-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Leveraging Genetics to Advance Type 2 Diabetes Prevention
In this Perspective, Jose Florez discusses how information from genetics and genomics may be able to contribute to prevention of type 2 diabetes and predicting individual responses to behavioral and other interventions.
Published in the journal: . PLoS Med 13(7): e32767. doi:10.1371/journal.pmed.1002102
Category: Perspective
doi: https://doi.org/10.1371/journal.pmed.1002102Summary
In this Perspective, Jose Florez discusses how information from genetics and genomics may be able to contribute to prevention of type 2 diabetes and predicting individual responses to behavioral and other interventions.
Type 2 diabetes is a preventable disease. The pandemic that has reached 415 million people worldwide continues to grow, threatens to undermine the global economy, and is taking place on the back of a profound transformation in the modern lifestyle, which involves pervasive detrimental changes in diet and physical activity patterns [1]. Though the disease undoubtedly has a genetic component, the recent explosion in its prevalence is clearly driven by a dynamic environment operating on a relatively static genetic background. But this equation also has a favorable counterpoint: as a deleterious environment has driven diabetes prevalence into double percentage digits in just a few decades, so reverting this trend by the widespread introduction of healthy behavioral patterns should, in turn, reduce disease burden.
Lifestyle intervention strategies have been proven to be effective in randomized controlled trials. The Chinese Da Qing study [2], the Finnish Diabetes Prevention Study [3], the U.S. Diabetes Prevention Program (DPP) [4], and the Indian Diabetes Prevention Program [5] have consistently shown that intensive lifestyle modification is able to prevent, in some, and delay, in others, the incidence of diabetes, despite the high-risk metabolic profile of participants. Lifestyle interventions are successful across the full range of genetic risk (Fig 1) [6,7] and can also reduce cardiovascular mortality if given enough time [8]. However, many of the prediabetic participants continue to march toward diabetes, albeit at a slower pace [9], and deploying a successful lifestyle intervention across the entire population at risk (~52 million in North America) is a massive undertaking.
Fig. 1. An intensive lifestyle intervention, as deployed in the U.S. Diabetes Prevention Program (DPP), is effective regardless of genetic risk score for type 2 diabetes. 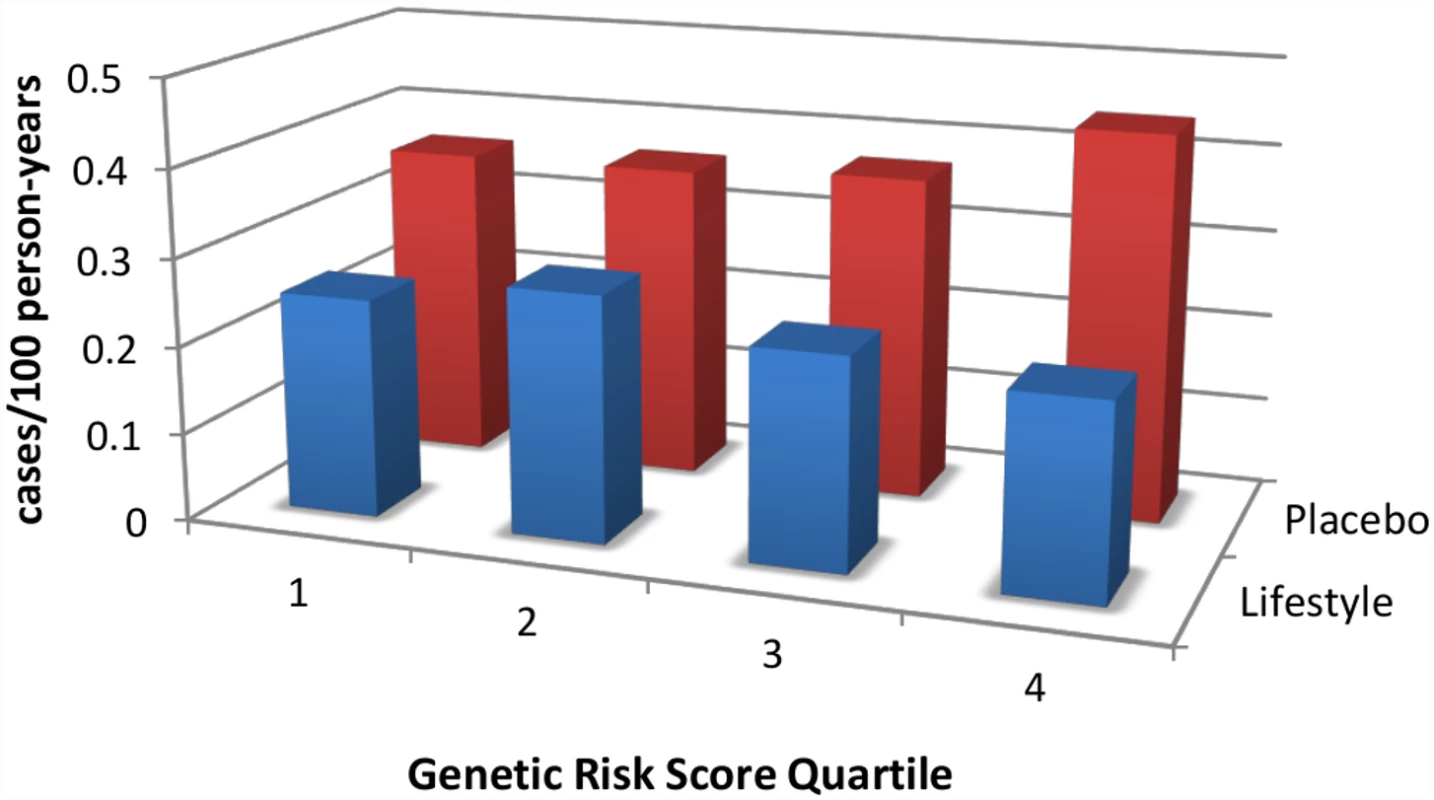
DPP participants were stratified by quartile of genetic risk constructed by adding risk alleles from 34 known type 2 diabetes-associated variants. Whereas the genetic risk score predicts diabetes incidence in the placebo arm (red bars), it does not do so in the lifestyle arm (blue bars); indeed, the intervention is highly effective in reducing diabetes incidence even in the quartile with the highest genetic risk. Data are from reference [11]. While logistically challenging, preventive strategies do make financial sense. An economic analysis of participants who adhered to the intervention in the DPP found that, over a ten-year period, intensive, individualized lifestyle modification costs less than US$5,000 per quality-adjusted life year [10], well within the expense range of other health interventions routinely embraced by high-income societies. Furthermore, preventive treatment with metformin was found to be cost-saving for those participants who adhered to treatment in the DPP study. However, these interventions would still involve a substantial investment, and governments are not necessarily in a position to provide such interventions to all their citizens at risk.
Identifying Individuals at Increased Risk
In a world of finite resources, it may make sense to prioritize those most likely to benefit. Therefore, great interest has emerged in whether nascent technologies can be used to identify individuals at risk of future type 2 diabetes and/or predisposed to experience a favorable response to preventive strategies.
Among all available approaches for more detailed personalized profiling, genetics has led the way for a number of reasons. First, it is now possible to query millions of variants across the human genome in a single experiment with great precision, and to interpret the ensuing results with appropriate statistical rigor. Second, germline genetic variation is fixed in the individual, and thus need be measured only once in the person’s lifetime. Third, DNA is easily accessible and does not require the biopsy of type 2 diabetes-relevant tissues such as beta cells, which would be required for many other biological modalities.
Advances in genotyping and sequencing, as well as plummeting costs and widespread international collaboration, have led to the identification of over 100 genetic variants associated with type 2 diabetes or related glycemic traits [12]. Most of them are common and shared across populations and, with a few exceptions [13,14], tend to have a small effect on risk (odds ratios 1.1–1.4; Fig 2). Combined genetic risk scores composed by the weighted sum of the risk alleles at these loci have been tested for their ability to predict diabetes in individuals, above and beyond the information provided by clinical risk factors. However, the available clinical prediction tools are good, they include variables (such as fasting glucose or body mass index) that already capture genetic information, and the combined effect of these genetic variants is modest; thus, genetic data do not seem to add much to clinical information in type 2 diabetes prediction [15–17]. The addition of metabolomic biomarkers only offers marginal additional benefit [18], and the incremental genetic information only reclassifies a few individuals into more accurate categories of risk. Genotype scores perform slightly better in younger subgroups or those with longer follow-up [15,16].
Fig. 2. Chronological listing of type 2 diabetes-associated loci, plotted by year of definitive publication and approximate effect size. 
By convention, the gene name closest to the index variant is given. Loci identified via the candidate gene approach are shown in red, loci identified via agnostic genome-wide association approaches are shown in light blue, loci identified by exome sequencing are shown in green, and loci identified by whole-genome sequencing are shown in purple. TCF7L2 (shown in dark blue) was discovered by dense fine-mapping under a linkage signal. TBC1D4 (shown in orange) was identified by exome sequencing of a locus found to be associated with a diabetes-related quantitative trait. Gene names that have asterisks (*) indicate initial discovery by association with quantitative glycemic traits; gene names that have a number sign (#) denote identification in population isolates. These results suggest that, if genetic prediction tools composed of common genetic variants were to be deployed to enhance prediction and target subgroups for preventive interventions, they would need to be combined with other risk factors that capture the global risk profile of an individual. As an illustration of this concept, Claudia Langenberg and colleagues used the EPIC-Interact study to demonstrate that, for individualized risk prediction, absolute risk, and not relative risk, is what matters. They stratified participants by their genetic risk scores as well as by their obesity categories (obese, overweight or normal weight). Obese individuals in the lowest genetic risk quartile were much more likely to develop type 2 diabetes than normal-weight individuals in the highest genetic risk quartile (Fig 3), indicating that if only genetics had been used for risk stratification, the individuals at highest risk would not have been targeted for intervention [19].
Fig. 3. The relative effect of genetic risk and obesity on future type 2 diabetes. 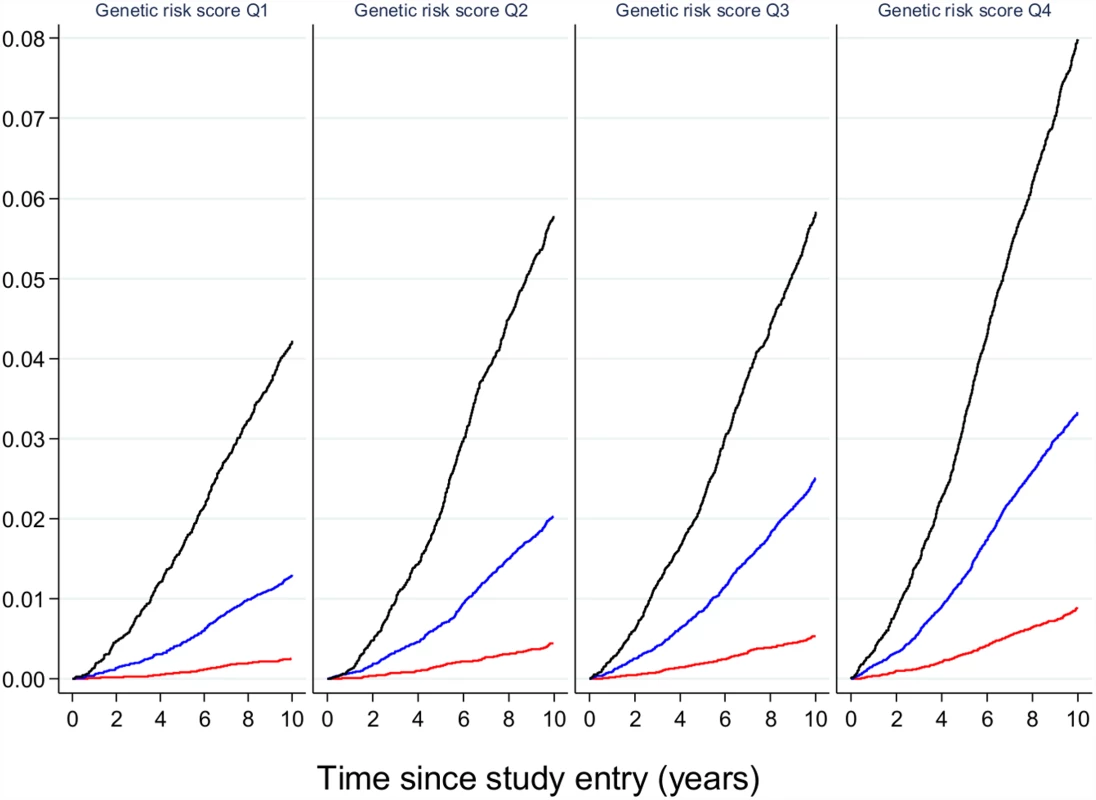
Participants in the EPIC-Interact study were stratified by quartiles of genetic risk and by strata of adiposity (obese in black, overweight in blue, and normal weight in red). The likelihood of developing type 2 diabetes is shown, indicating that obese participants with the lowest genetic burden have a higher absolute risk of diabetes than lean or overweight participants with the highest genetic burden. Image is from Langenberg et al. [19]. While this is the case for common genetic variants of small effects shared across populations, a potential exception might be made for variants that have much stronger effects. Explorations that target a lower frequency of the allelic spectrum have led to the identification of loci that are unique to a specific ethnic group but are present at appreciable frequencies in members of that group. For example, whole-exome sequencing in ~4,000 people of Latino descent uncovered the coding variant E508K in the gene HNF1A, which increases diabetes risk 5-fold [13]. Though complete loss of function in the transcription factor HNF1A causes maturity-onset diabetes of the young (MODY) type 3, the E508K allele only attenuates transcriptional activity by ~60%; as a result, the clinical phenotype of carriers is indistinguishable from regular type 2 diabetes, which would not alert the clinician to a mutation in a known MODY gene. The variant is carried by ~2% of Mexicans with type 2 diabetes, and, given its large impact on diabetes risk, it may help to target carriers (identified because they have a prediabetic phenotype, or they are relatives of a diabetic carrier, or they have undergone population screening when available) for more intensive surveillance and prevention. Similarly, Ida Moltke and colleagues identified a nonsense variant in the gene TBC1D4, which is present in 17% of Inuit individuals in Greenland [14]. Loss of function of its protein product interferes with insulin-stimulated Akt-induced glucose uptake in skeletal muscle, and the variant is thus associated with higher two-hour glucose and a 10-fold increased risk of type 2 diabetes. The clinician caring for this population would do well in using oral glucose tolerance tests to diagnose prediabetes and diabetes, to ensure that the 2-hour glucose measure is considered in diagnostics, and carriers might be better treated with an insulin sensitizer for either diabetes prevention or treatment.
Identifying Individuals More Likely to Respond to Intervention
While the value of genetic testing in type 2 diabetes prediction is limited, it may have a more useful role in the selection of patients more likely to respond to intervention. Though many of these observations require independent replication, the DPP Research Group has shown that variation in the obesity-associated gene MC4R modifies the ability of the lifestyle intervention to induce weight loss [20], the missense type 2 diabetes-associated variant P446L in GCKR modifies the effect of the intensive lifestyle intervention on triglyceride concentrations [21], and a genetic risk score for lipid traits also modifies the response to lifestyle intervention on LDL cholesterol concentrations and small LDL particle number [22]. A combined meta-analysis of the DPP (in prediabetic participants) and the Look AHEAD trial (in participants with established type 2 diabetes) has shown that genotype at the obesity-associated gene MTIF3 also predicts the degree of weight loss after a lifestyle intervention [23].
With regard to metformin, several genetic variants seem to modify metformin’s ability to reduce diabetes incidence [10]. One that has been shown to have an effect on metformin response in an independent cohort [24] is a polymorphism in the gene SLC47A1, which encodes the metformin transporter MATE1. The extent to which genetic loci associated with glycemic response in patients with established type 2 diabetes will exert a similar effect on diabetes prevention awaits more detailed analyses, although preliminary evidence for the top locus unearthed in a genome-wide association study of metformin response [25] indicates that the two traits may not have perfectly overlapping genetic architectures [26]. That is to say, the genetic variants that modify glycemic response to metformin in a person with established diabetes may not necessarily influence metformin’s ability to prevent diabetes and vice versa, in that the two processes may act via related but separate molecular pathways at different stages of disease progression.
Conclusion
In summary, while type 2 diabetes represents one of the most serious threats to global public health in the 21st century, strategies exist to stem its spread. To rationalize deployment of diabetes prevention strategies in a cost-efficient manner, it may help to stratify the population into groups at highest risk or most likely to benefit. Genetic prediction does not seem to provide much additional information beyond traditional clinical predictors in identifying those at increased risk, with the potential exception of some variants with strong effects that are more prevalent in specific ethnic groups; thus, if included, genetic predictors should always be considered in conjunction with other markers to obtain an overall estimate of risk. Whether the suggestive evidence that genetic predictors may help stratify response to preventive interventions is eventually translated into clinical practice awaits the completion of well-powered clinical trials.
Zdroje
1. International Diabetes Federation. IDF Diabetes Atlas. 7 ed: International Diabetes Federation; 2015.
2. Pan XR, Li GW, Hu YH, Wang JX, Yang WY, An ZX, et al. Effects of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care. 1997;20(4):537–544. 9096977.
3. Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344(18):1343–1350. 11333990.
4. The Diabetes Prevention Program Research Group. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346(6):393–403. 11832527.
5. Ramachandran A, Snehalatha C, Mary S, Mukesh B, Bhaskar AD, Vijay V. The Indian Diabetes Prevention Programme shows that lifestyle modification and metformin prevent type 2 diabetes in Asian Indian subjects with impaired glucose tolerance (IDPP-1). Diabetologia. 2006;49(2):289–297. 16391903.
6. Hivert MF, Jablonski KA, Perreault L, Saxena R, McAteer JB, Franks PW, et al. Updated genetic score based on 34 confirmed type 2 diabetes loci is associated with diabetes incidence and regression to normoglycemia in the Diabetes Prevention Program. Diabetes. 2011;60(4):1340–1348. 21378175; PubMed Central PMCID: PMC3064108. doi: 10.2337/db10-1119.
7. Hivert MF, Christophi CA, Franks PW, Jablonski KA, Ehrmann DA, Kahn SE, et al. Lifestyle and metformin ameliorate insulin sensitivity independently of the genetic burden of established insulin resistance variants in Diabetes Prevention Program participants. Diabetes. 2016;65(2):520–526. Epub 2015/11/04. doi: 10.2337/db15-0950 26525880; PubMed Central PMCID: PMC4747453.
8. Li G, Zhang P, Wang J, An Y, Gong Q, Gregg EW, et al. Cardiovascular mortality, all-cause mortality, and diabetes incidence after lifestyle intervention for people with impaired glucose tolerance in the Da Qing Diabetes Prevention Study: a 23-year follow-up study. Lancet Diabetes Endocrinol. 2014;2(6):474–480. Epub 2014/04/16. doi: 10.1016/s2213-8587(14)70057-9 24731674.
9. Knowler WC, Fowler SE, Hamman RF, Christophi CA, Hoffman HJ, Brenneman AT, et al. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet. 2009;374(9702):1677–1686. Epub 2009/11/03. doi: 10.1016/s0140-6736(09)61457-4 19878986; PubMed Central PMCID: PMC3135022.
10. Herman WH, Edelstein SL, Ratner RE, Montez MG, Ackermann RT, Orchard TJ, et al. Effectiveness and cost-effectiveness of diabetes prevention among adherent participants. Am J Manag Care. 2013;19(3):194–202. Epub 2013/04/03. 23544761; PubMed Central PMCID: PMC3985133.
11. Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, Hanson RL, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the Diabetes Prevention Program. Diabetes. 2010;59(10):2672–2681. Epub 2010/08/05. doi: 10.2337/db10-0543 20682687; PubMed Central PMCID: PMC3279522.
12. Mohlke KL, Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum Mol Genet. 2015;24(R1):R85–92. doi: 10.1093/hmg/ddv264 26160912; PubMed Central PMCID: PMC4572004.
13. Estrada K, Aukrust I, Bjorkhaug L, Burtt NP, Mercader JM, Garcia-Ortiz H, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311(22):2305–2314. Epub 2014/06/11. doi: 10.1001/jama.2014.6511 24915262; PubMed Central PMCID: PMC4425850.
14. Moltke I, Grarup N, Jorgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512(7513):190–193. doi: 10.1038/nature13425 25043022.
15. Meigs JB, Shrader P, Sullivan LM, McAteer JB, Fox CS, Dupuis J, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359(21):2208–2219. 19020323; PubMed Central PMCID: PMC2746946. doi: 10.1056/NEJMoa0804742.
16. Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, Tuomi T, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359(21):2220–2232. 19020324. doi: 10.1056/NEJMoa0801869.
17. Vassy JL, Hivert MF, Porneala B, Dauriz M, Florez JC, Dupuis J, et al. Polygenic type 2 diabetes prediction at the limit of common variant detection. Diabetes. 2014;63(6):2172–2182. Epub 2014/02/13. doi: 10.2337/db13-1663 24520119; PubMed Central PMCID: PMC4030114.
18. Walford GA, Porneala BC, Dauriz M, Vassy JL, Cheng S, Rhee EP, et al. Metabolite traits and genetic risk provide complementary information for the prediction of future type 2 diabetes. Diabetes Care. 2014;37(9):2508–2514. Epub 2014/06/21. doi: 10.2337/dc14-0560 24947790; PubMed Central PMCID: PMC4140156.
19. Langenberg C, Sharp SJ, Franks PW, Scott RA, Deloukas P, Forouhi NG, et al. Gene-lifestyle interaction and type 2 diabetes: the EPIC interact case-cohort study. PLoS Med. 2014;11(5):e1001647. Epub 2014/05/23. doi: 10.1371/journal.pmed.1001647 24845081; PubMed Central PMCID: PMC4028183.
20. Pan Q, Delahanty LM, Jablonski KA, Knowler WC, Kahn SE, Florez JC, et al. Variation at the melanocortin 4 receptor gene and response to weight-loss interventions in the Diabetes Prevention Program. Obesity (Silver Spring). 2013;21(9):E520–526. doi: 10.1002/oby.20459 23512951.
21. Pollin TI, Jablonski KA, McAteer JB, Saxena R, Kathiresan S, Kahn SE, et al. Triglyceride response to an intensive lifestyle intervention is enhanced in carriers of the GCKR Pro446Leu polymorphism. J Clin Endocrinol Metab. 2011;96(7):E1142–E1147. 21525158; PubMed Central PMCID: PMC3205512. doi: 10.1210/jc.2010-2324.
22. Pollin TI, Isakova T, Jablonski KA, de Bakker PI, Taylor A, McAteer J, et al. Genetic modulation of lipid profiles following lifestyle modification or metformin treatment: The Diabetes Prevention Program. PLoS Genet. 2012;8(8):e1002895. 22951888; PubMed Central PMCID: PMC3431328.
23. Papandonatos GD, Pan Q, Pajewski NM, Delahanty LM, Peter I, Erar B, et al. Genetic predisposition to weight loss and regain with lifestyle intervention: Analyses from the Diabetes Prevention Program and the Look AHEAD randomized controlled trials. Diabetes. 2015;64(12):4312–4321. Epub 2015/08/09. doi: 10.2337/db15-0441 26253612; PubMed Central PMCID: PMC4657576.
24. Becker ML, Visser LE, van Schaik RHN, Hofman A, Uitterlinden AG, Stricker BHC. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: A preliminary study. Diabetes. 2009;58(3):745–749. doi: 10.2337/db08-1028 19228809.
25. Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, Tavendale R, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet. 2011;43(2):117–120. Epub 2010/12/28. ng.735 [pii] doi: 10.1038/ng.735 21186350; PubMed Central PMCID: PMC3030919.
26. Florez JC, Jablonski KA, Taylor A, Mather K, Horton E, White NH, et al. The C allele of ATM rs11212617 does not associate with metformin response in the Diabetes Prevention Program. Diabetes Care. 2012;35(9):1864–1867. 22751958; PubMed Central PMCID: PMC3425006.
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2016 Číslo 7- Není statin jako statin aneb praktický přehled rozdílů jednotlivých molekul
- Moje zkušenosti s Magnosolvem podávaným pacientům jako profylaxe migrény a u pacientů s diagnostikovanou spazmofilní tetanií i při normomagnezémii - MUDr. Dana Pecharová, neurolog
- Magnosolv a jeho využití v neurologii
- S prof. Vladimírem Paličkou o racionální suplementaci kalcia a vitaminu D v každodenní praxi
- Biomarker NT-proBNP má v praxi široké využití. Usnadněte si jeho vyšetření POCT analyzátorem Afias 1
-
Všechny články tohoto čísla
- The Clinical and Public Health Challenges of Diabetes Prevention: A Search for Sustainable Solutions
- Screening for Dysglycemia: Connecting Supply and Demand to Slow Growth in Diabetes Incidence
- Diabetes: A Cinderella Subject We Can’t Afford to Ignore
- Prescribing Exercise and Lifestyle Training for High Risk Women in Pregnancy and Early Post-partum—Is It Worth It?
- Population Approaches to Prevention of Type 2 Diabetes
- First-Year Evaluation of Mexico’s Tax on Nonessential Energy-Dense Foods: An Observational Study
- Effects of Saturated Fat, Polyunsaturated Fat, Monounsaturated Fat, and Carbohydrate on Glucose-Insulin Homeostasis: A Systematic Review and Meta-analysis of Randomised Controlled Feeding Trials
- Leveraging Genetics to Advance Type 2 Diabetes Prevention
- Cardiometabolic Risk Factor Changes Observed in Diabetes Prevention Programs in US Settings: A Systematic Review and Meta-analysis
- Risks and Population Burden of Cardiovascular Diseases Associated with Diabetes in China: A Prospective Study of 0.5 Million Adults
- Dietary Diversity, Diet Cost, and Incidence of Type 2 Diabetes in the United Kingdom: A Prospective Cohort Study
- Detecting Dysglycemia Using the 2015 United States Preventive Services Task Force Screening Criteria: A Cohort Analysis of Community Health Center Patients
- Exercise Training and Weight Gain in Obese Pregnant Women: A Randomized Controlled Trial (ETIP Trial)
- Associations between Recreational and Commuter Cycling, Changes in Cycling, and Type 2 Diabetes Risk: A Cohort Study of Danish Men and Women
- Association of Plasma Phospholipid n-3 and n-6 Polyunsaturated Fatty Acids with Type 2 Diabetes: The EPIC-InterAct Case-Cohort Study
- Engagement, Retention, and Progression to Type 2 Diabetes: A Retrospective Analysis of the Cluster-Randomised "Let's Prevent Diabetes" Trial
- Obesity and Life Expectancy with and without Diabetes in Adults Aged 55 Years and Older in the Netherlands: A Prospective Cohort Study
- Supported Telemonitoring and Glycemic Control in People with Type 2 Diabetes: The Telescot Diabetes Pragmatic Multicenter Randomized Controlled Trial
- Mothers after Gestational Diabetes in Australia (MAGDA): A Randomised Controlled Trial of a Postnatal Diabetes Prevention Program
- Cycling and Diabetes Prevention: Practice-Based Evidence for Public Health Action
- Consumption of Meals Prepared at Home and Risk of Type 2 Diabetes: An Analysis of Two Prospective Cohort Studies
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Mothers after Gestational Diabetes in Australia (MAGDA): A Randomised Controlled Trial of a Postnatal Diabetes Prevention Program
- Consumption of Meals Prepared at Home and Risk of Type 2 Diabetes: An Analysis of Two Prospective Cohort Studies
- Obesity and Life Expectancy with and without Diabetes in Adults Aged 55 Years and Older in the Netherlands: A Prospective Cohort Study
- Effects of Saturated Fat, Polyunsaturated Fat, Monounsaturated Fat, and Carbohydrate on Glucose-Insulin Homeostasis: A Systematic Review and Meta-analysis of Randomised Controlled Feeding Trials
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



