-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Chronic Wasting Disease in Bank Voles: Characterisation of the Shortest Incubation Time Model for Prion Diseases
In order to assess the susceptibility of bank voles to chronic wasting disease (CWD), we inoculated voles carrying isoleucine or methionine at codon 109 (Bv109I and Bv109M, respectively) with CWD isolates from elk, mule deer and white-tailed deer. Efficient transmission rate (100%) was observed with mean survival times ranging from 156 to 281 days post inoculation. Subsequent passages in Bv109I allowed us to isolate from all CWD sources the same vole-adapted CWD strain (Bv109ICWD), typified by unprecedented short incubation times of 25–28 days and survival times of ∼35 days. Neuropathological and molecular characterisation of Bv109ICWD showed that the classical features of mammalian prion diseases were all recapitulated in less than one month after intracerebral inoculation. Bv109ICWD was characterised by a mild and discrete distribution of spongiosis and relatively low levels of protease-resistant PrPSc (PrPres) in the same brain regions. Despite the low PrPres levels and the short time lapse available for its accumulation, end-point titration revealed that brains from terminally-ill voles contained up to 108,4 i.c. ID50 infectious units per gram. Bv109ICWD was efficiently replicated by protein misfolding cyclic amplification (PMCA) and the infectivity faithfully generated in vitro, as demonstrated by the preservation of the peculiar Bv109ICWD strain features on re-isolation in Bv109I. Overall, we provide evidence that the same CWD strain was isolated in Bv109I from the three-cervid species. Bv109ICWD showed unique characteristics of “virulence”, low PrPres accumulation and high infectivity, thus providing exceptional opportunities to improve basic knowledge of the relationship between PrPSc, neurodegeneration and infectivity.
Published in the journal: . PLoS Pathog 9(3): e32767. doi:10.1371/journal.ppat.1003219
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003219Summary
In order to assess the susceptibility of bank voles to chronic wasting disease (CWD), we inoculated voles carrying isoleucine or methionine at codon 109 (Bv109I and Bv109M, respectively) with CWD isolates from elk, mule deer and white-tailed deer. Efficient transmission rate (100%) was observed with mean survival times ranging from 156 to 281 days post inoculation. Subsequent passages in Bv109I allowed us to isolate from all CWD sources the same vole-adapted CWD strain (Bv109ICWD), typified by unprecedented short incubation times of 25–28 days and survival times of ∼35 days. Neuropathological and molecular characterisation of Bv109ICWD showed that the classical features of mammalian prion diseases were all recapitulated in less than one month after intracerebral inoculation. Bv109ICWD was characterised by a mild and discrete distribution of spongiosis and relatively low levels of protease-resistant PrPSc (PrPres) in the same brain regions. Despite the low PrPres levels and the short time lapse available for its accumulation, end-point titration revealed that brains from terminally-ill voles contained up to 108,4 i.c. ID50 infectious units per gram. Bv109ICWD was efficiently replicated by protein misfolding cyclic amplification (PMCA) and the infectivity faithfully generated in vitro, as demonstrated by the preservation of the peculiar Bv109ICWD strain features on re-isolation in Bv109I. Overall, we provide evidence that the same CWD strain was isolated in Bv109I from the three-cervid species. Bv109ICWD showed unique characteristics of “virulence”, low PrPres accumulation and high infectivity, thus providing exceptional opportunities to improve basic knowledge of the relationship between PrPSc, neurodegeneration and infectivity.
Introduction
Chronic wasting disease (CWD) of cervids belongs to the family of transmissible spongiform encephalopathies (TSE) or prion diseases, a group of fatal neurodegenerative pathologies affecting animals and humans. They are characterised by spongiform changes, gliosis and the deposition in the brain of a post-translational misfolded isoform (PrPSc) of the host-encoded cellular prion protein (PrPc). Prion diseases also include Creutzfeldt-Jakob disease (CJD) of humans, scrapie of sheep and goats and bovine spongiform encephalopathy (BSE) of cattle.
CWD is the only prion disease known to affect free-ranging wild animals. It was first described in the United States in the late 1960s [1]. Currently, CWD has been diagnosed in farmed and free-ranging cervids in several areas of North America [2] and South Korea, where it was accidentally imported from Canada [3]. The disease has been documented in Rocky Mountain elk (Cervus elaphus nelsoni), mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and moose (Alces alces) [4].
Epidemiological evidence indicates that CWD spreads naturally with relative efficiency and recent trend analyses suggest that its prevalence is increasing [5].
The development of laboratory models for CWD has long been hampered by the very inefficient transmission of CWD to wild-type mice [6]. Significant progress was made by the generation of transgenic mice over-expressing cervid PrP [6], [7], [8], [9], [10], [11]. CWD has also been transmitted, albeit less efficiently, to transgenic mice over-expressing mouse [12] or Syrian hamster PrP, as well as to different hamster species [13]. Recently, CWD was successfully transmitted to different species of North American wild rodents [14] and ferrets [15].
In recent years, with the aim of developing new animal models for prion diseases, we have studied the susceptibility of the bank vole (Myodes glareolus) to a wide range of human and animal prion diseases. Two lines of voles, one homozygous for methionine and the other for isoleucine at codon 109 of PrP – here designated Bv109M and Bv109I, respectively – were investigated. Bv109M was shown to be susceptible to sporadic and genetic CJD [16], [17], sheep scrapie [18], [19], mouse - and hamster-adapted scrapie strains [20], [21], [22], cattle and sheep BSE [16], [22] and atypical BSE [22]. Overall susceptibility of Bv109I was found to be comparable to that of the methionine-carrying line, although with differences depending on the specific prion disease (unpublished data).
In the present study, we investigated the susceptibility of bank voles to CWD. To this purpose, we inoculated Bv109M and Bv109I with various CWD sources and found that CWD replicated faster in Bv109I compared to Bv109M. We then focused on the Bv109I line, which showed unprecedented short survival times upon CWD adaptation. We describe a thorough characterization of Bv109I-adapted CWD, showing that in spite of their short survival time, high infectious titres accumulate in the brains of affected animals. Furthermore, we provide evidence that Bv109I-adapted CWD can be easily and faithfully replicated in vitro by serial automated PMCA (saPMCA).
Results
Bank voles are susceptible to CWD isolates from three cervid species
All CWD isolates transmitted with 100% attack rate and short survival times in both Bv109I and Bv109M (Table 1). In some groups survival times were not uniform. Indeed, while most of the voles inoculated were clinically affected between 150 and 350 days post-infection (d.p.i.), a minor proportion of voles showed much longer survival times (Figure S1). Interestingly, Bv109I replicated CWD faster than Bv109M, with 60–100 days shorter median survival times (Figure S1). Conversely, SS7B, a scrapie isolate previously shown to replicate efficiently in Bv109M [19], showed a 60 days longer median survival time in Bv109I. Overall, these data demonstrate that the 109 methionine/isoleucine variation influences the susceptibility of voles in a strain dependent fashion and that Bv109I is a fast model for CWD replication. We thus further investigated the transmission features of CWD in the bank vole line expressing isoleucine at codon 109.
Tab. 1. Survival times of Bv109I and Bv109M after primary transmission of CWD. 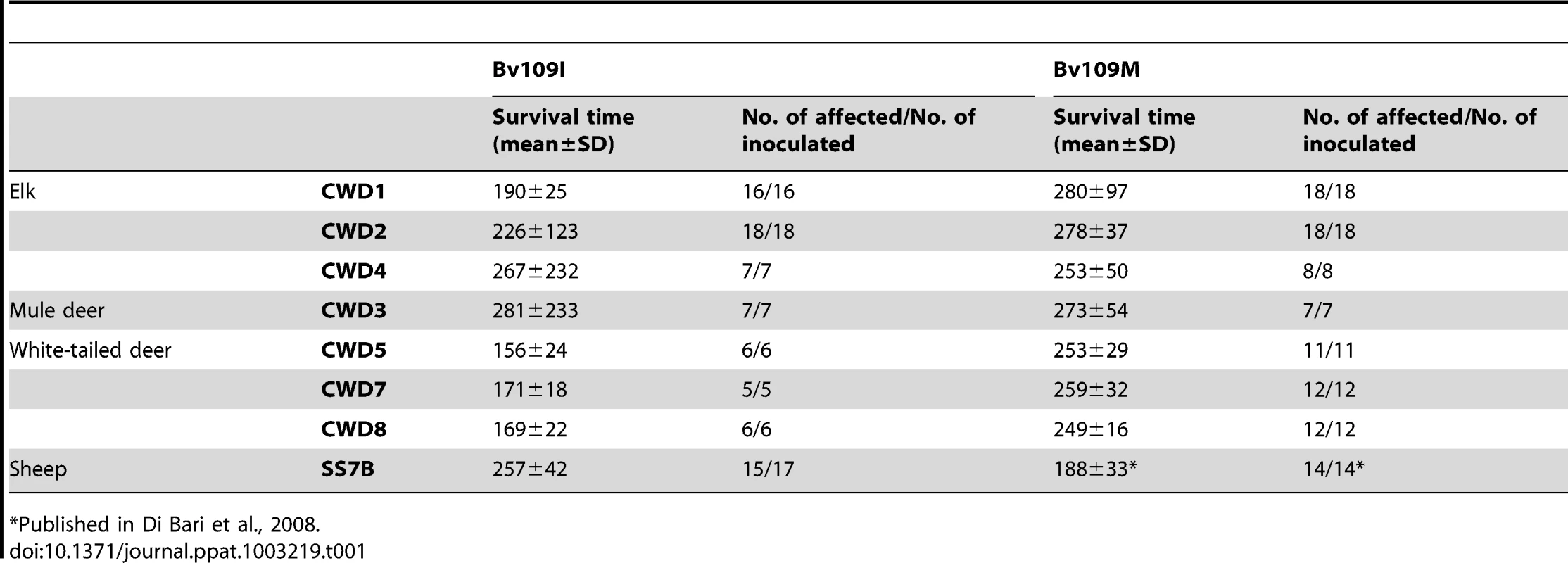
Published in Di Bari et al., 2008. Transmission features of CWD isolates in Bv109I
CWD isolates transmitted in Bv109I with mean survival times ranging from 156 to 281 d.p.i. (Table 1). All isolates induced a similar clinical picture. The onset of disease was characterised by slight behavioural alterations; hypoactivity and hyporeactivity were sometimes observed along with the disappearance of the typical behaviour of hiding under the sawdust lining the cage. Dorsal kyphosis, slight ataxia of the forelimb and occasional head bobbing (upward movements of the head) accompanied the progression of the disease. Overall, clinical signs were faint compared with those previously observed in voles infected with classical scrapie [19] and occasionally animals were found dead because of the rapid progression to terminal stage. The three isolates from white-tailed deer gave the shortest survival times, ranging from 156 to 171 d.p.i., while the survival times were slightly longer with isolates from elk and mule deer. As mentioned above, individual voles showing a late clinical onset were observed in CWD2, with two outliers at 414 and 656 d.p.i., in CWD3 (outlier at 805 d.p.i.) and CWD4 (792 d.p.i.) (Figure S1).
All infected voles showed moderate spongiform changes at brain histopathology, accompanied by gliosis and neuronal loss (Figure 1). All groups had a similar distribution of spongiform degeneration, which mainly involved the superior colliculus, the thalamus and, to a lesser extent, the cerebral cortex (Figure 2A). At the cortical level, spongiosis preferentially involved the V and VI deep layers (data not shown). Interestingly, spongiosis was not observed in the cerebral cortex of four voles surviving longer than 400 d.p.i. (Figure S2), although they showed spongiform degeneration in the mesencephalon and diencephalon with a distribution similar to that of the other infected voles (data not shown).
Fig. 1. Histopathological and immunohistochemical analysis of Bv109I inoculated with Bv109ICWD. 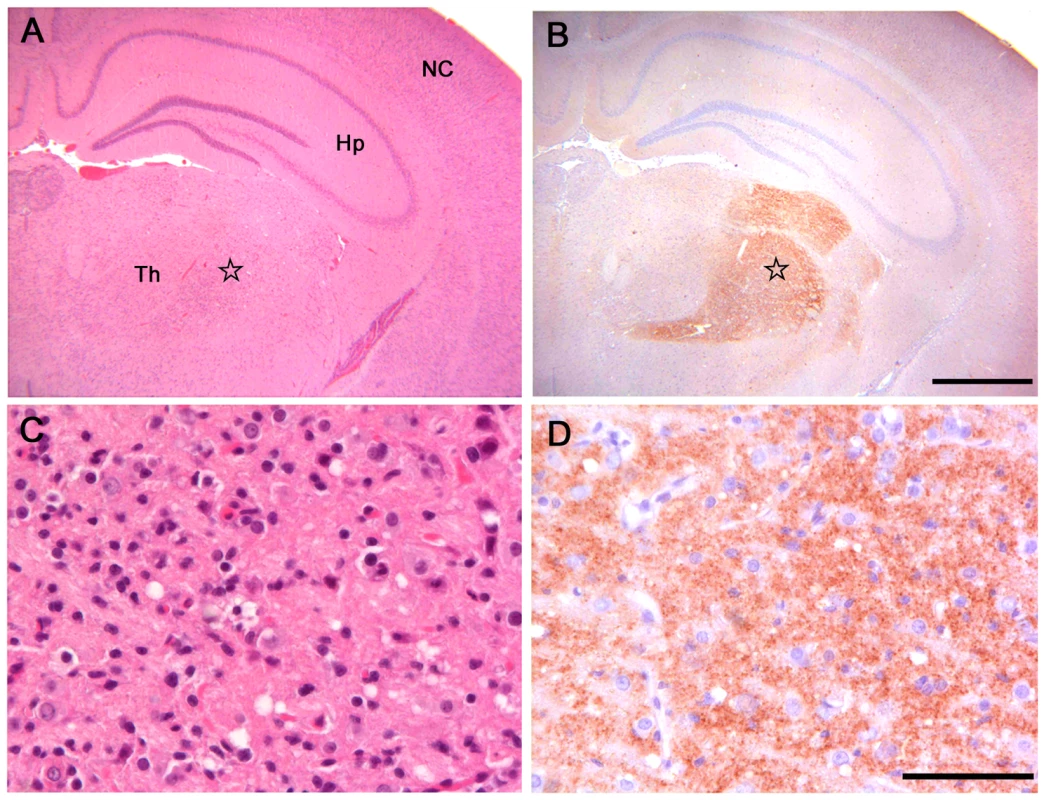
(A) Serial sections showing neocortex (NC), hippocampus (Hp) and thalamus (Th), stained by haematoxylin and eosin and (B) by immunohistochemistry for PrPSc with SAF84 mAb - bar 500 µm. (C) and (D): magnification of the ventral thalamic nucleus (star) from (A) and (B), respectively - bar 50 µm. Fig. 2. Lesion profiles of Bv109I infected with CWD following primary transmission and third passage. 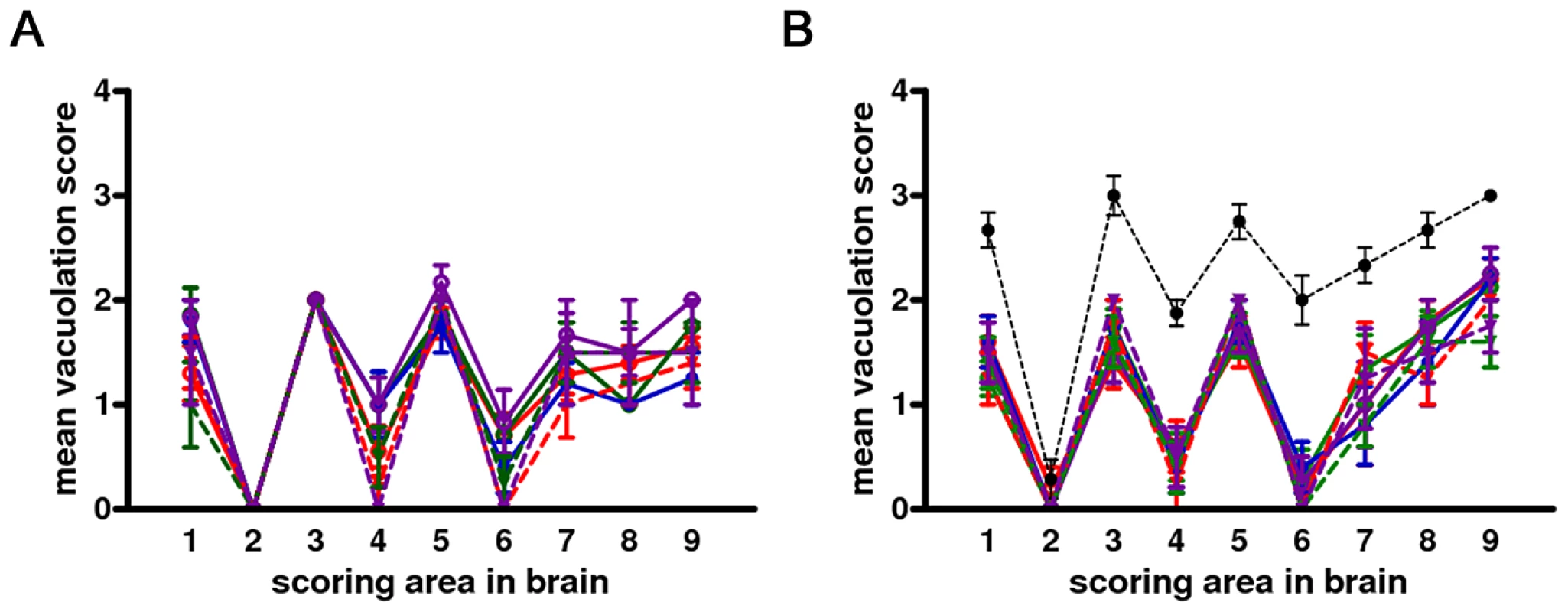
Pathological phenotypes observed after primary transmission and third passage are shown in (A) and (B), respectively. CWD1, 2 and 4 from elk, are shown in red, green and violet lines with open circles, respectively; CWD3 from mule deer, in blue line and closed circles; CWD5, 6 and 7 from W-T deer in red, green and violet dashed lines and closed triangles, respectively. Vole-adapted sheep scrapie is shown in black dashed line and closed circles. Brain-scoring areas are: medulla (1), cerebellum (2), superior colliculus (3), hypothalamus (4), thalamus (5), hippocampus (6), septum (7), retrosplenial and adjacent motor cortex (8), cingulate and adjacent motor cortex (9). PrPSc was readily detected in all infected voles by IHC, PET-blot and WB. IHC showed punctate PrPSc deposition (Figure 1) restricted to specific areas. The thalamus, substantia nigra, geniculate and vestibular nuclei were the most involved areas. The hippocampus was mostly spared, though some fibres of CA2 were occasionally found positive. Overall, a topographical correlation between vacuolation, neuronal loss and PrPSc deposition was observed (Figure 1).
Western blot analysis showed the typical three-band PrPres pattern, which was similar in all CWD-infected voles (Figure 3). Compared with elk and deer PrPres, bank vole CWD PrPres showed a lower molecular mass of the unglycosylated band (Figure 3A), similar to that reported in prairie voles infected with mule deer CWD [23]. In keeping with the discrete brain distribution of PrPSc observed by PET-blot or IHC, the overall level of PrPres detected in brain homogenates was much lower than that usually found in vole-adapted strains [16], [19], [22]. Direct comparison with Bv109I-adapted SS7B showed that PrPres levels were at least 30 fold lower in Bv109I-adapted CWD (Figure S3). In addition, the migration pattern was different from that previously reported in bank vole TSEs, being intermediate between scrapie and BSE (Figure 3B).
Fig. 3. Western blot analysis of the PrPres level in the brain of CWD-affected voles. 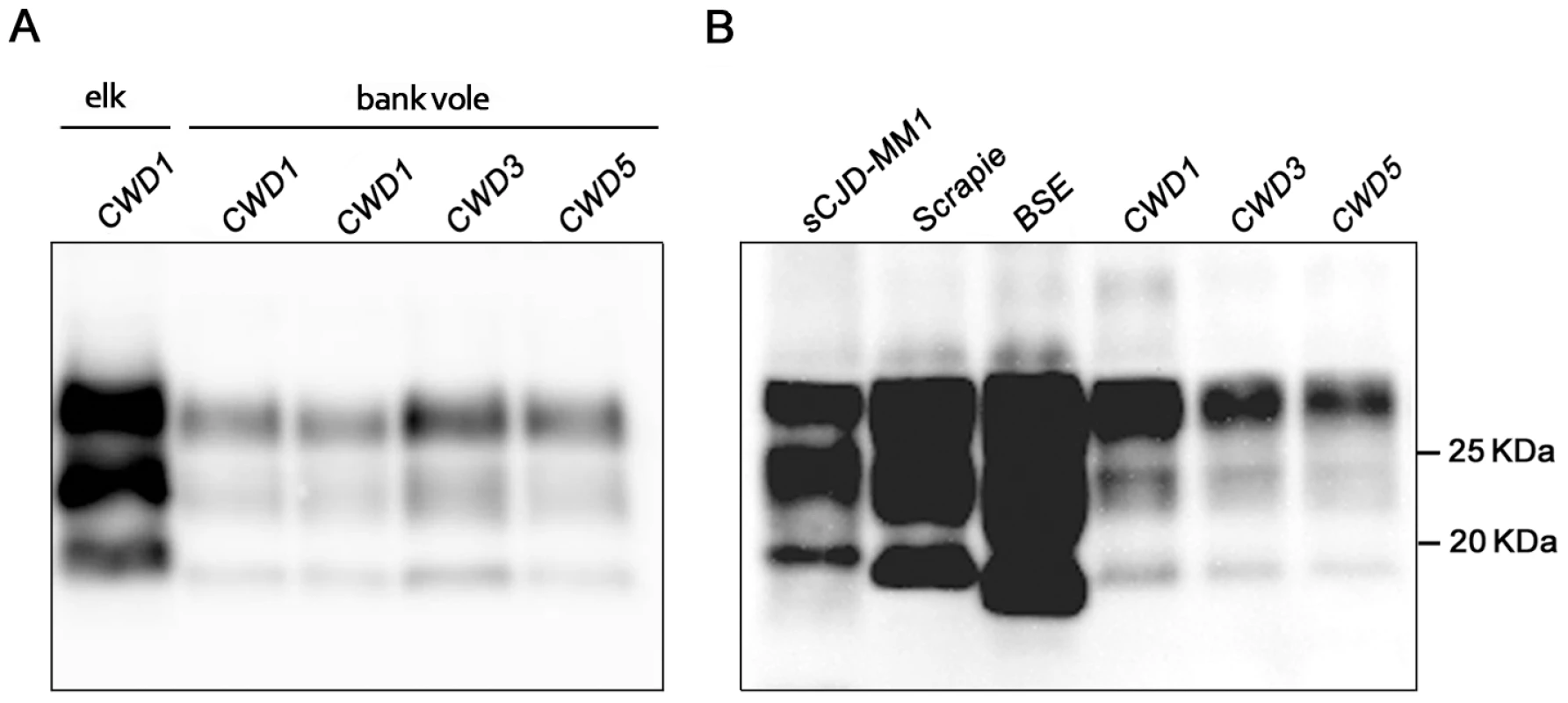
(A) Immunoblot of proteinase K–resistant PrPSc (PrPres) from one original elk isolate (CWD1) and from voles infected with CWD1, 3 and 5. (B) Comparison of PrPres amount and glycoprofile between Bv109I-adapted CWD and Bv109M-adapted sporadic CJD, sheep scrapie and cattle BSE, at third passage. Membranes were probed with SAF84. Molecular size markers are shown in kilodaltons on the right. All the twelve control challenged voles, culled at the end of the experiment (810 d.p.i.) or found dead for intercurrent disease (two voles, at 550 and 721 d.p.i.), were found PrPSc negative by Western blot analysis.
CWD adapts to Bv109I with strikingly short survival times
In order to study the adaptation of CWD prions in Bv109I and to compare more closely the biological properties of the various CWD isolates, vole-passaged CWD isolates were further propagated in Bv109I. On the second passage we observed a dramatic decrease in the incubation times, with behavioural alterations already evident at 25–28 d.p.i.. Following a short clinical phase all CWD isolates produced strikingly short mean survival times of between 34 and 44 d.p.i. (Table 2). Survival times were stable or slightly shorter on third passage, with all Bv109I-adapted CWD strains giving a survival time of ∼35 d.p.i. (Table 2).
Tab. 2. Survival times of Bv109I and Bv109M after subsequent passages of CWD. 
The number of animals in each group is reported in brackets. The survival times of voles used for vole-to-vole transmissions, indicated as ‘donors’, are shown. The neuropathological phenotype after the second and subsequent passages was similar to that observed on primary transmission (Figure 2B), with lesion profiles showing a slight increase in cortical involvement (Figure 2B). The pattern of brain PrPSc distribution by PET-blot was the same in all groups, characterised by PrPSc deposition mainly in the thalamus, substantia nigra, geniculate nuclei, vestibular nuclei and the deep layers of the cerebral cortex (Figure 4). Bv109I-adapted SS7B showed a marked neurodegeneration in all areas considered in lesion profiles, with the exception of the cerebellar cortex. PET-blot showed abundant PrPSc, distributed throughout the brain. Prominent immunolabelling was observed in the cortices, septal nucleus, hippocampus, thalamus, superior colliculi, geniculate nuclei and medulla oblongata.
Fig. 4. Regional distribution, by PET-blot, of PrPres following the third passage of CWD1, CWD3, CWD5 and sheep scrapie in Bv109I. 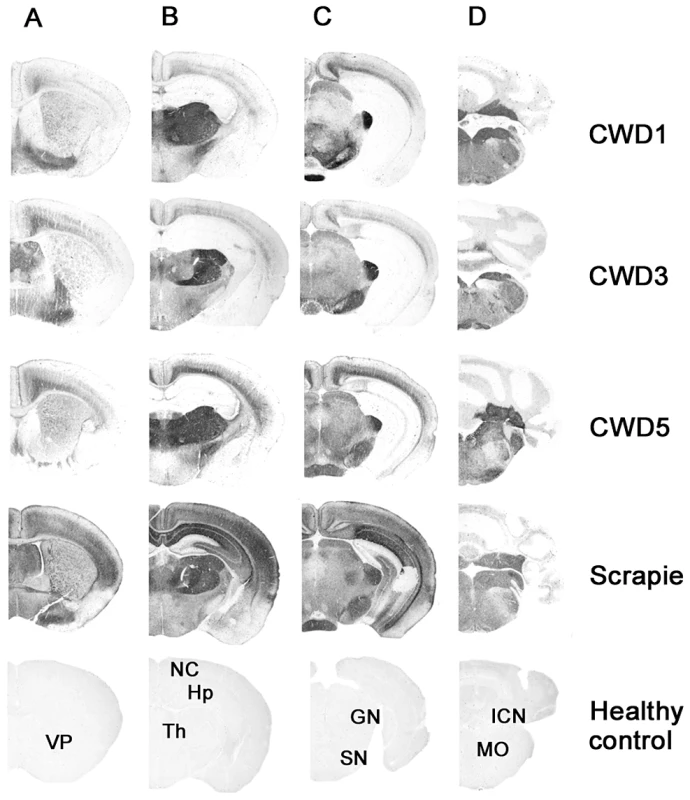
Coronal sections of the forebrain representing: telencephalon (A), diencephalon (B), midbrain (C) and hindbrain (D). In the lower part of the figure, the labelled coronal sections of negative control brain from 150 days old Bv109I are shown: VP, ventral pallidum; NC, neocortex; Hp, hippocampus; Th, thalamus; GN, geniculate nuclei; SN, substantia nigra; ICN, interposed cerebellar nucleus; MO, medulla oblongata. Overall these data suggest that the same strain, designated Bv109ICWD, was isolated from elk, mule deer and white-tailed deer.
Strain characterisation from Bv109I outliers suggests the isolation of more than one prion component in primary transmission
Two voles, designated CWD3outlier and CWD4outlier, which developed clinical signs after unusually long times of 805 and 792 d.p.i. at primary transmission of CWD3 and CWD4 were selected for supplementary strain typing. Second passages from CWD3outlier and CWD4outlier gave much longer survival times compared with CWD3 and CWD4 (Table 2). At subsequent passages the survival time decreased progressively and CWD3outlier and CWD4outlier were fully adapted only at the fourth passage (Table 2). Interestingly, neuropathological assessment at the second (data not shown) and third (Figure S2) passages showed much less severe spongiform changes in the cortex and superior colliculus than Bv109ICWD at ∼35 d.p.i.. By the fourth passage, the lesion profiles converged to Bv109ICWD (Figure S2) and the survival time decreased to ∼35 d.p.i.. These data suggest that the Bv109ICWD strain was eventually isolated also from CWD3outlier and CWD4outlier, although it required four subsequent passages to emerge. The deviant neuropathological profile observed in CWD3outlier and CWD4outlier at first passage was propagated for at least two vole-to-vole passages, suggesting that a second CWD strain was isolated in these outliers, which was progressively outcompeted by the extreme rapidity of the Bv109ICWD strain.
Characterisation of Bv109ICWD
The infectious titre of Bv109ICWD was determined by endpoint titration. Increasing survival times were observed with 10−2 to 10−5 dilutions, while dilutions higher than 10−5 also produced a decreasing attack rate (Figure 5). The infectious titre of Bv109ICWD was 108,4 i.c. ID50 U g−1. The vast majority of diseased animals succumbed within ∼100 d.p.i., and only 3 out of 44 voles developed the disease later on, with unequivocal clinical signs (Figure 5). Thus Bv109ICWD undergoes very fast replication kinetics, allowing high prion titres to accumulate in a very short time.
Fig. 5. End-point titration of Bv109ICWD infectivity in Bv109I. 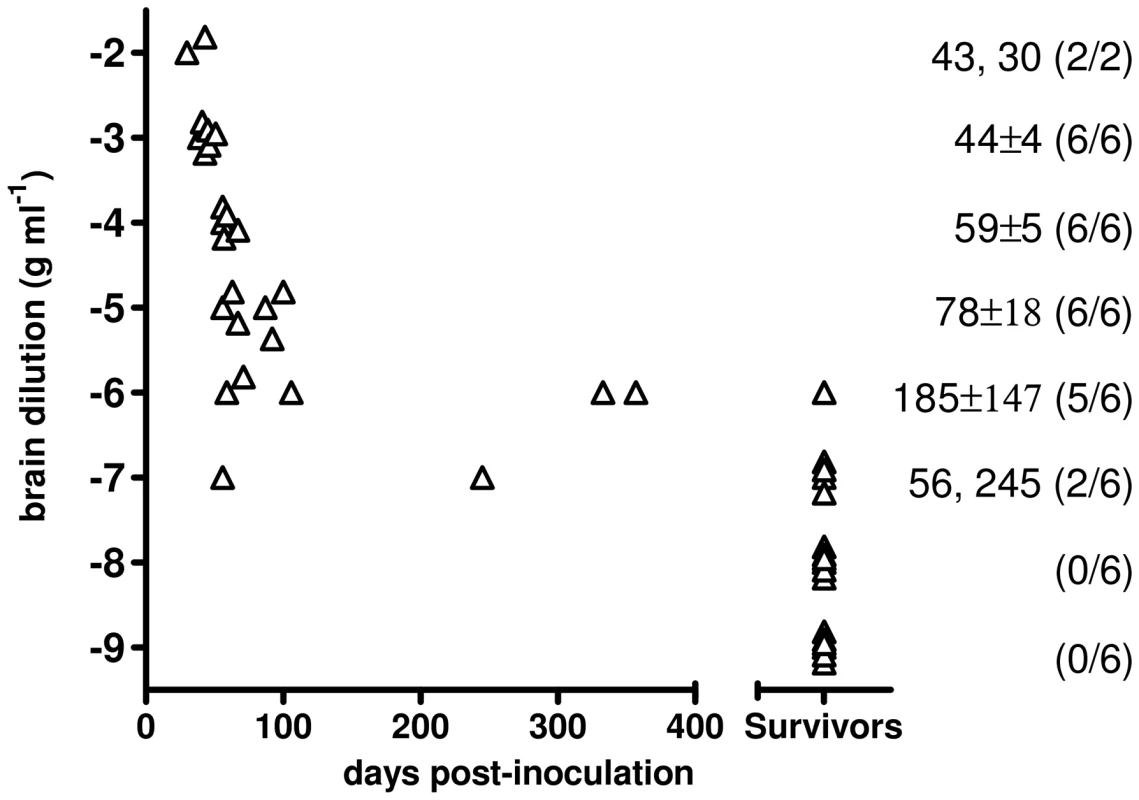
Triangles represent individual survival times. Voles that did not show signs of infection after 450 d.p.i. are plotted in compressed form after the x axes break point. The mean survival times (days ± standard deviation) and the numbers of diseased/inoculated voles are indicated on the right of each chart. To analyse the potential of Bv109ICWD to be amplified in vitro, we performed PMCA experiments using 1/100 mixtures of Bv109ICWD as seed and brain homogenates from Bv109I voles as substrate. As already shown with other vole species [24], Bv109ICWD was efficiently amplified already at the first PMCA round (amplified products from rounds 4 to 13 are shown in Figure S4). In order to verify whether Bv109ICWD infectivity was efficiently propagated during saPMCA and to study its biological properties in comparison with the original Bv109ICWD, we performed saPMCA experiments for 15 successive PMCA rounds so that the original seed was diluted 1016-fold. This dilution was such that only the newly generated PrPSc was theoretically present in the final PMCA product. Groups of nine voles were inoculated with 10-fold dilutions of either in vitro-generated Bv109ICWD (designated Bv109ICWDPMCA) or the original Bv109ICWD used as seed. Bv109ICWDPMCA induced terminal disease in 42±2 d.p.i., a survival time similar to that of the control Bv109ICWD inoculum (38±2 d.p.i.). The second passage of Bv109ICWDPMCA produced disease in 34±2 d.p.i.. The clinical phenotype and the lesion profile of animals infected with Bv109ICWDPMCA, after primary transmission and second passage, were indistinguishable from the control group (Figure 6A). The type of PrPSc deposition and the pattern of PrPSc distribution in the brain, analysed by IHC (not shown) and PET-blot (Figure 6B), were also the same in Bv109ICWDPMCA and Bv109ICWD. Overall, Bv109ICWD produced in vitro was highly infectious and faithfully maintained the properties of the original Bv109ICWD strain.
Fig. 6. Biological properties of in vitro-generated Bv109ICWD (Bv109ICWDPMCA). 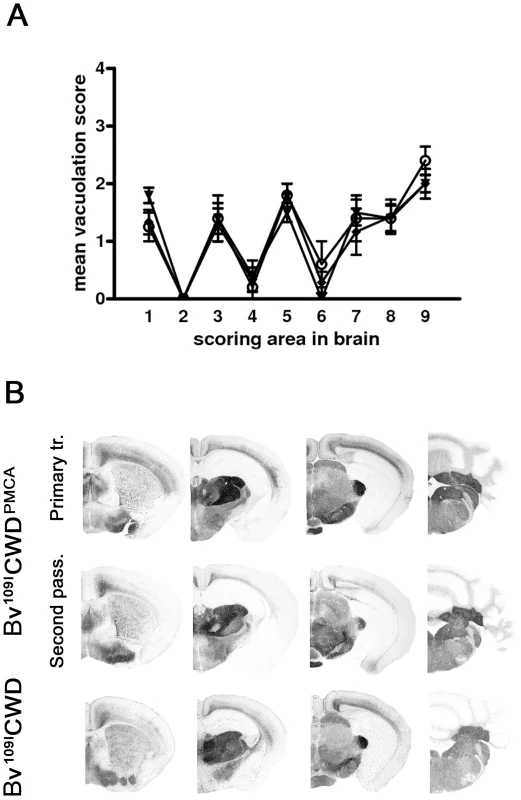
A) Lesion profiles of Bv109I infected with Bv109ICWDPMCA following primary transmission (open circles) and second passage (closed diamonds), compared with Bv109I infected with Bv109ICWD (triangles). Brain-scoring positions are described in Figure 2. B) Regional distribution, by PET-blot with SAF84 mAb, of PrPres in Bv109I infected with Bv109ICWDPMCA following primary transmission and second passage compared with the control group (Bv109ICWD). Brain areas are as in Figure 4. Discussion
In the present study we investigated the susceptibility of Bv109I and Bv109M to seven CWD isolates from three deer species and found that these animal models are highly permissive to CWD, showing 100% attack rate and mean survival times between 156 and 281 d.p.i.. A deepened transmission and characterization study of CWD was carried out in Bv109I. The susceptibility of this model appeared comparable to that of transgenic mice expressing cervid PrP [6], [7], [9], [10], [11]. The reasons for such a high susceptibility are unclear but apparently not related to a different expression of PrPC. As a matter of fact, its expression level in Bv109I is comparable to that of mouse and hamster (Figure S5). The dramatic drop in survival time with vole-to-vole sub-passages suggests that CWD still encounters a high transmission barrier in Bv109I, implying that Bv109I permissiveness to CWD was not due to the absence of transmission barrier, as observed in transgenic mice expressing cervid PrP. Using Bv109M voles we have previously shown that this species is permissive to a variety of human and animal prion diseases. Studies aimed at investigating the molecular basis of the susceptibility of bank voles to foreign prions and their selective strain preferences suggested that two asparagine residues at positions 150 and 170, specific to vole PrP, might play a role [18], [22], [25]. Interestingly, cervid PrP also has asparagine at residue 170, suggesting that sequence identity at codon 170 might facilitate the transmission of CWD to Bv109I. This interpretation is supported by the relative ease of transmission of CWD to meadow voles [14], which also have asparagine at positions 150 and 170, as well as by studies in MoPrP170N, 174T transgenic mice [24], [26] and by in vitro amplification of CWD by PMCA [27].
A striking feature of Bv109I-adapted CWD was the short incubation time of less than one month. In an earlier work, we showed that bank voles and related rodent species have peculiarly short survival times after infection with adapted prions, and presumably support equally fast prion replication kinetics, possibly due to the previously mentioned 150N–170N PrP residues [22]. This was also observed in vitro using Bv109M brain homogenates as substrate for PMCA-driven prion replication [28]. Notwithstanding this, our previous and on-going studies with Bv109M have not shown evidence of ultra-fast strains such as Bv109ICWD, and the fastest strains observed so far in voles show incubation times of ∼2 months [19, unpublished data). Transmission studies of CWD to Bv109M have not yet completed but we have observed that also CWD adapts to Bv109M with survival times longer than Bv109ICWD (60–100 d.p.i.) (unpublished data). The short survival time of Bv109ICWD is also unprecedented when compared with those found in transgenic mouse models, in which PrP overexpression greatly fosters prion diseases [6], [7], [8], [9], [10], [11]. Indeed, the fastest rodent models reported so far, i.e. Tga20 [29], Tg52NSE [30], Tg7 [31], Tg4053 [32] and Tg338 mice [33], express several-fold higher PrP levels compared with wild-type mice and have incubation periods at least twice as long as Bv109ICWD. Interestingly, it was recently shown that TgS3581 mice overexpressing vole PrP encoding for Isoleucine at position 109, undergo spontaneous prion disease and that it adapts to the same model with mean survival time of 35 days [34]. These findings suggest that the presence of Isoleucine at position 109 of the vole PrP plays a specific role in determining the short survival time of Bv109ICWD
In the present work we provide evidence that all the hallmarks of TSEs were recapitulated within one month in bank vole CWD. The finding that neurodegeneration and PrPSc deposition showed a discrete brain distribution, involving specific neuronal populations such as those in the medulla and thalamus, might suggest that Bv109ICWD replication primarily involves the so called clinical target areas (CTAs) which, once colonised by prions, trigger the clinical signs and death of the animals [35], [36]. A recent study showed that shorter time periods were needed to initiate the clinical phase when the 127S scrapie strain primarily targeted CTAs, as in intraperitoneally-inoculated Tg338 mice, compared with intracerebrally-inoculated mice [37]. This was accompanied by comparatively low levels of PrPres and infectivity in the brain of ip-inoculated mice. In Bv109ICWD we also observed unusually low levels of PrPres, compared with those observed in most of the vole-adapted prion strains (Figure 3). However, by endpoint titration we found unexpected high prion titres in Bv109ICWD, 108,4 i.c. ID50 U g−1, similar to those usually observed in standard hamster and mouse-adapted scrapie strains, whose incubation times are 3–10 times longer than Bv109ICWD. This implies that Bv109ICWD undergoes extraordinarily fast replication kinetics in Bv109I brain.
Several observations suggest that prion infectivity and toxicity might be uncoupled [38], [39], [40] and these observations are currently incorporated in a general model of prion replication and toxicity [41]. According to this model, neurotoxicity is mediated by a lethal PrP species, PrPL, which is distinct from PrPSc, but its formation is catalysed during the autocatalytic replication of PrPSc. Neurotoxicity may require a critical PrPL concentration to be reached, which would depend on the kinetics of prion propagation. The relative levels of toxicity and infectivity are governed by the ratio of the initial rate of PrPC conversion (which leads to the production of PrPL) to the rate of its maturation into PrPSc. Thus fast prion replication in Bv109ICWD might have triggered the production of high levels of PrPL in short time periods, leading to rapid disease onset and animal death. The low levels of PrPSc and the fast replication kinetics observed in Bv109ICWD are consistent with this interpretation.
A recent work showed that in mice inoculated with the RML scrapie strain the concentration of PrPC did not affect the overall level of prion infectious titres at terminal disease, while it was directly related to the incubation time, suggesting that the production of PrPL is directly proportional to PrPC concentrations [42]. Our observations with Bv109ICWD, i.e. the unusually short survival time and the high prion infectious titre in a model that does not overexpress PrPC, suggest that the kinetics of prion propagation and toxicity are governed by mechanisms that cannot be interpreted solely on the basis of the amount of available substrate (PrPC).
The unique and easily distinguishable features of Bv109ICWD prompted us to pursue its in vitro propagation by saPMCA. Bv109ICWD PrPSc was indeed easily propagated in vitro, which allowed us to produce Bv109ICWDPMCA PrPSc, theoretically devoid of any PrPSc formed in vivo. Bv109ICWDPMCA was highly infectious and faithfully reproduced the peculiar phenotype of Bv109ICWD. These findings confirm that CWD prions can be generated in vitro, as already demonstrated by others using transgenic mice expressing cervid PrP [43], [44], [45], [46] and prairie voles [47]. Furthermore, given the unique characteristics of this strain, it is extremely unlikely that their faithful maintenance during saPMCA could have occurred by chance and our results represent a convincing confirmation of other studies that have already demonstrated the ability of PMCA to replicate prion strains faithfully [44], [45], [47], [48], [49].
Elk, mule deer and white-tailed deer are the species most affected by CWD. The homogeneous and peculiar phenotypes observed in Bv109I inoculated with CWD isolates from these three cervid species indicate that the same CWD strain was isolated from all species. Interestingly, Bv109ICWD was isolated not only from natural cases of disease in elk (CWD1, 2 and 4) and mule deer (CWD3), but also from white-tailed deer experimentally inoculated with CWD-affected white-tailed deer, mule deer and elk (CWD5, 7 and 8, respectively). Along with previous findings in transgenic mice expressing cervid PrP [6], [9], and in keeping with the ease of indirect horizontal transmission of CWD [50], these data suggest that the same CWD strain circulates among different cervid species and maintains its characteristics following interspecies transmission. Recently, a large transmission study with elk and mule deer isolates provided substantial evidence for two prevalent CWD prion strains and suggested that individual CWD inocula might contain mixtures of the two prion strains [11]. Interestingly, we found similar evidence in at least two of the seven inocula investigated, derived from elk and mule deer, although we were unable to stabilize two different Bv109I-adapted CWD strains. Indeed, on primary transmission of CWD2, CWD3 and CWD4 inocula we observed voles that developed clinical signs after unusually long times, showing a slightly different neuropathological profile from that of voles with shorter survival times. A sub-passage in Bv109I of two of these outliers induced a survival time of 150–160 d.p.i. on second passage, compared with 35–45 d.p.i. observed with all other sub-passages. Such a long survival time might have depended on a low infectious titre in the brain from outlier voles, although they showed levels of PrPres similar to the other voles of their groups. However this hypothesis is excluded when these results are compared with the survival times observed in the endpoint titration experiment, which showed that even 10−5 dilution of Bv109ICWD had a mean survival time <80 d.p.i.. In addition, the slightly deviant lesion profile observed in outlier voles was preserved for 2 sub-passages, before both the survival time and the neuropathological profile converged with the fast Bv109ICWD strain. Overall, our findings strongly suggest that a second CWD strain was propagated in vole outliers, which was progressively outcompeted by the extremely rapid Bv109ICWD strain. The “evanescent” presence of a second strain might be also interpreted, according to the quasi species nature proposed for prions [51], as the progressive elimination of the less fitting conformers occurred following interspecies transmission.
Here we demonstrate the high susceptibility of Bv109I to CWD, which adds Bv109I to the portfolio of animal models useful for the study of CWD strains. The unique properties of Bv109ICWD provide exceptional opportunities to improve basic knowledge of the relationship between PrPSc, neurodegeneration and infectivity. The short survival time of Bv109ICWD, coupled with its high infectious titre, offers useful advantages for titration studies, while its unique clinico-pathological phenotype makes Bv109ICWD one of the best options for studies aimed at investigating strain fidelity in experimental conditions.
Materials and Methods
Ethics statement
Bv109I and Bv109M were obtained from the breeding colony at the Istituto Superiore di Sanità (ISS). The research protocol, approved by the Service for Biotechnology and Animal Welfare of the ISS and authorised by the Italian Ministry of Health, adhered to the guidelines contained in the Italian Legislative Decree 116/92, which transposed the European Directive 86/609/EEC on Laboratory Animal Protection.
Samples and transmission experiments
Brain tissues from CWD affected elk (n = 3), mule deer (n = 1) and white-tailed deer (n = 3) were used for primary transmissions. Two isolates from elk, CWD1 and CWD2, were provided by the United States Department of Agriculture (Dr. A.L. Jenny) and carried the wild type PRNP genotype. The third elk isolate, CWD4, was heterozygous methionine/leucine at codon 132. It was provided by the University of Wyoming (Dr. J.E. Jewel) as the mule deer isolate, CWD3, which was homozygous for serine at codon 225. The three isolates from white-tailed deer derived from an intracerebral experimental challenge [52]. All deer were heterozygous glycine/serine at codon 96 and were challenged with a white-tailed deer CWD isolate (CWD5, code 654 in the original paper), a mule deer CWD (CWD7, code 643) or a elk CWD (CWD8, code 677). Brain sample from a healthy elk was also inoculated as negative control.
An Italian scrapie isolate from Sarda sheep (SS7B) carrying the ARQ/ARQ genotype previously characterised into Bv109M [19], was used for comparison.
Tissues were homogenised at 10% (wt/vol) in phosphate buffered saline (PBS) and stored at −80°C. The PrPres amount estimated by Western blot was comparable in all the homogenates (data not shown).
Groups of eight-week-old Bv109I and Bv109M were inoculated intracerebrally with 20 µ of homogenate into the left cerebral hemisphere, under ketamine anaesthesia (ketamine 0.1 µg/g). The animals were examined twice a week until neurological signs appeared, after which they were examined daily. Diseased animals were culled with carbon dioxide at the terminal stage of the disease, but before neurological impairment was such as to compromise their welfare, in particular, their ability to drink and feed adequately. Survival time was calculated as the interval between inoculation and culling or death. After that, the brain was cut parasagittally into two parts. The smaller portion was stored at −80°C and the larger one was fixed in formalin.
The inocula for the second and third passages were prepared, as 10% wt/vol homogenates in PBS, using the brain of one of the first animals of each group that developed the disease. Supplementary second and subsequent passages from two voles that showed much longer survival times compared with the rest of their groups (outliers) were also performed.
Titration of Bv109ICWD
The specific infectivity of Bv109ICWD was assessed in Bv109I by end-point titration using tenfold dilutions of brain homogenates from the third passage of CWD1 and calculated as ID50 U g−1 according to the Spearman and Kärber method [53]. Survivors were animals that survived until the end of the experiment (450 d.p.i.) with no sign of infection, as assessed by Western blot. The transmission rate was calculated as the ratio between voles confirmed positive by Western blot and the number of voles inoculated, excluding animals culled for intercurrent disease before 30 d.p.i..
Histopathology
Histology, immunohistochemistry (IHC) and PET-blot analysis were performed on formalin-fixed tissues as previously described [16], [19]. Briefly, brains were trimmed at standard coronal levels, embedded in paraffin wax and cut at 6 µm for haematoxylin and eosin staining, immunohistochemistry and PET-blot. Sections were randomly mixed and coded for blind pathological assessment. For the construction of lesion profiles, vacuolar changes were scored in nine grey-matter areas of the brain on haematoxylin and eosin stained sections [16], [54], [55]. Vacuolation scores were derived from the examination of at least six voles per group. PET-blot and IHC were performed as described in Di Bari et al. [19], [26] using the SAF84 mAb.
Western blot
CWD inocula (10% w/v homogenates in PBS) were added to PBS/sarcosyl up to a final 2% sarcosyl (Sigma) concentration and incubated for 20 min at room temperature before digestion with proteinase K (200 µg/ml) for 1 hour at 37°C with gentle shaking. Protease treatment was stopped with 3 mM PMSF.
Brain homogenates (10% w/v) from individual voles were prepared in 100 mM Tris-HCl, pH 7.4, containing 2% sarcosyl, incubated for 20 min at room temperature and then digested with proteinase K (50 µg/ml) for 1 hour at 37°C with gentle shaking. Protease treatment was stopped with 3 mM PMSF.
Electrophoresis and Western blotting were performed as previously described [16]. Briefly, samples were denatured by adding NuPage LDS Sample Buffer (Invitrogen, Carlsbad, California, United States) and NuPage Sample Reducing Agent (Invitrogen), and heated for 10 min at 90°C. After centrifugation at 10,000 g for 5 min each sample was loaded onto 12% bis-Tris polyacrylamide gels (Invitrogen). After electrophoresis and Western blotting on PVDF membranes (Immobilon-P; Millipore, Bedford, MA, USA), the blots were processed by SNAP i.d. Protein Detection System (Millipore) in accordance with the manufacturer's instructions. Vole PrPres was detected using monoclonal antibodies SAF84 (1.2 µg/ml; epitope at amino acids 163–169 of the bank vole PrP sequence) and 12B2 (2.4 µg/ml; epitope at amino acids 89–93 of the sheep PrP sequence). Horseradish peroxidase-conjugated anti-mouse immunoglobulin (Pierce Biotechnology, Rockford, Illinois, United States) was used as secondary antibody (1∶13,000). The membranes were developed using an enhanced chemiluminescence method (SuperSignal Femto, Pierce). The chemiluminescence signal was detected using the VersaDoc imaging system (Bio-Rad) and was quantified by QuantityOne software (Bio-Rad).
Serial automated protein misfolding cyclic amplification
A vole brain from the third passage of CWD1 was homogenised at 10% w/v in PBS and divided into two aliquots. The first aliquot was used as a seed for saPMCA while the second was used for bioassays. Substrates from Bv109I voles were prepared as 10% brain homogenates in conversion buffer (PBS, 0.15 M NaCl and 1% Triton X) and saPMCA was performed as previously described [28] by diluting 1∶100 the seed into Bv109I substrate followed by one round of PMCA using a Misonix 3000 Sonicator, with the following settings: 20 seconds of sonication every 30 minutes for 24 hours (48 cycles of incubation/sonication); 200–250 watts (potency 8) sonication power. For successive rounds of saPMCA, the product of the previous round was diluted 1∶10 into fresh substrate and subjected to a further PMCA round. This procedure was repeated 14 times to reach a 10−15 final dilution of the initial CWD infected brain homogenate. The detailed protocol for saPMCA has been published elsewhere [56], [57], [58]. After each round, PrPres was evaluated by Western blot (Figure S4). Given the risk of contamination that is intrinsic for an ultrasensitive technique as PMCA [28], several healthy Bv109I brain homogenates were processed, as negative controls, in each round. Before each successive round, the products of the previous round were analysed by Western blot to confirm the amplification of Bv109ICWD and the negativity of controls.
Supporting Information
Zdroje
1. Williams ES & YoungS (1980) Chronic Wasting Disease of captive mule deer: a spongiform encephalopathy. J Wildl Disease 16 : 89–98.
2. WilliamsES, MillerMW, KreegerTR, KahnRH, ThorneET (2002) Chronic wasting disease of deer and elk: A review with recommendations for management. Journal of Wildlife Management 66 : 551–563.
3. SohnHJ, KimJH, ChoiKS, NahJJ, JooYS, et al. (2002) A case of chronic wasting disease in an elk imported to Korea from Canada. J Vet Med Sci 64 : 855–8.
4. SigurdsonCJ (2008) A prion disease of cervids: chronic wasting disease. Vet Res 39 : 41.
5. WalshDP, MillerMW (2010) A weighted surveillance approach for detecting Chronic Wasting Disease foci. J of Wildlife Diseases 46 : 118–135.
6. BrowningSR, MasonGL, SewardT, GreenM, EliasonGA, et al. (2004) Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J Virol 78 : 13345–50.
7. KongQ, HuangS, ZouW, VanegasD, WangM, et al. (2005) Chronic wasting disease of elk: transmissibility to humans examined by transgenic mouse models. J Neurosc 25 : 7944–9.
8. LaFauciG, CarpRI, MeekerHC, YeX, KimJI, et al. (2006) Passage of chronic wasting disease prion into transgenic mice expressing Rocky Mountain elk (Cervus elaphus nelsoni) PrPC. J Gen Virol 87 : 3773–80.
9. TamgüneyG, GilesK, Bouzamondo-BernsteinE, BosquePJ, MillerMW, et al. (2006) Transmission of elk and deer prions to transgenic mice. J Virol 80 : 9104–14.
10. Meade-WhiteK, RaceB, TrifiloM, BossersA, FavaraC, et al. (2007) Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. J Virol 81 : 4533–9.
11. AngersRC, KangHE, NapierD, BrowningS, SewardT, et al. (2010) Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 328 : 1154–8.
12. SigurdsonCJ, MancoG, SchwarzP, LiberskiP, HooverEA, et al. (2006) Strain fidelity of chronic wasting disease upon murine adaptation. J Virol 80 : 12303–11.
13. RaymondGJ, RaymondLD, Meade-WhiteKD, HughsonAG, FavaraC, et al. (2007) Transmission and adaptation of chronic wasting disease to hamsters and transgenic mice: evidence for strains. J Virol 81 : 4305–14.
14. HeiseyDM, MickelsenNA, SchneiderJR, JohnsonCJ, JohnsonCJ, et al. (2010) Chronic wasting disease (CWD) susceptibility of several North American rodents that are sympatric with cervid CWD epidemics. J Virol 84 : 210–5.
15. PerrottMR, SigurdsonCJ, MasonGL, HooverEA (2012) Evidence for distinct chronic wasting disease (CWD) strains in experimental CWD in ferrets. J Gen Virol 93 : 212–21.
16. NonnoR, Di BariMA, CardoneF, VaccariG, FazziP, et al. (2006) Efficient Transmission and Characterization of Creutzfeldt–Jakob Disease Strains in Bank Voles. PLoS Pathog 2 : 12.
17. ZanussoG, PoloA, FarinazzoA, NonnoR, CardoneF, et al. (2007) Novel prion protein conformation and glycotype in Creutzfeldt-Jakob disease. Arch Neur 64 : 595–9.
18. PieningN, NonnoR, Di BariMA, WalterS, WindlO, et al. (2006) Conversion efficiency of bank vole prion protein in vitro is determined by residues 155 and 170, but does not correlate with the high susceptibility of bank voles to sheep scrapie in vivo. J Biol Chem 281 : 9373–84.
19. Di BariMA, ChianiniF, VaccariG, EspositoE, ConteM, et al. (2008) The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie. J Gen Virol 89 : 2975–85.
20. CartoniC, SchininàME, MarasB, NonnoR, VaccariG, et al. (2005) Identification of the pathological prion protein allotypes in scrapie-infected heterozygous bank voles (Clethrionomys glareolus) by high-performance liquid chromatography-mass spectrometry. J Chromatogr A 1081 : 122–6.
21. CartoniC, SchininàME, MarasB, NonnoR, VaccariG, et al. (2007) Quantitative profiling of the pathological prion protein allotypes in bank voles by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 849 : 302–306.
22. AgrimiU, NonnoR, Dell'OmoG, Di BariMA, ConteM, et al. (2008) Prion protein amino acid determinants of differential susceptibility and molecular feature of prion strains in mice and voles. PLoS Pathog 25: e1000113.
23. KurtTD, SeeligDM, SchneiderJR, JohnsonCJ, TellingGC, et al. (2011) Alteration of the chronic wasting disease species barrier by in vitro prion amplification. J Virol 85 : 8528–37.
24. SigurdsonCJ, NilssonKP, HornemannS, MancoG, Fernández-BorgesN, et al. (2010) A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest 120 : 2590–9.
25. Agrimi U, Di Bari MA, Pirisini L, Marcon S, D'Agostino C, et al.. (2009) Biological typing of prion strains. Comparison of animal and human prion isolates following transmission to bank vole (Myodes glareolus) [Abstract]. In Proceedings of the 3th International Symposium on The New Prion Biology: Basis Science, Diagnosis And Therapy; 2–4 April 2009, Venice, Italy.
26. SigurdsonCJ, Joshi-BarrS, BettC, WinsonO, MancoG, et al. (2011) Spongiform encephalopathy in transgenic mice expressing a point mutation in the β2-α2 loop of the prion protein. J Neurosci 31 : 13840–7.
27. KurtTD, TellingGC, ZabelMD, HooverEA (2009) Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology 387 : 235–43.
28. CossedduGM, NonnoR, VaccariG, BucalossiC, Fernandez-BorgesN, et al. (2011) Ultra-efficient PrPSc amplification highlights potentialities and pitfalls of PMCA technology. PLoS Pathog 7: e1002370.
29. ThackrayAM, KleinMA, AguzziA, BujdosoR (2002) Chronic subclinical prion disease induced by low-dose inoculum. J Virol 76 : 2510–7.
30. DemaimayR, RaceR, ChesebroB (1999) Effectiveness of polyene antibiotics in treatment of transmissible spongiform encephalopathy in transgenic mice expressing Syrian hamster PrP only in neurons. J Virol 73 : 3511–3.
31. PrusinerSB, ScottM, FosterD, PanKM, GrothD, et al. (1990) Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 16 : 673–86.
32. SupattaponeS, MuramotoT, LegnameG, MehlhornI, CohenFE, et al. (2001) Identification of two prion protein regions that modify scrapie incubation time. J Virol 75 : 1408–13.
33. ThackrayAM, HopkinsL, LockeyR, SpiropoulosJ, BujdosoR (2012) Propagation of ovine prions from “poor” transmitter scrapie isolates in ovine PrP transgenic mice. Exp Mol Pathol 92 : 167–74.
34. WattsJC, GilesK, StöhrJ, OehlerA, BhardwajS, et al. (2012) Spontaneous generation of rapidly transmissible prions in transgenic mice expressing wild-type bank vole prion protein. Proc Natl Acad Sci USA 109 : 3498–503.
35. KimberlinRH, HallSM, WalkerCA (1983) Pathogenesis of mouse scrapie. Evidence for direct neural spread of infection to the CNS after injection of sciatic nerve. J Neurol Sci 61 : 315–25.
36. KimberlinRH, WalkerCA, FraserH (1989) The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J Gen Virol 70 : 2017–25.
37. LangevinC, AndréolettiO, Le DurA, LaudeH, BéringueV (2011) Marked influence of the route of infection on prion strain apparent phenotype in a scrapie transgenic mouse model. Neurobiol Dis 41 : 219–25.
38. HillAF, JoinerS, LinehanJ, DesbruslaisM, LantosPL, et al. (2000) Species-barrier-independent prion replication in apparently resistant species. Proc Natl Acad Sci USA 97 : 10248–53.
39. RaceR, RainesA, RaymondGJ, CaugheyB, ChesebroB (2001) Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: analogies to bovine spongiform encephalopathy and variant Creutzfeldt-Jakob disease in humans. J Virol 75 : 10106–12.
40. HillAF, CollingeJ (2003) Subclinical prion infection. Trends Microbiol 11 : 578–84.
41. CollingeJ, ClarkeAR (2007) A general model of prion strains and their pathogenicity. Science 318 : 930–936.
42. SandbergMK, Al-DoujailyH, SharpsB, ClarkeAR, CollingeJ (2011) Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature 470 : 540–2.
43. KurtTD, PerrottMR, WiluszCJ, WiluszJ, SupattaponeS, et al. (2007) Efficient in vitro amplification of chronic wasting disease PrPres. J Virol 81 : 9605–8.
44. GreenKM, CastillaJ, SewardTS, NapierDL, JewellJE, et al. (2008) Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathog 4: e1000139.
45. MeyerettC, MichelB, PulfordB, SprakerTR, NicholsTA, et al. (2008) In vitro strain adaptation of CWD prions by serial protein misfolding cyclic amplification. Virology 382 : 267–76.
46. JohnsonCJ, AikenJM, McKenzieD, SamuelMD, PedersenJA (2012) Highly efficient amplification of chronic wasting disease agent by protein misfolding cyclic amplification with beads (PMCAb). PLoS One 7: e35383.
47. CastillaJ, SaáP, HetzC, SotoC (2005) In vitro generation of infectious scrapie prions. Cell 121 : 195–206.
48. CastillaJ, Gonzalez-RomeroD, SaáP, MoralesR, De CastroJ, et al. (2008) Crossing the species barrier by PrPSc replication in vitro generates unique infectious prions. Cell 134 : 757–68.
49. ShikiyaRA, BartzJC (2011) In vitro generation of high-titer prions. J Virol 85 : 13439–42.
50. MillerMW, WilliamsES, HobbsNT, WolfeLL (2004) Environmental sources of prion transmission in mule deer. Emerg Infect Dis 10 : 1003–6.
51. WeissmannC, LiJ, MahalSP, BrowningS (2011) Prions on the move. EMBO Rep 12 : 1109–17.
52. HamirAN, RichtJA, MillerJM, KunkleRA, HallSM, et al. (2008) Experimental transmission of chronic wasting disease (CWD) of elk (Cervus elaphus nelsoni), white-tailed deer (Odocoileus virginianus), and mule deer (Odocoileus hemionus hemionus) to white-tailed deer by intracerebral route. Vet Pathol 45 : 297–306.
53. HamiltonMA, RussoRC, ThurstonRV (1977) Trimmed Spearman-Karber method for estimating median lethal concentrations in toxicity bioassays. Environ Sci Technol 11 : 714–719.
54. FraserH, DickinsonAG (1968) The sequential development of the brain lesions of scrapie in three strains of mice. J Comp Pathol 78 : 301–311.
55. Di Bari MA, Nonno R, Agrimi U (2012) The mouse model for scrapie: inoculation, clinical scoring, and histopathological techniques. In: Sigurdsson E M, Calero M, Gasset M, editors. Amyloid Proteins, Methods in Molecular Biology, Vol 849. New York, NY: Humana Press. pp 453–471.
56. Castilla J, Saa' P, Soto C (2004) Cyclic amplification of prion protein misfolding. In: Lehmann S, Grassi J, editors. Methods and Tools in Bioscience and Medicine Techniques in Prion Research. Basel, Switzerland: Birkhauser Verlag. pp 198–213.
57. CastillaJ, SaáP, MoralesR, AbidK, MaundrellK, SotoC (2006) Protein misfolding cyclic amplification for diagnosis and prion propagation studies. Methods Enzymol 412 : 3–21.
58. Saa' P, Castilla J, Soto C (2004) Cyclic amplification of protein misfolding and aggregation. In: Sigurdsson EM, editor. Amyloid Proteins: Methods and Protocols. New York, NY: Humana Press. pp 53–65.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Surviving the Heat of the Moment: A Fungal Pathogens Perspective
- Redefining the Immune System as a Social Interface for Cooperative Processes
- Post-Treatment HIV-1 Controllers with a Long-Term Virological Remission after the Interruption of Early Initiated Antiretroviral Therapy ANRS VISCONTI Study
- Rational Engineering of Recombinant Picornavirus Capsids to Produce Safe, Protective Vaccine Antigen
- Influenza Virus Aerosols in Human Exhaled Breath: Particle Size, Culturability, and Effect of Surgical Masks
- Glycomic Analysis of Human Respiratory Tract Tissues and Correlation with Influenza Virus Infection
- Evolution of Virulence in Emerging Epidemics
- Monomeric Nucleoprotein of Influenza A Virus
- The Enterovirus 71 A-particle Forms a Gateway to Allow Genome Release: A CryoEM Study of Picornavirus Uncoating
- HIV Restriction by APOBEC3 in Humanized Mice
- Transcriptional Responses of Praziquantel Exposure in Schistosomes Identifies a Functional Role for Calcium Signalling Pathway Member CamKII
- Genome-wide Determinants of Proviral Targeting, Clonal Abundance and Expression in Natural HTLV-1 Infection
- TIM-3 Does Not Act as a Receptor for Galectin-9
- Chronic Wasting Disease in Bank Voles: Characterisation of the Shortest Incubation Time Model for Prion Diseases
- Escapes Fumagillin Control in Honey Bees
- Generators of Phenotypic Diversity in the Evolution of Pathogenic Microorganisms
- Plasminogen Controls Inflammation and Pathogenesis of Influenza Virus Infections via Fibrinolysis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Escapes Fumagillin Control in Honey Bees
- TIM-3 Does Not Act as a Receptor for Galectin-9
- HIV Restriction by APOBEC3 in Humanized Mice
- Influenza Virus Aerosols in Human Exhaled Breath: Particle Size, Culturability, and Effect of Surgical Masks
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



