-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Secret Life of Viral Entry Glycoproteins: Moonlighting in Immune Evasion
article has not abstract
Published in the journal: . PLoS Pathog 9(5): e32767. doi:10.1371/journal.ppat.1003258
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003258Summary
article has not abstract
Survival of infection with Ebola virus (EBOV) depends on the ability of the host to mount early and strong immune responses [1], [2]. However, given that EBOV cases are associated with 40%–90% human mortality, EBOV has developed intricate solutions to human immunological defenses. Enveloped viruses, like EBOV, must deposit their genetic material within a cell to ensure their propagation. The roles of viral envelope glycoproteins in mediating virus attachment to host cells and catalyzing the subsequent fusion of the viral and host plasma membranes have been well described (reviewed in [3]). Given the limited number of genes in EBOV and other viruses, it stands to reason that these conformationally labile glycoproteins are also involved in more than just the initial steps of a productive infection. There is strong evidence that viral entry glycoproteins (GP) are modulators of host antiviral defenses (Table 1). In this article, we discuss our current structural understanding of the functions of envelope entry glycoproteins in immune evasion using EBOV as our example.
Tab. 1. Viral entry glycoprotein-mediated immune evasion strategies in other viral families. 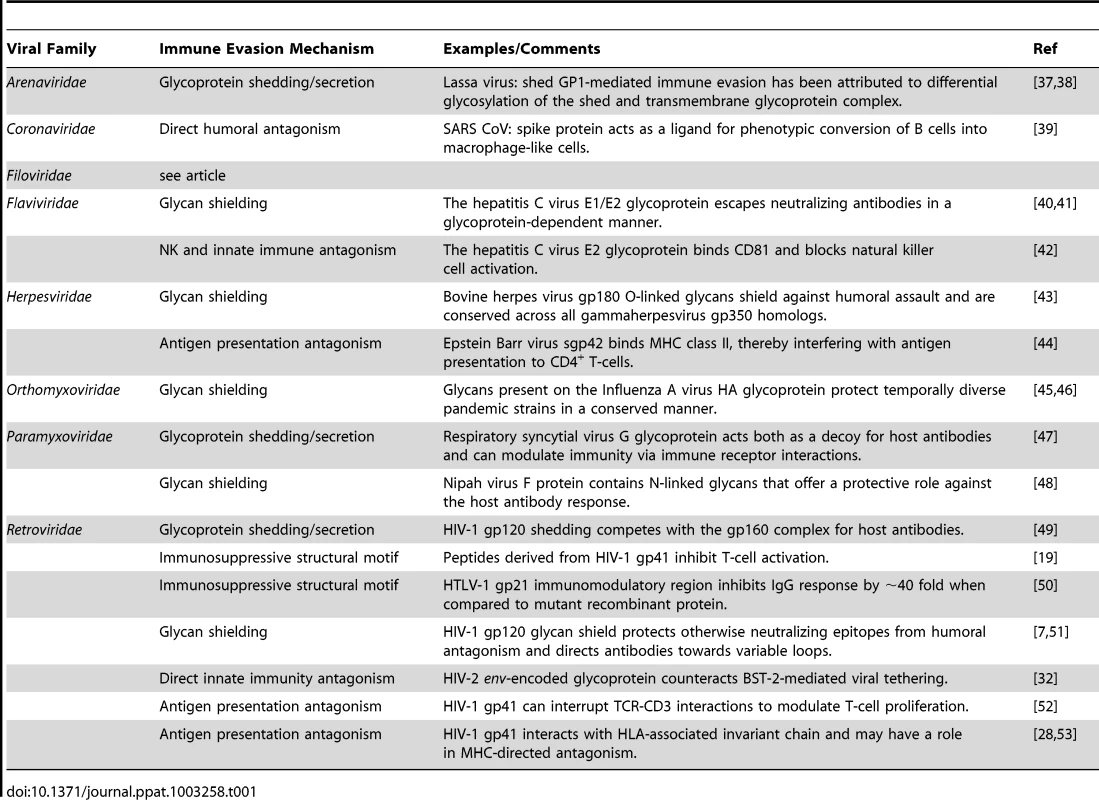
How Does Glycosylation of Ebola Virus Envelope Proteins Facilitate Immune Evasion?
In EBOV, four variants of the envelope glycoprotein are synthesized as a result of transcriptional stuttering or post-translational processing (Figure 1A). About 25% of transcripts from the GP gene produce the virion-attached or envelope spike GP that is important for entry. The surface of the envelope GP is covered with N - and O-linked glycans. Depending on the EBOV species, the envelope GP contains 11–18 N-linked glycan sites. Furthermore, EBOV GP contains an unstructured ∼150-residue mucin-like domain that is heavily modified with O-linked glycans (∼80 sites) [4]. The N-linked glycans are a heterogeneous mixture of ∼60 different species of high-mannose, hybrid, and complex oligosaccharides, while the O-linked glycans consist of primarily smaller trisaccharide structures (core 2) that contain varying amounts of sialic acids [5].
Fig. 1. Ebola virus glycoproteins. 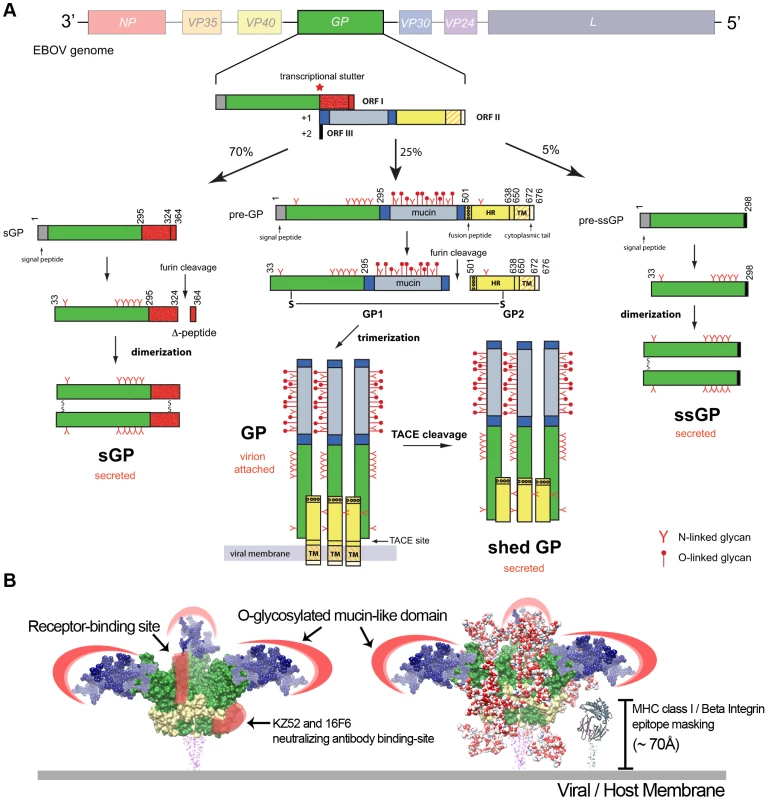
(A) Processing of EBOV glycoproteins. The EBOV genome contains seven genes (3′-NP-VP35-VP40-GP-VP30-VP24-L-5′), but nine proteins are produced due to editing events in the GP gene. The GP gene primary transcript encodes for a ∼110 kDa, dimeric secreted GP (pre-sGP). Furin cleavage of pre-sGP produces mature sGP and a secreted Δ-peptide. Transcriptional stuttering results in the production of the envelope-attached GP and a small, secreted GP (ssGP). The GP is the only virally encoded protein on the EBOV surface and is cleaved by furin to form a disulfide-linked GP1-GP2 heterodimer, which then assembles as trimers on the virus surface. GP1 contains the receptor-binding site for host cell attachment, while GP2 contains a helical heptad-repeat (HR) region, transmembrane anchor (TM), and a 4-residue cytoplasmic tail. A cleavage at the membrane-proximal external region by the tumor necrosis factor-α converting enzyme (TACE) releases the shed GP. The first 295 residues of ssGP, sGP, and GP are common, but each protein has a different C-terminus, leading to different functions. (B) Epitope masking by EBOV glycoproteins. Molecular surface of EBOV GP subunits (PDB code: 3CSY) are shown in green (GP1) and yellow (GP2). Complex-type N-linked glycans are modeled onto the EBOV GP surface as red/white spheres to reveal a heavy glycan layer that buries much of the GP surface, including the receptor-binding site; only a small patch at the base of the GP is accessible (KZ52/16F6 antibody-binding site). The O-linked glycosylated mucin-like domain (blue) is modeled onto EBOV GP, and thought to form an extended structure that provides another glycan layer of protection to the virus. EBOV GP is estimated to be ∼150 Å in height. Given the size and shape of EBOV GP, smaller cellular surface proteins, such as MHC class I and β-integrins (∼70 Å in height), may be sterically blocked. Epitope masking is a major mechanism of viral immune evasion. Modeling of the EBOV GP core structure reveals a surface covered in oligosaccharides (Figure 1B). The dense clustering of glycans creates an unfavorable environment for the interaction of otherwise neutralizing antibodies. Moreover, critical regions on EBOV GP, such as the receptor-binding site, are hidden under layers of glycan. No antibodies have been identified to target the receptor-binding site, however a number of neutralizing antibodies have been generated against the more variable mucin-like domain [6]. The mucin-like domain is not necessary for EBOV entry [4]. Essentially, the EBOV GP glycans direct the immune system to produce antibodies against highly variable or dispensable regions on the viral surface. This also occurs in hosts infected with HIV-1; nonbroadly neutralizing antibodies are generated against the variable V1/V2/V3 loops [7]. In mice, removal of the mucin-like domain of the EBOV GP leads to the production of cross-species antibodies directed at the conserved glycoprotein core structure [8]. A small area near the base of the EBOV GP core is available to immune surveillance (Figure 1B). This nonglycosylated patch on GP is conserved in both Zaire and Sudan EBOV species, and the neutralizing antibodies KZ52 and 16F6-1 bind to this hotspot [9]. However, given its close proximity to the viral membrane and the density of GP spikes on the surface, it is not clear how accessible this epitope is.
EBOV GP also has the unique ability to mask the function of host cellular proteins important in response to viral pathogens. Transient expression of EBOV GP results in low detectable levels of various cell surface proteins such as major histocompatibility complex (MHC) class I proteins and several members of the β-integrin family [10], [11]. Initially, it was thought that EBOV GP downregulated expression or degraded these proteins from the cell surface. However, MHC class I and β-integrins are not removed from the cell surface. Rather, the mucin-like domain of EBOV GP provides a “glycan umbrella” that shields surface epitopes and inhibits surface protein recognition [11]–[13] (Figure 1B). This represents a novel mechanism of disrupting immune function that does not involve downregulation or degradation of surface proteins.
What Roles Do Shed Viral Glycoproteins Play in Immune Evasion?
The shedding or secretion of soluble viral glycoproteins exemplifies another viral strategy of humoral misdirection. Many enveloped viruses, including EBOV, Lassa, respiratory syncytial, herpes simplex, and HIV-1, generate free glycoproteins that act as either “antibody sinks” or decoys of host immunity (Table 1). EBOV-infected cells secrete two glycoproteins (secreted GP and shed GP) into an infected person's sera [14], [15]. Most of the transcripts (70%) for the GP gene encode the 110-kDa, dimeric, secreted GP (sGP) (Figure 1A). A cleavage at the membrane-proximal external region by the tumor necrosis factor-α converting enzyme (TACE) releases the trimeric glycoprotein, termed shed GP. In 5% of the transcripts, insertion of two adenosines produces a small 298-residue secreted GP (ssGP), of unknown function. The first 295 amino acids of sGP are common with the envelope GP, but due to transcriptional stuttering, the sGP C-terminus forms different disulfide linkages leading to a homodimeric rather than a trimeric assembly (Figure 1A). As a result, sGP lacks regions found in GP that have been shown to be important in the neutralization of the virus [9]. sGP and shed GP likely compete with virion-attached GP for antibody binding [16]. Most of the antibodies derived from EBOV survivors or macaques are directed towards sGP rather than the virion-attached GP [17], [18]. Antibodies that bind to sGP or shed GP are likely nonneutralizing, and those neutralizing antibodies that cross-react between sGP and GP may be absorbed by the much more abundant sGP. In a guinea pig model of EBOV infection, shed GP inhibits the neutralizing activity of EBOV antibodies [15].
How Do Viral Glycoproteins Actively Suppress Host Immunity?
In a seminal paper, Cianciolo et al. described immunomodulation by a synthetic peptide derived from the fusion subunit of the HIV-1 envelope glycoprotein [19]. The immunomodulatory region (IR) is comprised of a disulfide-linked loop situated between the heptad-repeat regions of the fusion subunit, and similar structures have been identified in numerous retroviruses and filoviruses (Figure 2A). Point mutations introduced into the IR of HIV-1 gp41 abrogate the modulation of host cytokine expression in vitro and increase antibody responses in rats immunized with mutant protein [20]. Synthetic peptides derived from the IR regions of GP2 of Ebola and Marburg viruses inhibit the expression of IFN-γ, IL-2, and IL-10, lower CD4+ and CD8+ cell activation, and increase immune cell apoptosis [21]. The fusion domain from Moloney murine leukemia virus expressed on various tumor cell lines facilitates xenograph immune evasion and natural killer cell antagonism [22]. Interestingly, the human endogenous retrovirus-derived syncytin-2 retains the immunomodulatory activity associated with the viral envelope glycoproteins, but the closely related syncytin-1 differs in the IR region, ablating this function [23]. These retrovirus-derived syncytin proteins are implicated in both cell–cell fusion during placental development and in maternal–fetal tolerance, clearly pointing to a role in immune evasion [24]. Available structures of the post-fusion glycoprotein subunit show that the disulfide-bonded immunomodulatory motif exists as a conformationally conserved region at the apex of the fusion subunit, with residues identified by mutagenesis as important for immunosuppression displayed outwards (Figure 2B). Interestingly, in the SIV fusion subunit the same region is not found at the apex but rather on the central helical heptad-repeat region. One possible target of the HIV-1 gp41 IR is CD74, a type II single-pass transmembrane protein that, among other functions, chaperones MHC class II dimers from the endoplasmic reticulum to the MHC class II compartment (MIIC) for antigen loading [25], [26], and is also implicated in MHC class I cross-presentation [27]. Recently, it was determined that the ectodomain of human CD74 binds to residues corresponding to a region adjacent to the conserved HIV-1 gp41 IR. When peripheral blood mononuclear cells (PBMCs) were incubated with recombinant post-fusion HIV-1 gp41, an increase in phosphorylated ERK occurred. Furthermore, this activation was inhibited in a dose-dependent manner by treatment with soluble recombinant CD74 ectodomain [28]. The activation of the ERK/MAPK pathway via high levels of the CC-chemokine RANTES (or other exogenous signals) is responsible for increased infectivity of HIV-1 [29]. In support of these findings, siRNA knockdown of CD74 effectively curbs HIV-1 infectivity [30]. Although these works are stimulating, more extensive research is required to generate a complete description of viral fusion glycoprotein-associated immunosuppression. Like HIV-1, the host targets of the immunomodulatory motif found in other species of virus remain poorly defined and await further studies.
Fig. 2. Structural conservation of the viral glycoprotein immunomodulatory region. 
The immunomodulatory region is approximately 20 amino acids long and is found within numerous retroviruses and filoviruses. In each case presented here, the experimentally defined immunomodulatory region is rendered in yellow and residues that are necessary for the observed immunomodulatory activity are depicted as spheres. (A) Head-on view of viral glycoproteins exhibiting a conserved three-fold pinwheel structure. (B) Side-view illustrating the differences in possible interaction faces between the lentiviral gp41 immunomodulatory region and a representative retrovirus, HTLV-1. The outward positioning of the important immunomodulatory residues shown for HTLV-1 gp21 can be observed in all available retroviral and filoviral post-fusion glycoprotein structures except SIV gp41. The PDB codes for the fusion glycoproteins are as follows: EBOV GP2, 2EBO; MARV GP2, 4G2K; HTLV-1 gp21, 1MG1; syncytin-2, 1Y4M; SIV gp41, 2EZO. What Are the Innate Restriction Strategies Targeted toward Viral Glycoproteins?
Host strategies for viral restriction are not limited to the humoral arm of the immune system. The interferon-α-induced innate viral restriction factor BST-2 (also called tetherin and CD317) is a common target of viral glycoprotein modulation [31]. As viruses bud from the cell surface, they are coated with a membrane derived from the host cell. As a result, host BST-2 is incorporated in the membrane of the nascent virion and forms a protein tether to prevent viral release. This nonspecific restriction factor potentially plays a protective role against infections due to retroviruses, filoviruses, arenaviruses, flaviviruses, rhabdoviruses, and orthomyxoviruses (Table 1).
EBOV and HIV-2 both downregulate BST-2 by interactions mediated through their respective viral glycoproteins [32], whereas HIV-1 makes use of the accessory protein Vpu to achieve this same outcome. Some viruses degrade BST-2 or sequester it in intracellular compartments. For example, the HIV-2 envelope glycoprotein appears to sequester the constitutively endocytosed BST-2 in transferrin-positive endosomes [33]. Recent studies have shown that EBOV GP does not remove BST-2 from the cellular surface [34] or sequester it in intracellular sites or lipid rafts [35]. EBOV GP interferes with BST-2-mediated virion capture independently of the mucin-like domain, and neither an engineered form of GP lacking the transmembrane domain nor the dimeric sGP antagonize BST-2 restriction [32]. HIV-1 Vpu interacts with BST-2 via a helical interface found within the transmembrane domains of the two proteins [36]. Accordingly, it may be worthwhile to explore the role of the transmembrane domain of EBOV GP in BST-2 antagonism.
Perspectives
Viruses have developed remarkable mechanisms to inhibit the adaptive and innate immune systems of their hosts. Clearly, viral entry glycoproteins play critical roles in these activities. However, many of these roles and biological pathways are poorly defined. With new infectious diseases emerging and classical viral diseases reemerging, closer examination of viral entry glycoproteins as targets for preventative or therapeutic strategies is warranted.
Zdroje
1. SanchezA, LukwiyaM, BauschD, MahantyS, SanchezAJ, et al. (2004) Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: cellular responses, virus load, and nitric oxide levels. J Virol 78 : 10370–10377.
2. QiuX, AudetJ, WongG, PilletS, BelloA, et al. (2012) Successful treatment of Ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Transl Med 4 : 138ra181.
3. WhiteJM, DelosSE, BrecherM, SchornbergK (2008) Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol 43 : 189–219.
4. JeffersSA, SandersDA, SanchezA (2002) Covalent modifications of the Ebola virus glycoprotein. J Virol 76 : 12463–12472.
5. RitchieG, HarveyDJ, StroeherU, FeldmannF, FeldmannH, et al. (2010) Identification of N-glycans from Ebola virus glycoproteins by matrix-assisted laser desorption/ionisation time-of-flight and negative ion electrospray tandem mass spectrometry. Rapid Commun Mass Spectrom 24 : 571–585.
6. WilsonJA, HeveyM, BakkenR, GuestS, BrayM, et al. (2000) Epitopes involved in antibody-mediated protection from Ebola virus. Science 287 : 1664–1666.
7. PantophletR, BurtonDR (2006) GP120: target for neutralizing HIV-1 antibodies. Annu Rev Immunol 24 : 739–769.
8. OuW, DelisleJ, JacquesJ, ShihJ, PriceG, et al. (2012) Induction of Ebola virus cross-species immunity using retrovirus-like particles bearing the Ebola virus glycoprotein lacking the mucin-like domain. Virol J 9 : 32.
9. DiasJM, KuehneAI, AbelsonDM, BaleS, WongAC, et al. (2011) A shared structural solution for neutralizing Ebola viruses. Nat Struct Mol Biol 18 : 1424–1427.
10. SimmonsG, Wool-LewisRJ, BaribaudF, NetterRC, BatesP (2002) Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J Virol 76 : 2518–2528.
11. ReynardO, BorowiakM, VolchkovaVA, DelpeutS, MateoM, et al. (2009) Ebolavirus glycoprotein GP masks both its own epitopes and the presence of cellular surface proteins. J Virol 83 : 9596–9601.
12. LeeJE, FuscoML, HessellAJ, OswaldWB, BurtonDR, et al. (2008) Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454 : 177–182.
13. FrancicaJR, Varela-RohenaA, MedvecA, PlesaG, RileyJL, et al. (2010) Steric shielding of surface epitopes and impaired immune recognition induced by the Ebola virus glycoprotein. PLoS Pathog 6: e1001098 doi:10.1371/journal.ppat.1001098.
14. SanchezA, YangZY, XuL, NabelGJ, CrewsT, et al. (1998) Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol 72 : 6442–6447.
15. DolnikO, VolchkovaV, GartenW, CarbonnelleC, BeckerS, et al. (2004) Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J 23 : 2175–2184.
16. ItoH, WatanabeS, TakadaA, KawaokaY (2001) Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J Virol 75 : 1576–1580.
17. DruarC, SainiSS, CossittMA, YuF, QiuX, et al. (2005) Analysis of the expressed heavy chain variable-region genes of Macaca fascicularis and isolation of monoclonal antibodies specific for the Ebola virus' soluble glycoprotein. Immunogenetics 57 : 730–738.
18. MaruyamaT, ParrenPW, SanchezA, RensinkI, RodriguezLL, et al. (1999) Recombinant human monoclonal antibodies to Ebola virus. J Infect Dis 179 Suppl 1: S235–239.
19. CiancioloGJ, CopelandTD, OroszlanS, SnydermanR (1985) Inhibition of lymphocyte proliferation by a synthetic peptide homologous to retroviral envelope proteins. Science 230 : 453–455.
20. MorozovVA, MorozovAV, SemaanM, DennerJ (2012) Single mutations in the transmembrane envelope protein abrogate the immunosuppressive property of HIV-1. Retrovirology 9 : 67.
21. YaddanapudiK, PalaciosG, TownerJS, ChenI, SariolCA, et al. (2006) Implication of a retrovirus-like glycoprotein peptide in the immunopathogenesis of Ebola and Marburg viruses. FASEB J 20 : 2519–2530.
22. MangeneyM, HeidmannT (1998) Tumor cells expressing a retroviral envelope escape immune rejection in vivo. Proc Natl Acad Sci U S A 95 : 14920–14925.
23. MangeneyM, RenardM, Schlecht-LoufG, BouallagaI, HeidmannO, et al. (2007) Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci U S A 104 : 20534–20539.
24. DupressoirA, LavialleC, HeidmannT (2012) From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33 : 663–671.
25. MoldenhauerG, HenneC, KarhausenJ, MollerP (1999) Surface-expressed invariant chain (CD74) is required for internalization of human leucocyte antigen-DR molecules to early endosomal compartments. Immunology 96 : 473–484.
26. CresswellP (1994) Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol 12 : 259–293.
27. BashaG, OmilusikK, Chavez-SteenbockA, ReinickeAT, LackN, et al. (2012) A CD74-dependent MHC class I endolysosomal cross-presentation pathway. Nat Immunol 13 : 237–245.
28. ZhouC, LuL, TanS, JiangS, ChenYH (2011) HIV-1 glycoprotein 41 ectodomain induces activation of the CD74 protein-mediated extracellular signal-regulated kinase/mitogen-activated protein kinase pathway to enhance viral infection. J Biol Chem 286 : 44869–44877.
29. ChangTL, GordonCJ, Roscic-MrkicB, PowerC, ProudfootAE, et al. (2002) Interaction of the CC-chemokine RANTES with glycosaminoglycans activates a p44/p42 mitogen-activated protein kinase-dependent signaling pathway and enhances human immunodeficiency virus type 1 infectivity. J Virol 76 : 2245–2254.
30. DunnSJ, KhanIH, ChanUA, ScearceRL, MelaraCL, et al. (2004) Identification of cell surface targets for HIV-1 therapeutics using genetic screens. Virology 321 : 260–273.
31. NeilSJ, ZangT, BieniaszPD (2008) Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451 : 425–430.
32. KaletskyRL, FrancicaJR, Agrawal-GamseC, BatesP (2009) Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A 106 : 2886–2891.
33. LauD, KwanW, GuatelliJ (2011) Role of the endocytic pathway in the counteraction of BST-2 by human lentiviral pathogens. J Virol 85 : 9834–9846.
34. LopezLA, YangSJ, HauserH, ExlineCM, HaworthKG, et al. (2010) Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84 : 7243–7255.
35. LopezLA, YangSJ, ExlineCM, RengarajanS, HaworthKG, et al. (2012) Anti-tetherin activities of HIV-1 Vpu and Ebola virus glycoprotein do not involve removal of tetherin from lipid rafts. J Virol 86 : 5467–5480.
36. SkaskoM, WangY, TianY, TokarevA, MunguiaJ, et al. (2012) HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J Biol Chem 287 : 58–67.
37. BrancoLM, GroveJN, MosesLM, GobaA, FullahM, et al. (2010) Shedding of soluble glycoprotein 1 detected during acute Lassa virus infection in human subjects. Virol J 7 : 306.
38. BrancoLM, GarryRF (2009) Characterization of the Lassa virus GP1 ectodomain shedding: implications for improved diagnostic platforms. Virol J 6 : 147.
39. ChiangSF, LinTY, ChowKC, ChiouSH (2010) SARS spike protein induces phenotypic conversion of human B cells to macrophage-like cells. Mol Immunol 47 : 2575–2586.
40. KongL, GiangE, RobbinsJB, StanfieldRL, BurtonDR, et al. (2012) Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci U S A 109 : 9499–9504.
41. HelleF, VieyresG, ElkriefL, PopescuCI, WychowskiC, et al. (2010) Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol 84 : 11905–11915.
42. CrottaS, StillaA, WackA, D'AndreaA, NutiS, et al. (2002) Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J Exp Med 195 : 35–41.
43. MachielsB, LétéC, GuillaumeA, MastJ, StevensonPG, et al. (2011) Antibody evasion by a gammaherpesvirus O-glycan shield. PLoS Pathog 7: e1002387 doi:10.1371/journal.ppat.1002387.
44. RessingME, van LeeuwenD, VerreckFA, KeatingS, GomezR, et al. (2005) Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J Virol 79 : 841–852.
45. WeiCJ, BoyingtonJC, DaiK, HouserKV, PearceMB, et al. (2010) Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci Transl Med 2 : 24ra21.
46. DasSR, PuigbòP, HensleySE, HurtDE, BenninkJR, et al. (2010) Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog 6: e1001211 doi:10.1371/journal.ppat.1001211.
47. BukreyevA, YangL, FrickeJ, ChengL, WardJM, et al. (2008) The secreted form of respiratory syncytial virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J Virol 82 : 12191–12204.
48. AguilarHC, MatreyekKA, FiloneCM, HashimiST, LevroneyEL, et al. (2006) N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol 80 : 4878–4889.
49. MooreJP, SodroskiJ (1996) Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol 70 : 1863–1872.
50. Schlecht-LoufG, RenardM, MangeneyM, LetzelterC, RichaudA, et al. (2010) Retroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc Natl Acad Sci U S A 107 : 3782–3787.
51. McLellanJS, PanceraM, CarricoC, GormanJ, JulienJP, et al. (2011) Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480 : 336–343.
52. CohenT, CohenSJ, AntonovskyN, CohenIR, ShaiY (2010) HIV-1 gp41 and TCRα trans-membrane domains share a motif exploited by the HIV virus to modulate T-cell proliferation. PLoS Pathog 6: e1001085 doi:10.1371/journal.ppat.1001085.
53. AshkenaziA, FaingoldO, KaushanskyN, Ben-NunA, ShaiY (2013) A highly conserved sequence associated with the HIV gp41 loop region is an immunomodulator of antigen-specific T cells in mice. Blood 121 : 2244–2252.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 5- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Volatile Metabolites of Pathogens: A Systematic Review
- Behind Closed Membranes: The Secret Lives of Picornaviruses?
- DNA from Skeletal Remains from the 6 Century AD Reveals Insights into Justinianic Plague
- Experimental Evolution of Pathogenesis: “Patient” Research
- Bile Acid Recognition by the Germinant Receptor, CspC, Is Important for Establishing Infection
- Malaria Parasite cGMP-dependent Protein Kinase Regulates Blood Stage Merozoite Secretory Organelle Discharge and Egress
- The Secret Life of Viral Entry Glycoproteins: Moonlighting in Immune Evasion
- Uracil DNA Glycosylase Counteracts APOBEC3G-Induced Hypermutation of Hepatitis B Viral Genomes: Excision Repair of Covalently Closed Circular DNA
- Differences in Gastric Carcinoma Microenvironment Stratify According to EBV Infection Intensity: Implications for Possible Immune Adjuvant Therapy
- Polyphosphate and Its Diverse Functions in Host Cells and Pathogens
- Activation of the NLRP3 Inflammasome by IAV Virulence Protein PB1-F2 Contributes to Severe Pathophysiology and Disease
- Plant Virus Ecology
- Proving Lipid Rafts Exist: Membrane Domains in the Prokaryote Have the Same Properties as Eukaryotic Lipid Rafts
- Structural and Functional Basis for Inhibition of Erythrocyte Invasion by Antibodies that Target EBA-175
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Malaria Parasite cGMP-dependent Protein Kinase Regulates Blood Stage Merozoite Secretory Organelle Discharge and Egress
- The Secret Life of Viral Entry Glycoproteins: Moonlighting in Immune Evasion
- Structural and Functional Basis for Inhibition of Erythrocyte Invasion by Antibodies that Target EBA-175
- DNA from Skeletal Remains from the 6 Century AD Reveals Insights into Justinianic Plague
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



