 S. epidermidis, and 12 other CoNS strains (Staphylococcus capitis, Staphylococcus lugdunensis, Staphylococcus warneri, Staphylococcus hominis) in SNM3 was compared.
All S. aureus grew in SNM3 liquid cultures without long lag-phases and reached their highest densities after about 12.5 hours (Fig. 2B). When serial dilutions of the same strains were spotted on SNM3 agar, which should correspond better to the sessile lifestyle on the nasal epithelial surface than liquid SNM3, identical CFUs as on complex medium agar (basic medium, BM) were found for all S. aureus except for two strains (Fig. 3A). This indicates that SNM3 offers efficient growth conditions for the vast majority of nasal S. aureus strains. In contrast, most of the S. epidermidis and other CoNS grew in liquid SNM3 only after long lag-phases and often with much longer generation times compared to S. aureus (Fig. 2B). Moreover, only a very small percentage of cells (one to ten out of a million viable cells) of the S. epidermidis and other CoNS strains were able to form colonies on SNM3 agar (Fig. 3A). This property appeared to be a stable trait because sub-cultivation of such outgrowing clones resulted in much higher numbers of colonies on SNM3 compared to the parental clones (data not shown). Hence, most S. aureus appear to be metabolically well adapted to life in the human nose, whereas the vast majority of CoNS exhibited arrested growth with only a small minority of cells starting multiplication on SNM3 agar. These differences reflect recent findings that the nose of permanent S. aureus carriers usually contains substantially lower numbers of CoNS than S. aureus [7], [31].
Fig. 3. Colony formation abilities on SNM agar of S. aureus, S. epidermidis and other CoNS strains isolated from human nares. 
(A) Serial dilutions of all nasal isolates grown in BM overnight were spotted on BM or 3× SNM (SNM3) agar. The y-axis shows the percentage of cells forming colonies on SNM3 compared to BM. The influence of enhanced nutrient concentrations in 5×, 10×, and 20× SNM (B) or of 10–100 µM methionine in SNM3 (C) on the colony-forming ability was analyzed with a subset of strains shown in Figure 3A. S. aureus is shown as filled circles, S. epidermidis as triangles and other coagulase-negative staphylococci (CoNS) as open circles. The horizontal bar indicates the median of each group of strains. Statistically significant differences calculated by the ANOVA Kruskal-Wallis test with Post-test Dunns and 95% confidence intervals are indicated: *, p≤0.05; **, p≤0.01; ***, p≤0.001. To investigate if S. aureus is simply better adapted to dilute nutrient concentrations than CoNS, a selection of strains, whose ability to form colonies on SNM3 was in the median range, was tested for colony formation on SNM agar with three, five, ten, and twenty-fold concentrated nutrients. As shown in Figure 3B, concentrating nutrients in SNM agar plates ten to twenty-fold improved the outgrowth of most of the tested S. epidermidis and some other CoNS strains, but only some strains reached similar growth capacities as S. aureus, while most formed colonies at several magnitudes lower numbers than S. aureus even at the highest nutrient concentrations. Thus, CoNS appear to be much less capable of adapting to diluted nutrient concentrations than S. aureus and exhibit enormous intra-species variation in their capacities to grow on SNM3 agar, even when nutrient concentrations were strongly increased.
Expression of S. aureus marker genes in SNM3 is similar to that in the nose
Two recent studies have described if and how efficiently critical S. aureus genes are transcribed during nasal colonization of humans or cotton rats [17], [18]. In order to evaluate if growth in SNM3 leads to similar transcriptional profiles as found during in vivo colonization, we compared marker gene expression of S. aureus USA300 actively growing in SNM3 or BM by quantitative RT-PCR (qRT-PCR) (Fig. 4). Bacteria grown to stationary phase in BM (BM-stat) were also included because this growth condition has recently been used to assess intranasal expression of relevant S. aureus marker genes [17], [18]. The global virulence regulator RNAIII, which responds to the concentration of a secreted agr autoinducer peptide (quorum sensing), and the agr-controlled psmß have been shown to be only moderately expressed in the nose [17], [18], and artificial induction of the RNAIII transcript reduces the nasal colonization capacity of S. aureus in the cotton rat model [20]. These previous findings corresponded to the low expression of RNAIII and psmß in BM and SNM3 compared to expression in BM stat. Also, the keratin-binding adhesin gene clfB was expressed in SNM3 at a similar level as in growing BM cultures, corresponding to the expression profile found recently in human noses [17]. Moreover, expression of the iron-regulated isdA and the lytic transglycosylase gene sceD, found to be up-regulated in the nose compared to BM or BM-stat cultures [17], [18], was also enhanced in SNM3. Taken together, these data indicate that the composition of SNM3 provides suitable conditions for in vitro simulation of S. aureus growth and gene expression in the nasal habitat.
Fig. 4. Relative gene expression of selected genes of S. aureus USA300 LAC in BM and SNM. 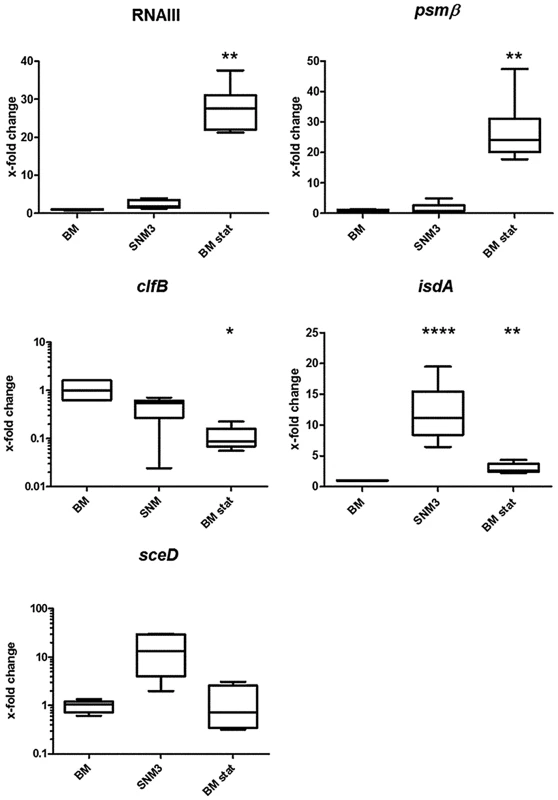
Gene expression was assayed by qRT-PCR for at least 6 independent cultures growing in BM or SNM3 or in stationary phase BM cultures (BM stat) each. Analyzed transcripts represent RNAIII, phenol soluble modulins β1 and β2 (psm), clumping factor B (clfB), iron-regulated surface determinant A (isdA), and a lytic transglycosylase (sceD). The upper and lower box limits and the horizontal lines within the boxes represent 25 and 75% percentiles and the means, respectively. The whiskers of the plots indicate minimum and maximum values. Statistically significant differences vs. BM calculated by the unpaired, two-tailed Student's t test with Welch's correction are indicated: *, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001. Global S. aureus gene expression in SNM3 reveals an important role of amino acid anabolism during nasal colonization
SNM3-grown S. aureus cultures enabled us to monitor global gene expression under conditions reflecting nasal colonization and to compare these with expression profiles from previous studies, which have usually used S. aureus grown in complex media. RNA from S. aureus USA300, actively growing either in SNM3 or BM, was hybridized to Affymetrix microarrays and analyzed with respect to basic cellular and metabolic pathways (data deposited under GEO Series accession number GSE43712). Multivariate data analysis was used to show differences or similarities between the transcriptomic data. Principal component analysis (PCA) confirmed that the three biological replicates performed for each of the two conditions led to very reproducible results, with substantial differences in SNM3 or BM-derived transcription profiles and a PCA mapping value of 77.7% (Fig. 5A).
Fig. 5. Differences in the transcriptome of S. aureus USA300 grown in SNM3 and complex medium (BM). 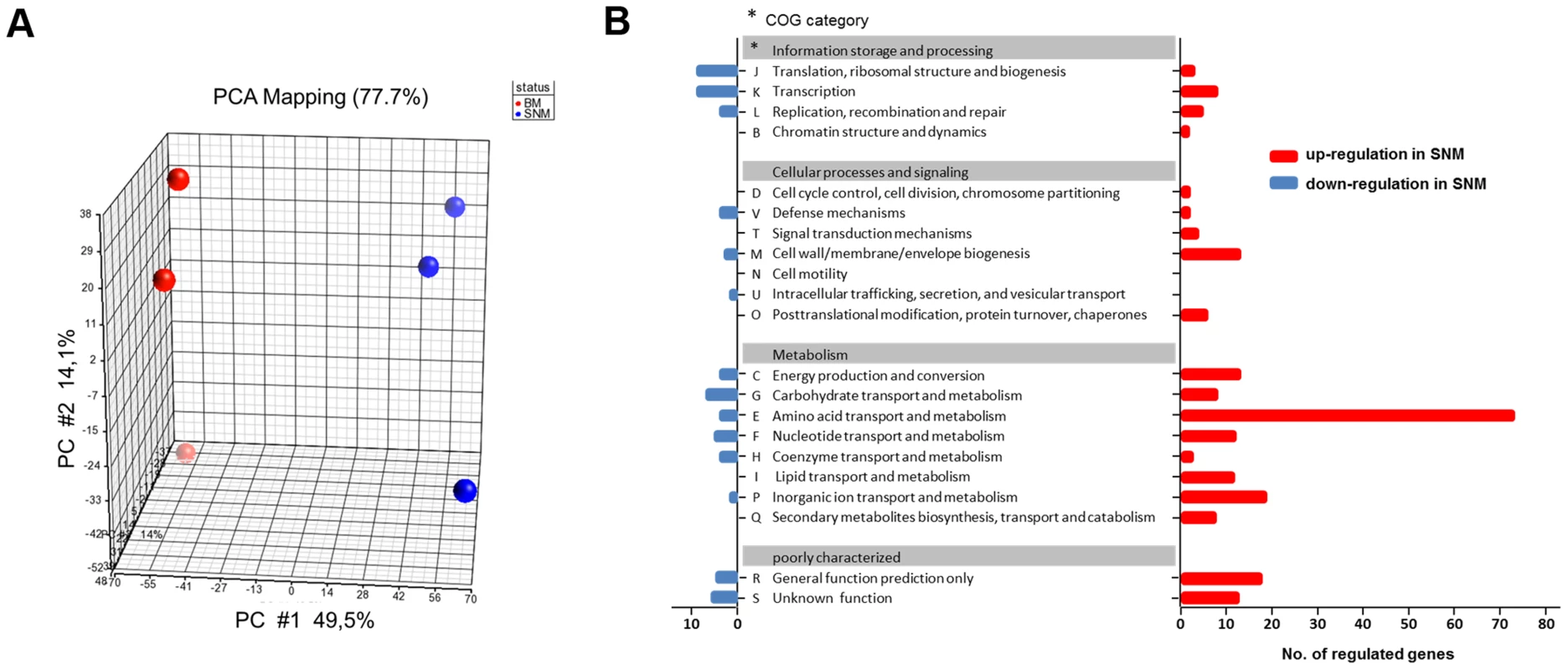
(A) PCA mapping of transcriptome profiles from BM compared to SNM3 cultures from microarray experiments. Each sphere represents an individual GeneChip result with the plotted location based upon the correlation of each sample relative to the others. Results from three independent BM and SNM cultures are compared. (B) Functional classes of genes (COG, Clusters of Orthologous Groups of proteins) detected in the microarray analysis, which were more than two-fold up- (red) or down-regulated (blue) and p<0.05 in SNM3 compared to complex medium (BM) are shown. The x-axis indicates the number of differentially regulated genes in each COG subgroup. The genes of subgroups E, M, and P are listed in detail in the supplementary Figure S2. Expression of 521 signals corresponding to 341 genes differed more than two-fold upon growth in SNM3 vs. BM with p-values below 0.05 (Fig. 5B). Thus, complex media represent a very artificial situation for S. aureus that differs profoundly from colonization-related conditions. A total of about 12.6% of the genes categorized in the “Clusters of Orthologous Groups of proteins” (COG) database [32] as being involved in “cellular processes and signalling” (Fig. 5B) showed more than two-fold expression differences between the two growth conditions, with 22 genes being up - and only six down-regulated. Most pronounced in this group was the two - to threefold higher expression of various capsule biosynthesis genes (functional category M) in SNM3 compared to BM (Supplementary Fig. S2). This finding is in agreement with the crucial role of the capsule in S. aureus nasal colonization recently shown in a rodent model [33]. The up-regulated signals also included the genes for the iron-regulated IsdC surface protein and the osmoprotectant system OpuCC and OpuD. In the group of genes categorized for “information storage and processing” 10.6% of the genes were differentially expressed, 13 genes were up - and 20 down-regulated including various transcriptional regulators.
As expected, most of the major differences were found among genes governing the central “metabolism” (26.6% of all genes assigned to this group), in which 54% of all genes from the functional group E (“amino acid transport and metabolism”) differed more than twofold in expression between the chosen growth conditions. Here, 79 genes were up - and only three down-regulated in SNM3 compared to BM (Supplementary Fig. S2). Significant up-regulation of amino acid biosynthesis genes was observed for glutamate, histidine, lysine, valine, leucine, isoleucine and methionine in SNM3 compared to BM. Remarkably, all of these amino acids, except for methionine and isoleucine, are integral components of SNM3. Whereas isoleucine can easily be generated from threonine, methionine has to be synthesized from aspartate, which was also not detectable in nasal secretions and therefore was not included in SNM3. Via O-acetyl-L-homoserine, aspartate reacts in a transsulfuration reaction with cysteine to form L-cystathionine and L-homocysteine (Fig. 6). The genes for cystathionine-γ-synthase (metI, SAUSA300_0360) and cystathionine-β-lyase (metC, SAUSA300_0359), whose gene products are responsible for these enzymatic reactions, were 26 - to 32-fold up-regulated and exhibited the strongest up-regulation of all genes in SNM3. Among the methionine-biosynthetic genes those for MetE (5-methyltetrahydropteroyltriglutamate-homocysteine S-methyl-transferase) and its homologue MetF SAUSA300_0358 (bifunctional homocysteine S-methyltransferase), responsible for the conversion of L-homocysteine to L-methionine, were 13 to 14-fold up-regulated. The strong up-regulation of two L-methionine ABC-transport systems (SAUSA300_0435-0437 and SAUSA300_0796-0798) underscored the importance of methionine supply during growth in SNM3. In addition to the biosynthetic operons of the above mentioned amino acids, many ABC-type dipeptide/oligopeptide (opp genes) as well as metal ion transporters (especially for iron-siderophores), were strongly up-regulated in SNM3 (Supplementary Fig. S2, functional category P). The presence of iron-limiting conditions in SNM3 was reflected by distinct up-regulation of the operon for biosynthesis of staphyloferrin B (also called staphylobactin, sbnABCDEFGHI; SAUSA300_0118 to SAUSA300_0126), a potent siderophore facilitating the extraction of iron from human transferrin [34]. The ferric uptake regulator (Fur) controls the expression of iron-regulated genes via binding to a consensus sequence in the promoter regions in staphylococci [35]. Besides the sbn operon additional Fur-regulated genes and operons, as listed at the RegPrecise regulon site (http://regprecise.lbl.gov/RegPrecise/regulon.jsp?regulon_id=6608), exhibited slightly to moderately altered expression in SNM3 compared to BM. These included the isd genes (iron-regulated surface determinant system; also called sir; staphylococcal iron-regulated proteins) and those for the ferritin storage protein, the TatAC system and the ferrichrome ABC-transporter SstABCD (supplementary Figure S3).
Fig. 6. Schematic representation of the methionine-biosynthetic pathway of S. aureus USA300 (adapted from KEGG pathway database). 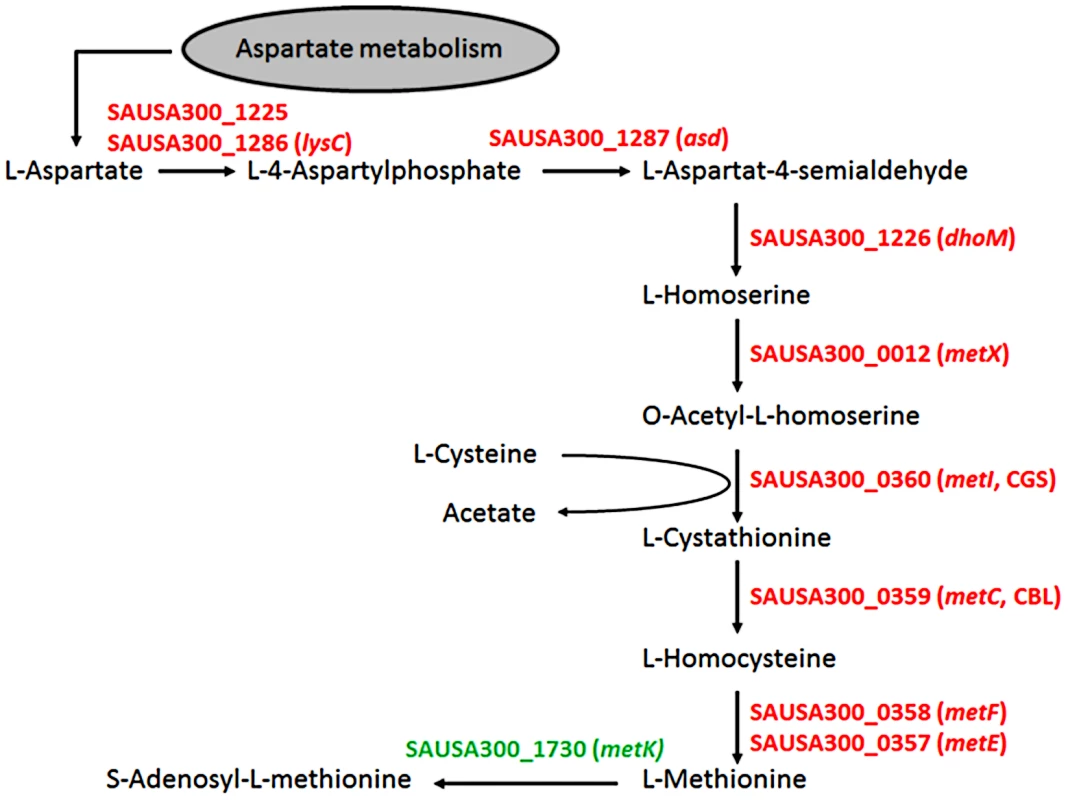
All genes shown in red were up-regulated in SNM3 compared to BM as determined by microarray analysis, with SAUSA300_0357 to SAUSA300_0360 exhibiting the strongest effects (13 to 32-fold). The metK gene, shown in green, was expressed at similar levels in BM and SNM3. The cystathionine-γ-synthase (MetI) represents the target for DL-propargylglycin. To evaluate the validity of the microarray results, gene expression of selected S. aureus USA300 genes, which were strongly up-regulated in SNM3, was reinvestigated by qRT-PCR. Besides metI (methionine biosynthesis; SAUSA300_0360) and the methionine transporter gene metN (SAUSA300_0435), expression was analyzed for hisC (histidine biosynthesis; SAUSA300_2610), aspartate kinase (SAUSA300_1225), oligopeptide transporter oppB (SAUSA300_0201) and staphyloferrin B biosynthesis gene sbnC (SAUSA300_0120). As shown in Figure 7, all of the investigated genes exhibited significant up-regulation in SNM3 compared to BM, thereby confirming the microarray data.
Fig. 7. Validation of relative gene expression of selected genes up-regulated in the microarrays by qRT-PCR. 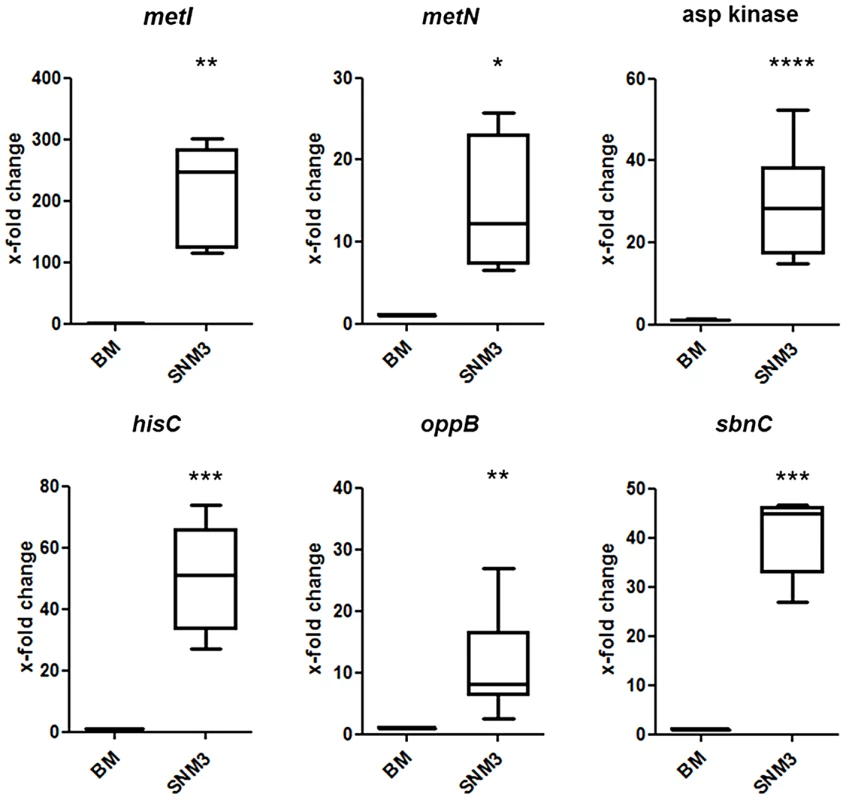
Results from at least six independent BM or SNM3 cultures each are depicted as the x-fold change of expression of the respective genes in SNM3 vs. BM. Analyzed transcripts represent cysteine-γ-synthase (metI), a methionine transporter (metN), aspartate kinase, histidine biosynthesis gene (hisC), oligopeptide permease (oppB), and staphyloferrin B biosynthesis gene (sbnC). The upper and lower box limits and the horizontal lines within the boxes represent 25 and 75% percentiles and the means, respectively. The whiskers of the plots indicate minimum and maximum range. Significant differences calculated by the unpaired, two-tailed Student's t test with Welch's correction are indicated: * p≤0.05; **p≤0.01; *** p≤0.001; **** p≤0.0001. The methionine-biosynthetic pathway represents a potential target for antimicrobial strategies
The strong expression of methionine-biosynthetic genes prompted us to investigate if the absence of methionine is significantly limiting the growth of S. aureus USA300 in SNM3. However, the step-wise increase of methionine in SNM3 had no effect on colony formation on SNM3 agar (Fig. 3C) and only a marginal impact on S. aureus growth in liquid SNM3 (data not shown). Hence, methionine limitation does not compromise S. aureus growth, probably as a result of efficient ways to synthesize methionine. We also investigated if methionine limitation might be a reason for the inefficient outgrowth of CoNS on SNM3 agar. For this purpose, we analyzed colony formation of various nasal CoNS isolates and the laboratory strain S. epidermidis 1457 on SNM3 agar with increasing methionine concentrations (Fig. 3C). However, even high methionine amounts of 100 µM increased colony formation of S. epidermidis and CoNS strains only slightly, and only a small minority of strains benefitted strongly from the addition of methionine. This finding suggests that the inability of CoNS to thrive on SNM3 agar is probably the result of complex differences in the metabolic or regulatory properties of S. aureus and CoNS.
In order to analyze the importance of the capacity of S. aureus to synthesize methionine, the gene of the methionine-biosynthetic enzyme MetI was inactivated in S. aureus Newman. The resulting mutants showed no growth defects in complex medium. However, they were completely unable to grow in SNM3, while addition of methionine restored growth of the mutants, thereby demonstrating a crucial role of MetI under colonization-related conditions (Supplementary Fig. S4). Antimicrobial targets for new decolonization drugs are urgently needed because of increasingly emerging mupirocin resistant S. aureus [23]. The synthetic compound DL-propargylglycine has been shown to inhibit the bacterial cytathionine-γ-synthase MetI leading to a block of methionine biosynthesis [36]. We hypothesized that it may have antimicrobial activity against S. aureus under conditions where MetI plays a crucial role. DL-propargylglycine hardly affected growth of S. aureus USA300 in BM (minimal inhibitory concentration, MIC>10 mg/ml), whereas it inhibited growth of all tested S. aureus strains in SNM3 (Table 3). These data suggest that MetI inhibitors might in fact be useful to limit the growth of S. aureus in vivo. Supplementation of SNM3 with methionine abrogated the antimicrobial activity of DL-propargylglycine (MIC>10 mg/ml) confirming that DL-propargylglycine inhibits S. aureus growth by blocking an indispensable step of methionine biosynthesis.
Tab. 3. MIC values of DL-propargylglycine for S. aureus strains in complex medium (BM) and SNM3. 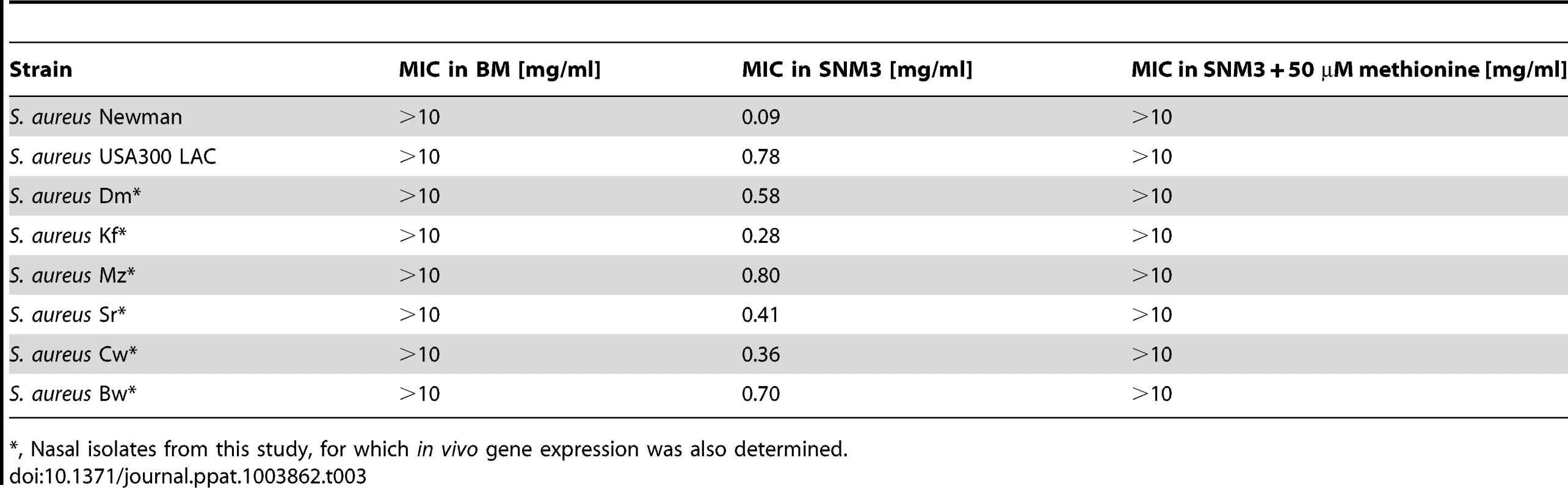
, Nasal isolates from this study, for which in vivo gene expression was also determined. metI is strongly expressed during S. aureus colonization of the human nose and is crucial for efficient nasal colonization of cotton rats
In order to analyze if the critical genes found to be up-regulated by S. aureus USA300 in SNM3 vs. BM exhibit similar expression profiles during nasal colonization, their transcription was measured> by qRT-PCR in nasal swabs from six documented S. aureus carriers. Their corresponding nasal S. aureus strains were subsequently grown in SNM3 and BM for RNA isolation, and all samples were analyzed by qRT-PCR (Fig. 8). For some samples expression of certain genes was not detectable, possibly as a result of sequence variation at primer binding sites or low RNA concentrations.
Fig. 8. Relative expression of selected genes in nasal swabs from six S. aureus carriers (in vivo) compared to growth of the corresponding strains in BM or SNM3. 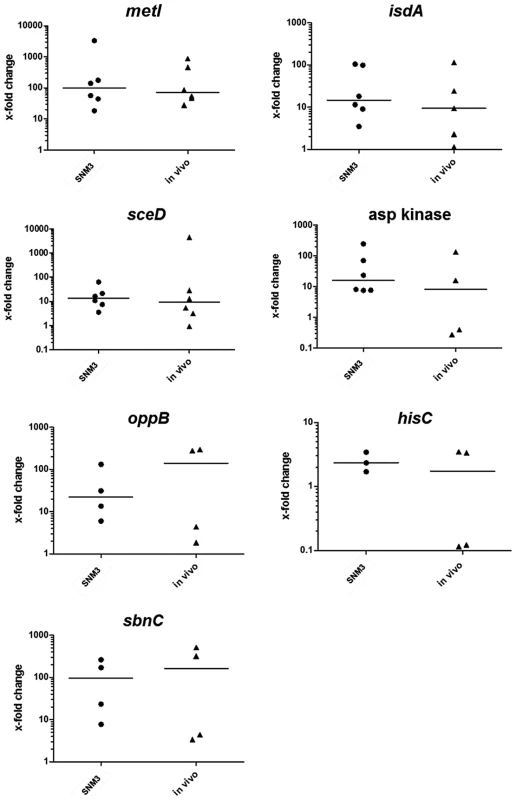
The relative gene expression levels in nasal swabs (in vivo) and SNM3 cultures measured by qRT-PCR were compared with mRNA level of the corresponding strain in BM, which was defined as 1. Each symbol per group (SNM3 or in vivo) represents an independent nasal isolate. The means per group are indicated as horizontal lines. Less than six data points per group are presented for some of the genes since some samples yielded no expression signals. Analyzed transcripts represent cysteine-γ-synthase (metI), iron-regulated surface determinant A (isdA), a lytic transglycosylase (sceD), aspartate kinase, oligopeptide permease (oppB), histidine biosynthesis gene (hisC), and staphyloferrin B biosynthesis gene (sbnC). The expression patterns in SNM3 and in vivo were overall very similar, and none of the analyzed genes exhibited significant differences in vivo and in SNM3 compared to BM. Gene expression in the six nasal isolates was generally more variable in vivo than in SNM3, suggesting that some parameters of the nasal living conditions may vary to a certain extent between donors. In most of the samples isdA, sceD, and oppB expression analysis confirmed up-regulation in vivo and in SNM3 compared to BM. While all strains that yielded detectable qRT-PCR signals for aspartate kinase and hisC were consistently up-regulated in SNM3 compared to BM, this was only found in some of the in vivo samples. This variability indicates that in vivo expression of these genes depends on the individual host. Expression of sbnC was high in SNM3 and in vivo compared to complex medium, confirming that iron-limiting conditions were indeed present under both conditions. Notably, expression of metI was strongly up-regulated in SNM3 compared to BM in all six strains (median about 100-fold, Fig. 8), and the in vivo metI expression was nearly the same as in SNM3.
Cotton rats have been shown to be a suitable model for S. aureus nasal colonization [11], [37]. When S. aureus Newman and the isogenic ΔmetI mutant were used to inoculate the noses of cotton rats, a strongly reduced colonization capacity of the mutant was observed compared to the parental strain (Figure 9). Thus, MetI has a critical role during nasal colonization, and the methionine-biosynthetic pathway may include previously unrecognized targets for new antimicrobial strategies.
Fig. 9. Cotton rat nasal colonization model. 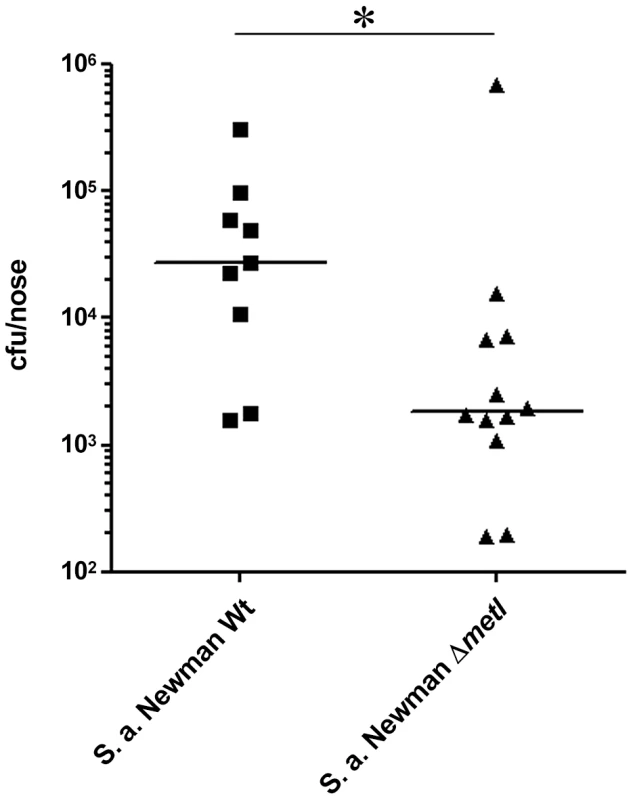
Bacterial numbers were determined six days after intranasal inoculation of cotton rats with 1×108 colony forming units (CFU) of S. aureus (Newman). After six days the noses were dissected and bacterial CFUs were enumerated on S. aureus-selective HighChrome agar. Statistical analysis was performed by D'Agostino & Pearson omnibus normality test and a subsequent Mann Whitney test. Significant differences between groups are indicated by one (P<0.05) asterisk (*). Discussion
The metabolomics analysis of nasal secretions reveals that the human nose represents an environment with rather limited nutrient availability. The concentrations of glucose and amino acids are substantially lower in nasal secretions compared to human plasma of healthy individuals (glucose about 0.04–1 mM vs. 4–8 mM; amino acids about 0.65–2.2 mM vs. 2.6 to 4.3 mM, respectively) [38]. The sputum covering lung epithelia of cystic fibrosis (CF) patients, which are frequently infected by S. aureus [39], [40], contains similar concentrations of glucose and even higher concentrations of free amino acids than plasma (1.3 to 4.5 mM and 4.7 to 24.7 mM, respectively [41]. These differences in nutrient availability suggest that S. aureus requires different metabolic activities during colonization of the human nose or infection of sterile tissues. It is interesting to note that lactate, an abundant compound on skin with concentrations around 2.5 mM [42] and a product of S. aureus energy metabolism [43], was undetectable in nasal secretions. Accordingly, metabolites in the nasal habitat differ from those on skin, and S. aureus metabolism does not seem to affect much the nasal metabolome. Our data indicate that the concentration of many nutrients in nasal secretions varies between donors, but none of the differences could be associated with the S. aureus carrier status. Thus, factors other than nutrient supply, such as differences in epithelial immunity or ligands for S. aureus adhesins, may be responsible for the predisposition to the S. aureus carriage status.
Various chemically defined media have been developed over the past decades with the purpose to achieve maximal growth and high capsule or protein expression in S. aureus [44]–[47]. In contrast, our aim was to simulate the in vivo situation in the human nose, to use a representative synthetic nasal medium for comparing colonization capacities of different staphylococcal strains and to elucidate colonization-related bacterial metabolic processes. Expression profiles of representative S. aureus genes upon growth in the artificial nasal medium SNM3 corresponded well to those found in the human nose indicating that SNM3 provides suitable conditions for simulating S. aureus growth during nasal colonization. Of note, global transcriptomes of bacteria from SNM3 and complex media differed extensively indicating that complex media can hardly reflect in vivo-related gene expression. In accord with this finding, transcriptome data of S. aureus grown in human blood and serum have also yielded major differences compared to previously described expression profiles from complex-media grown bacteria [48], [49]. Thus, SNM3 will allow to investigate bacterial metabolic processes during nasal colonization in detail and to monitor S. aureus competition with other resident bacteria under in vivo-like conditions. Nevertheless, results obtained with SNM3 in genome-wide in vitro experiments should be validated in in vivo models.
In accordance with the limited nasal nutrient supply, the human nasal microbiome has been shown to be much less complex [31], [50] than those of the upper or lower digestive tract, where bacteria are in regular contact with ingested food [30], [51]. Hence, bacteria in the human nose can be expected to compete fiercely for available nutrients and depend on mechanisms allowing them to thrive even with very low amounts of nutrients. In line with this notion all tested nasal S. aureus isolates grew well in SNM3, and almost all viable cells formed colonies on SNM3 agar. S. aureus is obviously adapted to growth in moist environments with very dilute nutrients, while human skin is usually dry except for atopic dermatitis patients, whose skin structure and permeability is severely perturbed [52]. In contrast, CoNS colonize healthy skin as their major habitat [53], which is in accordance with our finding that most of the tested CoNS isolates had major problems to form colonies on SNM3 agar and grew in liquid SNM3 only after long lag phases. It has remained unclear if CoNS use the human nose as a preferred or only as a transient habitat. While permanent S. aureus carriers are usually colonized by one specific single clone, multiple S. epidermidis strains can be found per nose and the pattern of strains is highly variable over time [54]. Our results indicate that CoNS are usually much less adapted to growth in nasal secretions than S. aureus. In line with our data a recent study has found a negative association between the colonisation with S. aureus and the abundance of S. epidermidis. It has been speculated that these bacterial species compete with each other during colonization of the nares [31]. Our data supports the notion that most S. aureus strains can overgrow CoNS because of better metabolic adaptation. In addition to differences in the abilities to utilize dilute nutrients, competition between S. aureus and CoNS may also involve different capacities to produce bacteriocins such as lantibiotics [55], [56], to induce and resist host antimicrobial peptides such as defensins [57], which have been detected in nasal secretions [58], and to produce factors that compromise the ability of other staphylococci to adhere to epithelial surfaces such as the S. epidermidis Esp protease [59].
Metabolomics analyses have been proposed to facilitate the identification of new antimicrobial targets and have recently helped to define the staphylococcal pyruvate dehydrogenase as a target for a new class of organobismuth antimicrobials [60]. We demonstrate here that a combined metabolomics and transcriptomics approach can lead to the identification of targets that are specifically important during in vivo-like conditions. Gene expression analysis of S. aureus grown in SNM3, or from in vivo samples, revealed that various amino acid biosynthesis operons are up-regulated during colonization. This implies a general importance of several anabolic pathways under such conditions.
Since SNM3 does not contain isoleucine, it can be assumed that the global transcription repressor CodY plays an important role for the up-regulation of a number of genes under the conditions used for microarray experiments, especially those for isoleucine biosynthesis [61], [62]. It has recently been shown that CodY also influences S. aureus metICFE-mdh expression, which is essentially regulated by a T-box riboswitch that recognizes uncharged initiator tRNAfMet [63]. Despite the obvious influence of CodY on the gene expression pattern no clear signs of stringent response, like down-regulation of ribosomal protein rpsL, could be detected [64].
The absence of methionine in nasal secretions and SNM3 was reflected by the strong up-regulation of S. aureus methionine import and, most conspicuously, methionine biosynthesis genes in SNM3 and in the noses of human volunteers. In a similar approach, metabolomics analysis of sputum from CF patients has recently allowed to develop a synthetic sputum medium, leading to the unexpected finding that Pseudomonas aeruginosa depends on L-alanine as carbon source during CF lung infections. Furthermore, the high concentrations of aromatic amino acids in CF-sputum and the corresponding synthetic medium have been implicated in high-level production of pyocyanin [41], [65]. This cytotoxic respiratory inhibitor is important in competition of P. aeruginosa with S. aureus in the CF lung, since the latter is strongly inhibited because of its pyocyanin-sensitive cytochrome bd oxidase [66].
The methionine biosynthetic enzymes fulfil important criteria for antimicrobial drug targets, because they are absent from human cells that need to take up methionine from exogenous sources. However, such enzymes have hardly been regarded as antimicrobial targets before, because their importance has probably been missed when growing bacteria in complex media in antimicrobial screening programs. Nevertheless, this pathway has recently been proposed as a potential staphylococcal “Achilles heel” [63]. In our study the synthetic compound DL-propargylglycine, described as an inhibitor of cystathionine-γ-synthase (MetI), which is unique to microorganisms and plants [36], [67] was used. The activity of DL-propargylglycine against S. aureus and the inability of metI mutants to grow in SNM3 underscore the potential use of MetI or other methionine-biosynthetic enzymes as targets for new nasal decolonisation drugs. These drugs could be alternatives to eradicate e.g. mupirocin-resistant S. aureus. In agreement with this notion, the S. aureus metI gene was strongly upregulated in human noses, and the metI mutant was compromised in nasal colonization of cotton rats. However, DL-propargylglycin itself does not seem to be suitable as a drug, because it also inhibits the human cystathionine-γ-lyase. This enzyme converts exogenous methionine to cysteine via the transsulfuration pathway [68], [69], leading to reduced production of the gaseous messenger molecule hydrogen sulfide with various eminent consequences [70]. While the rather low activity against S. aureus and the severe impact on mammalian metabolism precluded the use of DL-propargylglycine in animal models, derivatives or new compounds with higher selectivity and activity against bacterial MetI or other methionine-biosynthetic enzymes may become promising lead substances for the development of new antimicrobial drugs. Notably, all these enzymes were significantly up-regulated in SNM3 (Fig. 6).
Materials and Methods
Ethics statement
The nasal secretion study and sample collection procedures were approved by the clinical ethics committee of the University of Tuebingen (No. 109/2009 BO2) and informed written consent was obtained from all volunteers. Secretion samples and nasal swabs were taken exclusively from healthy adults.
Strains and growth conditions
The staphylococcal laboratory strains used in this study are S. aureus USA300 LAC [71], S. aureus Newman [72], and S. epidermidis 1457 [73]. Beside these characterised strains a set of 87 staphylococcal isolates from nasal swabs from 37 healthy volunteers were used. The collection of nasal isolates includes 18 S. aureus, 57 S. epidermidis, six S. capitis, three S. lugdunensis, two S. warneri and one S. hominis strain. Identification was accomplished according to an established scheme [74] by sequencing of a variable ca. 931-bp PCR fragment of the glyceraldehyde-3-phosphate dehydrogenase gene (gap), amplified with primer pair gap-F and gap-R (Table S1). Ambiguities were clarified by additional sequencing of a variable part of the dnaJ gene, amplified with primer pair dnaJ-F and dnaJ-R (Table S1).
BM (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% glucose and 0.1% K2HPO4, pH 7.2) was used as standard complex medium. The composition of the chemically defined medium SNM, corresponding to nasal secretions, is listed in Tables 1 and 2. The content of inorganic ions in human nasal secretions listed in Table 1 has been published earlier [24], [25] and the described buffer, salt and co-factor concentrations were included in SNM as listed in Table 2. In contrast to the published data calcium was omitted from the medium, because it led to precipitates. Trace elements and cofactors were added from 1000-fold concentrated stock solutions. The iron-complexing agent 2, 2′-bipyridine (Merck, Darmstadt, Germany) was added at a final concentration of 200 µM. SNM3 contained the same concentrations of inorganic salts, urea, trace elements, and cofactors as SNM while amino acids, organic acids, and glucose as listed in Table 1 were threefold concentrated. BM and SNM agar plates contained 1.5% agar. For growth curves bacteria from overnight cultures grown in BM were centrifuged, washed with PBS, and diluted in SNM3 to an initial OD600 nm of 0.02 in microtiter plates (MTP) and grown in a TECAN Infinite 200 PRO reader (Tecan Group Ltd., Switzerland) with shaking (180 rpm) at 37°C with continuous measurement of optical densities. For MIC determination 24-well MTP plates, essentially inoculated as described above, were grown for 48 hours at 37°C and 160 rpm. For the calculation of growth, inoculation density was subtracted from final optical density and the MICs, defined as the concentration of DL-propargylglycine (Sigma-Aldrich, Taufkirchen, Germany) at which 75% growth inhibition occurred, were calculated. For monitoring bacterial growth in three to 20-fold SNM cultures 12-ml medium in 100-ml buffled Erlenmeyer flasks were inoculated with an SNM3 over-night culture to an initial OD578 nm of 0.02 and incubated with vigorous shaking at 160 rpm at 37°C, until the final OD578 nm was determined after 90 h growth. For RNA isolation all media were inoculated with SNM3 overnight cultures, grown at 37°C with vigorous shaking at 160 rpm in 250-ml buffled Erlenmeyer flasks.
For continuous cultures sterile fresh medium was added from a reservoir to the growing culture with a peristaltic pump. A second pump was used to remove consumed medium, including bacteria, from the culture. 100 ml SNM in 500-ml glass bottles (Schott) were inoculated to an OD578 nnm of 0.02 and incubated with shaking at 140 rpm at 37°C with a sterile filter in the bottle lid to allow aeration. About 4 ml fresh medium were added and simultaneously removed from the culture per hour resulting in exchange of the complete culture volume within approximately 24 hours. Non-continuous batch cultures were performed in the same way, except that continuous medium exchange was omitted.
Collection of nasal secretions and metabolite analysis
Nasal secretions were taken with the help of slight vacuum suction with suction catheters (model 14 Ch, Bicakcilar Healthcare Products, Turkey) mounted on sputum collection traps (P. J. Dahlhausen & Co.GmbH, Germany). After short centrifugation the samples were immediately frozen at −80°C. Metabolites from frozen nasal secretions were extracted with methanol/chloroform/water 4/4/2.85 after addition of internal standards (ribitol and norvaline), samples were vortexed twice for 10 s and then centrifuged (4°C, 10 min, 13000 rpm). Supernatants were transferred to new glass vials and dried by lyophilisation. Dry samples were derivatized for GC-MS analyses and measured according to a previously described method [75]. Briefly, metabolites were identified by matching retention time and identification ion of pure chemical standards, measured under the same conditions. Ratios of peak areas to the respective internal standard (ribitol) were used for absolute and relative quantification. Calibration curves of pure substances were measured over a wide range of concentrations under the same conditions and were used for the calculation of the total, micromolar metabolite concentration.
Because arginine is unstable when derivatized for GC-MS, its content was determined by HPLC using ortho-phthaldehyde (OPA) pre-column derivatization. OPA was diluted to a final concentration of 1 mg/ml with 1 M sodium-borate-buffer, pH 9.0. Nasal secretion samples were sonicated for 20 s in a sonication water bath to reduce viscosity and diluted 1∶1 with distilled water. Subsequently, each sample (6 µl) was mixed with 1.5 µl OPA for 90 s and immediately injected and separated on an Agilent 1200 series HPLC-system using a Grom-SIL OPA-3 (5 µm), 4.0×150 mm column (Grace Davison, Lokeren, Belgium). A linear gradient from 100% buffer A (25 mM sodium-phosphate buffer, 0.7% tetrahydrofuran, pH 7.2) to 100% buffer B (50% buffer A, 35% methanol, 15% acetonitrile) in 24 min was run at a flow rate of 1.1 ml/min. Arginine was detected at 450 nm and quantified against a standard calibration curve with arginine concentrations of 10 µM, 100 µM, 1 mM, and 10 mM. Data were analyzed by Agilent ChemStation software.
Relative abundance is given in cases were no dilution series of pure standard compounds was measured. Data analysis was performed within the GC-MS software Chemstation (Vers. E.02.00 Service Pack 2, Agilent). Statistical analysis was accomplished with Aabel 3.0.4 (Gigawiz) and SIMCA P+ 12.0.1, for principal component analysis (PCA), and for partial least square (PLS) analysis log-transformed peak areas with UV scaling were used.
RNA isolation and amplification
For qRT-PCR 40-ml cultures were inoculated to an optical density of 0.02 at 578 nm (ca. 2×107 CFU/ml) and grown for three hours. RNA was isolated by a modified protocol of Bhagwat et al. adapted to large culture volumes [76]. Briefly, cells were immediately killed and RNA was stabilized by the addition of 1/9 vol. of an ice-cold 9∶1 ethanol∶phenol solution (equilibrated with Tris/EDTA-buffer (TE); Applichem, Darmstadt, Germany). After 5 min on ice cells were harvested (20 min, 4500×g, 4°C), resuspended in 1 ml TRIZOL solution (Invitrogen - Life Technologies Corporation, Darmstadt, Germany), and lysed with 0.5 ml zirconia-silica beads (Karl Roth GmbH, Karlsruhe, Germany; 0.1 mm-diameter) in a high-speed benchtop homogenizer (FastPrep-24, MP Biomedicals, Germany). Subsequently, RNA was isolated as described in the instructions provided by the manufacturer of the RNA isolation kit (ExpressArt RNAready, AmpTec GmbH, Germany). In order to get rid of potential RT-PCR inhibitors, a first washing step with 0.5 ml ‘Inhibitor removal buffer’ was applied (High Pure PCR Template Preparation Kit, Roche Applied Science). DNA was removed by on-column DNAse treatment according to the ExpressArt RNAready protocol.
RNA for microarrays was isolated from 100-ml cultures, inoculated to an OD578 nm of 0.005 and grown until OD578 nm of 0.02. RNA was stabilized as described above by ethanol∶phenol addition. After centrifugation, the cell pellet was washed with 2 ml ice-cold ethanol∶acetone (1∶1) to remove phenol traces. A second washing and stabilisation step was applied by resuspending the cells in 2 ml RNAprotect bacteria reagent (QIAGEN GmbH, Germany)∶TE-buffer (2∶1). After centrifugation cells were lysed in 100 µl TE buffer containing 10 µl lysostaphin (2 mg/ml, Genmedics GmbH, Germany) by incubation for 3 min at room temperature. After the addition of 350 µl RLT buffer (RNeasy Kit, QIAGEN GmbH, Germany) cells were lysed with 0.2 ml zirconia-silica beads as described above and RNA was purified with the RNeasy Kit according to the manufacturer's instructions.
RNA for qRT-PCR from nasal swabs was isolated essentially as described above for in vitro cultures. The cotton swabs (MSP Schmeiser, Horb, Germany) were soaked in sterile PBS and used to carefully wipe the anterior nares of volunteers and afterwards directly resuspended in 1 ml TRIZOL. For confirming the carrier status additional swabs were taken, resuspended in PBS, and serial dilutions were plated on BM agar. The presence of S. aureus was confirmed with the Slidex Staph Plus latex agglutination test (bioMérieux Deutschland GmbH, Nürtingen, Germany) according to the manufacturer's instructions.
Since some of the volunteers had quite low numbers of S. aureus in their nose (<1.000 S. aureus/swab), the isolated RNA amounts were sometimes not sufficient for qRT-PCR. In such cases RNA was amplified once with the ‘Bacterial Nano mRNA amplification Kit’ (AmpTec GmbH, Hamburg, Germany) as described by the manufacturer.
Reverse transcription and quantitative real-time PCR (qRT-PCR)
Relative quantification of various transcripts was performed as described previously [18]. Briefly, isolated RNA from cultures and nasal swabs was transcribed into complementary DNA using SuperScriptIII Reverse Transcriptase (Invitrogen) and 200 ng of random hexamer primers (Fermentas, St. Leon-Rot, Germany). For relative quantification standards were generated by PCR with the primers listed in Table S1 with S. aureus USA300 genomic DNA as template. All agarose gel-purified PCR products were used in 10-fold serial dilutions. For the genes metN and sbnC PauI (Fermentas) -digested and purified chromosomal DNA of S. aureus USA300 was used as standard. Complementary DNA was diluted 1∶10 and quantitative real-time PCR (qRT-PCR) was performed with primers listed in Supplementary Table S1, using the LightCycler instrument (LightCycler 480, Roche) in combination with SYBR Green I (QuantiFast SYBR Green PCR Kit, QIAGEN). Master mixes were prepared according to the manufacturer's instructions. Relative gene expression level was calculated by the method of Pfaffl with PCR efficiency correction [77].
Microarray analysis
5 µg of RNA from three biological replicates per condition were applied to GeneChip microarrays (Affymetrix) and processed according to the manufacturer's protocol. The biological replicates yielded highly reproducible expression profiles.
The GeneChip S. aureus genome array was provided by MFTServices (www.mftservices.de), a Core Lab provider authorized by Affymetrix Inc. (Santa Clara, CA). GeneChip hybridization, washing, staining, and scanning were performed as described by the manufacturer. The images were processed with Expression Console (Affymetrix). The raw data from the array scans were normalized by median-centering genes for each array, followed by log transformation. Expressed genes were identified using Affymetrix GeneChip Operating Software (GCOS, Ver.1.1). To identify genes that are differentially expressed in treated samples compared to controls, the Partek software version 6.6 was used. To select the differentially expressed genes, we used threshold values of ≥2.0 - and ≤−2.0-fold change between the conditions. The false discovery rate (FDR) significance level with Benjamini-Hochberg was <5%. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [78] and are accessible through GEO Series accession number GSE43712 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43712).
Construction of a cystathionine-γ-synthase mutant and colonisation of cotton rat nares
For the construction of a markerless mutant of S. aureus Newman, the flanking regions of metI were amplified with primer pairs cgsF1 up/cgsF1 down or cgsF2 up/cgsF2 down. After digestion of PCR product F1 with EcoRI/BglII and of PCR product F2 with BglII/NheI the two fragments were ligated into the previously described vector pBASE6 [64], digested with EcoRI and NheI and used to transform E. coli DC10B [79]. The resulting plasmid pBASE6-ΔmetI was isolated and directly transferred to S. aureus Newman, where the homologous recombination process resulted in metI mutants, which were confirmed by PCR analysis.
For the colonisation of cotton rats spontaneous streptomycin-resistant mutants of S. aureus Newman wild type and ΔmetI were selected on BM agar plates with 500 µg/ml streptomycin. The cotton rat model was used as described earlier [11]. Cotton rats were anesthetized and instilled intranasally with 10 µl of 1×108 colony-forming units (CFU) of S. aureus. Six days after bacterial instillation the animals were euthanized and noses were removed surgically. The noses were vortexed in 1 ml of PBS containing 0.5% Tween for 30 s. Samples were plated on appropriate agar plates (B-medium, sheep blood containing 250 µg/ml streptomycin and HiCrome Aureus Agar (Fluka)) and the bacterial CFU was determined. All animals received drinking water with 2.5 mg/ml streptomycin continuously, starting three days prior to the experiment to reduce the natural nasal flora. All animal experiments were conducted in accordance with German laws after approval (protocol T1/10) by the local authorities (Regierungspraesidium Tuebingen).
Supporting Information
Zdroje
1. LowyFD (1998) Staphylococcus aureus infections. N Engl J Med 339 : 520–532.
2. MilesAA (1941) Some problems of wound infection. The Lancet 238 : 507–510.
3. KrismerB, PeschelA (2011) Does Staphylococcus aureus nasal colonization involve biofilm formation? Future Microbiol 6 : 489–493.
4. WeidenmaierC, GoerkeC, WolzC (2012) Staphylococcus aureus determinants for nasal colonization. Trends Microbiol 20 : 243–250.
5. OttoM (2009) Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat Rev Microbiol 7 : 555–567.
6. GouldJC, McKE (1954) The carriage of Staphylococcus pyogenes var. aureus in the human nose. J Hyg (Lond) 52 : 304–310.
7. van BelkumA, VerkaikNJ, de VogelCP, BoelensHA, VerveerJ, et al. (2009) Reclassification of Staphylococcus aureus nasal carriage types. J Infect Dis 199 : 1820–1826.
8. von EiffC, BeckerK, MachkaK, StammerH, PetersG (2001) Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med 344 : 11–16.
9. WertheimHF, VosMC, OttA, van BelkumA, VossA, et al. (2004) Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364 : 703–705.
10. van BelkumA, MellesDC, NouwenJ, van LeeuwenWB, van WamelW, et al. (2009) Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol 9 : 32–47.
11. WeidenmaierC, Kokai-KunJF, KristianSA, ChanturiyaT, KalbacherH, et al. (2004) Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med 10 : 243–245.
12. WeidenmaierC, PeschelA (2008) Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6 : 276–287.
13. O'BrienLM, WalshEJ, MasseyRC, PeacockSJ, FosterTJ (2002) Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol 4 : 759–770.
14. WertheimHF, WalshE, ChoudhurryR, MellesDC, BoelensHA, et al. (2008) Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med 5: e17.
15. ClarkeSR, AndreG, WalshEJ, DufreneYF, FosterTJ, et al. (2009) Iron-regulated surface determinant protein A mediates adhesion of Staphylococcus aureus to human corneocyte envelope proteins. Infect Immun 77 : 2408–2416.
16. MulcahyME, GeogheganJA, MonkIR, O'KeeffeKM, WalshEJ, et al. (2012) Nasal Colonisation by Staphylococcus aureus Depends upon Clumping Factor B Binding to the Squamous Epithelial Cell Envelope Protein Loricrin. PLoS Pathog 8: e1003092.
17. BurianM, WolzC, GoerkeC (2010) Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS One 5: e10040.
18. BurianM, RautenbergM, KohlerT, FritzM, KrismerB, et al. (2010) Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J Infect Dis 201 : 1414–1421.
19. ClarkeSR, WiltshireMD, FosterSJ (2004) IsdA of Staphylococcus aureus is a broad spectrum, iron-regulated adhesin. Mol Microbiol 51 : 1509–1519.
20. PynnonenM, StephensonRE, SchwartzK, HernandezM, BolesBR (2011) Hemoglobin promotes Staphylococcus aureus nasal colonization. PLoS Pathog 7: e1002104.
21. BrownSA, PalmerKL, WhiteleyM (2008) Revisiting the host as a growth medium. Nat Rev Microbiol 6 : 657–666.
22. BodeLG, KluytmansJA, WertheimHF, BogaersD, Vandenbroucke-GraulsCM, et al. (2010) Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med 362 : 9–17.
23. PatelJB, GorwitzRJ, JerniganJA (2009) Mupirocin resistance. Clin Infect Dis 49 : 935–941.
24. VanthanouvongV, RoomansGM (2004) Methods for determining the composition of nasal fluid by X-ray microanalysis. Microsc Res Tech 63 : 122–128.
25. LorinMI, GaerlanPF, MandelID (1972) Quantitative composition of nasal secretions in normal subjects. J Lab Clin Med 80 : 275–281.
26. MahRA, FungDY, MorseSA (1967) Nutritional requirements of Staphylococcus aureus S-6. Appl Microbiol 15 : 866–870.
27. BiswasL, BiswasR, NerzC, OhlsenK, SchlagM, et al. (2009) Role of the twin-arginine translocation pathway in Staphylococcus. J Bacteriol 191 : 5921–5929.
28. NouwenJL, OttA, Kluytmans-VandenberghMF, BoelensHA, HofmanA, et al. (2004) Predicting the Staphylococcus aureus nasal carrier state: derivation and validation of a “culture rule”. Clin Infect Dis 39 : 806–811.
29. AasJA, PasterBJ, StokesLN, OlsenI, DewhirstFE (2005) Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43 : 5721–5732.
30. O'HaraAM, ShanahanF (2006) The gut flora as a forgotten organ. EMBO Rep 7 : 688–693.
31. FrankDN, FeazelLM, BessesenMT, PriceCS, JanoffEN, et al. (2010) The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5: e10598.
32. TatusovRL, KooninEV, LipmanDJ (1997) A genomic perspective on protein families. Science 278 : 631–637.
33. KiserKB, Cantey-KiserJM, LeeJC (1999) Development and characterization of a Staphylococcus aureus nasal colonization model in mice. Infect Immun 67 : 5001–5006.
34. CheungJ, BeasleyFC, LiuS, LajoieGA, HeinrichsDE (2009) Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol Microbiol 74 : 594–608.
35. XiongA, SinghVK, CabreraG, JayaswalRK (2000) Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology 146 (Pt 3) 659–668.
36. JohnstonM, JankowskiD, MarcotteP, TanakaH, EsakiN, et al. (1979) Suicide inactivation of bacterial cystathionine gamma-synthase and methionine gamma-lyase during processing of L-propargylglycine. Biochemistry 18 : 4690–4701.
37. Kokai-KunJF (2008) The Cotton Rat as a Model for Staphylococcus aureus nasal colonization in humans: cotton rat S. aureus nasal colonization model. Methods Mol Biol 431 : 241–254.
38. NassetES, HealdFP, CallowayDH, MargenS, SchneemanP (1979) Amino acids in human blood plasma after single meals of meat, oil, sucrose and whiskey. J Nutr 109 : 621–630.
39. GoerkeC, WolzC (2010) Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int J Med Microbiol 300 : 520–525.
40. GossCH, MuhlebachMS (2011) Review: Staphylococcus aureus and MRSA in cystic fibrosis. J Cyst Fibros 10 : 298–306.
41. PalmerKL, AyeLM, WhiteleyM (2007) Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189 : 8079–8087.
42. PetersenLJ (1999) Interstitial lactate levels in human skin at rest and during an oral glucose load: a microdialysis study. Clin Physiol 19 : 246–250.
43. FerreiraMT, MansoAS, GasparP, PinhoMG, NevesAR (2013) Effect of oxygen on glucose metabolism: utilization of lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS One 8: e58277.
44. MillerRD, FungDY (1973) Amino acid requirements for the production of enterotoxin B by Staphylococcus aureus S-6 in a chemically defined medium. Appl Microbiol 25 : 800–806.
45. OnoueY, MoriM (1997) Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int J Food Microbiol 36 : 77–82.
46. StringfellowWT, DassyB, LiebM, FournierJM (1991) Staphylococcus aureus growth and type 5 capsular polysaccharide production in synthetic media. Appl Environ Microbiol 57 : 618–621.
47. TaylorD, HollandKT (1989) Amino acid requirements for the growth and production of some exocellular products of Staphylococcus aureus. J Appl Bacteriol 66 : 319–329.
48. MalachowaN, WhitneyAR, KobayashiSD, SturdevantDE, KennedyAD, et al. (2011) Global changes in Staphylococcus aureus gene expression in human blood. PLoS One 6: e18617.
49. den ReijerPM, Lemmens-den ToomN, KantS, SnijdersSV, BoelensH, et al. (2013) Characterization of the Humoral Immune Response during Staphylococcus aureus Bacteremia and Global Gene Expression by Staphylococcus aureus in Human Blood. PLoS One 8: e53391.
50. Wos-OxleyML, PlumeierI, von EiffC, TaudienS, PlatzerM, et al. (2010) A poke into the diversity and associations within human anterior nare microbial communities. ISME J 4 : 839–851.
51. DewhirstFE, ChenT, IzardJ, PasterBJ, TannerAC, et al. (2010) The human oral microbiome. J Bacteriol 192 : 5002–5017.
52. ChoSH, StricklandI, BoguniewiczM, LeungDY (2001) Fibronectin and fibrinogen contribute to the enhanced binding of Staphylococcus aureus to atopic skin. J Allergy Clin Immunol 108 : 269–274.
53. WiderstromM, WistromJ, SjostedtA, MonsenT (2012) Coagulase-negative staphylococci: update on the molecular epidemiology and clinical presentation, with a focus on Staphylococcus epidermidis and Staphylococcus saprophyticus. Eur J Clin Microbiol Infect Dis 31 : 7–20.
54. HuL, UmedaA, AmakoK (1995) Typing of Staphylococcus epidermidis colonizing in human nares by pulsed-field gel electrophoresis. Microbiol Immunol 39 : 315–319.
55. BastosMC, CeottoH, CoelhoML, NascimentoJS (2009) Staphylococcal antimicrobial peptides: relevant properties and potential biotechnological applications. Curr Pharm Biotechnol 10 : 38–61.
56. BierbaumG, SahlHG (2009) Lantibiotics: mode of action, biosynthesis and bioengineering. Curr Pharm Biotechnol 10 : 2–18.
57. WankeI, SteffenH, ChristC, KrismerB, GotzF, et al. (2011) Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Invest Dermatol 131 : 382–390.
58. ColeAM, DewanP, GanzT (1999) Innate antimicrobial activity of nasal secretions. Infect Immun 67 : 3267–3275.
59. IwaseT, UeharaY, ShinjiH, TajimaA, SeoH, et al. (2010) Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465 : 346–349.
60. BirkenstockT, LiebekeM, WinstelV, KrismerB, GekelerC, et al. (2012) Exometabolome analysis identifies pyruvate dehydrogenase as a target for the antibiotic triphenylbismuthdichloride in multiresistant bacterial pathogens. J Biol Chem 287 : 2887–2895.
61. MajerczykCD, DunmanPM, LuongTT, LeeCY, SadykovMR, et al. (2010) Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192 : 2861–2877.
62. PohlK, FrancoisP, StenzL, SchlinkF, GeigerT, et al. (2009) CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191 : 2953–2963.
63. SchoenfelderSM, MarincolaG, GeigerT, GoerkeC, WolzC, et al. (2013) Methionine Biosynthesis in Staphylococcus aureus Is Tightly Controlled by a Hierarchical Network Involving an Initiator tRNA-Specific T-box Riboswitch. PLoS Pathog 9: e1003606.
64. GeigerT, FrancoisP, LiebekeM, FraunholzM, GoerkeC, et al. (2012) The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog 8: e1003016.
65. PalmerKL, MashburnLM, SinghPK, WhiteleyM (2005) Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J Bacteriol 187 : 5267–5277.
66. VogguL, SchlagS, BiswasR, RosensteinR, RauschC, et al. (2006) Microevolution of cytochrome bd oxidase in Staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol 188 : 8079–8086.
67. MarcotteP, WalshC (1975) Active site-directed inactivation of cystathionine gamma-synthetase and glutamic pyruvic transaminase by propargylglycine. Biochem Biophys Res Commun 62 : 677–682.
68. RaoAM, DrakeMR, StipanukMH (1990) Role of the transsulfuration pathway and of gamma-cystathionase activity in the formation of cysteine and sulfate from methionine in rat hepatocytes. J Nutr 120 : 837–845.
69. SteegbornC, ClausenT, SondermannP, JacobU, WorbsM, et al. (1999) Kinetics and inhibition of recombinant human cystathionine gamma-lyase. Toward the rational control of transsulfuration. J Biol Chem 274 : 12675–12684.
70. PaulBD, SnyderSH (2012) H(2)S signalling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol 13 : 499–507.
71. KazakovaSV, HagemanJC, MatavaM, SrinivasanA, PhelanL, et al. (2005) A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med 352 : 468–475.
72. DuthieES, LorenzLL (1952) Staphylococcal coagulase; mode of action and antigenicity. Journal of General Microbiology 6 : 95–107.
73. MackD, SiemssenN, LaufsR (1992) Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun 60 : 2048–2057.
74. GhebremedhinB, LayerF, KonigW, KonigB (2008) Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J Clin Microbiol 46 : 1019–1025.
75. LiebekeM, DorriesK, ZuhlkeD, BernhardtJ, FuchsS, et al. (2011) A metabolomics and proteomics study of the adaptation of Staphylococcus aureus to glucose starvation. Mol Biosyst 7 : 1241–1253.
76. BhagwatAA, PhadkeRP, WheelerD, KalantreS, GudipatiM, et al. (2003) Computational methods and evaluation of RNA stabilization reagents for genome-wide expression studies. J Microbiol Methods 55 : 399–409.
77. PfafflMW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
78. EdgarR, DomrachevM, LashAE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 : 207–210.
79. MonkIR, ShahIM, XuM, TanMW, FosterTJ (2012) Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite MovementČlánek Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi ScreenČlánek IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed ChildrenČlánek Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive SubjectsČlánek Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 1- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- HIV-1 Accessory Proteins Adapt Cellular Adaptors to Facilitate Immune Evasion
- Ranaviruses: Not Just for Frogs
- Effectors and Effector Delivery in
- Plasmacytoid Dendritic Cell Dynamics Tune Interferon-Alfa Production in SIV-Infected Cynomolgus Macaques
- Lu/BCAM Adhesion Glycoprotein Is a Receptor for Cytotoxic Necrotizing Factor 1 (CNF1)
- A Substrate-Fusion Protein Is Trapped inside the Type III Secretion System Channel in
- Parvovirus-Induced Depletion of Cyclin B1 Prevents Mitotic Entry of Infected Cells
- Red Blood Cell Invasion by : Structural Basis for DBP Engagement of DARC
- NsrR, GadE, and GadX Interplay in Repressing Expression of the O157:H7 LEE Pathogenicity Island in Response to Nitric Oxide
- Loss of Circulating CD4 T Cells with B Cell Helper Function during Chronic HIV Infection
- TREM-1 Deficiency Can Attenuate Disease Severity without Affecting Pathogen Clearance
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- Glutamate Utilization Couples Oxidative Stress Defense and the Tricarboxylic Acid Cycle in Phagosomal Escape
- Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite Movement
- Recovery of an Antiviral Antibody Response following Attrition Caused by Unrelated Infection
- Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi Screen
- Absence of Intestinal PPARγ Aggravates Acute Infectious Colitis in Mice through a Lipocalin-2–Dependent Pathway
- Induction of a Stringent Metabolic Response in Intracellular Stages of Leads to Increased Dependence on Mitochondrial Metabolism
- CTCF and Rad21 Act as Host Cell Restriction Factors for Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Lytic Replication by Modulating Viral Gene Transcription
- Gammaherpesviral Gene Expression and Virion Composition Are Broadly Controlled by Accelerated mRNA Degradation
- The Arabidopsis Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes
- Inflammatory Stimuli Reprogram Macrophage Phagocytosis to Macropinocytosis for the Rapid Elimination of Pathogens
- Alphavirus Mutator Variants Present Host-Specific Defects and Attenuation in Mammalian and Insect Models
- Phosphopyruvate Carboxylase Identified as a Key Enzyme in Erythrocytic Carbon Metabolism
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Electron Tomography of HIV-1 Infection in Gut-Associated Lymphoid Tissue
- Characterisation of a Multi-ligand Binding Chemoreceptor CcmL (Tlp3) of
- Single Cell Stochastic Regulation of Pilus Phase Variation by an Attenuation-like Mechanism
- Cell Tropism Predicts Long-term Nucleotide Substitution Rates of Mammalian RNA Viruses
- Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive Subjects
- RNA-seq Analysis of Host and Viral Gene Expression Highlights Interaction between Varicella Zoster Virus and Keratinocyte Differentiation
- Kaposi's Sarcoma Associated Herpesvirus Tegument Protein ORF75 Is Essential for Viral Lytic Replication and Plays a Critical Role in the Antagonization of ND10-Instituted Intrinsic Immunity
- DAMP Molecule S100A9 Acts as a Molecular Pattern to Enhance Inflammation during Influenza A Virus Infection: Role of DDX21-TRIF-TLR4-MyD88 Pathway
- Variable Suites of Non-effector Genes Are Co-regulated in the Type III Secretion Virulence Regulon across the Phylogeny
- Reengineering Redox Sensitive GFP to Measure Mycothiol Redox Potential of during Infection
- Preservation of Tetherin and CD4 Counter-Activities in Circulating Alleles despite Extensive Sequence Variation within HIV-1 Infected Individuals
- KSHV 2.0: A Comprehensive Annotation of the Kaposi's Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features
- Nutrient Limitation Governs Metabolism and Niche Adaptation in the Human Nose
- Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
- Investigation of Acetylcholine Receptor Diversity in a Nematode Parasite Leads to Characterization of Tribendimidine- and Derquantel-Sensitive nAChRs
- Intranasal Vaccination Promotes Detrimental Th17-Mediated Immunity against Influenza Infection
- -Mediated Inhibition of Iron Export Promotes Parasite Replication in Macrophages
- Variation in RNA Virus Mutation Rates across Host Cells
- A Single Amino Acid in the Stalk Region of the H1N1pdm Influenza Virus HA Protein Affects Viral Fusion, Stability and Infectivity
- Group B Engages an Inhibitory Siglec through Sialic Acid Mimicry to Blunt Innate Immune and Inflammatory Responses
- Synthesis and Biological Properties of Fungal Glucosylceramide
- HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets
- Recruitment of PfSET2 by RNA Polymerase II to Variant Antigen Encoding Loci Contributes to Antigenic Variation in
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Enhancing Virus-Specific Immunity by Combining Therapeutic Vaccination and PD-L1 Blockade in Chronic Hepadnaviral Infection
- Suppression of Interferon Lambda Signaling by SOCS-1 Results in Their Excessive Production during Influenza Virus Infection
- Inflammation Fuels Colicin Ib-Dependent Competition of Serovar Typhimurium and in Blooms
- Host-Specific Enzyme-Substrate Interactions in SPM-1 Metallo-β-Lactamase Are Modulated by Second Sphere Residues
- STING-Dependent Type I IFN Production Inhibits Cell-Mediated Immunity to
- From Scourge to Cure: Tumour-Selective Viral Pathogenesis as a New Strategy against Cancer
- Lysine Acetyltransferase GCN5b Interacts with AP2 Factors and Is Required for Proliferation
- Narrow Bottlenecks Affect Populations during Vertical Seed Transmission but not during Leaf Colonization
- Targeted Cytotoxic Therapy Kills Persisting HIV Infected Cells During ART
- Murine Gammaherpesvirus M2 Protein Induction of IRF4 via the NFAT Pathway Leads to IL-10 Expression in B Cells
- iNKT Cell Production of GM-CSF Controls
- Malaria-Induced NLRP12/NLRP3-Dependent Caspase-1 Activation Mediates Inflammation and Hypersensitivity to Bacterial Superinfection
- Detection of Host-Derived Sphingosine by Is Important for Survival in the Murine Lung
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



