-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Molecular Basis of Increased Serum Resistance among Pulmonary Isolates of Non-typeable
Non-typeable Haemophilus influenzae (NTHi), a common commensal of the human pharynx, is also an opportunistic pathogen if it becomes established in the lower respiratory tract (LRT). In comparison to colonizing isolates from the upper airway, LRT isolates, especially those associated with exacerbations of chronic obstructive pulmonary disease, have increased resistance to the complement - and antibody-dependent, bactericidal effect of serum. To define the molecular basis of this resistance, mutants constructed in a serum resistant strain using the mariner transposon were screened for loss of survival in normal human serum. The loci required for serum resistance contribute to the structure of the exposed surface of the bacterial outer membrane. These included loci involved in biosynthesis of the oligosaccharide component of lipooligosaccharide (LOS), and vacJ, which functions with an ABC transporter encoded by yrb genes in retrograde trafficking of phospholipids from the outer to inner leaflet of the cell envelope. Mutations in vacJ and yrb genes reduced the stability of the outer membrane and were associated with increased cell surface hyrophobicity and phospholipid content. Loss of serum resistance in vacJ and yrb mutants correlated with increased binding of natural immunoglobulin M in serum as well as anti-oligosaccharide mAbs. Expression of vacJ and the yrb genes was positively correlated with serum resistance among clinical isolates. Our findings suggest that NTHi adapts to inflammation encountered during infection of the LRT by modulation of its outer leaflet through increased expression of vacJ and yrb genes to minimize recognition by bactericidal anti-oligosaccharide antibodies.
Published in the journal: . PLoS Pathog 7(1): e32767. doi:10.1371/journal.ppat.1001247
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001247Summary
Non-typeable Haemophilus influenzae (NTHi), a common commensal of the human pharynx, is also an opportunistic pathogen if it becomes established in the lower respiratory tract (LRT). In comparison to colonizing isolates from the upper airway, LRT isolates, especially those associated with exacerbations of chronic obstructive pulmonary disease, have increased resistance to the complement - and antibody-dependent, bactericidal effect of serum. To define the molecular basis of this resistance, mutants constructed in a serum resistant strain using the mariner transposon were screened for loss of survival in normal human serum. The loci required for serum resistance contribute to the structure of the exposed surface of the bacterial outer membrane. These included loci involved in biosynthesis of the oligosaccharide component of lipooligosaccharide (LOS), and vacJ, which functions with an ABC transporter encoded by yrb genes in retrograde trafficking of phospholipids from the outer to inner leaflet of the cell envelope. Mutations in vacJ and yrb genes reduced the stability of the outer membrane and were associated with increased cell surface hyrophobicity and phospholipid content. Loss of serum resistance in vacJ and yrb mutants correlated with increased binding of natural immunoglobulin M in serum as well as anti-oligosaccharide mAbs. Expression of vacJ and the yrb genes was positively correlated with serum resistance among clinical isolates. Our findings suggest that NTHi adapts to inflammation encountered during infection of the LRT by modulation of its outer leaflet through increased expression of vacJ and yrb genes to minimize recognition by bactericidal anti-oligosaccharide antibodies.
Introduction
The mucosal surface of the human nasopharynx is serially colonized by different strains of Haemophilus influenzae [1]. When host factors allow this opportunistic pathogen to gain access to the normally sterile parts of the respiratory tract, inflammatory diseases such as otitis media, sinusitis or pneumonia may result [2]. Widespread immunization against encapsulated strains with the type b polysaccharide has greatly reduced the incidence of invasive disease by H. influenzae in children. However, non-typeable strains (NTHi), which do not express a capsule, remain amongst the most common etiologic agents of localized infectious diseases of the airway in all age groups [3]. The damaged airways in adults with chronic obstructive pulmonary disease (COPD) are especially susceptible, and identification of a newly acquired NTHi isolate in sputum is temporally associated with exacerbations of disease symptoms and decline in pulmonary function [4], [5]. COPD ranks as the fourth leading cause of death in the US and is rapidly becoming recognized as a public health problem of similar proportions in other parts of the world [6], [7].
Characteristics of the organism that allow it to transition from its commensal state in the upper airway and survive the inflammatory milieu of the lower respiratory tract (LRT) and elsewhere are poorly understood. In particular, during early infection this predominantly extracellular pathogen will be exposed to increasing levels of natural (i.e. pre-existing) antibody and complement produced locally or extravasated from serum. For encapsulated H. influenzae, the thick polysaccharide coat protects the organism from recognition by immunoglobulin, the activation of complement and complement-dependent bactericidal activity. For gram-negative bacteria, the exposed surface is its outer membrane, an asymmetric lipid bilayer consisting of an outer leaflet of lipid A attached to a polysaccharide (LPS) and an inner leaflet of phospholipid [8], [9], [10]. For H. influenzae, LPS is referred to as a lipooligosaccharide (LOS) because of its more limited number of attached sugars. There is marked strain to strain heterogeneity in the presence and linkages of these sugars and oligosaccharide epitopes these residues generate indicating that antigenic variation may contribute to immune evasion by NTHi [11], [12]. Structural features of the surface oligosaccharide that inhibit complement-dependent killing have been analyzed extensively [13], [14], [15], [16]. The expression of many of these oligosaccharide components is controlled by highly repetitive DNA sequences and, as a consequence of slipped stranded mispairing, the expression of oligosaccharide structures is turned on and off at high frequency [13], [17]. While this would predict that the presence of bactericidal antibody and complement would select for variants with increased resistance, many of these structures decorating the surface oligosaccharide are present on both serum sensitive and resistant isolates. Therefore, our current understanding does not fully account for why only some NTHi are serum resistant and how this phenotype correlates with the pathogenicity of the species.
In this study, we addressed whether increased resistance to the complement-mediated bactericidal activity of normal human serum is a characteristic of isolates from the LRT. We then used a whole genomic approach to identify the genes required for the expression of serum resistance among these isolates. We describe an important role for genes involved in trafficking of phospholipids in evading natural antibody and the expression of serum resistance by NTHi.
Results
Lung isolates have increased serum resistance and decreased binding of natural IgM
Collections of recent clinical isolates maintained with minimal in vitro passage were compared for their ability to survive following a 60 min incubation in 5% normal human serum (NHS). Bactericidal activity was complement-dependent, since killing was not observed in controls using heat-inactivated serum. Sputum isolates from the lower respiratory tract (LRT) (n = 22) were significantly more serum resistant than colonizing strains (n = 25) cultured from the upper respiratory tract (Fig. 1A). Among the LRT isolates, those obtained at the time of a COPD exacerbation were the most serum resistant. Next, we examined whether differences in serum resistance correlated with the binding of immunoglobulin present in normal human serum as measured by flow cytometry. There was no difference between serum resistant and serum sensitive isolates in binding of IgG (Fig. 1B1). In contrast, the serum sensitive strains bound significantly more IgM than serum resistant strains (Fig. 1B2). There was no difference between serum resistant and serum sensitive strains in killing by baby rabbit serum (2.5%), which lacks natural antibody to H. influenzae, as a source of complement (Fig. 1C). Addition of IgM, but not IgG, purified from NHS to baby rabbit serum significantly enhanced killing of serum sensitive, but not serum resistant isolates (Fig. 1C). Together these results demonstrate 1) an association between serum resistance and the ability of NTHi to infect the LRT and 2) that resistant isolates bind less natural, bactericidal IgM.
Fig. 1. Characterization of clinical isolates. 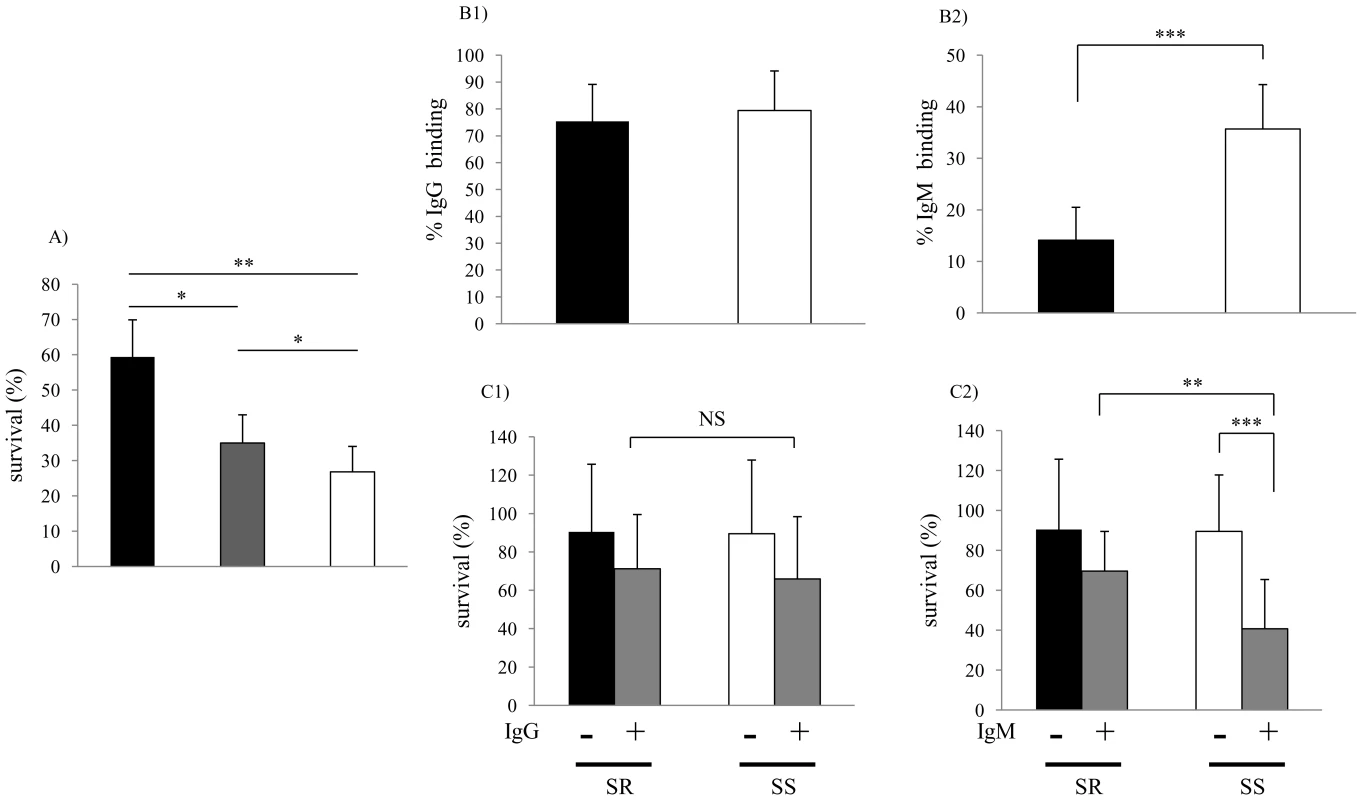
(A) Comparison of serum sensitivity between lower and upper respiratory tract isolates. Survival was determined over 60 min in 5% normal human serum and expressed relative to controls in which complement was inactivated. Groups included lower respiratory tract isolates from patients with chronic obstructive pulmonary disease (COPD) at the time of clinical exacerbation (black bars; n = 11); isolates from patients with COPD during clinically stable periods (grey bars; n = 11); and upper respiratory tract colonizing strains (white bars; n = 25). Values are the mean of three determinations in triplicate ± SEM. (B) Comparison of antibody binding in serum resistant (>50% survival in pooled NHS, black bars; n = 10) and serum sensitive (<50% survival in pooled NHS, white bars; n = 10) isolates. B1 and B2 show percent of IgG and IgM bound following incubation in 5% heat-inactivated NHS as determined by flow cytometry, respectively. (C) Serum IgM contributes to bactericidal killing of clinical isolates of NTHi. Strains were incubated with or without IgG (C1, 0.25 µg/ml) or IgM (C2, 0.07μg/ml) purified from NHS for 60 min in 2.5% baby rabbit serum as a complement source. Percent survival was calculated by viable counts with and without antibody. The mean values of two independent experiments in triplicate are shown ± SD, *P<0.05, **P<0.01, ***P<0.001. Serum sensitive (SS), Serum resistant (SR). Genetic basis of increased serum resistance
In order to identify the complete set of genes required for serum resistance in NTHi, we screened mariner transposon mutants generated in strain R2866, a previously described serum resistant isolate for which the whole genome sequence was available, for increased susceptibility to NHS [18]. A total of 6912 mutants were individually screened to provide ∼4-fold representation of open reading frames. Genomic DNA from candidates showing <10% survival was back transformed into the parent, R2866, and these back transformants were tested to confirm that the insertion mutation conferred a serum sensitive phenotype. Sixty serum sensitive mutants (representing 0.87% of the total strains screened) were identified and for these the mariner insertion site was defined. We focused on the genes (13 total and 12 of ‘known’ function) for which there was more than a single ‘hit’(Table 1). Eight loci, including lgtC, galE, waaQ, lic2A, lex2B, lpsA, yhxB and galU, function in the biosynthesis of the surface oligosaccharide and these were not considered further [19]. The most striking effect on serum resistance (Fig. 2A) was observed with mutations in HI0718 (encoding VacJ, a putative lipoprotein), and in a separate operon with ‘hits’ in HI1083, HI1085, and HI1086 (encoding orthologs of other gram-negative species; YrbB, a putative NTP binding protein; YrbD, an ABC transporter periplasmic protein; and YrbE, an ABC transporter permease, respectively). VacJ, YrbD and YrbE each share homology (>60% sequence identity) with members of the E. coli Mla transport system, which have been proposed to function in preventing phospholipid accumulation in the outer leaflet and thereby maintain the lipid asymmetry and the barrier function of the gram-negative outer membrane [20]. A double mutant in vacJ and the ABC transporter gene yrbE had a similar serum sensitive phenotype to that of each mutant, which confirmed that these may act in the same pathway. The effect on vacJ is unlikely to be caused by a polar effect of the insertion mutation, since a mutant in the immediate downstream gene, HI0719, maintained serum resistance (data not shown). There was no effect of vacJ or yrb genes on LOS or outer membrane protein profiles as assessed following separation using tricine gel electrophoresis followed by silver staining (data not shown).
Fig. 2. Characterization of vacJ and yrb mutants. 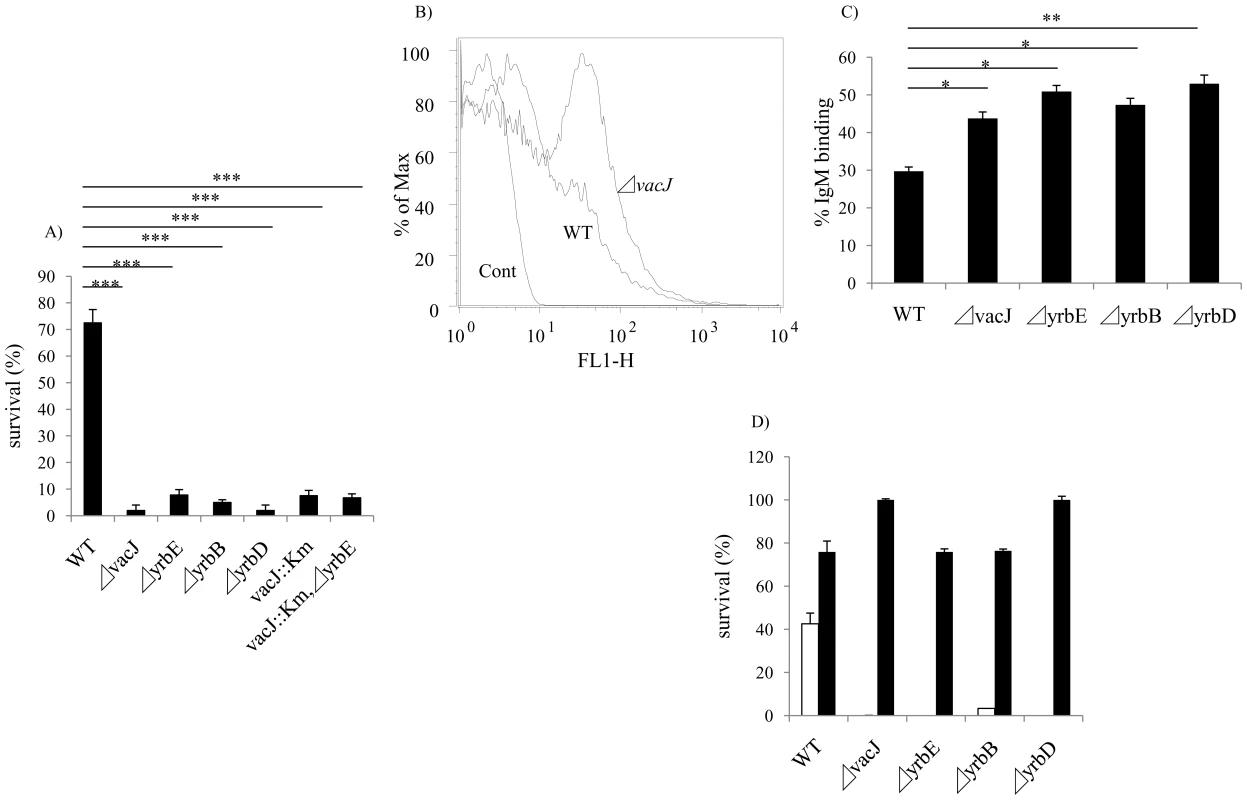
(A) Effect of mutations in vacJ and genes of the yrb ABC transporter on serum resistance of strain R2866. Survival was determined over 60 min in 5% normal human serum and expressed relative to controls in which complement was inactivated. (B) Representative histogram comparing the binding, as measured by fluorescence intensity (x-axis), of total IgM purified from normal human serum to parent strain (WT) or vacJ by flow cytometry. Control performed without IgM (C) Percent IgM binding for each mutant was determined by calculating the percentage of 50,000 events with an increase in mean fluorescence intensity following incubation in 5% heat-inactivated normal human serum compared to no serum controls. (D) Survival of mutants in 10% normal human serum in the presence (black bars) or absence (white bars) of Mg-EGTA to inhibit the classical pathway of complement activation. Values represent two independent experiments in triplicate ± SD. *P<0.05, **P<0.01, ***P<0.001. Tab. 1. List of sites with multiple transposon insertions affecting serum resistance in strain R2866. 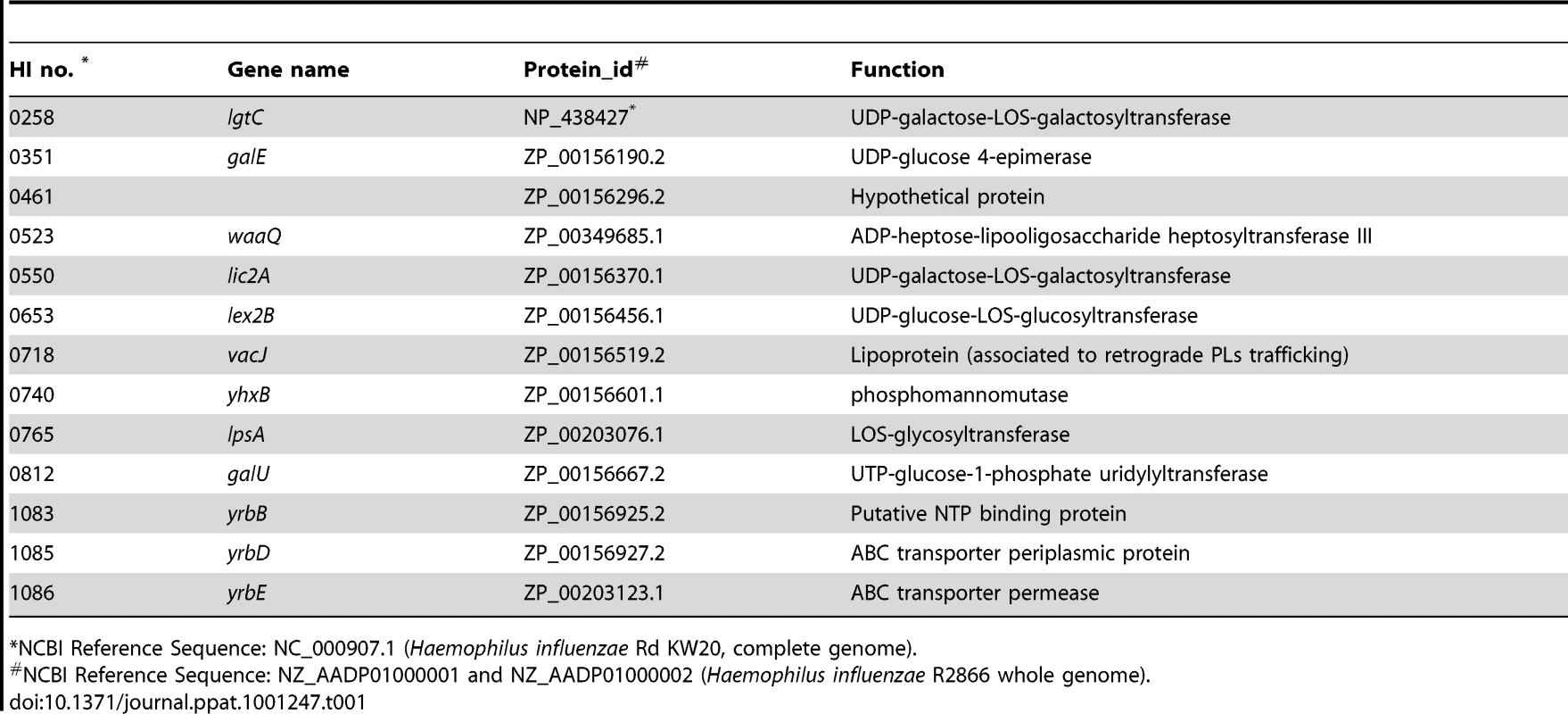
*NCBI Reference Sequence: NC_000907.1 (Haemophilus influenzae Rd KW20, complete genome). vacJ and yrb ABC transporter genes contribute to serum resistance and IgM binding
After incubation in baby rabbit serum, no significant difference was observed in the binding of rabbit complement factor 3 between wild type and vacJ mutant (data not shown) indicating a requirement for antibody in the differential susceptibility of the mutants. To determine whether vacJ and yrbE, B and D mediate serum resistance by affecting antibody binding, we compared the deposition of IgG and IgM purified from NHS by flow cytometry. No detectible effect of these mutations on the binding of IgG was observed (data not shown). In contrast, there was a significant increase in the binding of IgM to the mutants (Fig. 2B and 2C). To determine if increased binding of IgM was sufficient to account for loss of serum resistance of the mutants, IgM purified from human serum was used with 2.5% baby rabbit serum as a complement source. Under these conditions each of the mutants was more susceptible to IgM dependent killing (data not shown). To define the complement pathway affected, the survival of the mutants was studied in the presence of Mg-EGTA buffer, which inhibits the classical pathway. When the classical pathway was inhibited, a significant increase in the survival of each mutant was observed (Fig. 2D). The requirement for the classical pathway of complement showed that the anti-bacterial effect of IgM was not caused by agglutination. Together these results demonstrated that vacJ and yrbE, B and D are needed for serum resistance in R2866 by limiting the binding of natural IgM that promotes killing via the classical pathway of complement activation.
vacJ and yrb ABC transporter genes affect binding of anti-LOS antibody
Next, we considered the target of bactericidal, natural IgM affected by mutations in vacJ and yrb ABC transporter genes. We performed FACS analysis to compare the binding of murine mAbs 4C4 and TEPC-15, which bind specifically to LOS components, Galα1-4Gal and phosphorylcholine, respectively. Since these are both phase variable LOS epitopes, we first enriched for mAb 4C4 or TEPC-15 positive cells by colony immunoblotting. Mutations in vacJ and the yrb ABC transporter genes significantly increased the binding of mAbs 4C4 and TEPC-15 (Fig. 3A and 3B). Furthermore, mAb 4C4 was bactericidal in the presence of normal mouse serum as a source of complement and each mutant was significantly more sensitive compared to the parent strain. (Fig. 3C). We then investigated whether vacJ affects the antibody binding to cell surface proteins by flow cytometry in two different ways. First, we compared binding of the mAb 7B11 to an exposed epitope on outer membrane protein P2 on strain H782 [21]. However, the mutation in vacJ in strain H782 did not alter the binding of mAb 7B11 (data not shown). Second, we labeled exposed lysine residues on cell surface proteins by treating whole bacteria with the fluorescent dye Cy5. There was no difference in the levels of bound Cy5 between wild type and vacJ mutant (data not shown). Our data suggested that vacJ affects binding of antibody to exposed LOS but not outer membrane protein epitopes. Since mAb 4C4 is IgG and TEPC-15 is IgA, the effect on antibody binding and killing was not specific to IgM. This suggests that our observations about natural antibody in NHS could be because the bactericidal antibody targeting LOS is predominantly IgM.
Fig. 3. Effect of vacJ and yrb mutants on antibody binding and bactericidal activity. 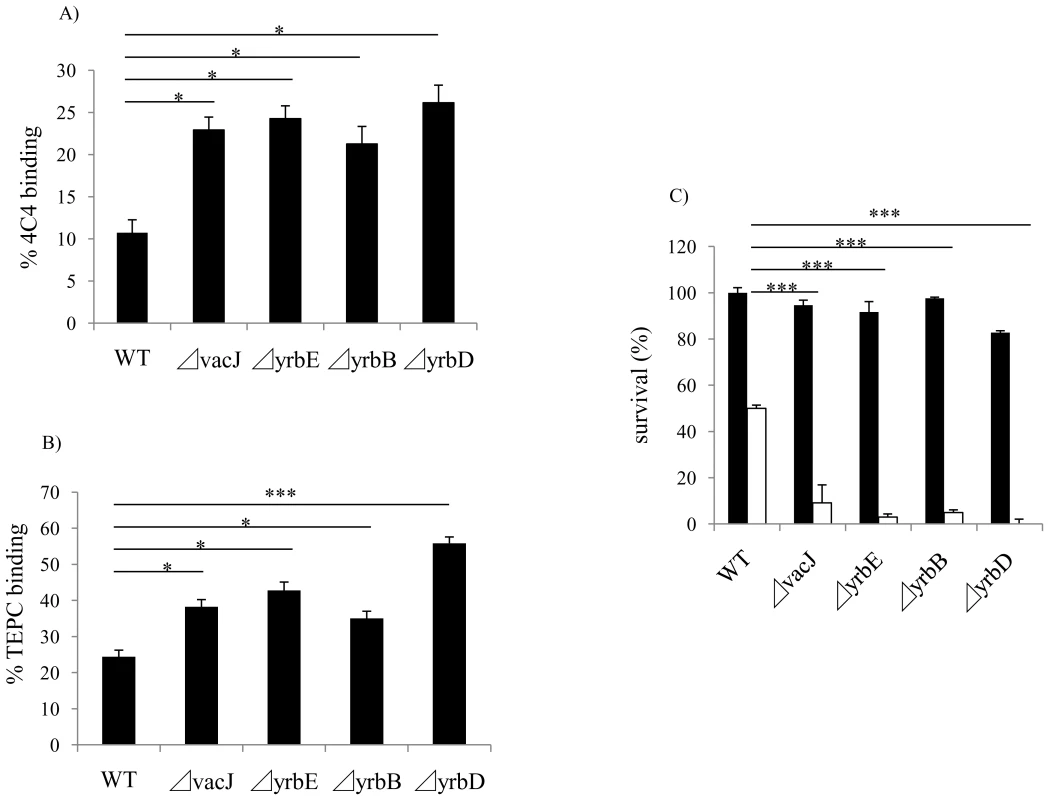
Binding of (A) mAb 4C4 to the LOS structure Galα1-4Gal or (B) mAb TEPC-15 to the LOS structure phosphorylcholine was compared by flow cytometry for the mutants indicated. Percent mAb binding for each mutant was determined by calculating the percentage of 50,000 events with an increase in mean fluorescence intensity compared to no primary antibody controls. (C) Effect of mutations in vacJ and genes of the yrb ABC transporter on the bactericidal effect of mAb 4C4. Bactericidal assays were performed with (white bars) or without (black bars) mAb with 5.0% normal mouse serum as a complement source. Values represent two independent experiments in triplicate ± SD. *P<0.05, ***P<0.001. vacJ and yrb ABC transporter genes affect outer membrane stability
To test whether vacJ and yrb ABC transporter genes affect the integrity of the outer membrane, we analyzed the sensitivity of the mutants to small antimicrobial compounds, including vancomycin (MW 1449), novobiocin (MW 613), bacitracin (MW 1423), and polymyxin-B (MW 1302). There was no difference in sensitivity to these compounds compared to the parent strain suggesting that the outer membrane barrier of the mutants is largely intact (data not shown). We then addressed the stability of the outer membrane by the addition of EDTA, which chelates divalent cations and compromises the outer leaflet by interrupting intermolecular associations between LOS phosphate groups. A concentration of 25 mM EDTA had no effect on the parent strain, but resulted in a >3-log decrease in viability for each of the mutants (Fig. 4A). Similarly, the mutants were sensitive to the detergent deoxycholate at a concentration 4-fold lower than that required to inhibit growth of the parent strain (data not shown). Our observations of increased sensitivity to EDTA and deoxycholate demonstrated that the stability of the outer membrane was impaired in the mutants. Next, we compared the physical properties of the outer leaflet of the mutants by measuring the rate of uptake of 1-N-phenylnaphthylamine (NPN), a probe that changes fluorescence upon transfer from a hydrophilic to hydrophobic environment. We predicted that higher phospholipid content would increase the hydrophobic character of the cell surface of the mutants. As shown in Fig. 4B, vacJ and yrbE mutants had more rapid NPN uptake than the wild type strain, demonstrating that both mutants have increased surface hydrophobicity. In addition, we directly compared the content of surface exposed phospholipids by treating whole cells with phospholipase C and then detecting released diacylglycerol using thin layer chromatography (Fig. 4C). In comparison to the parent strain, amounts of released diacylglycerol were increased in vacJ (spot density increased 219%) and yrbE (spot density increased 143%) mutants, showing that the amount of surface phospholipid accessible to phospholipase C treatment was increased in the mutants. The similar results for each mutant provided further evidence that vacJ and yrb genes act in the same pathway and were consistent with their previously proposed function in E. coli in excluding phospholipids from the outer leaflet described [20]. Together our findings suggested that maintaining the asymmetry of the outer leaflet by the exclusion of phospholipids is important in limiting recognition of surface oligosaccharide epitopes by antibody.
Fig. 4. Effect of mutations in vacJ and genes of the yrb ABC transporter on outer membrane characteristics. 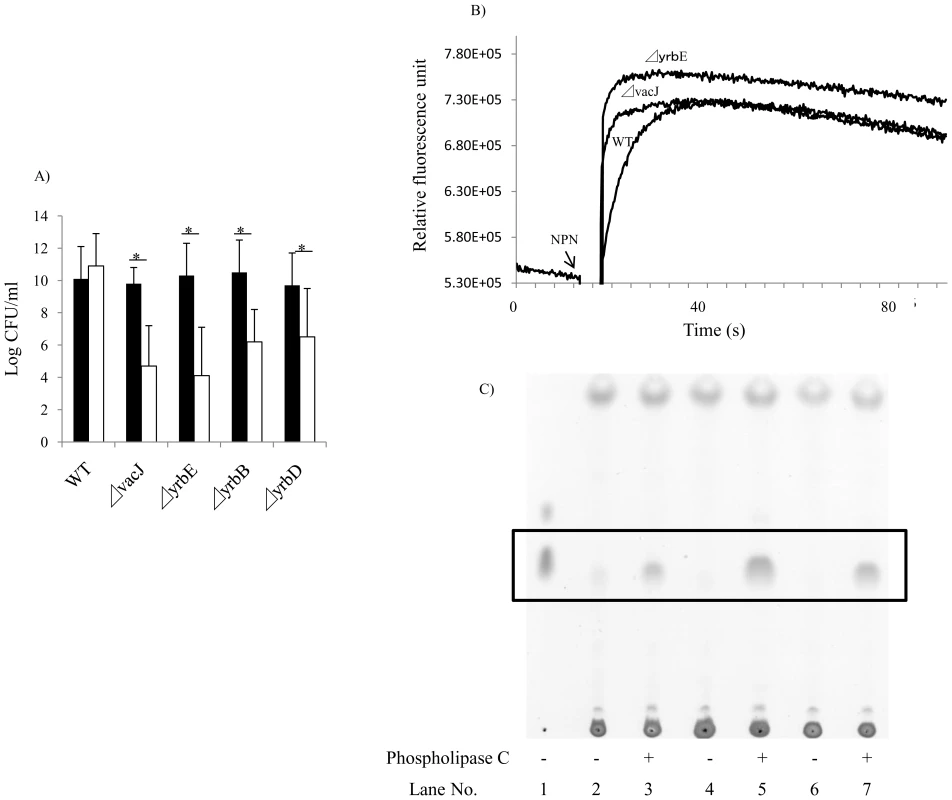
(A) To compare outer membrane stability, following overnight culture of the strain indicated, viable counts were obtained after incubation at 37°C for 4 h in the presence (white bars) or absence (black bars) of 25 mM EDTA. Values represent two independent experiments in triplicate ± SD. *P<0.05. (B) To compare surface hydrophobicity, the rate of uptake of membrane permeant 1-N-phenylnaphthylamine (NPN) was monitored by fluorescence. NPN was added at the time indicated and a representative experiment shown. (C) To compare amounts of surface phospholipids, diacylglycerol (boxed area) released by phospholipase C treatment of whole bacteria was detected by thin layer chromatography. Lane 1; diacylglycerol (standard), Lane 2 and 3; R2866 (wild type), Lane 4 and 5; 32F2 (vacJ mutant), Lane 6 and 7; 69G3 (yrbE mutant). Expression of vacJ and yrb ABC transporter genes correlates with serum resistance in clinical isolates
To investigate the relationship between outer leaflet stability and serum resistance in NTHi, we compared the sensitivity of serum resistant and serum sensitive clinical isolates to EDTA. As shown in Fig. 5A, serum resistant isolates were significantly more resistant to EDTA than serum sensitive isolates. We then determined whether differences in serum resistance and resistance to EDTA correlated with the expression of vacJ and yrb genes by qRT-PCR. As shown in Fig. 5B, vacJ expression was ∼5-fold higher in serum resistant strains compared to serum sensitive isolates. In addition, yrbE and yrbD expression was ∼3-fold higher in serum resistant compared to serum sensitive isolates (Fig. 5C and 5D). To further investigate the relationship between serum resistance and vacJ expression, we serially passaged strain H725, a serum sensitive clinical isolate, in 2.5% NHS to select for variants with an increasing capacity to resist the bactericidal effect of serum. With each passage H725 became more serum resistant and this adaptation was associated with a stepwise increase in vacJ expression and resistance to EDTA (Fig. 6A–C). Our data suggest that among clinical isolates differences in serum resistance correlate with the level of expression of vacJ and its effect on stability of the outer leaflet of NTHi.
Fig. 5. Membrane stability and vacJ and yrb expression among clinical isolates. 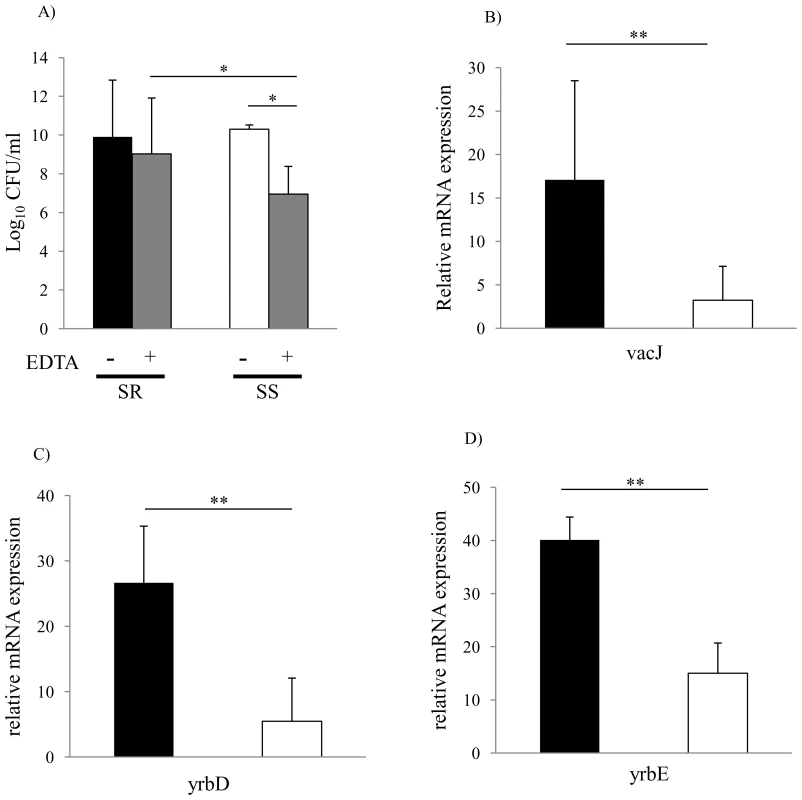
(A) Outer membrane stability of serum sensitive (n = 31) compared to serum resistant (n = 16) clinical isolates. Following overnight culture, viable counts were obtained after incubation at 37°C for 4 h with or without 25 mM EDTA. Values represent two independent experiments in triplicate ± SD. *P<0.05, Serum sensitive (SS), Serum resistant (SR). (B) Relative expression of vacJ mRNA, (C) yrbD and (D) yrbE by qRT-PCR in serum sensitive (white bar, n = 7) and serum resistant isolates (black bar, n = 7). Error bars indicate SD, **P<0.01. Fig. 6. Repeated serum treatment selects for serum resistance, increased vacJ expression, and increased outer membrane stability. 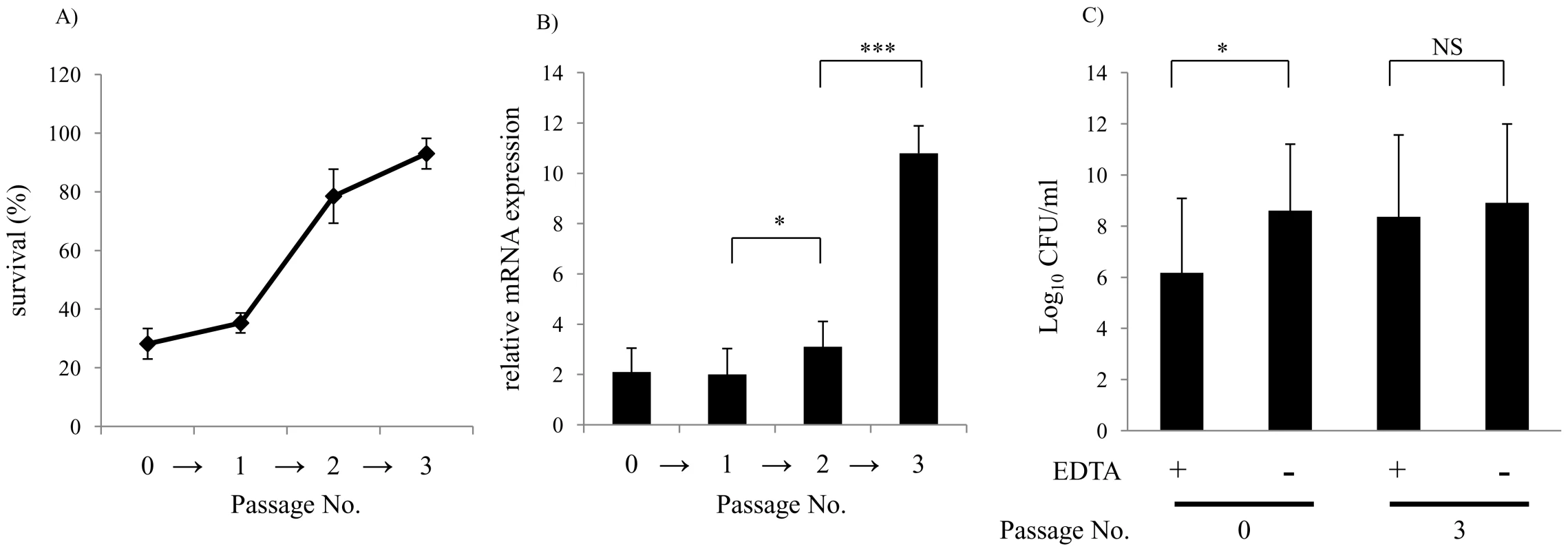
(A) Serum sensitive clinical isolate H725 was treated three times in 2.5% NHS and survival quantified after the passage indicated. (B) Following each passage in the bactericidal assay survivors were tested for relative expression of vacJ mRNA by qRT-PCR and (C) survival in the presence of 25 mM EDTA. Values represent two independent experiments in triplicate ± SD.*P<0.05, ***P<0.001. Discussion
Although generally co-existing in a commensal relationship with its host, NTHi is able to survive the robust inflammatory response it induces in normally sterile sites in the respiratory tract such as the lung. Since humans are serially colonized beginning early in childhood, prior exposure to NTHi, as well as to other microbial species that induce cross reactivity, provides an abundant source of natural antibody [22], [23]. Individuals with defects in generating antibody are particularly susceptible to recurrent respiratory tract infection with NTHi [24]. To survive the inflammatory response in the LRT, this pathogen has to evade the effects of the host's pool of pre-existing antibody, which when bound to the bacterial surface activates complement and induces lytic killing. In this report we used a serum killing assay to show that survival of NTHi isolated from the LRT is associated with increased resistance to the complement-dependent bactericidal effect of antibody. Additional evidence in support of this conclusion is the finding that serum resistance was highest among the isolates obtained at the time of COPD exacerbations when clinical signs of inflammation, such as increased production of sputum, are more pronounced. Although antibodies of other isotypes can be bactericidal, we found that most of the natural bactericidal antibody present in NHS is IgM. Our findings demonstrate that serum killing correlates with binding of natural IgM followed by activation of the classical pathway of complement. Observations on the prominence of anti-LOS IgM in the bactericidal activity of NHS correlate with previous reports looking at natural bactericidal antibody present in animal sera [25]. Findings in the current study are also consistent with prior reports from this laboratory using a mouse model of airway infection showing that complement and natural antibody protect the host from NTHi [26].
Previous analysis of serum resistance in NTHi has been limited by the marked heterogeneity within and between strains. This is largely caused by the rapid variation in the expression of surface antigens targeted by natural antibody and demonstrates the ability of the organism to escape antibody-dependent, complement-mediated killing. Several specific oligosaccharide structures that contribute to serum resistance have been described. Some strains express siayltransferases that use serum CMP-NANA to cap the oligosacchaide with sialic acid, which inhibits the activation of complement through the alternative pathway [16]. The disaccharide Galα1-4Gal, which mimics the human Pk blood group antigen, blocks recognition by anti-LOS antibodies [13]. The oligosaccharide structures requiring the galactosyltransferase LgtC affects expression of Galα1-4Gal modulating deposition of C4b and activation of the classical pathway [15]. These oligosaccharide decorations are each variably expressed within and between strains. Our finding that 8 of the 13 genes necessary for the expression of serum resistance in a highly serum resistant NTHi strain function in biosynthesis of the surface oligosaccharide highlights its central role in this phenotype and the pathogenicity of the organism. In addition, we demonstrate that the oligosaccharide is the major target of bactericidal human antibody. This may explain the predominance of natural IgM in targeting the NTHi oligosaccharide, since antibody to LPS antigens is dominated by IgM generated by B-1 cells [27]. For example, a large proportion of B-1 cells generate IgM reactive with phosphorylcholine [28].
The selection for a more serum resistant phenotype in the LRT correlated with increased expression of vacJ and yrb genes, which alter cell surface characteristics and thereby limit the binding of bactericidal antibody. The identification of VacJ and an ABC transporter with related function as necessary for the expression of serum resistance shows that surface characteristics other than structural components of the oligosaccharide contribute to serum resistance of NTHi. The lipoprotein VacJ was previously identified as a virulence determinant contributing to intracellular survival by Shigella flexneri [29]. The group of Silhavy proposed that VacJ acts with the Mla ABC transporter to maintain the lipid asymmetry of the outer membrane by recycling phospholipids from the outer leaflet back to the inner leaflet [20]. In E. coli, mutants lacking these genes were more sensitive to the presence of SDS plus EDTA added to solid media, but not to antibiotics that need to access targets in the periplasmic space. These observations indicated that 1) stability of the outer leaflet generated through intermolecular bridging of LPS by divalent cations was compromised by the accumulation of phospholipids and 2) the permeability barrier of the outer leaflet remains largely intact in the mutants. We observed similar characteristics of mutants in vacJ and Mla ABC transporter homologs in NTHi. In addition, we provided direct evidence that vacJ and yrb genes function in determining key characteristics of the cell surface (i.e. its hydrophobicity) and that these genes affect amounts of surface exposed phosholipid. The increase in antibody binding and killing seen in these mutants provides a new insight into how these physical characteristics of the outer membrane contribute to serum resistance. Our results suggest that the intramolecular forces bridging LOS molecules also serve to limit binding of antibodies to the oligosaccharide. Thus, when phospholipids are more thoroughly excluded (high expression of vacJ and yrb genes), LOS molecules are more tightly packed and accessibility of LOS epitopes is restricted (serum resistance). Whereas when phospholipids accumulate in the outer leaflet (low expression of vacJ and yrb genes), intermolecular associations of LOS molecules are interrupted allowing for increased access of oligosaccharide epitopes recognized by bactericidal antibodies (serum sensitivity).
Interestingly, the expression of vacJ and yrb genes, which are required for serum resistance, is variable within and between NTHi strains. Serum resistant isolates demonstrated increased levels of transcription compared to serum sensitive isolates and serial passage of a sensitive isolate in serum selected for increased resistance and resulted in a higher level of vacJ expression. Thus, differences in transcription of vacJ are another factor accounting for the marked differences in serum resistance that characterizes this species. It is somewhat surprising that for many clinical isolates levels of vacJ expression is low enough to affect the stability of the outer membrane. This suggests that some level of phospholipid accumulation in the outer leaflet is tolerated by NTHi and implies that there must be an advantage to a less stable, more hydrophobic outer membrane, particularly during colonization when restricting recognition by antibody may be less critical for survival. It appears, however, that this fitness advantage is lost during infection of the lung when increased vacJ and yrb gene expression is selected for. Because NTHi resides in the respiratory tract where it is not exposed to the detergent effect of bile in the intestine, the physical requirements of its outer membrane may be different from the paradigm described in the classic studies based on enteric bacteria [30].
In conclusion, we show that resistance to the bactericidal effect of immunoglobulin together with complement correlates with the ability of NTHi to infect the human LRT. Analysis of a serum resistant isolate revealed that genes contributing to the biosynthesis of its surface oligosaccharide are required for this phenotype. In addition, we describe a novel mechanism for serum resistance whereby NTHi limits the binding of bactericidal anti-LOS antibody by increasing the exclusion of surface phospholipids.
Materials and Methods
Bacterial strains and growth conditions
Strains used in this study are listed in Table 2. COPD strains. Strains were isolated from expectorated sputum samples as part of a prospective study at the Buffalo VA Medical Center (4). COPD exacerbation strains fulfilled the following criteria: 1) First isolation of the strain in an adult with COPD based on molecular typing of isolates recovered from monthly sputum cultures; 2) NTHi is the only potential pulmonary pathogen isolated in the sputum sample; 3) simultaneous onset of clinical symptoms of an exacerbation (increased sputum volume, increased sputum purulence and increased shortness of breath compared to baseline symptoms). Non-exacerbation strains were from patients with COPD during clinically stable periods who fulfilled criteria 1 and 2 but symptoms unchanged from baseline upon acquisition of the strain. Upper respiratory tract strains. 25 NTHi strains were isolated from throat cultures of 25 healthy children attending 17 different day care centers [31], [32]. Each isolate had a different pulse-field gel electrophoresis pattern and was confirmed to be H. influenzae (and not non-hemolytic H. haemolyticus) based on previous criteria [1], [33]. The absence of bexA and bexB confirmed that the isolates were not capsule-negative variants.
Tab. 2. The list of strains, plasmids and primers used in this study. 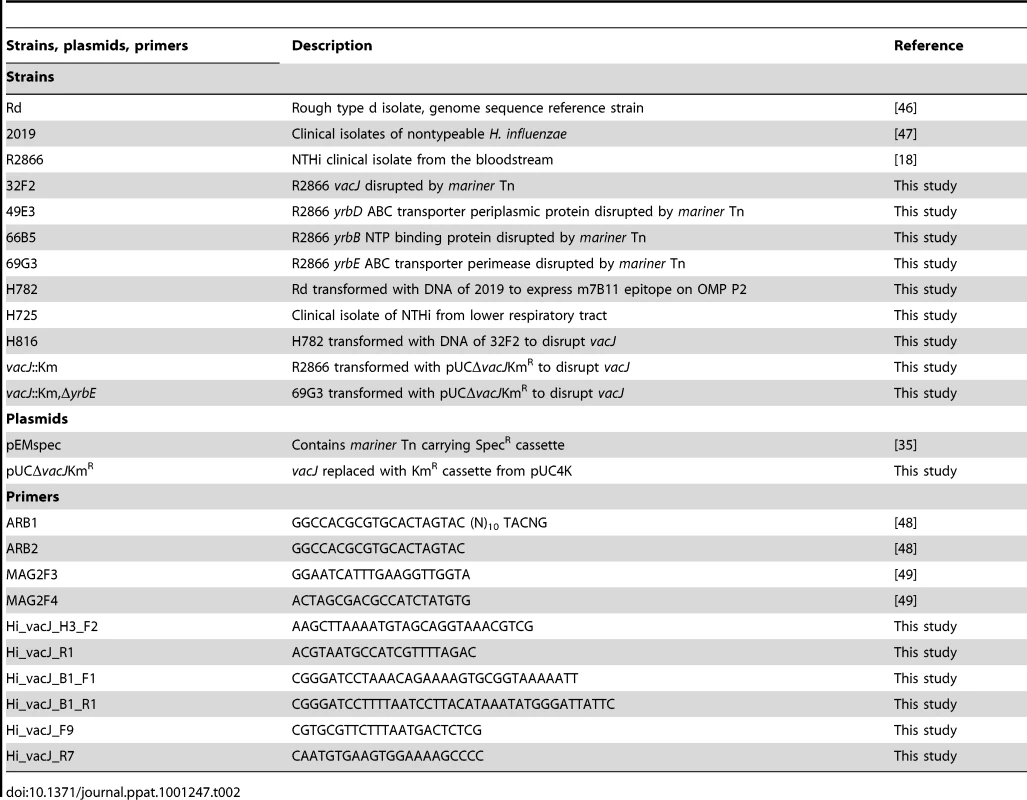
Strains were routinely grown at 37°C in brain heart infusion broth (Becton Dickinson) supplemented with 2% Fildes enrichment (Remel) and 20 µg/ml β-NAD hydrate (NAD; Sigma). H782 was created from strain Rd by transformation with a PCR product from outer membrane protein P2 locus of strain 2019 followed by screening for expression of the surface epitope recognized by mAb 7B11 epitope [21].
Ethics statement
The strains from adults with COPD were obtained from subjects enrolled in a study at the Buffalo VA Medical Center that was approved by the IRB of the VA Western NY Healthcare System. All subjects provided written informed consent. Strains collected from healthy children in day care, under protocols reviewed by IRB Health at the University of Michigan, were deemed EXEMPT on the basis of: EXEMPTION #4 (45 CFR 46.101(b)(4)), because the data were collected and analyzed without personal identifiers attached to the bacterial isolates.
In vitro transposon mutagenesis, DNA transformation and transposon library construction
Mariner mutants of strain R2866 were created by in vitro transposon mutagenesis as previously described [34]. Briefly, in vitro transposition reactions were carried out on NTHi genomic DNA treated with purified MarC9 transposase and pEMspec [35]. To repair gaps, reactions were ethanol-precipitated and resuspended in the gap repair buffer [50 mM Tris (pH 7.8)], 10 mM MgCl2, 1 mM DTT, 100 nM dNTP, and 50 ng of BSA] and then treated with T4 DNA polymerase (Invitrogen) and E. coli DNA ligase (Invitrogen) [36]. DNA was transformed into competent R2866 by the method of Herriott et al [37] and transformants where selected for on sBHI agar (1%) plates containing spectinomycin (100 µg/ml).
Identification of serum sensitive transposon mutants
To identify NTHi genes essential for serum resistance, mariner transposon mutants were screened in a 96-well serum bactericidal assay. Following growth from single colonies in 200 µl sBHI, mutants were diluted to 105 CFU/ml and 10 µl of the culture solution was added to 90 µl consisting of 55 µl PBS, 30μl Hank's buffer (Ca2+, Mg2+) and 5 µl of normal human serum (NHS). Serum was obtained from a single donor for the initial screen to minimize variability in screening large numbers of mutants. Bacteria were incubated at 37°C for 1 h with shaking before the reaction was stopped at 4°C. Controls included serum from the same donor treated at 56°C for 30 min to inactivate complement. Mutants showing >90% killing in the primary screen were further tested to minimize false positives. Genomic DNA was used to back transform competent R2866 and three spectinomycin resistant colonies were picked and rescreened in the bactericidal assay. The site of the transposon insertion was determined for mutants in which 3/3 back transformants were serum sensitive.
Arbitrary primed PCR and nucleotide sequence
The primers used for inverse PCR are listed in Table 2. The first round of PCR was performed in a final volume of 50 µl. Primers (0.5 µmol/reaction mixture) ARB1 paired with mag2F3 (transposon) were used in PCR under the following conditions: 1 cycle of 95°C for 8 min; 6 cycles of 95°C for 30 s, 30°C for 30 s, and 72°C for 1.5 min; 30 cycles of 95°C for 30 s, 45°C for 30 s, and 72°C for 2 min; and 72°C for 5 min. A second round of PCR was performed with primers (0.5 µmol/reaction) ARB2 paired with the internal mag2F4 (transposon) primer under the following conditions: 1 cycle of 95°C for 8 min, 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 1.5 min, followed by 72°C for 10 min. PCR products were purified with the QIAGEN PCR cleanup kit and then used for nucleotide sequence analysis.
Construction of ΔvacJ::KmR mutant
The vacJ gene and flanking regions were amplified from strain R2866 using primers Hi_vacJ_H3_F2 and Hi_vacJ_R1 and cloned into pCR2.1TOPO vector. The entire vacJ gene was deleted by inverse PCR using Hi_vacJ_B1_F1 and Hi_vacJ_B1_R1 primers introducing a BamHI site. The resulting ΔvacJ fragment was subcloned into pUC19 and the kanamycin-resistance cassette from pUC4K was then inserted using BamHI creating pUCΔvacJ::KmR. This plasmid was used to transform strains R2866 and 69G3 strains creating single and double ΔvacJ::KmR mutants. Disruption of vacJ was confirmed by PCR using primers vacJ_ F9 and Hi_vacJ_R7 located outside of the originally cloned region.
Serum bactericidal assays
To test clinical isolates in bactericidal assays, serum was collected, pooled and stored at -80°C from 5 healthy adult volunteers. Assays were performed with 20 µl of a suspension of midlog phase organisms (OD620 0.3–0.4) diluted to 105 CFU/ml in Hank's buffer with Ca2+ and Mg2+ (GIBCO, Auckland, New Zealand), 10 µl of NHS, 110 µl of PBS and 60 µl of Hank's buffer with Ca2+ and Mg2+. After incubation for 60 min at 37°C with rotation, the assay was stopped by cooling to 4°C and dilutions were made for quantitative culture. To inhibit the classical pathway of complement, veronal buffer (pH 7.4) containing 70 mM NaCl, 140 mM glucose, 0.1% gelatin, 7 mM MgCl2, and 10 mM Mg-EGTA was substituted for Hank's buffer [38]. To calculate the percent survival, viable counts were compared to controls in which complement activity had been eliminated by heat-inactivation at 56°C for 30 min. IgG was removed from NHS using a protein G column according to the manufacturer's instructions (GE Healthcare, Uppsala, Sweden). The flow-through of IgG column was used for purification of IgM. Purified IgM was obtained by using IgM purification column according to the manufacturer's instructions (GE Healthcare, Uppsala, Sweden). The concentration of purified IgG and IgM were determined by the Micro BCA protein assay (Pierce Chemical Co.) and a titrated enzyme-linked immunosorbent assay (ELISA). Where indicated, three-four week old baby rabbit serum was used as a complement source (Pel-Freez Biologicals, Rogers, AR).
Colony immunoblotting
Colonies lifted onto nitrocellulose were immunoblotted to separate phase variants as previously described [39]. Briefly, the LOS and ChoP epitope on colonies lifted onto nitrocellulose were detected using mAbs 4C4 against Galα1-4Gal [40] and TEPC-15 (Sigma) against phosphorylcholine followed by alkaline phosphatase–conjugated anti – mouse IgG and IgA, respectively. Single colonies of a uniform phenotype were selected and confirmed by repeated immunoblotting.
Labeling of surface proteins
200 µl of a suspension of midlog phase organisms, R2866 and vacJ mutant, (OD620 0.5) was pelleted, washed twice with 1 ml of 10 mM carbonate buffer (pH 8.5), and then resuspended in 200 µl of buffer (10 mM carbonate, 1 M urea). Surface-exposed lysine residues were labeled with 10 µl of Cy5 (400 pmol, GE Healthcare) on ice in the dark for 20 min [41]. Reactions were stopped by addition of 20 µl of 10 mM lysine. The resulting pellets were washed twice with 500 µl of carbonate buffer, centrifuged, and resuspended in 200μl 1% BSA. Cy5 binding on bacteria was analyzed by flow cytometry with 10,000 events analyzed per sample.
Deposition of complement factor 3
R2866 and vacJ mutant were grown to an OD620 ∼0.5. The bacterial suspension (200 µl) was pelleted and resuspended in Hank's buffer with Ca2+ and Mg2+ (GIBCO, Auckland, New Zealand) with 5% fetal calf serum (HyClone) and incubated with 5μl of baby rabbit sera for 1°h. Bacteria were pelleted and resuspended in Hanks' buffer plus 5% fetal calf serum containing a 1 : 100 dilution of a FITC-conjugated polyclonal goat anti-rabbit C3 antibody (MP Biomedical Cappel, Irvine, CA) for 60 min at 4°C in dark and analyzed by flow cytometry.
Quantitative real-time RT-PCR
Total cellular RNA was extracted from mid-log phase grown NTHi clinical isolates by using the RNeasy mini Kit (QIAGEN). To eliminate genomic DNA, samples were incubated with 20 U of RNase-free DNase (QIAGEN) for 20 min at 25°C using the RNeasy columns, according to the manufacturer's instructions. 1 to 1.5 µg RNA was used for reverse transcription in a 20 µl reaction with the high-capacity cDNA reverse transcription kit (Applied Biosystems) together with random primers and 20 U RNase inhibitor (Promega). 1 µl of cDNA from this reaction was used as template with 0.5 µM primers (vacJ-F: 5′-TCCGTGGGCATTAGTGAAAT-3′; vacJ-R:5′-AATTCTGCATTATTGAGATTTTTCG-3′; yrbD-F:5′-TACTGTGACGGCAACTTTCG-3′; yrbD-R:5′-AATCGCGATGCTTACTTTCG-3′; yrbE-F:5′-TCGTGTTAATCGATTTTTCTGC-3′; yrbE-R:5′ - CAGGGCCTAATTCTCGTAAAAG-3′) and SYBR Green PCR Master Mix in a 20-μl reaction (Applied Biosystems). Standard runs of the reactions on fast optical 96-well reaction plates (Applied Biosystems) were carried out using the StepOnePlus Real-Time PCR system (Applied Biosystems). The gyrA gene (primers gyr-F: 5′-GCGTGTTGTGGGTGATGTAA-3′; and gyr-R: 5′ - GTTGTGCCATACGAACGATG-3′) was used as the internal standard gene for RNA quantity normalization [42]. Quantitative comparison was obtained through the ΔΔCT method as described at http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_041435.pdf.
Antibody binding assays
200 µl of mid-logarithmic-phase bacterial cells (OD620 0.5) were pelleted and resuspended in 200 µl of Hanks' buffer without Ca2+ and Mg2+ (Gibco) supplemented with 5% fetal calf serum (HyClone). Primary antibodies (1 : 200 for heat inactivated NHS, 1 : 50 for mAb 4C4, 1 : 1000 for mAb TEPC-15, and 1 : 100 for mAb 7B11) were added to reaction and then incubated at 37°C for 60 min. Bacteria were pelleted and incubated with Hanks' buffer without Ca2+ and Mg2+ plus 5% fetal calf serum containing a 1 : 200 dilution of the appropriate secondary antibody; goat anti-human IgG-FITC conjugate (Sigma), goat anti-human IgM FITC conjugate (Sigma), anti-mouse IgG-FITC conjugate (Sigma), anti-mouse IgA-FITC conjugate (Sigma) for 60 min at 4°C in the dark. Reaction mixtures were then washed and resuspended in 200 µl of PBS containing 1% bovine serum albumin and 0.5% paraformaldehyde. A total of 50,000 cells were collected for each sample. All samples were subjected in full volume to flow cytometry analysis on a BD FACS Calibur flow cytometer (BD Biosciences), and groups were compared using FlowJo software (Tree Star).
Growth inhibition assays
Ethylenediaminetetraacetate (EDTA) was added to 500 µl of an overnight culture in sBHI broth (25 mM final concentration) and incubated at 37°C for 4 h under aeration prior to obtaining viable counts. The minimum inhibitory concentration of vancomycin, novobiocin, bacitracin, polymyxin-B and deoxycholate was determined by broth microdilution method. Briefly, 96-well plates containing 2-fold dilution of chemical were prepared in sBHI and wells were inoculated with ∼5×105 cfu/ml of overnight bacterial cultures. Viable counts were determined after incubation for 18–24 h at 37°C.
NPN uptake assay
1-N-phenylnaphthylamine (NPN) (Sigma) uptake assay was performed as described previously [43]. Briefly, bacteria were grown to OD620 0.5 in sBHI broth. Pellets were washed and resuspended in 5 mmol/L HEPES buffer (pH 7.2). NPN was dissolved in acetone for a 500 µmol/L stock solution and diluted to 40 µmol/L in HEPES buffer. The NPN solution (50 µl) was added after 15 s to the bacterial suspension (150 µl) for a final NPN concentration of 10 µmol/L. Fluorescence was monitored for a total of 90 s using the PTi fluorescence system (Photon Technology International), with excitation at 350 nm, emission at 420 nm, and slit width of 2 nm.
Phospholipase C treatment
Phospholipase C treatment and thin layer chromatography (TLC) were performed as described previously [44]. 1 L of each bacterial culture were grown in sBHI broth to OD620 0.5 and centrifuged ×5000 g for 20 min. Pellets were washed once and resuspended in a sucrose-PBS-MgCl2 buffer [0.4M sucrose, 1×PBS, 15 mM MgCl2, (pH 7.5)] to a final volume of 1.8 ml. 200 µl aliquots of bacterial suspension were treated with 5u of phospholipase C from Bacillus cereus (Sigma) for 20 min at 37°C. Reactions were stopped by adding 600 µl CHCl3-methanol 1 : 2 (v/v). The lipids were extracted by the method of Bligh and Dyer [45]. Lipid samples were normalized to protein content and separated by TLC on glass backed silica gel 60 plates (Merck) with CHCl3-Acetone 94 : 4 (v/v) solvent system and were stained by iodine. The standard was 10 µg of 1,2-diglycerides from pig liver (Serdary Research Lab.).
Statistical analysis
All data were analyzed using StatView software (Abacus Concepts, Cary, NC). The significance of differences between or among groups was examined using ANOVA followed by Tukey or Dunnett post-tests.
Zdroje
1. MukundanD
EcevitZ
PatelM
MarrsC
GilsdorfJ
2007 Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J Clin Microbiol 45 3207 3217
2. MurphyT
2003 Respiratory infections caused by non-typeable Haemophilus influenzae. Curr Opin Infect Dis 16 129 134
3. TurkDC
1984 The pathogenicity of Haemophilus influenzae. J Med Microbiol 18 1 16
4. SethiS
EvansN
GrantBJ
MurphyTF
2002 New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med 347 465 471
5. BandiV
ApicellaMA
MasonE
MurphyTF
SiddiqiA
2001 Nontypeable Haemophilus influenzae in the lower respiratory tract of patients with chronic bronchitis. Am J Respir Crit Care Med 164 2114 2119
6. PauwelsR
BuistA
CalverleyP
JenkinsC
HurdS
2001 Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med 163 1256 1276
7. MurrayC
LopezA
1997 Alternative projections of mortality and disability by cause 1990-2020: global burden of disease study. Lancet 349 1498 1504
8. NikaidoH
2003 Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67 593 656
9. JiaW
El ZoeibyA
PetruzzielloTN
JayabalasinghamB
SeyedirashtiS
2004 Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem 279 44966 44975
10. KamioY
NikaidoH
1976 Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15 2561 2570
11. SwordsW
JonesP
ApicellaM
2003 The lipo-oligosaccharides of Haemophilus influenzae: an interesting array of characters. J Endotoxin Res 9 131 134
12. SchwedaE
RichardsJ
HoodD
MoxonE
2007 Expression and structural diversity of the lipopolysaccharide of Haemophilus influenzae: implication in virulence. Int J Med Microbiol 297 297 306
13. WeiserJN
PanN
1998 Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol Microbiol 30 767 775
14. ErwinAL
AllenS
HoDK
BonthuisPJ
JarischJ
2006 Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum. Infect Immun 74 6226 6235
15. HoDK
RamS
NelsonKL
BonthuisPJ
SmithAL
2007 lgtC expression modulates resistance to C4b deposition on an invasive nontypeable Haemophilus influenzae. J Immunol 178 1002 1012
16. FigueiraM
RamS
GoldsteinR
HoodD
MoxonE
2007 Role of complement in defense of the middle ear revealed by restoring the virulence of nontypeable Haemophilus influenzae siaB mutants. Infect Immun 75 325 333
17. WeiserJN
LoveJM
MoxonER
1989 The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell 59 657 665
18. WilliamsBJ
MorlinG
ValentineN
SmithAL
2001 Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect Immun 69 695 705
19. HoodD
DeadmanM
AllenT
MasoudH
MartinA
1996 Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol Microbiol 22 951 965
20. MalinverniJC
SilhavyTJ
2009 An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 106 8009 8014
21. HaaseE
CampagnariA
SarwarJ
SheroM
WirthM
1991 Strain specific and immunodominant surface epitopes of the P2 porin protein of nontypeable Haemophilus influenzae. Infect Immun 59 1278 1284
22. OchsenbeinAF
FehrT
LutzC
SuterM
BrombacherF
1999 Control of early viral and bacterial distribution and disease by natural antibodies. Science 286 2156 2159
23. FadenH
DuffyL
WilliamsA
KrystofikD
WolfJ
1996 Epidemiology of nasopharyngeal colonization with nontypeable Haemophilus influenzae in the first two years of life. Acta Otolaryngol Suppl 523 128 129
24. OksenhendlerE
GérardL
FieschiC
MalphettesM
MouillotG
2008 Infections in 252 patients with common variable immunodeficiency. Clin Infect Dis 46 1547 1554
25. ErwinA
BrewahY
CouchenourD
BarrenP
BurkeS
2000 Role of lipopolysaccharide phase variation in susceptibility of Haemophilus influenzae to bactericidal immunoglobulin M antibodies in rabbit sera. Infect Immun 68 2804 2807
26. ZolaTA
LysenkoES
WeiserJN
2009 Natural antibody to conserved targets of Haemophilus influenzae limits colonization of the murine nasopharynx. Infect Immun 77 3458 3465
27. ReidR
ProdeusA
KhanW
HsuT
RosenF
1997 Endotoxin shock in antibody-deficient mice: unraveling the role of natural antibody and complement in the clearance of lipopolysaccharide. J Immunol 159 970 975
28. BoesM
ProdeusA
SchmidtT
CarrollM
ChenJ
1998 A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. J Exp Med 188 2381 2386
29. SuzukiT
MuraiT
FukudaI
TobeT
YoshikawaM
1994 Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol 11 31 41
30. NikaidoH
VaaraM
1985 Molecular Basis of Bacterial Outer Menbrane Permeability. Microbiol Rev 49 1 32
31. St SauverJ
MarrsC
FoxmanB
SomselP
MaderaR
2000 Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg Infect Dis 6 622 630
32. FarjoR
FoxmanB
PatelM
ZhangL
PettigrewM
2004 Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr Infect Dis J 23 41 46
33. McCreaK
XieJ
LaCrossN
PatelM
MukundanM
2008 Relationships of non-typeable Haemophilus influenzae with hemolytic and non-hemolytic Haemophilus haemolyticus. J Clin Microbiol 46 406 416
34. LampeDJ
ChurchillME
RobertsonHM
1996 A purified mariner transposase is sufficient to mediate transposition in vitro. Embo J 15 5470 5479
35. MartinB
PrudhommeM
AlloingG
GranadelC
ClaverysJP
2000 Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol 38 867 878
36. AkerleyB
RubinE
CamilliA
LampeD
RobertsonH
1998 Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci U S A 95 8927 8932
37. HerriottRM
MeyerEM
VogtM
1970 Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol 101 517 524
38. MatsushitaM
OkadaH
1986 Alternative complement pathway activation by C4b deposited during classical pathway activation. J Immunol 136 2994 2998
39. WeiserJN
ShchepetovM
ChongST
1997 Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect Immun 65 943 950
40. GuligPA
PatrickCC
HermanstorferL
McCrackenGHJr
HansenEJ
1987 Conservation of epitopes in the oligosaccharide portion of the lipooligosaccharide of Haemophilus influenzae type b. Infect Immun 55 513 520
41. KhemiriA
GallandA
VaudryD
Chan Tchi SongP
VaudryH
2008 Outer-membrane proteomic maps and surface-exposed proteins of Legionella pneumophila using cellular fractionation and fluorescent labelling. Anal Bioanal Chem 390 1861 1871
42. GiufreM
CarattoliA
CardinesR
MastrantonioP
CerquettiM
2008 Variation in expression of HMW1 and HMW2 adhesins in invasive nontypeable Haemophilus influenzae isolates. BMC Microbiol 8 83
43. Martinez de TejadaG
MorivónI
1993 The outer membranes of Brucella spp. are not barriers to hydrophobic permeants. J Bacteriol 175 5273 5275
44. GoldfineH
JohnstonN
BishopD
1982 Ether phospholipid asymmetry in Clostridium butyricum. Biochem Biophys Res Commun 108 1502 1507
45. BlighE
DyerW
1959 A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37 911 917
46. FleischmannRD
AdamsMD
WhiteO
ClaytonRA
KirknessEF
1995 Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269 496 512
47. PhillipsNJ
ApicellaMA
GriffissJM
GibsonBW
1992 Structural characterization of the cell surface lipooligosaccharides from a nontypable strain of Haemophilus influenzae. Biochemistry 31 4515 4526
48. MerrellDS
HavaDL
CamilliA
2002 Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol 43 1471 1491
49. HavaDL
CamilliA
2002 Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45 1389 1406
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 1- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Salivary Gland NK Cells Are Phenotypically and Functionally Unique
- Genetic Epidemiology of Tuberculosis Susceptibility: Impact of Study Design
- Early Target Cells of Measles Virus after Aerosol Infection of Non-Human Primates
- Multiple Plant Surface Signals are Sensed by Different Mechanisms in the Rice Blast Fungus for Appressorium Formation
- Biofilm Development on by Is Facilitated by Quorum Sensing-Dependent Repression of Type III Secretion
- Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus
- Molecular Basis of Increased Serum Resistance among Pulmonary Isolates of Non-typeable
- A Helminth Immunomodulator Exploits Host Signaling Events to Regulate Cytokine Production in Macrophages
- HCMV Spread and Cell Tropism are Determined by Distinct Virus Populations
- Characteristics of the Earliest Cross-Neutralizing Antibody Response to HIV-1
- Induction of a Peptide with Activity against a Broad Spectrum of Pathogens in the Salivary Gland, following Infection with Dengue Virus
- Structural Basis for the Recognition of Cellular mRNA Export Factor REF by Herpes Viral Proteins HSV-1 ICP27 and HVS ORF57
- Identification and Characterization of the Host Protein DNAJC14 as a Broadly Active Flavivirus Replication Modulator
- Dual-Use Research and Technological Diffusion: Reconsidering the Bioterrorism Threat Spectrum
- Pathogenesis of the 1918 Pandemic Influenza Virus
- A Cardinal Role for Cathepsin D in Co-Ordinating the Host-Mediated Apoptosis of Macrophages and Killing of Pneumococci
- Critical Role of IRF-5 in the Development of T helper 1 responses to infection
- The Pel Polysaccharide Can Serve a Structural and Protective Role in the Biofilm Matrix of
- Selective C-Rel Activation via Malt1 Controls Anti-Fungal T-17 Immunity by Dectin-1 and Dectin-2
- Imaging Single Retrovirus Entry through Alternative Receptor Isoforms and Intermediates of Virus-Endosome Fusion
- Aerosols Transmit Prions to Immunocompetent and Immunodeficient Mice
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dual-Use Research and Technological Diffusion: Reconsidering the Bioterrorism Threat Spectrum
- Pathogenesis of the 1918 Pandemic Influenza Virus
- Critical Role of IRF-5 in the Development of T helper 1 responses to infection
- A Cardinal Role for Cathepsin D in Co-Ordinating the Host-Mediated Apoptosis of Macrophages and Killing of Pneumococci
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



