-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Fresh Look at the Origin of , the Most Malignant Malaria Agent
From which host did the most malignant human malaria come:
birds, primates, or rodents? When did the transfer occur? Over the last half century, these have been some of the questions up for debate about the origin of Plasmodium falciparum, the most common and deadliest human malaria parasite, which is responsible for at least one million deaths every year. Recent findings bring elements in favor of a transfer from great apes, but are these evidences really solid? What are the grey areas that remain to be clarified? Here, we examine in depth these new elements and discuss how they modify our perception of the origin and evolution of P. falciparum. We also discuss the perspectives these new discoveries open.
Published in the journal: . PLoS Pathog 7(2): e32767. doi:10.1371/journal.ppat.1001283
Category: Review
doi: https://doi.org/10.1371/journal.ppat.1001283Summary
From which host did the most malignant human malaria come:
birds, primates, or rodents? When did the transfer occur? Over the last half century, these have been some of the questions up for debate about the origin of Plasmodium falciparum, the most common and deadliest human malaria parasite, which is responsible for at least one million deaths every year. Recent findings bring elements in favor of a transfer from great apes, but are these evidences really solid? What are the grey areas that remain to be clarified? Here, we examine in depth these new elements and discuss how they modify our perception of the origin and evolution of P. falciparum. We also discuss the perspectives these new discoveries open.Introduction
In the genus Plasmodium, four species are traditionally regarded as human parasites: Plasmodium malariae, Plasmodium ovale, Plasmodium vivax, and Plasmodium falciparum. These species are remotely related to each other, suggesting that adaptation to humans has occurred several times independently during the history of the genus. It is still unclear, however, when these associations began and from where they came [1]. The origin of P. falciparum, in particular, continues to be a highly debated topic.
Early molecular phylogenetic studies on the genus Plasmodium showed that P. falciparum clustered with two avian parasites rather than with those infecting mammals, thus suggesting that P. falciparum was the result of a transfer from birds to humans [2], [3]. According to the authors, this transfer took place at the beginning of agricultural development, when the human habitat was settled about 10,000 years ago. This result was quickly questioned, though, due to the small number of ingroup taxa considered for the phylogenetic analyses and the use of 18S rDNA sequences, which have proved their weakness in studies on Haemosporidia phylogeny (e.g., [4]).
Subsequent analyses demonstrated that the closest sister taxon of P. falciparum was Plasmodium reichenowi, a parasite isolated from a chimpanzee. Escalante and Ayala [5] suggested that these two parasites diverged at the time of the divergence between humans and chimpanzees. According to their results, P. falciparum did not directly originate from an avian malarial parasite. Nevertheless, the P. falciparum/P. reichenowi pair still was considered as a sister lineage of the parasites from birds and lizards [5].
The results of subsequent studies were contradictory. Some of them concluded that the P. falciparum/P. reichenowi clade was included in the mammalian group, being closer to the rodent or the primate Plasmodium [6]–[8]; others reached the opposite conclusion by showing that this group clustered more closely with parasites form birds [9]–[16]. Part of the confusion concerning the origin of P. falciparum arose because of biases in the representation of certain taxa, the small number of loci analysed and/or improper rooting. In 2008, the study of Martinesen et al. [4] did not suffer from these shortcomings and quite conclusively placed P. falciparum basal to all other mammalian malaria parasites. However, the origin of P. falciparum within this group of pathogens still remained unknown. Finally, it is only very recently, by adding more taxa from primates (in particular great apes), that the origin of P. falciparum was firmly established.
Great Apes Are Hosts of Higher Plasmodium Diversity Than Previously Thought
In one year, the known diversity of Plasmodium species infecting great apes and belonging to the P. falciparum lineage has burst. Until May 2009, only one species, P. reichenowi, was known to be phylogenetically a sister lineage of P. falciparum. P. reichenowi was isolated from a chimpanzee that was captured near Lake Edwards (Democratic Republic of Congo) [17], likely belonging to the Pan troglodytes schweinfurthii subspecies. Until recently, this isolate was the only great ape parasite genetically characterized [18].
In May 2009, Ollomo et al. [19] published the complete mitochondrial genome of a new, not yet described species of Plasmodium that circulates in great apes and belongs to the P. falciparum/P. reichenowi lineage. This species was discovered in two wild-borne chimpanzees kept as pets in villages of Gabon and was called Plasmodium gaboni in reference to the country where it was found. Based on the hypothesis that P. falciparum and P. reichenowi diverged about 6 million years (MY) ago, the authors proposed that P. gaboni diverged from the P. falciparum/P. reichenowi lineage about 21 MY ago, leading them to conclude that the ancestor of this African great apes/human parasite clade (also known as the Laverania subgenus, [20]) could have been already present in hominid ancestors [19].
Three months later, Rich and colleagues [21] described the diversity of Plasmodium species that circulate among wild and wild-borne chimpanzees from Ivory Coast and Cameroon. From blood samples, the authors identified, using partial sequences of the Cytochrome B (CytB) gene but also some apicoplastic and nuclear sequences, eight Plasmodium isolates that they described as P. reichenowi. However, the CytB sequences showed a high polymorphism and some of them were actually very close genetically to the previously described sequences of P. gaboni [21]. The phylogenetic analysis carried out by the authors suggests that P. falciparum arose from a recent transfer from chimpanzees to humans that may have occurred as early as between 5,000 to 50,000 years ago. This finding challenges the conventional belief that P. falciparum and P. reichenowi diverged at the same time as their respective hosts (human and chimpanzee) between 4 and 7 MY ago.
In January 2010, Prugnolle and colleagues [22] published a paper in which, using new non-invasive methods based on the use of great apes fecal samples, they described the diversity of Plasmodium species that circulate in wild West African chimpanzees and also, for the first time, in gorillas (subspecies Gorilla gorilla gorilla and G. gorilla dielhi). Their study confirmed the presence of P. gaboni and P. reichenowi in wild chimpanzees (subspecies P. troglodytes troglodytes and P. troglodytes vellerosus) and reported the existence of still unknown Plasmodium genetic lineages in wild gorillas. One clusters with P. reichenowi and P. falciparum (they called this phylogenetic lineage P. GorB) and the other is a sister lineage of P. gaboni (they called it P. GorA). Their results confirm that chimpanzees are infected by a large diversity of Plasmodium species from which P. falciparum seems to have originated. In addition, the authors identified P. falciparum in wild gorillas, an unexpected result since P. falciparum is considered as strictly specific to humans. They did not find any genetic differences between the P. falciparum circulating in humans and in gorillas based on the partial CytB sequence studied, thus suggesting the possibility of a recent transfer from humans to primates.
In February 2010, Krief and collaborators [23] confirmed the large diversity of Plasmodium species infecting great apes from Central Africa. The originality of their work lies in the presence of isolates from chimpanzees from East Africa (subspecies P. t,. schweinfurthii) and from bonobos. By sequencing the whole Plasmodium mitochondrial genome (as well as some apicoplastic and nuclear genes), they identified two distinct parasite lineages in chimpanzees. One is located at the root between P. falciparum and P. reichenowi and was called Plasmodium billcollinsi. The other one was named Plasmodium billbrayi and is phylogenetically very close to P. gaboni. Interestingly, they also discovered P. falciparum in bonobos and found that the genetic diversity of its mitochondrial genome was higher than the overall diversity found in human P. falciparum, thus suggesting that P. falciparum might have originated from bonobos and was transferred to humans as early as 30,000 years ago. This interpretation challenges the hypothesis by Rich et al. [21] and Prugnolle et al. [22], according to which P. falciparum likely originated from a transfer from chimpanzees.
In May 2010, Duval and colleagues [24] investigated the diversity of Plasmodium species in chimpanzees and gorillas from Cameroon by sequencing two mitochondrial genes (CytB and Cox1). Their study confirmed the existence of the lineage P. GorB in western lowland gorillas and of P. gaboni and P. reichenowi in chimpanzees. They also confirmed the presence of P. falciparum–related parasites in gorillas and demonstrated, for the first time, the presence of P. falciparum in blood samples from two different chimpanzee subspecies (P. t. vellerosus and P. t. troglodytes) from sanctuaries.
Finally, very recently, in September 2010, Liu et al. [25] published a study on the diversity of Plasmodium species in African great apes based on a very large collection of fecal samples from three subspecies of chimpanzees (P. t. troglodytes, P. troglodytes ellioti [also known as P. t. vellerosus], and P. t. schweinfurthii), bonobos, and two subspecies of gorillas (subspecies G. gorilla gorilla and G. gorilla graueri), and a method of single template amplification allowing them to sequence mitochondrial, apicoplastic, and nuclear genes of Plasmodium isolates from mixed infections. From their results, they conclude the existence of six Plasmodium species belonging to the Laverania subgenus, three in chimpanzees (they called them C1–C3) and three in gorillas (referred to as G1–G3). All these species were previously described, but by the greater depth of sampling, this study gives a more definitive picture of the diversity of the Laverania species infecting great apes. In particular, this study confirms the existence of a large diversity of P. falciparum–related parasites in gorillas but does not find any in natural populations of chimpanzees or bonobos. This latter finding suggests a likely gorilla origin for human P. falciparum, in opposition to all theories previously proposed.
Update on the Diversity of Plasmodium Circulating in African Great Apes…
Thanks to the complementarity between the different studies, in terms of the geographic areas and host species and subspecies sampled, a clear picture of the partitions existing within the Laverania lineage now emerges. Figure 1 presents phylogenies of the Laverania subgenus obtained from partial CytB sequences extracted from several of the previously reported studies (methods used to construct the phylogeny are as those presented in Prugnolle et al. [22]; see also the Figure 1 legend for more details). Figure 2 represents the distribution of the different newly recognized Plasmodium lineages in the different subspecies of chimpanzees, gorillas, and bonobos, and Table 1 presents an historic overview of the different names given to these lineages.
Fig. 1. Phylogeny of the Laverania subgenus. 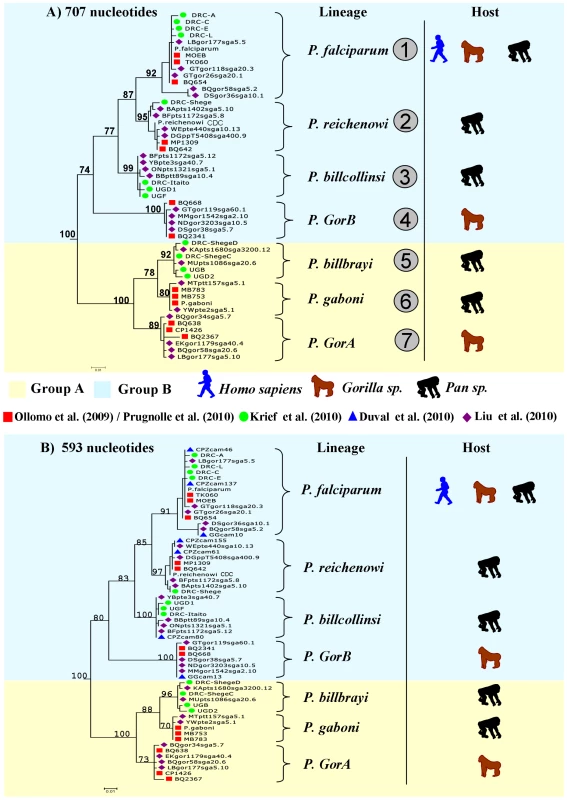
This phylogeny is based on partial CytochromeB sequences and including strains isolated and characterized in (A) Ollomo et al. [19], Prugnolle et al. [22], Krief et al. [23], and Liu et al. [25], and in (B) Ollomo et al. [19], Prugnolle et al. [22], Krief et al. [23], Duval et al. [24], and Liu et al. [25]. The phylogenies were produced using a maximum likelihood approach and robustness was tested using 100 bootstraps. Names of the lineages were given following their first denomination (see Table 1) except for P. billcollinsi, which was first named by Rich et al. [21] as P. reichenowi. Fig. 2. Distribution of the different subspecies of chimpanzees, bonobos, and gorillas in Africa and representation of the spread of the different <i>Plasmodium</i> species in these subspecies. 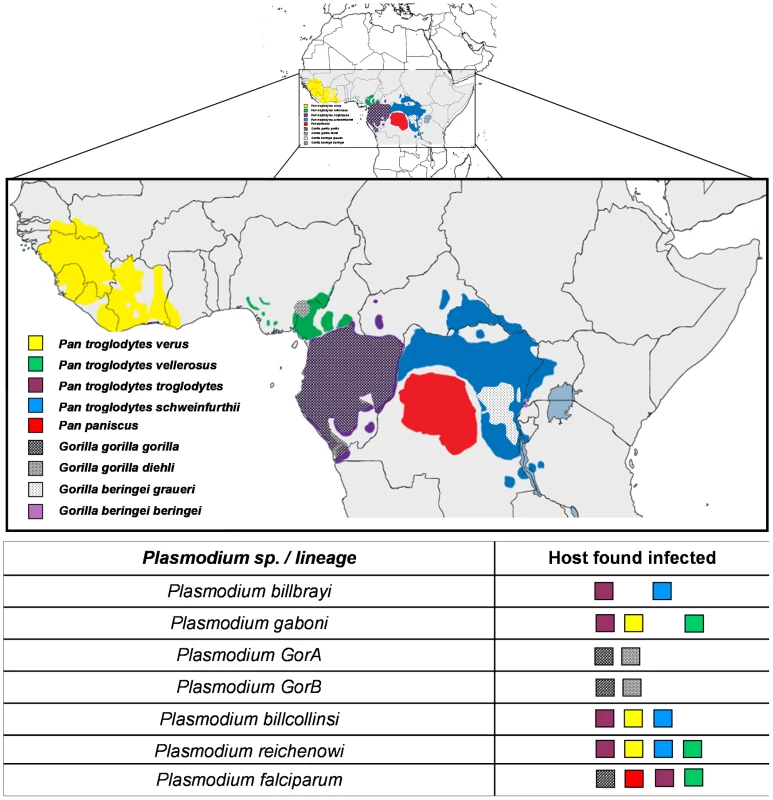
Tab. 1. Historic overview of the molecular descriptions and of the names given to the different lineages (seven lineages) of the Laverania subgenus. 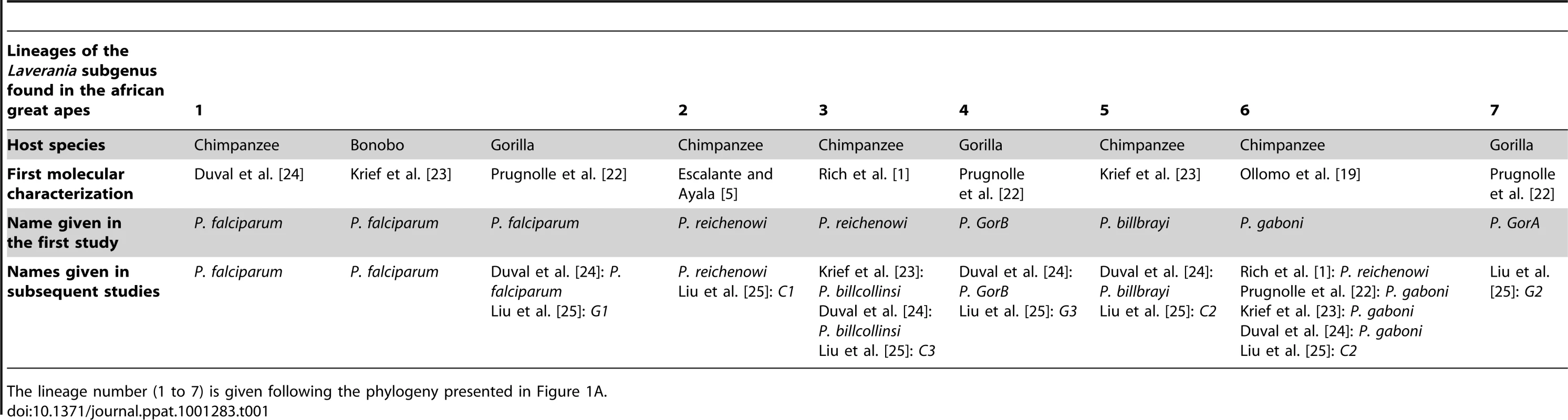
The lineage number (1 to 7) is given following the phylogeny presented in Figure 1A. As shown (Figure 1), the Laverania subgenus is subdivided in two main groups. We will refer to them as Group A and Group B. Group A is formed by two distinct and well-supported clades (Figure 1A and 1B). The first one is found in two gorilla subspecies, Gorilla gorilla gorilla and G. g. dielhi (Figure 2), and was originally described by Prugnolle et al. [22] as P. GorA (Table 1). The second clade is found in chimpanzees and is subdivided into two well-supported lineages (Figure 1A). The first includes the isolates called P. gaboni [19]; the other lineage is composed by P. billbrayi isolates [23] (Table 1). As shown in Figure 2, P. gaboni can infect at least three chimpanzee subspecies, P. troglodytes verus, P. t. vellerosus, and P. t. troglodytes, whereas P. billbrayi infects P. t. troglodytes and P. t. schweinfurthii.
Group B, the second main group of the Laverania clade, includes four distinct and well-supported lineages (Figure 1). The first is defined as P. GorB [22] (Table 1) and is constituted of isolates found in western lowland gorillas (subspecies G. g. gorilla and G. g. dielhi, Figures 1 and 2). The second main lineage, named P. billcollinsi, infects chimpanzees [23] (Table 1). As shown in Figure 2, to date, P. billcollinsi has been identified in three chimpanzee subspecies (P. t. verus, P. t. troglodytes, and P. t. schweinfurthii). The last two lineages are those of P. reichenowi and P. falciparum, respectively. As for the other Plasmodium species that infect chimpanzees, P. reichenowi is widespread and infects the four subspecies of chimpanzees (Figure 2). Until 2009, only one isolate (P. reichenowi CDC) was known, but now a lot of new isolates have been characterized in several different studies [21], [22], [24], [25]. Finally, as shown (Figures 1 and 3), gorillas [22], [25], bonobos [23], and chimpanzees [24] can also be infected by P. falciparum, although it used to be considered to be naturally strictly human specific.
Fig. 3. Sub-tree of the P. falciparum isolates. 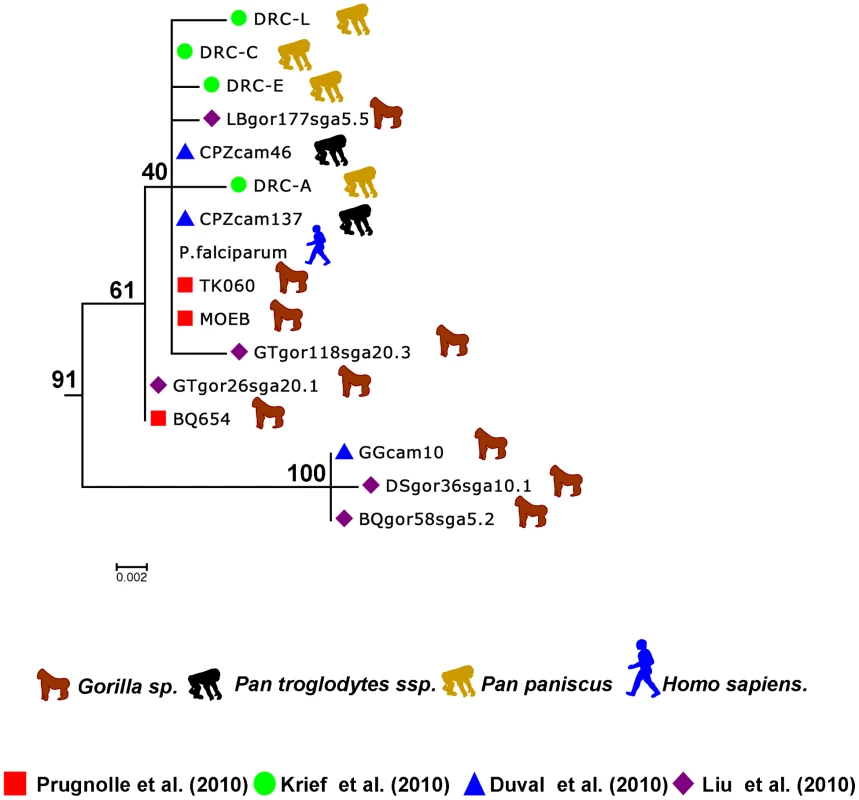
This sub-tree was extracted from the tree presented in Figure 1B and built using the data from Ollomo et al. [19], Prugnolle et al. [22], Krief et al. [23], Duval et al. [24], and Liu et al. [25]. …and on the Origin of P. falciparum
During the last year and as briefly described above, the proposed scenarios concerning the origin of P. falciparum have changed several times (origin in chimpanzees, bonobos, or gorillas) depending on the host species analyzed, the lineages of Plasmodium discovered, or the way data were analyzed.
Now it seems sure, in light of all the studies, that P. falciparum did not originate from a transfer from birds, lizards, or rodents but instead derived from some Plasmodium lineages evolving in African hominids. More precisely, the study by Liu and collaborators [25] (see also Figure 3) strongly suggests that P. falciparum is of gorilla origin and that all known strains circulating in humans nowadays have resulted from a single cross-transmission event from gorilla to human. The origin of the Laverania subgenus in itself remains an enigma, but we will not further discuss this issue here.
Although the scenario of a P. falcirparum gorilla origin is very likely in light of the data recently presented [25], we think nevertheless that more information should be gathered before being able to definitely conclude that P. falciparum really originated in gorillas. First of all, it is still impossible to definitely rule out some alternative scenarios. Among them, one possibility could be that P. falciparum diverged from P. reichenowi at the time of the divergence between chimpanzees and humans (as recently suggested by Hughes and Verra [26]) and then was transferred several times from archaic humans to gorillas over the history of the lineage. The small genetic diversity observed within the current human P. falciparum isolates (compared to the one observed in gorillas) being the result of a recent and rapid expansion of only a small fraction of the human P. falciparum diversity, at the moment, for instance, of the expansion of modern humans from a small east African source population (as recently proposed by [27]).
Second, and despite the very large number of individuals that were analyzed by Liu et al. [25], there is still a possibility that P. falciparum–related strains circulate in the other African great apes (chimpanzees and bonobos, as shown by Duval et al. [24] and Krief et al. [23]) but at lower prevalence [28]. Moreover, given the propensity of the genus Plasmodium to switch from one host to another—as exemplified by the transfer of P. knowlesi from macaques to humans [29] and P. vivax between humans and South American monkeys [30]—it would not be very surprising to find P. falciparum–related strains or other Laverania lineages infecting the other non-human primates in Africa. New data should be gathered to confirm or infirm this possibility.
Challenges for the Future
Causes of the Host Switch
Explaining why the host switch from gorilla to human occurred in the evolutionary history of the P. falciparum lineage will certainly be one of the greatest challenges for the near future.
The process of emergence of a disease into a new host can be schematized by four transition stages according to [31]. The first two stages are prerequisites for the emergence itself: (1) a human contact with the infectious agent and (2) the cross-species transmission. The two others are necessary for the development of the pandemicity: (3) sustained human-to-human transmission and (4) genetic adaptation to humans.
Describing these transition stages, or at least getting a better picture of what happened at these different stages, will require the involvement of different scientific disciplines, from anthropology to entomology, immunology, and genetics. The identification as well as the description of the ecology and the biology (in particular the trophic behavior and host preference) of the vectors of the different Laverania species may, in our mind, constitute a good start to understand the origin of the P. falciparum host transfer.
Dating the Events of Divergence
Dating the divergence time between the different species or lineages may help to understand the origin of P. falciparum and determine the factors that have led to speciation within the Laverania subgenus. For pathogens like those of the genus Plasmodium, speciation generally occurs through two main processes: 1) co-evolution with the hosts or 2) host switch. Estimating and comparing the Plasmodium divergence dates with those of the hosts should therefore allow to determine which process was involved. This implies, obviously, that the host phylogeny (be it the vertebrate or the vector phylogeny) has to be known or determined.
For parasites and soft organisms in general, fossil records are very rare if not absent. This implies that calibration of phylogenies is mostly based on speculation. The recent, aforementioned studies fell into this category. Ollomo et al. [19] proposed that P. gaboni diverged from the P. reichenowi/P. falciparum taxa about 21 MY ago based on the hypothesis that P. reichenowi/P. falciparum diverged between 4 and 7 MY ago. Krief et al. [23] considered other calibration points, as speculative as the one taken by Ollomo et al. [19]. They assumed, for example, that P. gonderi and macaque parasites co-diverged when Macaca branched from other Papionina, which led them to the conclusion that P. falciparum was transferred to humans from bonobos around 30,000 years ago.
In general, we advise readers to be cautious with time estimates and to give a second thought as to how these dates were obtained. In the absence of fossil calibration, one way to date the divergence between species is by using estimates of mutation rates [32]. The principle is that the time separating two sequences from a common ancestor (T) is a simple function of the substitution rate (r, which is equal to the mutation rate for neutral sites) and the observed divergence between sequences (d): T = d/2r. However, it is not easy to get good estimates of mutation rates. Ideally, they should be obtained for all the genes under study and for each species in the phylogeny because substitution rates may vary from one gene to another as well as among species [33]. Recently, Ricklefs and Outlaw [34] estimated the rate of substitution of CytB in Plasmodium species that infect birds and used this data to compute the divergence time of P. falciparum from P. reichenowi. They proposed a split around 2.5 MY ago, but the same previous remarks regarding the variation of substitution rates among unrelated lineages can be applied to this study.
Another possibility for calibrating a phylogeny is to use external calibration points such as well-documented events of vicariance caused by well-dated ancient geological or geographical events (e.g., [28]). In the case of Plasmodium, such an event could be, for example, the separation of Madagascar from the African continent (L. Duval, personal communication). Duval et al. [24] recently published the molecular description of a Plasmodium species isolated from a lemur of Madagascar (Plasmodium malagasy). Many studies have shown that the lemurs have been geographically isolated from other African primates for more than 55 MY [35], [36]. Because the island of Madagascar was always free from other non-human primate species, it should thus be possible to use this external calibration to date the other divergence events in the phylogeny.
Genome Evolution
All these recent discoveries open up the possibility to thoroughly study the evolution of Plasmodium species and their genome. Sequencing the new species' genome should indeed allow the analysis of lineage-specific evolution using comparative genomics, and hence, the identification of the genes responsible for the adaptation of these parasites to their specific hosts [37]. Genome comparison will advance our understanding of the differences in malaria pathology and the processes at work in the interaction with the vertebrate or the mosquito hosts. It is thus essential to rapidly complete the sequencing of the phylogenetically important Plasmodium species within the Laverania lineage, in order to enhance our knowledge on the functional genomics of the most malignant human malaria parasites and of the genetic adaptation that might have facilitated its transfer from gorilla to human.
Risk of Emergence: Human Invaded or Human Invader
The recent discovery of P. falciparum in bonobos [23], chimpanzees [24], and gorillas [22], [24] as well as P. ovale, P. malariae, and P. vivax in chimpanzees, bonobos, and gorillas [23], [24], [38], [39] highlights the risk of transfer of Plasmodium species from human to primates and vice versa. It is now urgent to identify the genetic and ecological factors that allow this group of pathogens to exploit a variety of host species. Notably, this feature should be of concern for the wildlife conservationist community, as recurrent release of human infectious diseases to great apes may accelerate their disappearance. Similarly, it is now important to systematically survey the presence of primate Plasmodium species in human populations, especially in those living in their vicinity (e.g., forest-dwelling populations) in order to evaluate if great apes may constitute a reservoir of Plasmodium for humans and the risk of emergence.
Basic Biology of the Laverania Species
Finally, it will be essential to gather information on the biology and ecology of the different species belonging to the Laverania lineage and on their interactions with the hosts. We think in particular that it will be of major interest to investigate their virulence against chimpanzees and gorillas and thus determine if they impose selective pressures on them. Today, our knowledge on this aspect of their biology is still very limited (see however [23], [40]). In parallel, it will also be interesting to document the response to infection of the hosts and determine, in particular, if they have evolved mechanisms of resistance.
Concluding Remarks
In conclusion, the recent data gathered from great apes in Africa have shown that a large diversity of Plasmodium species circulates among our relatives and have provided new insights into the evolutionary history of the malaria parasites of humans, particularly P. falciparum. These discoveries not only dramatically change our view on the evolution of the P. falciparum lineage, but also question the evolution and origin of the other human Plasmodium species (P. ovale, P. malariae, and P. vivax). This opens completely new areas of research and will certainly attract the attention of an all new community of scientists from various disciplines. Indeed, getting more information on the biology, ecology, and evolution of the different Plasmodium species infecting great apes will certainly help us to better understand and, therefore, fight against, the most virulent human malaria agent, P. falciparum.
Zdroje
1. RichSM
AyalaFJ
2006
Evolutionary origins of Human malaria parasites.
DronamrajuKR
AreseP
Malaria: genetic and evolutionary aspects
New York
Springer
2. WatersAP
HigginsDG
McCutchanTF
1991
Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts.
Proc Natl Acad Sci U S A
88
3140
3144
3. WatersAP
HigginsDG
McCutchanTF
1993
Evolutionary relatedness of some primate models of Plasmodium.
Mol Biol Evol
10
914
923
4. MartinsenES
PerkinsSL
SchallJJ
2008
A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches.
Mol Phylogenet Evol
47
261
273
5. EscalanteAA
AyalaFJ
1994
Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences.
Proc Natl Acad Sci U S A
91
11373
11377
6. AyalaFJ
EscalanteAA
RichSM
1999
Evolution of Plasmodium and the recent origin of the world populations of Plasmodium falciparum.
Parassitologia
41
55
68
7. LeclercMC
HugotJP
DurandP
RenaudF
2004
Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18S rDNA cladistic analysis.
Parasitology
129
677
684
8. PerkinsSL
SchallJJ
2002
A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences.
J Parasitol
88
972
978
9. EscalanteAA
AyalaFJ
1995
Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes.
Proc Natl Acad Sci U S A
92
5793
5797
10. EscalanteAA
FreelandDE
CollinsWE
LalAA
1998
The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome.
Proc Natl Acad Sci U S A
95
8124
8129
11. EscalanteAA
GoldmanIF
De RijkP
De WachterR
CollinsWE
1997
Phylogenetic study of the genus Plasmodium based on the secondary structure-based alignment of the small subunit ribosomal RNA.
Mol Biochem Parasitol
90
317
321
12. KedzierskiL
EscalanteAA
IseaR
BlackCG
BarnwellJW
2002
Phylogenetic analysis of the genus Plasmodium based on the gene encoding adenylosuccinate lyase.
Infect Genet Evol
1
297
301
13. KissingerJC
SouzaPC
SoarestCO
PaulR
WahlAM
2002
Molecular phylogenetic analysis of the avian malarial parasite Plasmodium (Novyella) juxtanucleare.
J Parasitol
88
769
773
14. McCutchanTF
KissingerJC
TourayMG
RogersMJ
LiJ
1996
Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications.
Proc Natl Acad Sci U S A
93
11889
11894
15. RathoreD
WahlAM
SullivanM
McCutchanTF
2001
A phylogenetic comparison of gene trees constructed from plastid, mitochondrial and genomic DNA of Plasmodium species.
Mol Biochem Parasitol
114
89
94
16. Vargas-SerratoE
CorredorV
GalinskiMR
2003
Phylogenetic analysis of CSP and MSP-9 gene sequences demonstrates the close relationship of Plasmodium coatneyi to Plasmodium knowlesi.
Infect Genet Evol
3
67
73
17. CoatneyGR
CollinsWE
WarrenM
ContacosPG
1971
The primate malarias.
Washington (D.C.)
US Government Printing Office
366
18. JeffaresDC
PainA
BerryA
CoxAV
StalkerJ
2007
Genome variation and evolution of the malaria parasite Plasmodium falciparum.
Nat Genet
39
120
125
19. OllomoB
DurandP
PrugnolleF
DouzeryE
ArnathauC
2009
A new malaria agent in African hominids.
PLoS Pathog
5
e1000446
doi:10.1371/journal.ppat.1000446
20. BrayRS
1963
The malaria parasites of anthropoids apes.
J Parasitol
49
888
891
21. RichSM
LeendertzFH
XuG
LeBretonM
DjokoCF
2009
The origin of malignant malaria.
Proc Natl Acad Sci U S A
106
14902
14907
22. PrugnolleF
DurandP
NeelC
OllomoB
AyalaFJ
2010
African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum.
Proc Natl Acad Sci U S A
107
1458
1463
23. KriefS
EscalanteAA
PachecoMA
MugishaL
AndreC
2010
On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from bonobos.
PLoS Pathog
6
e1000765
doi:10.1371/journal.ppat.1000765
24. DuvalL
FourmentM
NerrienetE
RoussetD
SadeuhSA
2010
African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus.
Proc Natl Acad Sci U S A
107
10561
10566
25. LiuW
LiY
LearnGH
RudicellRS
RobertsonJD
2010
Origin of the human malaria parasite Plasmodium falciparum in gorillas.
Nature
467
420
425
26. HughesAL
VerraF
2010
Malaria parasite sequences from chimpanzee support the co-speciation hypothesis for the origin of virulent human malaria (Plasmodium falciparum).
Mol Phylogenet Evol
7
135
143
27. TanabeK
MitaT
JombartT
ErikssonA
HoribeS
2010
Plasmodium falciparum accompanied the human expansion out of Africa.
Curr Biol
20
1283
1289
28. HolmesEC
2010
Malaria: The gorilla connection.
Nature
467
404
405
29. Cox-SinghJ
DavisTM
LeeKS
ShamsulSS
MatusopA
2008
Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening.
Clin Infect Dis
46
165
171
30. TaziL
AyalaFJ
2011
Unresolved direction of host transfer of Plasmodium vivax v. P. simium and P. malariae v. P. brasilianum.
Infect Genet Evol
11
209
221
31. ChildsJE
RichtJA
MackenzieJS
2007
Introduction: conceptualizing and partitioning the emergence process of zoonotic viruses from wildlife to humans.
ChildsJE
MackenzieJS
RichtJA
Wildlife and emerging zoonotic diseases: the biology, circumstances and consequences of cross species transmission
Berlin
Springer
30
32. GraurD
MartinW
2004
Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision.
Trends Genet
20
80
86
33. NabholzB
GleminS
GaltierN
2009
The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals.
BMC Evol Biol
9
54
34. RicklefsRE
OutlawDC
2010
A molecular clock for malaria parasites.
Science
329
226
229
35. PouxC
MadsenO
MarquardE
VieitesDR
de JongWW
2005
Asynchronous colonization of Madagascar by the four endemic clades of primates, tenrecs, carnivores, and rodents as inferred from nuclear genes.
Syst Biol
54
719
730
36. YangZ
YoderAD
2003
Comparison of likelihood and Bayesian methods for estimating divergence times using multiple gene Loci and calibration points, with application to a radiation of cute-looking mouse lemur species.
Syst Biol
52
705
716
37. PrugnolleF
McGeeK
KeeblerJ
AwadallaP
2008
Selection shapes malaria genomes and drives divergence between pathogens infecting hominids versus rodents.
BMC Evol Biol
8
223
38. DuvalL
NerrienetE
RoussetD
Sadeuh MbaSA
HouzeS
2009
Chimpanzee malaria parasites related to Plasmodium ovale in Africa.
PLoS ONE
4
e5520
doi:10.1371/journal.pone.0005520
39. HayakawaT
ArisueN
UdonoT
HiraiH
SattabongkotJ
2009
Identification of Plasmodium malariae, a human malaria parasite, in imported chimpanzees.
PLoS ONE
4
e7412
doi:10.1371/journal.pone.0007412
40. TarelloW
2005
A fatal Plasmodium reichenowi infection in a chimpanzee?
Revue de Médecine Vétérinaire
156
503
505
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance inČlánek The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding ModuleČlánek A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 2- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- A Fresh Look at the Origin of , the Most Malignant Malaria Agent
- In Situ Photodegradation of Incorporated Polyanion Does Not Alter Prion Infectivity
- Highly Efficient Protein Misfolding Cyclic Amplification
- Positive Signature-Tagged Mutagenesis in : Tracking Patho-Adaptive Mutations Promoting Airways Chronic Infection
- Charge-Surrounded Pockets and Electrostatic Interactions with Small Ions Modulate the Activity of Retroviral Fusion Proteins
- Whole-Body Analysis of a Viral Infection: Vascular Endothelium is a Primary Target of Infectious Hematopoietic Necrosis Virus in Zebrafish Larvae
- Inhibition of Nox2 Oxidase Activity Ameliorates Influenza A Virus-Induced Lung Inflammation
- STAT2 Mediates Innate Immunity to Dengue Virus in the Absence of STAT1 via the Type I Interferon Receptor
- Uropathogenic P and Type 1 Fimbriae Act in Synergy in a Living Host to Facilitate Renal Colonization Leading to Nephron Obstruction
- Elite Suppressors Harbor Low Levels of Integrated HIV DNA and High Levels of 2-LTR Circular HIV DNA Compared to HIV+ Patients On and Off HAART
- DC-SIGN Mediated Sphingomyelinase-Activation and Ceramide Generation Is Essential for Enhancement of Viral Uptake in Dendritic Cells
- Short-Lived IFN-γ Effector Responses, but Long-Lived IL-10 Memory Responses, to Malaria in an Area of Low Malaria Endemicity
- Induces T-Cell Lymphoma and Systemic Inflammation
- The C-Terminus of RON2 Provides the Crucial Link between AMA1 and the Host-Associated Invasion Complex
- Critical Role of the Virus-Encoded MicroRNA-155 Ortholog in the Induction of Marek's Disease Lymphomas
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Atypical/Nor98 Scrapie Infectivity in Sheep Peripheral Tissues
- Innate Sensing of HIV-Infected Cells
- BosR (BB0647) Controls the RpoN-RpoS Regulatory Pathway and Virulence Expression in by a Novel DNA-Binding Mechanism
- Compensatory Evolution of Mutations Restores the Fitness Cost Imposed by β-Lactam Resistance in
- Expression of Genes Involves Exchange of the Histone Variant H2A.Z at the Promoter
- The RON2-AMA1 Interaction is a Critical Step in Moving Junction-Dependent Invasion by Apicomplexan Parasites
- Epstein-Barr Virus Nuclear Antigen 3C Facilitates G1-S Transition by Stabilizing and Enhancing the Function of Cyclin D1
- Transcription and Translation Products of the Cytolysin Gene on the Mobile Genetic Element SCC Regulate Virulence
- Phosphatidylinositol 3-Monophosphate Is Involved in Apicoplast Biogenesis
- The Rubella Virus Capsid Is an Anti-Apoptotic Protein that Attenuates the Pore-Forming Ability of Bax
- Episomal Viral cDNAs Identify a Reservoir That Fuels Viral Rebound after Treatment Interruption and That Contributes to Treatment Failure
- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Relationship between Functional Profile of HIV-1 Specific CD8 T Cells and Epitope Variability with the Selection of Escape Mutants in Acute HIV-1 Infection
- The Genotype of Early-Transmitting HIV gp120s Promotes αβ –Reactivity, Revealing αβ/CD4 T cells As Key Targets in Mucosal Transmission
- Small Molecule Inhibitors of RnpA Alter Cellular mRNA Turnover, Exhibit Antimicrobial Activity, and Attenuate Pathogenesis
- The bZIP Transcription Factor MoAP1 Mediates the Oxidative Stress Response and Is Critical for Pathogenicity of the Rice Blast Fungus
- Entrapment of Viral Capsids in Nuclear PML Cages Is an Intrinsic Antiviral Host Defense against Varicella-Zoster Virus
- NS2 Protein of Hepatitis C Virus Interacts with Structural and Non-Structural Proteins towards Virus Assembly
- Measles Outbreak in Africa—Is There a Link to the HIV-1 Epidemic?
- New Models of Microsporidiosis: Infections in Zebrafish, , and Honey Bee
- The C-Terminal Domain of the Arabinosyltransferase EmbC Is a Lectin-Like Carbohydrate Binding Module
- A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- Secreted Bacterial Effectors That Inhibit Host Protein Synthesis Are Critical for Induction of the Innate Immune Response to Virulent
- Genital Tract Sequestration of SIV following Acute Infection
- Functional Coupling between HIV-1 Integrase and the SWI/SNF Chromatin Remodeling Complex for Efficient Integration into Stable Nucleosomes
- DNA Damage and Reactive Nitrogen Species are Barriers to Colonization of the Infant Mouse Intestine
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
- Targeted Disruption of : Invasion of Erythrocytes by Using an Alternative Py235 Erythrocyte Binding Protein
- Trivalent Adenovirus Type 5 HIV Recombinant Vaccine Primes for Modest Cytotoxic Capacity That Is Greatest in Humans with Protective HLA Class I Alleles
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genetic Mapping Identifies Novel Highly Protective Antigens for an Apicomplexan Parasite
- Type I Interferon Signaling Regulates Ly6C Monocytes and Neutrophils during Acute Viral Pneumonia in Mice
- Infections in Cells: Transcriptomic Characterization of a Novel Host-Symbiont Interaction
- The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



