-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural Competence
A critical step in building a functional nervous system is to generate neurons at the appropriate locations. Neural competence is acquired at the precursor stage with the expression of specific transcription factors. One such critical factor is Senseless (Sens), as precursors lacking Sens fail to develop to neurons. Here we describe the critical role of protein complex COP9 signalosome (CSN) that regulates Sens expression by integrating temporal and spatial information. This was studied in developing Drosophila wing tissues, in which the anterior wing margin develops neuron-innervated bristles, while the posterior wing margin develops non-innervated bristles. The CSN complex is required for the anterior-posterior difference in spatial patterning of neuron formation, and posterior cells lacking CSN develop innervated bristles like anterior cells. CSN accomplishes this by transforming the temporal hormonal ecdysone signaling from activation to repression of downstream target BR-Z1. As BR-Z1 itself is a transcription activator, repression of BR-Z1 in turn leads to repression of Sens in posterior wing margin, eventually terminating the neural competence. Repression of BR-Z1 expression requires the interaction between the CSN complex and the ecdysone receptors. Our results suggest a novel CSN-mediated regulation that converts temporal hormone signaling to the patterning of neurons at the right place.
Published in the journal: . PLoS Genet 10(11): e32767. doi:10.1371/journal.pgen.1004760
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004760Summary
A critical step in building a functional nervous system is to generate neurons at the appropriate locations. Neural competence is acquired at the precursor stage with the expression of specific transcription factors. One such critical factor is Senseless (Sens), as precursors lacking Sens fail to develop to neurons. Here we describe the critical role of protein complex COP9 signalosome (CSN) that regulates Sens expression by integrating temporal and spatial information. This was studied in developing Drosophila wing tissues, in which the anterior wing margin develops neuron-innervated bristles, while the posterior wing margin develops non-innervated bristles. The CSN complex is required for the anterior-posterior difference in spatial patterning of neuron formation, and posterior cells lacking CSN develop innervated bristles like anterior cells. CSN accomplishes this by transforming the temporal hormonal ecdysone signaling from activation to repression of downstream target BR-Z1. As BR-Z1 itself is a transcription activator, repression of BR-Z1 in turn leads to repression of Sens in posterior wing margin, eventually terminating the neural competence. Repression of BR-Z1 expression requires the interaction between the CSN complex and the ecdysone receptors. Our results suggest a novel CSN-mediated regulation that converts temporal hormone signaling to the patterning of neurons at the right place.
Introduction
Neural specification generates diverse neural cells that are located at exact positions necessary for specialized functions. It is well established that positional cues, such as extrinsic signals and intrinsic tissue-specific transcriptional factors, play key roles in neural specification and neuronal patterning. In Drosophila, the sensory bristles occupy stereotypical positions on the head, notum, abdomen, legs and anterior wing margins (AWMs). Patterning of sensory bristles is primarily defined by the spatially restricted expressions of bHLH proneural proteins Achaete (Ac) and Scute (Sc). High-level Ac and Sc expressions in the presumptive sensory organ precursors (SOPs) activate genetic programs for further specification and differentiation, leading to the generation of sensory bristles comprising neuron, sheath, hair, and socket cells [1], [2]. A key target of the proneural proteins, the Zinc finger protein Senseless (Sens), is turned on in smaller subsets of proneural cells, and the expression levels are further elevated in SOPs and SOP lineage cells [3]. Sens is required for the specifications of SOPs and SOP daughter cells pIIa [3], [4]. Misexpression of Sens in epithelial cells is sufficient to induce sensory organ formation in a process bypassing the requirement of proneural proteins, indicating that Sens plays a key role in sensory organ formation [4].
In addition to spatially restricted expression of transcription factors, specification of SOPs also depends on steroid hormonal ecdysone (20-hydroxyecdysone, 20E) signaling. For example, ecdysone signaling is required for the specification of chemosensory bristles at the AWM at late third instar larval stages [5]. Pulses of ecdysones set temporal boundaries in developmental processes like inducing molting and metamorphosis [6]–[10]. Ecdysone signaling is mediated through the heterodimeric complex between the ecdysone receptor EcR and the RXR ortholog Ultraspiracle (USP) that binds to the ecdysone response element (EcRE) [11], [12]. Ligand binding transforms the EcR/USP complex from a repressed to active state, leading to transcriptional activation of a hierarchy of target genes. Among them, broad (br) is the key early gene that mediates ecdysone signaling in the induction of metamorphosis. The br gene encodes four isoforms BR-Z1 to BR-Z4 through alternative splicing [13], [14]. In br mutants, responses of a large set of early and late genes to ecdysone signaling are abrogated [15], [16], and mutants die as wandering larvae unable to initiate puparium formation [17], [18]. Dysregulation of br also disrupts sensory neuron differentiation and suppresses adult abdominal cuticle formation [5], [19]. Therefore, tight regulation of br expression is crucial for development of various insect tissues.
The COP9 signalosome (CSN) is a highly conserved protein complex initially identified in Arabidopsis for suppression of photomorphogenesis [20]–[22]. Subsequent identification and characterization in mammalian cells, insects and yeast reveal that the CSN complex participates in diverse cellular and developmental processes (for reviews, see references [23] and [24]). The major CSN function is to deconjugate the ubiquitin-like peptide Nedd8 from cullins, the scaffolding proteins in cullin-RING ubiquitin ligases (CRLs) [25]. CSN-mediated deneddylation of cullins inactivates ubiquitin ligase activity and protects CRLs from turnover. Thus, cycling between neddylation and deneddylation maintains the physiological CRL activity [26], [27]. Additional CSN-associated activities such as deubiquitination and phosphorylation are linked to the maintenance of target protein stability [28]–[30].
Studies in Drosophila have revealed several CSN functions in development and adult physiology. Transcriptome analysis has shown a role for CSN4 and CSN5, two subunits of the CSN complex, in transcriptional repression of developmentally regulated genes [31]. CSN4 mutants show defect in larval molting while CSN5 mutations induce melanotic formation at larvae stages [32]. The CSN complex regulates protein degradation of CycE and Timeless during oogenesis and circadian rhythm, respectively [33], [34]. By regulating Cul1 and Cul3, the CSN complex has dual functions in dendrite morphogenesis [35], [36]. Interestingly, the CSN activity confers specifically the intermediate response in graded Hedgehog signaling [37]. In an RNAi-based screening to identify genes involved in Notch signaling, CSN subunits were identified to be essential in binary cell fate determination in sensory organ development [38].
The bristles along the anterior and posterior wing margin succumb to different fates; AWM bristles are innervated by neurons beneath the cuticle while posterior wing margin (PWM) bristles are non-innervated. Both types of bristles require transcription factors Sens and the bHLH protein Daughterless for precursor cell selection during third instar larval and prepupal stages [4]. However, it is still unclear how the neural competence of PWM bristles is suppressed to prevent formation of neurons and associated neural cells. We have identified a role of the CSN complex in inhibition of the neural competence of non-innervated bristles through Sens downregulation at the onset of pupal development. Activation of ecdysone signaling at the prepupa-to-pupa transition is required for pupal BR-Z1 suppression, which leads to Sens suppression. The CSN complex and ecdysone receptors act in the same genetic pathway and form protein complexes to block BR-Z1 expression in pupal wings. Thus, our results demonstrated that the CSN complex acts as a nexus to convert temporal hormone stimulation to spatial restriction of neural competence.
Results
Transformation of PWM bristles into innervated sensory bristles in CSN mutants
Innervated sensory bristles, including stout and slender mechanosensory and spaced chemosensory bristles, are located along the AWM of wild-type wings. These sensory bristles grow dome-shaped sockets at the base (Figure 1A), and are innervated by neurons [39]. Non-innervated bristles along the PWM, however, display only thin hairs without sockets (Figure 1B). In CSN4null and CSN5null mutant clones, PWM bristles adopted morphological characteristics of innervated sensory bristles. Mutant bristles had thicker hairs surrounded by sockets (arrowheads in Figure 1C, 1D). Some bristles had double hairs emerging from one large socket (arrows), reminiscent of the double hair/double socket phenotype caused by fate transformation of SOP lineage cells [40], [41]. Similar morphological changes of PWM bristles were also observed in wings expressing interference RNA (RNAi) by en-GAL4 to knockdown CSN1b, CSN2, CSN3, CSN6 and CSN7 in the posterior compartment (Figure 1F–H, and Figure S1A, S1B), suggesting that these CSN subunits function together as a holoenzyme to suppress formation of innervated bristles at the PWM.
Fig. 1. Transformation from non-innervated to innervated PWM bristles in CSN mutants. 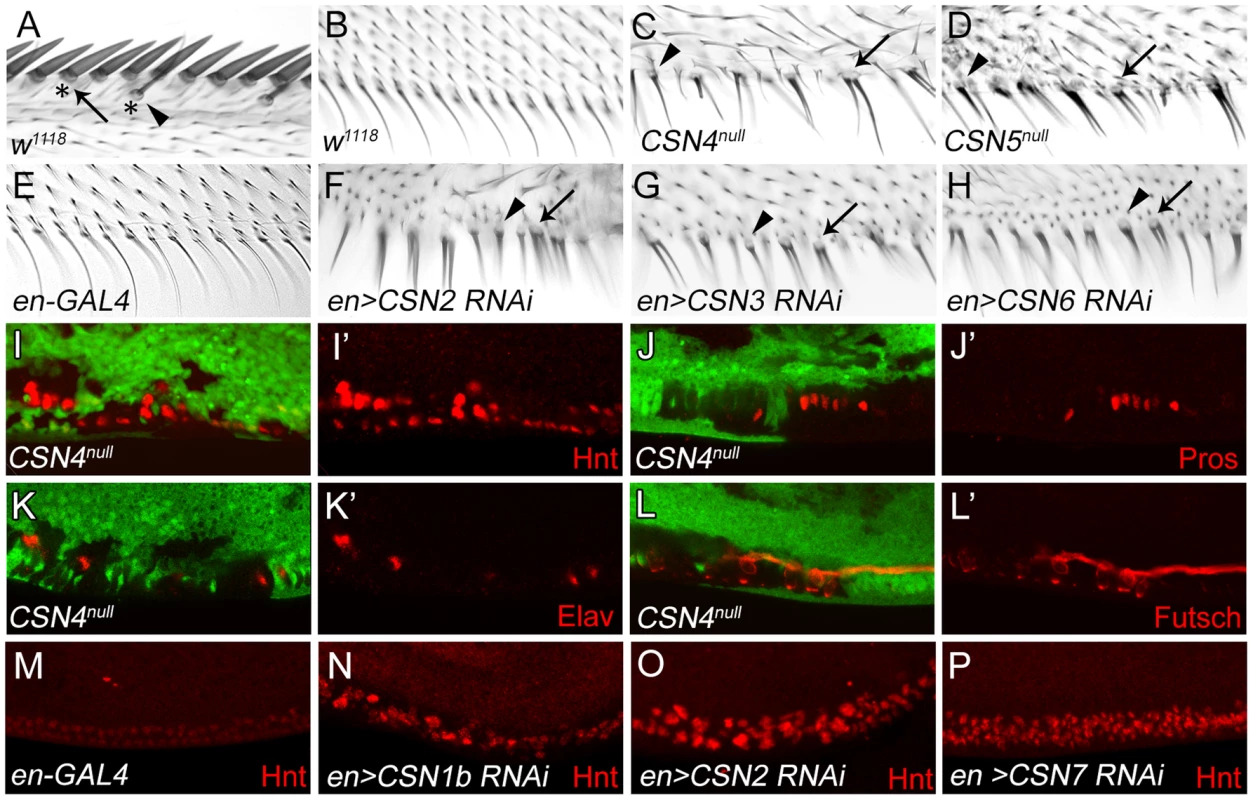
(A) AWM innervated bristles. Arrow and arrowhead indicate stout mechanosensory and chemosensory bristles, respectively. Also indicated are dome-shape sockets (asterisks) surrounding shafts. (B) PWM non-innervated bristles. (C–H) PWM bristles in CSN4null (C), CSN5null (D), en-GAL4 control (E), CSN2 RNAi (F), CSN3 RNAi (G) and CSN6 RNAi (H) driven by en-GAL4 driver. Arrowheads indicate single bristles with a thicker shaft and a socket. Arrows indicate single bristles with two shafts and one large socket. (I–L′) CSN4null clones, located at the PWM and marked by the absence of GFP, exhibited elevated Hnt expression 20–24 h APF (red in I, I′), and ectopic expression of Pros 24–28 h APF (red in J, J′), Elav 28–32 h APF (red in K, K′) and Futsch 28–32 h APF (red in L, L′). (I′, J′, K′, L′) Single-channel images. (M–P) Hnt (red) expressions 20–24 h APF at the PWM of en-GAL4 control (M), and CSN1b RNAi (N), CSN2 RNAi (O) and CSN7 RNAi (P) by en-GAL4. The altered developmental process of PWM bristles in CSN mutants was examined for the expression of nuclear protein Hindsight (Hnt)/Pebbled in bristle lineage cells [42]. In wild-type wing discs at 20–24 hours (h) after puparium formation (APF), clusters of AWM innervated bristle lineage cells had larger nuclei and expressed high levels of Hnt, while PWM non-innervated bristle cells, with smaller nuclei, expressed low levels of Hnt (Figure S1C–C″). In CSN4null and CSN5null PWM clones, cells with larger nuclei and high-level Hnt were observed (Figure 1I, 1I′ and Figure S1D, S1D′). Accumulations of Hnt in PWM cells were also detected in CSN1b, CSN2 and CSN7 knockdowns by en-GAL4 (Figure 1M–P). While non-innervated PWM bristles contain no neurons or sheath cells [4], [39], these two types of cells were detected in PWM clones for CSN4null, as shown by Prospero (Pros) expression in pIIb precursors and sheath cells [43] and Elav expression in neurons [44] (Figure 1J–K). These ectopic neurons extended axons as revealed by immunostaining for the microtubule-associated protein Futsch (Figure 1L, 1L′). Taken together, these morphological and molecular analyses indicate that the CSN inhibits the neural potential of PWM bristles.
The CSN suppresses Sens independent of proneural genes ac and sc
Overexpression of Sens in epithelial cells can induce innervated bristles [3]. As shown, overexpression of Sens by C96-GAL4 along the wing margin induced bristles with sockets around the PWM (Figure 2A, arrowheads). Concomitantly, high levels of Hnt expressions were also detected (Figure 2B), confirming that ectopic Sens expression induces innervated bristles at the PWM.
Fig. 2. The CSN suppresses Sens to inhibit neural differentiation of PWM bristles. 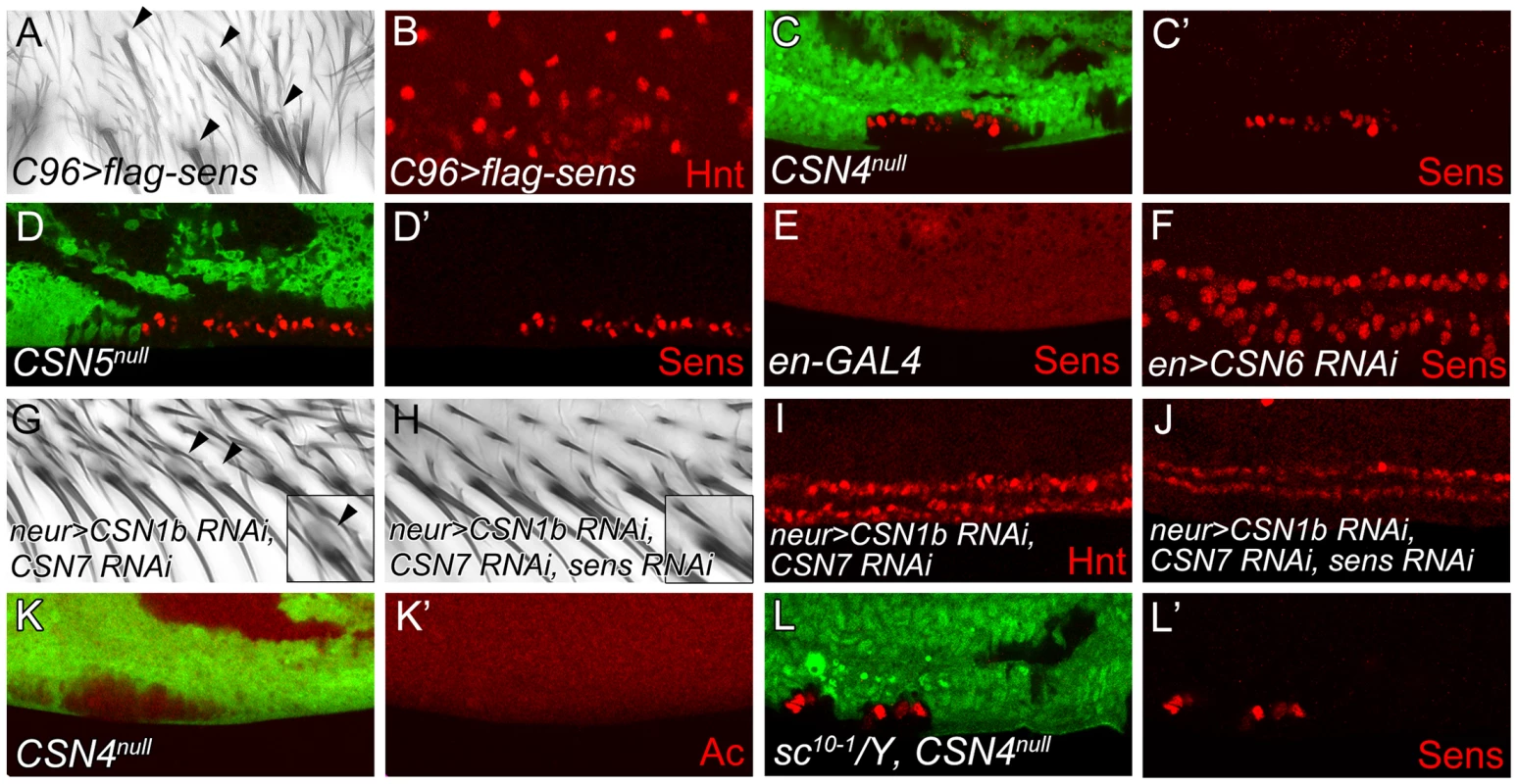
(A, B) Overexpression of sens in C96-GAL4-driven UAS-flag-sens resulted in ectopic innervated bristles at the PWM of adult wings (A) and ectopic Hnt-positive cells at the PWM of wing discs 20–24 h APF (B). (C–F) Sens (red) expression was detected in CSN4null clones (C, C′), CSN5null clones (D, D′) and CSN6 RNAi knockdown by en-GAL4 (E, F) at the PWM 20–24 h APF. (G, H) Innervated bristles with sockets (arrowheads) appeared at the PWM of neur-GAL4 double knockdown of CSN1b and CSN7 (G), which were no longer detected when sens was also knocked down (H). (Insets in G, H) Enlarged figures showing single bristle and socket (arrowhead). (I, J) Double knockdown of CSN1b and CSN7 by neur-GAL4 resulted in accumulation of higher-level Hnt at the PWM, which was suppressed by sens knockdown (J). (K, K′) Ac (red) was not expressed in CSN4null clones in wing disc 18–22 h APF. (L, L′) Ectopic Sens-positive cells were detected in PWM clones for CSN4null in sc10-1 null mutant disc 20–24 h APF. With the induction of innervated sensory bristles by high levels of Sens, we then examined whether Sens was upregulated in CSN mutants. In CSN4null and CSN5null PWM cells, the Sens levels were elevated (Figure 2C–D′). The elevation of Sens expression was also detected in CSN6, CSN2 and CSN3 knockdown cells, suggesting that CSN functions as a complex to suppress Sens expression (Figure 2E, 2F and Figure S2A, S2B). Next we addressed whether Sens is required for innervated bristle formation in CSN mutant cells at the PWM. Double knockdown of CSN1b and CSN7 in bristle lineage cells by neur-GAL4 resulted in socket formation in PWM bristles (Figure 2G), ectopic Sens expression (Figure S2C), and consistently, higher levels of Hnt in PWM cells with large nuclei (Figure 2I). When sens was knocked down simultaneously (Figure S2D), those features of innervated bristles in CSN mutants were suppressed. Socket morphology was no longer visible (Figure 2H), and most PWM cells had smaller nuclei and expressed lower levels of Hnt (Figure 2J). Thus, Sens induction in CSN mutant cells is required for the formation of innervated bristles at the PWM.
The proneural proteins Ac and Sc are specifically expressed in proneural clusters and precursors of AWM chemosensory bristles [45]. Formation of innervated sensory bristles in CSN mutant cells could be the consequence of ectopic proneural protein induction. However, Ac expression was not induced in CSN4null and CSN5null PWM cells. (Figure 2K, 2K′ and Figure S2E–F′). Also, removal of ac and sc in CSN4null clones still accumulated Sens in PWM cells (Figure 2L, 2L′). Thus, proneural proteins Ac and Sc are not required for Sens upregulation in CSN mutant cells at the PWM.
Sens accumulation is induced by BR-Z1 upregulation in CSN mutant cells
The Zinc-finger transcriptional factor BR-Z1 induces Sens expression in response to ecdysone signaling in chemosensory precursors [5]. Thus, we tested if the CSN also downregulates BR-Z1, leading to the suppression of Sens in PWM cells. As expected, the BR-Z1 levels were strongly elevated in CSN4null and CSN5null wing-disc cells 20–24 h APF (Figure 3A–B′). Elevated BR-Z1 levels were also detected in CSN2-knockdown cells (Figure S3A). Distinct from Sens regulation, CSN suppression of BR-Z1 was ubiquitous in all wing-disc cells, not restricted to the wing margin. Furthermore, forced expression of BR-Z1 in bristle lineage cells by neur-GAL4 elevated Sens and Hnt expression and induced Elav-positive neurons at the PWM (Figure 3C–E). In adult wings, all bristles at the PWM had dome-shape sockets (Figure 3F), suggesting that BR-Z1 promotes the formation of innervated bristles at the PWM. The br locus encodes three additional isoforms BR-Z2, BR-Z3 and BR-Z4, and functional redundancy was observed among these isoforms [46], [47]. We found that the activity to induce innervated bristles is not limited to BR-Z1, as overexpression of BR-Z3 also promoted Sens accumulation and innervated bristle formation at the PWM (Figure S3B, S3C).
Fig. 3. BR-Z1 upregulation is required for Sens upregulation at the PWM of CSN mutants. 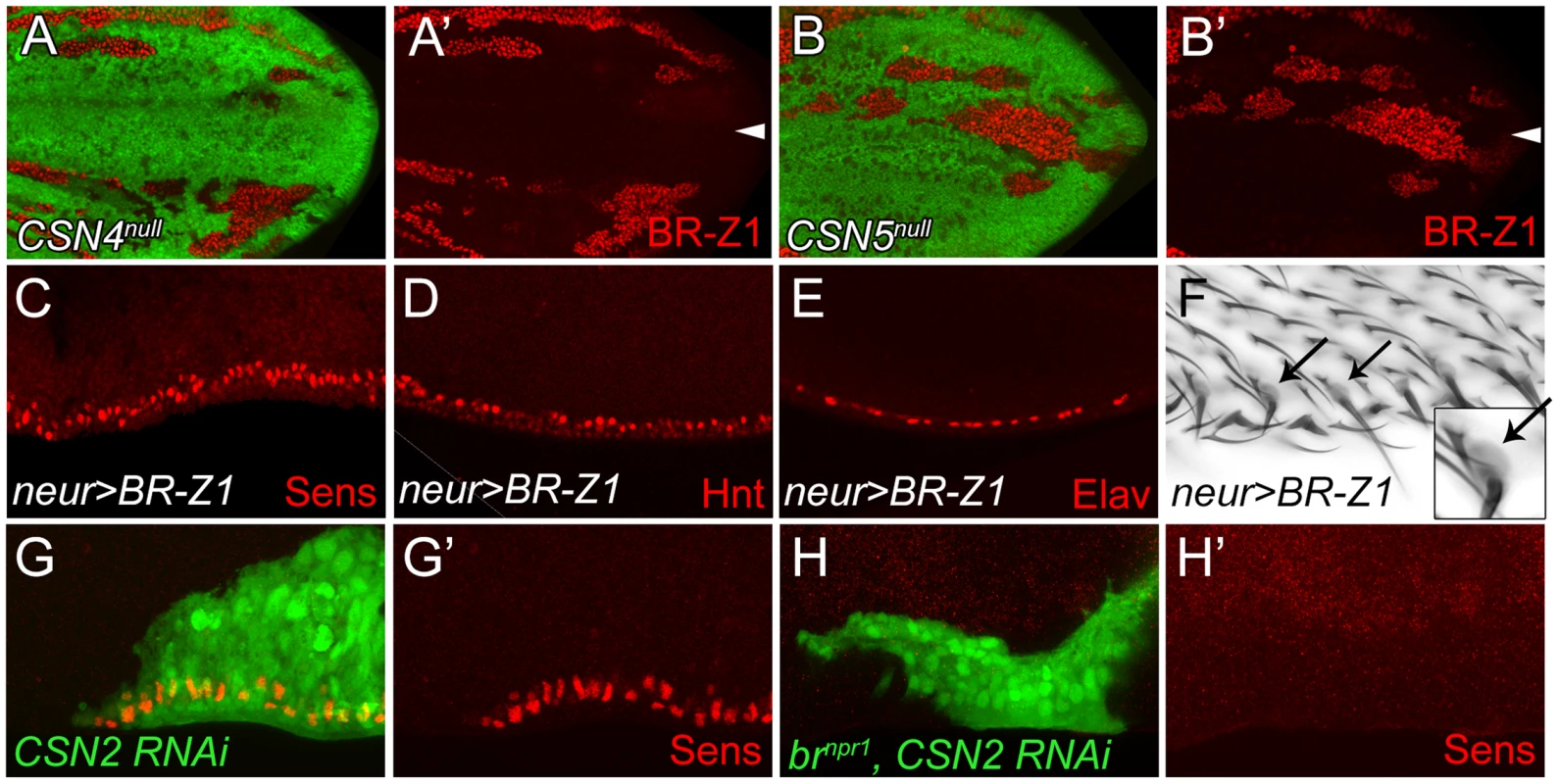
(A–B′) Upregulation of BR-Z1 (red) in CSN4null (A, A′) and CSN5null clones (B, B′) 20–24 h APF. White arrowheads indicate the anterior-posterior boundary. (C–F) Transient overexpression of BR-Z1 in bristle lineage cells via neur-GAL4 and Tub-GAL80ts. (C–E) Ectopic expression of Sens (red in C), accumulation of Hnt (red in D) and ectopic formation of Elav-positive neurons (red in E) were observed at the PWM 22–24 h APF. Prepupae grown at 18°C were shifted to 37°C for one hour at 8–10 h APF, and then incubated at 29°C until dissection. (F) Bristles with dome-shape sockets (indicated by arrows) were observed at the PWM. Inset: enlarged figure showing single bristle. The experiment was carried out similarly to (C–E) except for incubation at 29°C for 12 hours, and returning back to 18°C until eclosion. (G–H′) Knockdown of CSN2, in MARCM clones (green), induced Sens (red) upregulation at the PWM 20–24 h APF (G, G′), which was abolished in CSN2 RNAi brnpr1 double mutant MARCM clones (green) (H, H′). Lastly, we examined the role br plays in promotion of innervated bristle formation when CSN activity is abrogated. To completely inactivate all br isoforms to prevent functional redundancy, the br null allele, brnpr1, was used. In control clones, Sens expression was elevated in CSN2 RNAi cells (Figure 3G, 3G′). However, Sens elevation was no longer detected in CSN2 RNAi cells when br was simultaneously inactivated (Figure 3H, 3H′). These results unequivocally indicate that br is the critical factor required in CSN mutant cells to upregulate Sens expression.
CSN regulation of BR-Z1 and Sens levels is time-dependent
In response to ecdysone pulses, BR-Z1 expression is upregulated in wing-disc cells at late third-instar larval stages and peaked at the prepupal stage of 2–6 h APF (GFP-positive cells in Figure 4A–B′) [48]. BR-Z1 expression declined starting around 6–8 h APF and continued declining to almost undetectable level 24–28 h APF (GFP-positive cells in Figure 4C–F′). Quantification of immunofluorescent intensity following co-staining revealed that the BR-Z1 intensity was 1.6-fold of H3 intensity 2–6 h APF, but dramatically dropped to 0.07-fold of H3 24–28 h APF (dashed line in Figure 4G, and Figure S4A–B″), with the strongest reduction occurring between 12–14 h to 16–18 h APF.
Fig. 4. The CSN suppresses BR-Z1 expression after prepupa-to-pupa transition. 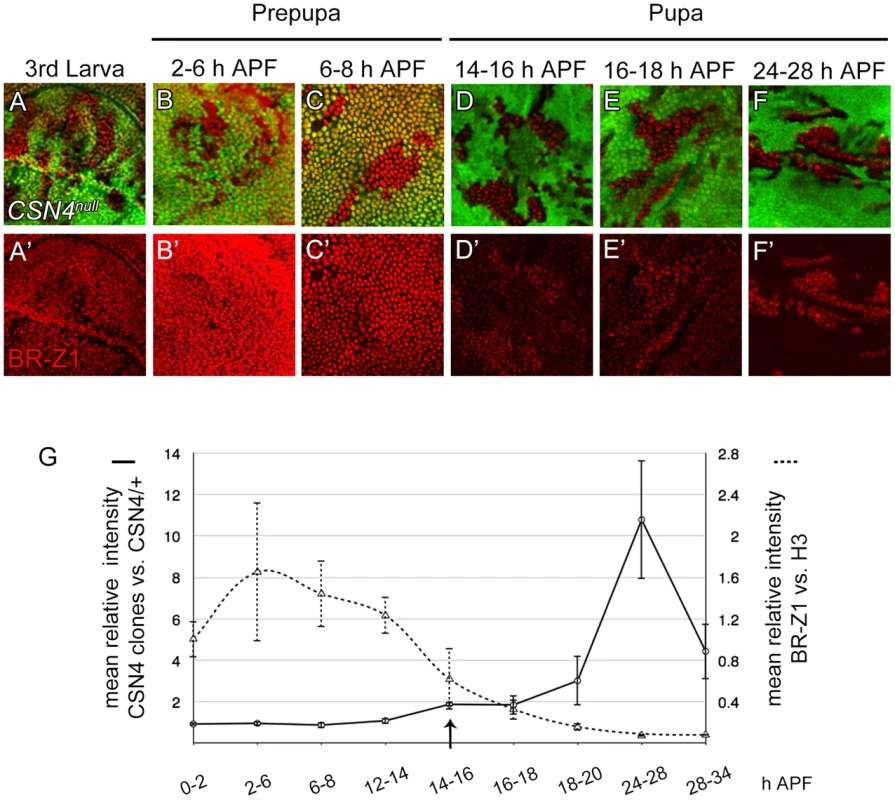
(A–F′) BR-Z1 (red) expression in CSN4null clones in wing discs from late third-instar larval to pupal stages 24–28 h APF. (A–C′) BR-Z1 expression was not affected in CSN4null clones at late third larval instar (A, A′) and prepupal stages (B–C′). (D–F′) BR-Z1 levels in CSN4null clones were upregulated at pupal stages. (G) Diagram showing the mean anti-BR-Z1 immunofluorescent intensity from 0 to 34 h APF. The dashed line represents the mean relative intensities of anti-BR-Z1/anti-H3 in wild-type w1118 discs. The solid line represents the relative mean intensity of anti-BR-Z1 in CSN4null cells/neighboring CSN4null/+ cells. The arrow marks the starting point when the BR-Z1 expression was significantly upregulated in CSN4null clones. Error bars represent the standard deviation (SD). Five w1118 wing discs were scored (N = 5) for each time point, and at least three CSN4null wing discs were scored (N≥3) for each time point, except for 12–14 h APF (N = 2). Strikingly, we found that the CSN initiates BR-Z1 suppression during this time period. From the late third-instar larval stage to 8 h APF, the BR-Z1 levels in CSN4null cells were comparable to neighboring heterozygous +/CSN4null cells, suggesting that CSN4 has no role in regulating BR-Z1 levels at these stages (Figure 4A–C′, solid line in 4G). However, while BR-Z1 continued to decline at the beginning of the prepupa-to-pupa transition, higher BR-Z1 levels in CSN4null clones were detected. Significant BR-Z1 upregulation in CSN4null cells was first detected 14–16 h APF (Figure 4D, 4D′, and arrow and solid line in 4G), two hours after the completion of prepupal development. The upregulation in CSN4null cells reached about two-fold of neighboring heterozygous cells 14–18 h APF and three-fold 18–20 APF, and the peak of a 10-fold increase was observed 24–28 h APF (Figure 4E–F′, solid line in 4G). The enhancement of BR-Z1 expression in CSN4null cells declined after 28 h APF (solid line in Figure 4G).
The activation of BR-Z1 expression in CSN mutants at pupal stages prefigures the timing in the upregulation of Sens. At the PWM, Sens was highly expressed in the precursors 6–8 h APF (Figure 5A) [4], declined to lower levels in lineage cells 16–18 h APF (Figure 5B), and became undetectable 20 h APF (Figure 5C). In CSN4null cells, while the Sens levels remained unchanged compared to neighboring control cells 6–8 h APF (Figure 5D, 5D′), Sens was upregulated at the pupal stage of 16–18 h APF (Figure 5E, 5E′), and the upregulation was maintained even 20–24 h APF (Figure 2C, 2C′). Taken together, the CSN downregulates BR-Z1 and Sens levels in a time-dependent manner, detected only after the prepupa-to-pupa transition.
Fig. 5. Temporal regulation of Sens expression by CSN4. 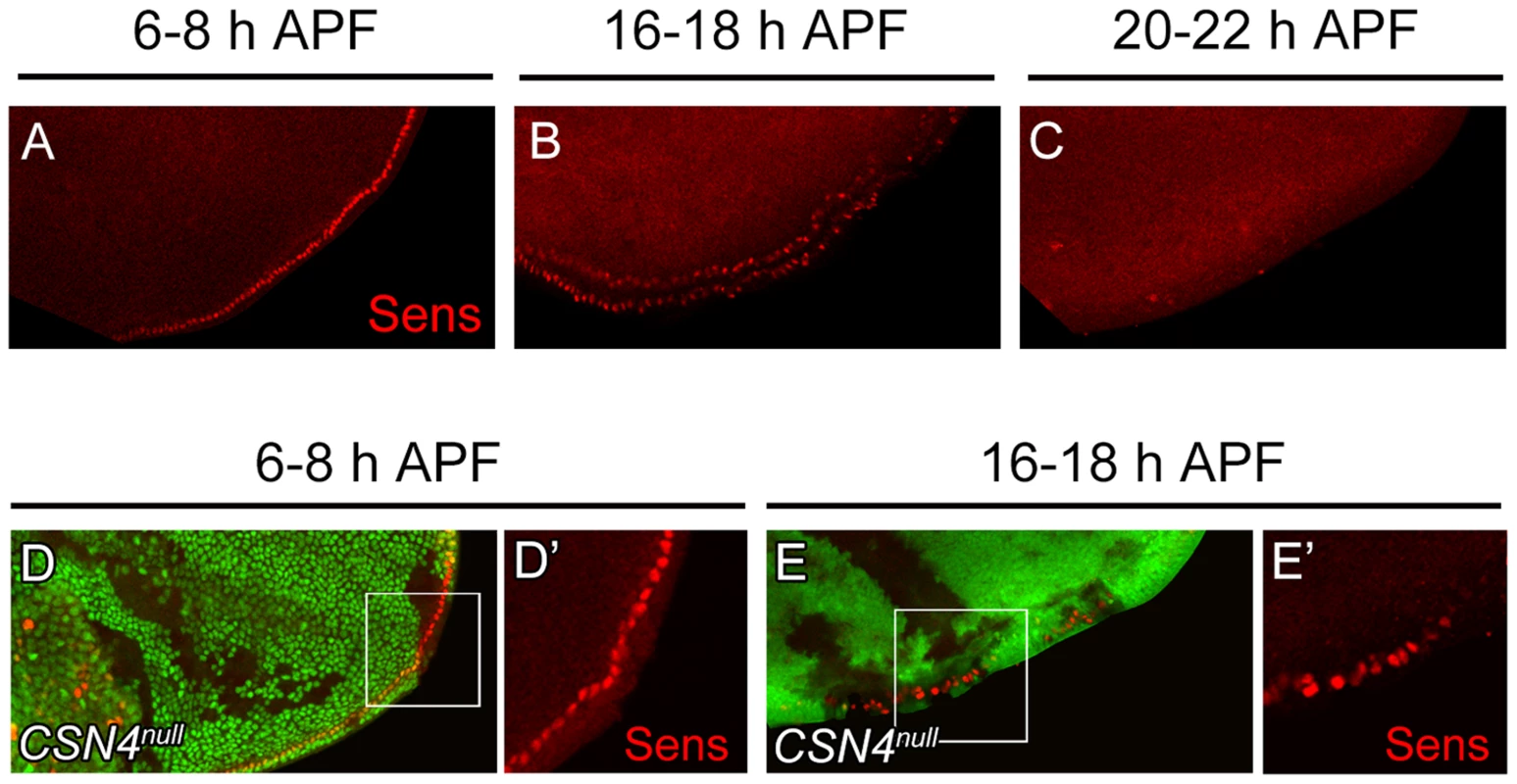
(A–C) Sens (red) expression in wild-type wing discs at the PWM. The expression gradually declined from 6–8 h APF (A) to 16–18 h APF (B), and was below detection 20–22 h APF (C). (D–E′) Sens (red) levels at the PWM 6–8 h APF were identical between CSN4null cells and neighboring CSN4null/+ cells (D, D′), and were upregulated in CSN4null clones 16–18 h APF (E, E′). Ecdysone receptor represses BR-Z1 after the prepupa-to-pupa transition
To test whether ecdysone receptor activity is also involved in the switch from CSN-resistant to CSN-sensitive BR-Z1 suppression after the prepupa-to-pupa transition, the transcriptional activity of EcR was inactivated by the expression of dominant-negative EcRB1ΔC655F645A (DN-EcR) [49]. In DN-EcR expression clones, BR-Z1 expression in late larval wing discs was abolished (Figure 6A, 6A′), indicating that EcR positively regulates BR-Z1 expression [50]. In contrast, BR-Z1 expression was strongly induced in DN-EcR clones 20–24 h APF (Figure 6B, 6B′), indicating that transcriptionally active EcR represses BR-Z1 expression after the prepupa-to-pupa transition.
Fig. 6. EcR regulation of BR-Z1 before and after the prepupa-to-pupa transition. 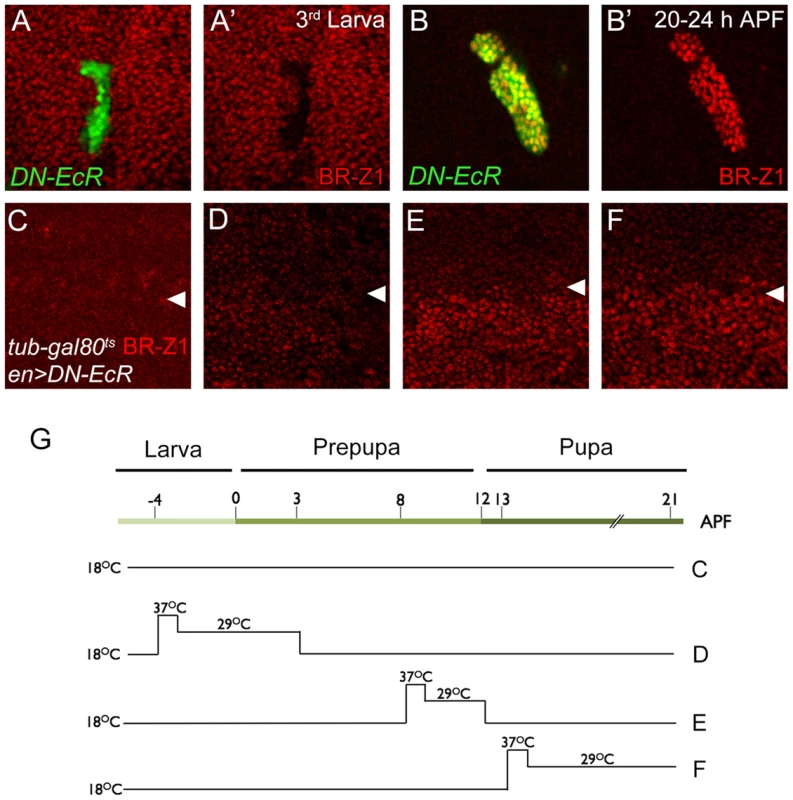
(A–B′) DN-EcR-expressing clones (green) displayed BR-Z1 (red) downregulation at third-instar larval stages (A, A′) and upregulation 20–24 h APF (B, B′). (C–F) BR-Z1 (red) expression detected in wing discs of en-GAL4>UAS-DN-EcR; Tub-GAL80ts/+ 21–22 h APF. White arrowheads indicate anterior-posterior boundaries. BR-Z1 expression in the posterior compartment (bottom) was unaffected when animals were incubated at 18°C from larva to pupa (C), or at 18°C except during larva-to-prepupa transition (−4 to 3 h APF) at non-permissive temperatures (37°C for one hour and 29°C for six hours) (D). BR-Z1 levels were upregulated in pupae incubated at non-permissive temperatures 8–12 h APF (E) or 13–21 h APF (F). (G) A diagram showing the temperature regimen in animals shown in Figure C–F. To further delineate the temporal regulation of BR-Z1 from activation to repression, DN-EcR was overexpressed by en-GAL4 in the posterior compartment, in combination with temperature-sensitive Tub-GAL80ts. At the permissive temperature throughout larval to pupal stages, en-GAL4-driven DN-EcR expression was blocked by GAL80, and BR-Z1 remained at low levels in both anterior and posterior compartments (Figure 6C, 6G). At the non-permissive temperature, inactivation of GAL80 allowed expression of DN-EcR in the posterior compartment. We found that expression of DN-EcR from 4 h before puparium formation to 3 h APF maintained BR-Z1 repression in pupal wing discs at 21 h APF (Figure 6D, 6G). However, the DN-EcR expression at 8–12 or 13–21 h APF induced BR-Z1 upregulation in the posterior compartment (Figure 6E, 6F, 6G). Thus, active EcR is required to suppress BR-Z1 expression in pupal wings at and after the prepupa-to-pupa transition.
CSN subunits associate with EcR to repress br transcription
EcR regulates BR-Z1 at the transcriptional level. To test whether the CSN also regulates BR-Z1 through transcriptional regulation, we measured the mRNA level for BR-Z1 in CSN mutants by semi-quantitative RT-PCR. While the BR-Z1 mRNA expression in en-GAL4 control pupal wing discs was undetected, expression of CSN2 RNAi by en-GAL4 induced the BR-Z1 mRNA level (Figure 7A). To test the transcriptional regulation by the CSN, the reporter EcRE-LacZ which contains seven tandem repeats of EcRE was used [51]. In CSN4null and CSN5null clones, the expression levels from EcRE-lacZ were also elevated in comparison to neighboring heterozygous cells in pupal wings (Figure 7B–C′). The regulation of EcRE-lacZ is specific to the pupal stage, as EcRE-LacZ expression was not elevated in CSN5null clones at the prepupal stage (Figure 7D, 7D′). Thus, the CSN-downregulated BR-Z1 expression is at the transcriptional level and after the prepupa-to-pupa transition.
Fig. 7. Association of CSN subunits and EcR in BR-Z1 repression. 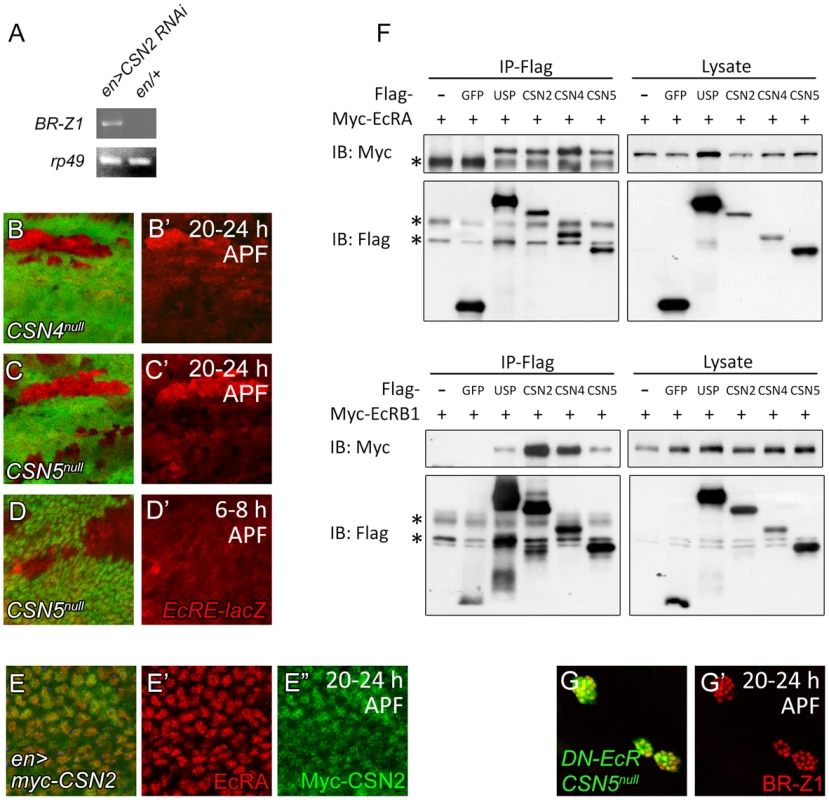
(A) The BR-Z1 mRNA level in wing discs 20–24 h APF, assayed by semi-quantitative RT-PCR, was upregulated in wing discs of CSN2 RNAi driven by en-GAL4 compared to en-GAL4/+ control. rp49 levels served as internal controls. (B–C′) EcRE-lacZ (red) expression was upregulated in CSN4null (B, B′) and CSN5null (C, C′) clones 20–24 h APF, but unaffected in CSN5null clones 6–8 h APF (D, D′). (E–E″) Both EcRA (red) and Myc-CSN2 (green) localized in nuclei in wing disc 20–24 h APF. (F) Western blots showing co-precipitations of Myc-EcRA (upper panel) or Myc-EcRB1 (lower panel) in Flag immunoprecipitates of Flag-USP, CSN2, CSN4 or CSN5 in S2 cell extract. Co-precipitation was not detected in Flag-GFP. * represents the non-specific bands. (G, G′) BR-Z1 levels (red) in double MARCM clones for CSN5null and DN-EcR (green) were not further elevated compared to CSN5null or DN-EcR single clones (Figure 3B, 6B). As a transcriptional regulator, EcRA was localized in nuclei in wing-disc cells (Figure 7E, 7E′). Examination of protein location of the Myc-tagged CSN2 and CSN4 subunits and the endogenous CSN5 subunit in wing disc cells showed that they primarily localized in nuclei as EcRA (Figure 7E, 7E″ and Figure S5A–B″). Thus, several CSN subunits and EcRA colocalize in nuclei, consistent with their roles in transcription regulation of BR-Z1.
We then performed co-immunoprecipitation to detect protein-protein interaction between CSN subunits and EcR. The S2 cells were co-transfected with expression plasmids for one of Flag-tagged CSN subunits and Myc-tagged EcRA or EcRB1. Both EcRA and EcRB1 were specifically detected in the Flag immunoprecipitates of CSN2, CSN4, CSN5 and the positive control USP, but not the negative control GFP (Figure 7F). The abilities of various subunits to associate with EcR suggest that the CSN interacts with EcR as protein complexes.
The results show that inactivating either the CSN or EcR is sufficient to derepress BR-Z1 expression at the pupal stage (Figure 3A–B′, 6B). We then examined the effect on BR-Z1 expression when both CSN and EcR were inactivated. In double mutants that expressed DN-EcR in CSN5null mutant clones, BR-Z1 was upregulated to the level comparable to that in single mutants of CSN5null or DN-EcR (Figure 7G, 7G′). This result supports the notion that the CSN and EcR function as protein complexes to repress BR-Z1 expression.
Nedd8 and multiple cullins are required to suppress BR-Z1 and Sens expressions
Deneddylation is coupled with neddylation to cycle cullins between two neddylation states for optimal CRL activities [52]. We first addressed whether the deneddylating activity of the CSN is required to repress BR-Z1 and Sens. Elevated BR-Z1 and Sens expressions in CSN5null cells were completely suppressed by the wild-type CSN5 transgene driven by MS1096-GAL4 in wing discs (Figure 8A, 8A′ 8C, 8C′). However, expression of the deneddylation-defective CSN5D148N mutant [26] failed to decrease BR-Z1 and Sens levels in CSN5null clones (Figure 8B, 8B′, 8D, 8D′). This result suggests the Nedd8 conjugation is involved in BR-Z1 and Sens regulations. Indeed, Nedd8AN015 null mutant clones located at the PWM of pupal wings also exhibited BR-Z1 and Sens upregulations (Figure 8E–E″).
Fig. 8. Deneddylation/neddylation and multiple cullins suppress BR-Z1 and Sens expressions at the PWM. 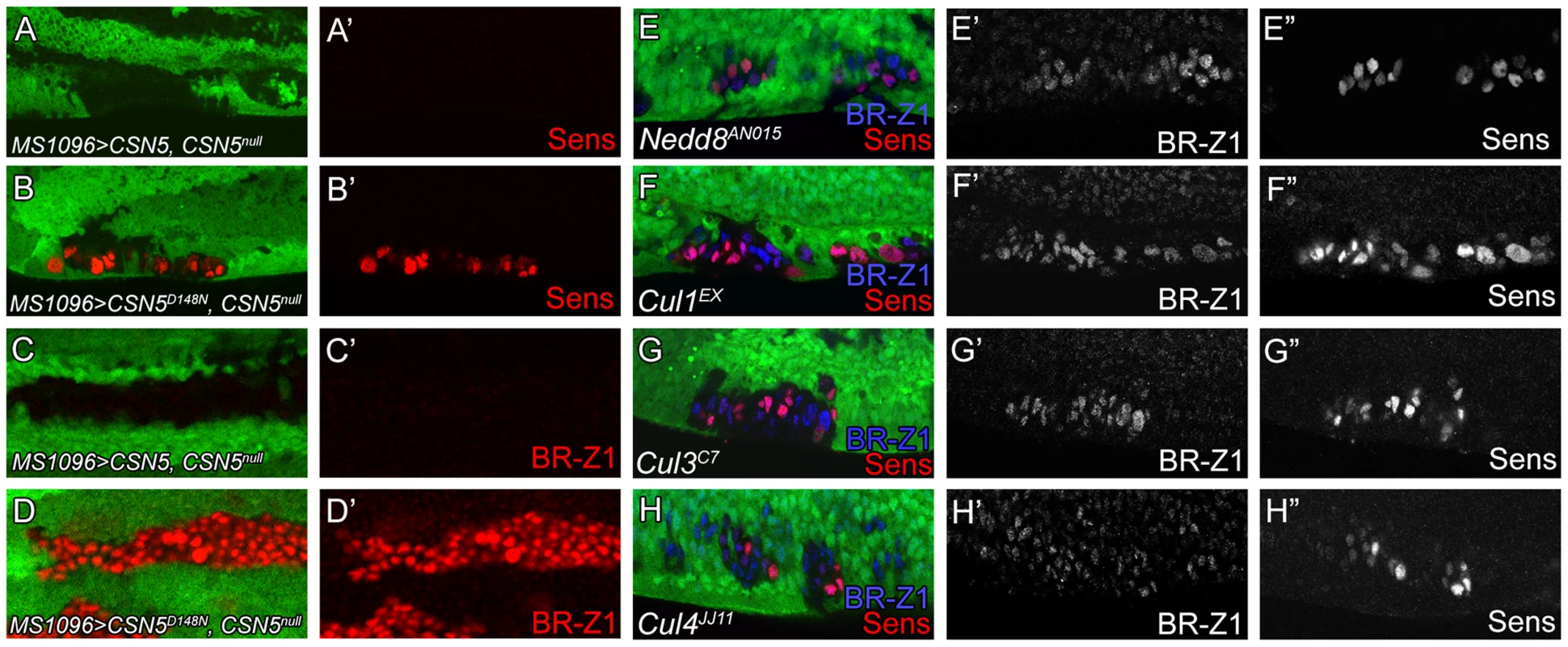
(A–H″) 20–24 h APF wing discs. (A–D′) Upregulation of Sens (red in A–B′) and BR-Z1 (red in C–D′) in PWM CSN5null clones were suppressed by wild-type CSN5 (A, A′, C, C′), but not deneddylation-defective CSN5D148N (B, B′, D, D′). (E–H) Upregulation of BR-Z1 (blue) and Sens (red) were observed in null clones of Nedd8AN015 (E), Cu11EX (F), Cul3C7 (G) and Cul4JJ11 (H) at the PWM 20–24 h APF, with a single channel for BR-Z1 (E′–H′) and Sens (E″–H″). We then addressed whether and which cullins are affected in CSN mutants for control of Sens and BR-Z1 suppression. Individual cullin proteins were inactivated by generating clones for the null alleles Cul1EX, Cul3C7 and Cul4JJ11. Interestingly, these Cul mutants showed upregulations of BR-Z1 and Sens in pupal wing discs of 20–24 h APF (Figure 8F–H″). Consistent with the pupa-specific regulation, BR-Z1 levels remained unchanged in the Cul1EX, Cul3C7 and Cul4JJ11 clones of late third instar larval wing discs (Figure S6A–C″). Therefore, our results indicate that cycling multiple cullin components between neddylation and deneddylation is required to completely suppress BR-Z1 and Sens in pupal PWM cells.
Discussion
To build the nervous system, spatial and temporal patterning cues have to be integrated to confer neural competence at specific locations of the developing tissues. We describe a critical role of the CSN, which by reading the temporal ecdysone signal cue, can determine neural competence at a specific location, the PWM. Interestingly, in response to ecdysone signaling, EcR activity is converted from transcription activation to repression of target genes, and the CSN-mediated regulation is involved in this conversion. Repression of the target gene br leads to the repression of BR-Z1-activated Sens, thus suppressing the neural differentiation of PWM bristles. We show that the CSN physically associates with EcR and represses BR-Z1 at the transcriptional level. Together with the genetic analyses between CSN subunits and EcR, our study indicates that the CSN integrates the temporal hormone signaling to endow a spatial neural repressive activity in sensory bristle patterning.
Suppression of neural competence of non-innervated bristle at the PWM
The difference between AWM and PWM bristles is that AWM bristles are innervated by sensory neurons to sense mechanical or chemical stimulations. PWM bristles with long and thin hair structures are non-innervated and without socket support, and they are non-functional in sense detection. During development, the transcription factor Sens is expressed dynamically at the wing margin. At the third-instar larval stage, Sens is initially expressed in both anterior and posterior margins. Expressions of proneural proteins Ac and Sc, which are limited to anterior cells, maintain Sens in developing chemosensory bristle cells [4]. At 6–8 h APF, Sens is re-activated in precursors of both anterior mechanosensory and posterior non-innervated bristles for precursor specification. At the pupal stage, however, Sens expression is suppressed in bristle lineage cells, thus preventing neural differentiation at PWM. Timely suppression of Sens expression plays a critical role in disruption of neural differentiation of PWM bristles, as continuing Sens expression allows PWM bristles to differentiate into innervated bristles, even after precursor specification.
While sens knockdown in bristle lineage cells prevented transformation of PWM non-innervated to innervated bristles in CSN mutants, it did not transform innervated bristles to non-innervated bristles at AWM (Figure S7A, S7B), indicating that diminishment of Sens levels in the lineage cells is not the sole determinant in non-innervated bristle formation, and additional mechanisms are activated to disrupt the differentiation of neurons and the associated neural cells at the PWM. It is shown that programmed cell death of the underlying neurons and sheath cells is one such mechanism. Suppression of programmed cell death, either by overexpression of anti-apoptotic protein p35 or by blocking Dmyb and Grim activities, induces ectopic neurons associated with PWM bristles [4], [53]. While overexpression of p35 prevented neuronal death (Figure S8B), we found that the morphology of PWM hairs was not affected and lacked associated sockets (). Thus, apoptosis has an effect only on internal neurons and sheath cells, while other mechanisms are required to prevent socket cell formation and to differentially regulate hair elongation at PWM.
Switch from activation to repression of ecdysone signaling target genes requires CSN deneddylation activity
Pulses of ecdysone provide temporal information to coordinate various developmental processes in insect development. Activation of ecdysone signaling in late larval and prepupal stages are required for larva-to-prepupa and prepupa-to-pupa transitions, respectively. In contrast to br activation in the larva-to-prepupa transition, br expression was thought to be low even with a large ecdysone pulse in the pupal stage [10], [19]. Our study indicates that this is indeed due to repression of br by ecdysone signaling, as shown by the expressions of BR-Z1 induced by dominant-negative EcR in pupal wings. The repressive regulation requires the input of the CSN, as BR-Z1 was also upregulated in pupal wing-disc cells of CSN4null and CSN5null mutants. Therefore, our results suggest that by integrating the CSN, ecdysone signaling activity plays a non-canonical role in the prepupa-to-pupa transition, and the br repression is critical to Sens suppression during PWM bristle development.
Neddylation is required for activating CRL activity both in vivo and in vitro [52], [54]–[56]. Deneddylation, while inactivating CRL in vitro, promotes CRL activity in vivo by protecting them from cellular degradation, thus maintaining a physiological pool of CRL for the next round of neddylation [26], [27]. Our results showed that repression of ecdysone signaling target genes requires cycling of Nedd8 substrates between neddylation and deneddylation states. Similar to CSN mutants, the protein levels of BR-Z1 and Sens were elevated in Nedd8 mutants. Also, CSN suppression of BR-Z1 and Sens depends on the deneddylating activity residing in the CSN5 subunit, as the deneddylation-defective CSN5 mutant failed to rescue the CSN5 phenotype. Thus, the requirement of neddylation and deneddylation is consistent with the involvement of multiple cullin proteins that are the major substrates of Nedd8 conjugation. Transcription regulation of ecdysone responsive genes requires multiple activators and repressors [57]–[61] whose protein activity and stabilities could be in turn regulated by multiple cullin-organized ubiquitin E3 ligases. Alternatively, these cullin ligases might act on the same set of targets, like the regulations of protein stability of Ci by multiple cullin ligases [54].
Our results on the requirement of the CSN holoenzyme in development of PWM bristles suggest different usages of CSN subunits in nuclear receptor signaling. A previous study showed that CSN4 but not CSN5 is required for larval molting [32]. In mammals, single CSN2 and CSN5 subunits interact with nuclear receptors to function as a co-repressor and a co-activator, respectively, in steroid hormone signaling [62]–[64]. Drosophila CSN2 physically interacts with several Drosophila hormone receptors including EcR [62]. In our study, seven out of the eight CSN subunits were shown to be required for Sens suppression and CSN2, CSN4 and CSN5 were shown to associate with EcR, strongly suggesting that the CSN associates with EcR as a complex. It has been shown that the CSN complex interacts with multiple cullins to protect them from self-ubiquitination and degradation [26], [65]. Thus, the association between EcR and the CSN holoenzyme might provide a platform for recruiting and maintaining multiple CRLs, composed of cullins and their associated components, in the proximity for efficient down-regulation of EcR-mediated transcription (Figure S9). Two models are proposed here for the CSN-dependent switch of EcRs from activation to repression of br (Figure S9). In model A, the switch depends on the induction of specific co-repressors upon the pulse of ecdysone at the prepupa-to-pupa transition. The co-repressor could then replace the co-activator, a step that could be facilitated by the nearby CRLs through the ubiquitination of co-activators for subsequent degradation, for example. In model B, the switch depends on the activation of specific CRLs by ecdysone signaling at the prepupa-to-pupa transition. The activation could be mediated through expression of the specific substrate receptors of CRLs, or phosphorylation of substrates to induce binding to substrate receptors. Once activated, these CRLs could inactivate co-activators to shut down br transcription. It is unlikely that the CSN-mediated pupa-specific BR-Z1 and Sens repression is through mechanisms involving changes in the CSN and EcR protein expression levels during the prepupa-to-pupa transition, as the expression of the catalytic subunit CSN5 and EcRA were expressed at comparable levels in wing discs at late third instar larval and pupal stages (Figure S10A–C, and S11A–F′). Also, the CSN is active in third-instar larvae for normal wing disc development [37]. Our finding that suppression of EcR target genes in pupae through the CSN and multiple cullins provide insights for a novel negative regulation of steroid hormone signaling in metazoa.
Materials and Methods
Fly strains and genetics
The following fly strains were used in this study. Mutant alleles: CSN5null, CSN4null [32], Nedd8AN015, Cul1EX [54], Cul3C7 [66], Cul4JJ11 [67], sc10-1 [68], brnpr1 [69], [70]. Reporter line: EcRE-LacZ (II, BL4516) [71]. GAL4 drivers: C96-GAL4 [72], en-GAL4 [73], neur-GAL4 [74] and MS1096-GAL4 [75]. UAS lines: UAS-BR-Z1, UAS-BR-Z3 [76], UAS-p35 (P[UAS-p35.HB]H3, BL6298) [77], UAS-EcRB1ΔC655F645A [49], UAS-CSN5, UAS-CSN5D148N [26], UAS-myc-CSN2 (this study), UAS-myc-CSN4 (this study), and UAS-3Xflag-sens (this study). The UAS-CSN1bRNAi (v34727), UAS-CSN2RNAi (v48044), UAS-CSN3RNAi (v101516), UAS-CSN6RNAi (v22308) and UAS-CSN7RNAi (v40691) were obtained from Vienna Drosophila RNAi Center (VDRC). The UAS-sensRNAi (NIG 32120R-2) was obtained from NIG-FLY stock center.
Clones were generated by FLP/FRT-mediated mitotic recombination [78]. The mutant or MARCM clones in developing tissues were identified by the absence or presence of GFP, respectively. For transient overexpression using GAL80ts, developing animals were grown at 18°C. Prepupae or pupae were then shifted to 37°C for one hour to rapidly inactivate GAL80ts to allow fast GAL4-activated gene expression, and then incubated at 29°C for several hours as indicated.
Immunofluorescence staining
Larvae and prepupae (0–10 h APF) were dissected in 1×PBS for isolating wing discs, which were then fixed in 4% paraformaldehyde for 15 minutes at room temperature (RT). For pupae 12–36 h APF, pupal cases were removed in 1×PBS and the whole pupae were fixed in 4% paraformaldehyde at RT for one hour. Internal pupal membranes on the surface of wing tissues were subsequently removed and the resulting wing discs were fixed again in 4% paraformaldehyde for 15 minutes. The following primary antibodies were used: mouse anti-Ac (DSHB, 1∶5) [79], mouse anti-ß-galactosidase 40-1a (DSHB, 1∶500) [80], mouse anti-BR-Z1 Z1.3C11.OA1 (DSHB, 1∶250) [48], mouse anti-EcRA 15G1a (DSHB, 1∶100) [81], mouse anti-Elav 9F8A9 (DSHB, 1∶500) [82], mouse anti-Futsch 22C10 (DSHB, 1∶500) [83], mouse anti-Hnt 1G9 (DSHB, 1∶25) [84], mouse anti-Pros MR1A (DSHB, 1∶100) [85], rabbit anti-c-Myc A-14 (Santa Cruz, 1∶500), mouse anti-Pol II CTD4H8 (Santa Cruz, 1∶500), rabbit anti-Histone H3 GTX122148 (GeneTex, 1∶500), rabbit anti-JAB1 (CSN5) (Sigma, 1∶250), and guinea pig anti-Sens (1∶2000) [3]. The anti-JAB1/CSN5 antibody was pre-cleaned by incubation of 10× volume antibody with CSN5null larval brain tissues at 4°C overnight.
Quantification of staining intensity
For assaying anti-BR-Z1 and anti-histone H3 immunostaining intensity, all images were obtained from the Zeiss LSM 510 Meta microscope with the same setting, and analyzed by Image J. For comparing BR-Z1 intensity in CSN4null clones and CSN4null/+ cells, the mean nuclear anti-BR-Z1 intensity per pixel in each wing disc was calculated from twenty randomly selected cells within CSN4null clones or the neighboring heterozygous tissue. For comparing BR-Z1 and histone H3 intensity, the mean intensity per pixel was obtained from the same twenty randomly selected cells in each w1118 wing discs. The mean intensity of CSN5 and Pol II in w1118 wing discs was measured using the same method.
Cell culture and plasmid construction
S2 cells were maintained in Schneider medium (Invitrogen) supplemented with 10% fetal bovine serum. Cells were transiently transfected with UAS-based expression plasmids together with driver pWA-GAL4 [86]. Transfection was carried out using Cellfectin II reagent (Invitrogen). The expression plasmids for CSN2, 4, 5, EcRA, and EcRB1 were constructed by cloning the ORFs into Drosophila gateway vectors pTFW or pTMW (obtained from Drosophila Genomics Resource Center, DGRC). The UAS-3Xflag-sens plasmid was constructed by cloning the 3Xflag-sens DNA into the pUAST vector.
Co-immunoprecipitation
The harvested cells were washed twice with ice-cold 1×PBS, and then lysed in mRIPA buffer [50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 5 mM EDTA (pH 8.0), 0.5% Triton X-100, 0.5% NP-40] supplemented with complete protease inhibitors (Roche). Lysates were diluted in mRIPA buffer to a final concentration of 5 ug/ul. 20 ul ANTI-FLAG M2 Affinity Gel (Sigma) was added to 1 mL lysate and incubated for overnight at 4°C. Associated protein complexes were analyzed by SDS-PAGE followed by western blotting. The primary antibodies used for western blots were mouse anti-Flag M2 (Sigma, 1∶10000) and mouse anti-c-Myc 9E10 (Santa Cruz, 1∶5000). Secondary antibody was goat anti-mouse HRP used at 1∶5000.
RT-PCR
Pupal wing discs were dissected in ice-cold 1×PBS. Total mRNA from thirty to forty pairs of pupal wing discs were extracted by TRIzol RNA Isolation Reagents (Invitrogen) and reverse-transcribed to cDNA by MMLV RT (PROSPEC). Expression of BR-Z1 mRNA was detected by the primer set: 5′—TGAAG AGGAG TGGTG ATTGA GCTGC—3′ and 5′—CCATC ACAAG TGCCT CCGGC ATC—3′. The mRNA for rp49 was used as the internal control.
Supporting Information
Zdroje
1. RomaniS, CampuzanoS, MacagnoER, ModolellJ (1989) Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes Dev 3 : 997–1007.
2. CubasP, de CelisJF, CampuzanoS, ModolellJ (1991) Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev 5 : 996–1008.
3. NoloR, AbbottLA, BellenHJ (2000) Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102 : 349–362.
4. Jafar-NejadH, TienAC, AcarM, BellenHJ (2006) Senseless and Daughterless confer neuronal identity to epithelial cells in the Drosophila wing margin. Development 133 : 1683–1692.
5. SchubigerM, CarreC, AntoniewskiC, TrumanJW (2005) Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development 132 : 5239–5248.
6. RiddifordLM (1993) Hormone receptors and the regulation of insect metamorphosis. Receptor 3 : 203–209.
7. HuetF, RuizC, RichardsG (1995) Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development 121 : 1195–1204.
8. ThummelCS (1995) From embryogenesis to metamorphosis: the regulation and function of Drosophila nuclear receptor superfamily members. Cell 83 : 871–877.
9. ThummelCS (1996) Flies on steroids–Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet 12 : 306–310.
10. ThummelCS (2001) Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell 1 : 453–465.
11. YaoTP, FormanBM, JiangZ, CherbasL, ChenJD, et al. (1993) Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 366 : 476–479.
12. King-JonesK, ThummelCS (2005) Nuclear receptors–a perspective from Drosophila. Nat Rev Genet 6 : 311–323.
13. DiBelloPR, WithersDA, BayerCA, FristromJW, GuildGM (1991) The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 129 : 385–397.
14. BayerCA, HolleyB, FristromJW (1996) A switch in broad-complex zinc-finger isoform expression is regulated posttranscriptionally during the metamorphosis of Drosophila imaginal discs. Dev Biol 177 : 1–14.
15. BelyaevaES, VlassovaIE, BiyashevaZM, KakpakovVT, RichardsG, et al. (1981) Cytogenetic analysis of the 2B3-4-2B11 region of the X chromosome of Drosophila melanogaster. II. Changes in 20-OH ecdysone puffing caused by genetic defects of puff 2B5. Chromosoma 84 : 207–219.
16. KarimFD, GuildGM, ThummelCS (1993) The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development 118 : 977–988.
17. KissI, BenczeG, FodorG, SzabadJ, FristromJW (1976) Prepupal larval mosaics in Drosophila melanogaster. Nature 262 : 136–138.
18. BelyaevaES, AizenzonMG, SemeshinVF, KissII, KoczkaK, et al. (1980) Cytogenetic analysis of the 2B3-4–2B11 region of the X-chromosome of Drosophila melanogaster. I. Cytology of the region and mutant complementation groups. Chromosoma 81 : 281–306.
19. ZhouX, RiddifordLM (2002) Broad specifies pupal development and mediates the ‘status quo’ action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development 129 : 2259–2269.
20. WeiN, DengXW (1992) COP9: a new genetic locus involved in light-regulated development and gene expression in arabidopsis. Plant Cell 4 : 1507–1518.
21. WeiN, ChamovitzDA, DengXW (1994) Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78 : 117–124.
22. ChamovitzDA, WeiN, OsterlundMT, von ArnimAG, StaubJM, et al. (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86 : 115–121.
23. WeiN, DengXW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19 : 261–286.
24. WeiN, SerinoG, DengXW (2008) The COP9 signalosome: more than a protease. Trends Biochem Sci 33 : 592–600.
25. LyapinaS, CopeG, ShevchenkoA, SerinoG, TsugeT, et al. (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292 : 1382–1385.
26. WuJT, LinHC, HuYC, ChienCT (2005) Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol 7 : 1014–1020.
27. WuJT, ChanYR, ChienCT (2006) Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol 16 : 362–369.
28. Bech-OtschirD, KraftR, HuangX, HenkleinP, KapelariB, et al. (2001) COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J 20 : 1630–1639.
29. UhleS, MedaliaO, WaldronR, DumdeyR, HenkleinP, et al. (2003) Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J 22 : 1302–1312.
30. WeeS, GeyerRK, TodaT, WolfDA (2005) CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol 7 : 387–391.
31. OronE, TullerT, LiL, RozovskyN, YekutieliD, et al. (2007) Genomic analysis of COP9 signalosome function in Drosophila melanogaster reveals a role in temporal regulation of gene expression. Mol Syst Biol 3 : 108.
32. OronE, MannervikM, RencusS, Harari-SteinbergO, Neuman-SilberbergS, et al. (2002) COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129 : 4399–4409.
33. DoronkinS, DjagaevaI, BeckendorfSK (2003) The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev Cell 4 : 699–710.
34. KnowlesA, KohK, WuJT, ChienCT, ChamovitzDA, et al. (2009) The COP9 signalosome is required for light-dependent timeless degradation and Drosophila clock resetting. J Neurosci 29 : 1152–1162.
35. DjagaevaI, DoronkinS (2009) Dual regulation of dendritic morphogenesis in Drosophila by the COP9 signalosome. PLoS One 4: e7577.
36. DjagaevaI, DoronkinS (2009) COP9 limits dendritic branching via Cullin3-dependent degradation of the actin-crosslinking BTB-domain protein Kelch. PLoS One 4: e7598.
37. WuJT, LinWH, ChenWY, HuangYC, TangCY, et al. (2011) CSN-mediated deneddylation differentially modulates Ci(155) proteolysis to promote Hedgehog signalling responses. Nat Commun 2 : 182.
38. Mummery-WidmerJL, YamazakiM, StoegerT, NovatchkovaM, BhaleraoS, et al. (2009) Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458 : 987–992.
39. HartensteinV, PosakonyJW (1989) Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development 107 : 389–405.
40. RhyuMS, JanLY, JanYN (1994) Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76 : 477–491.
41. PiH, WuHJ, ChienCT (2001) A dual function of phyllopod in Drosophila external sensory organ development: cell fate specification of sensory organ precursor and its progeny. Development 128 : 2699–2710.
42. PickupAT, LamkaML, SunQ, YipML, LipshitzHD (2002) Control of photoreceptor cell morphology, planar polarity and epithelial integrity during Drosophila eye development. Development 129 : 2247–2258.
43. ManningL, DoeCQ (1999) Prospero distinguishes sibling cell fate without asymmetric localization in the Drosophila adult external sense organ lineage. Development 126 : 2063–2071.
44. RobinowS, WhiteK (1988) The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol 126 : 294–303.
45. CousoJP, BishopSA, Martinez AriasA (1994) The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development 120 : 621–636.
46. BayerCA, von KalmL, FristromJW (1997) Relationships between protein isoforms and genetic functions demonstrate functional redundancy at the Broad-Complex during Drosophila metamorphosis. Dev Biol 187 : 267–282.
47. MugatB, BroduV, Kejzlarova-LepesantJ, AntoniewskiC, BayerCA, et al. (2000) Dynamic expression of broad-complex isoforms mediates temporal control of an ecdysteroid target gene at the onset of Drosophila metamorphosis. Dev Biol 227 : 104–117.
48. EmeryIF, BedianV, GuildGM (1994) Differential expression of Broad-Complex transcription factors may forecast tissue-specific developmental fates during Drosophila metamorphosis. Development 120 : 3275–3287.
49. CherbasL, HuX, ZhimulevI, BelyaevaE, CherbasP (2003) EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development 130 : 271–284.
50. SchubigerM, TrumanJW (2000) The RXR ortholog USP suppresses early metamorphic processes in Drosophila in the absence of ecdysteroids. Development 127 : 1151–1159.
51. KoelleMR, TalbotWS, SegravesWA, BenderMT, CherbasP, et al. (1991) The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67 : 59–77.
52. PintardL, KurzT, GlaserS, WillisJH, PeterM, et al. (2003) Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol 13 : 911–921.
53. RovaniMK, BrachmannCB, RamsayG, KatzenAL (2012) The dREAM/Myb-MuvB complex and Grim are key regulators of the programmed death of neural precursor cells at the Drosophila posterior wing margin. Dev Biol 372 : 88–102.
54. OuCY, LinYF, ChenYJ, ChienCT (2002) Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev 16 : 2403–2414.
55. WuK, ChenA, PanZQ (2000) Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J Biol Chem 275 : 32317–32324.
56. ReadMA, BrownellJE, GladyshevaTB, HotteletM, ParentLA, et al. (2000) Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol 20 : 2326–2333.
57. TsaiCC, KaoHY, YaoTP, McKeownM, EvansRM (1999) SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell 4 : 175–186.
58. BaiJ, UeharaY, MontellDJ (2000) Regulation of invasive cell behavior by taiman, a Drosophila protein related to AIB1, a steroid receptor coactivator amplified in breast cancer. Cell 103 : 1047–1058.
59. SedkovY, ChoE, PetrukS, CherbasL, SmithST, et al. (2003) Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426 : 78–83.
60. FrancisVA, ZorzanoA, TelemanAA (2010) dDOR is an EcR coactivator that forms a feed-forward loop connecting insulin and ecdysone signaling. Curr Biol 20 : 1799–1808.
61. SawatsubashiS, MurataT, LimJ, FujikiR, ItoS, et al. (2010) A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev 24 : 159–170.
62. DresselU, ThormeyerD, AltincicekB, PaululatA, EggertM, et al. (1999) Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol Cell Biol 19 : 3383–3394.
63. ChauchereauA, GeorgiakakiM, Perrin-WolffM, MilgromE, LoosfeltH (2000) JAB1 interacts with both the progesterone receptor and SRC-1. J Biol Chem 275 : 8540–8548.
64. PollyP, HerdickM, MoehrenU, BaniahmadA, HeinzelT, et al. (2000) VDR-Alien: a novel, DNA-selective vitamin D(3) receptor-corepressor partnership. FASEB J 14 : 1455–1463.
65. OlmaMH, RoyM, Le BihanT, SumaraI, MaerkiS, et al. (2009) An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. J Cell Sci 122 : 1035–1044.
66. OuCY, WangCH, JiangJ, ChienCT (2007) Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev Biol 308 : 106–119.
67. LinHC, WuJT, TanBC, ChienCT (2009) Cul4 and DDB1 regulate Orc2 localization, BrdU incorporation and Dup stability during gene amplification in Drosophila follicle cells. J Cell Sci 122 : 2393–2401.
68. CampuzanoS, CarramolinoL, CabreraCV, Ruiz-GomezM, VillaresR, et al. (1985) Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell 40 : 327–338.
69. StewartM, MurphyC, FristromJW (1972) The recovery and preliminary characterization of X chromosome mutants affecting imaginal discs of Drosophila melanogaster. Dev Biol 27 : 71–83.
70. KissI, BeatonAH, TardiffJ, FristromD, FristromJW (1988) Interactions and developmental effects of mutations in the Broad-Complex of Drosophila melanogaster. Genetics 118 : 247–259.
71. SchwedesC, TulsianiS, CarneyGE (2011) Ecdysone receptor expression and activity in adult Drosophila melanogaster. J Insect Physiol 57 : 899–907.
72. GustafsonK, BoulianneGL (1996) Distinct expression patterns detected within individual tissues by the GAL4 enhancer trap technique. Genome 39 : 174–182.
73. FietzMJ, JacintoA, TaylorAM, AlexandreC, InghamPW (1995) Secretion of the amino-terminal fragment of the hedgehog protein is necessary and sufficient for hedgehog signalling in Drosophila. Curr Biol 5 : 643–650.
74. BellaicheY, GhoM, KaltschmidtJA, BrandAH, SchweisguthF (2001) Frizzled regulates localization of cell-fate determinants and mitotic spindle rotation during asymmetric cell division. Nat Cell Biol 3 : 50–57.
75. MilanM, Diaz-BenjumeaFJ, CohenSM (1998) Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev 12 : 2912–2920.
76. ZhouX, ZhouB, TrumanJW, RiddifordLM (2004) Overexpression of broad: a new insight into its role in the Drosophila prothoracic gland cells. J Exp Biol 207 : 1151–1161.
77. ZhouL, SchnitzlerA, AgapiteJ, SchwartzLM, StellerH, et al. (1997) Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc Natl Acad Sci U S A 94 : 5131–5136.
78. XuT, RubinGM (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117 : 1223–1237.
79. SkeathJB, PanganibanG, SelegueJ, CarrollSB (1992) Gene regulation in two dimensions: the proneural achaete and scute genes are controlled by combinations of axis-patterning genes through a common intergenic control region. Genes Dev 6 : 2606–2619.
80. GotoS, HayashiS (1999) Proximal to distal cell communication in the Drosophila leg provides a basis for an intercalary mechanism of limb patterning. Development 126 : 3407–3413.
81. TalbotWS, SwyrydEA, HognessDS (1993) Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell 73 : 1323–1337.
82. O'NeillEM, RebayI, TjianR, RubinGM (1994) The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78 : 137–147.
83. ZipurskySL, VenkateshTR, TeplowDB, BenzerS (1984) Neuronal development in the Drosophila retina: monoclonal antibodies as molecular probes. Cell 36 : 15–26.
84. YipML, LamkaML, LipshitzHD (1997) Control of germ-band retraction in Drosophila by the zinc-finger protein HINDSIGHT. Development 124 : 2129–2141.
85. SpanaEP, DoeCQ (1995) The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121 : 3187–3195.
86. KanukaH, KuranagaE, TakemotoK, HiratouT, OkanoH, et al. (2005) Drosophila caspase transduces Shaggy/GSK-3beta kinase activity in neural precursor development. EMBO J 24 : 3793–3806.
Štítky
Genetika Reprodukční medicína
Článek Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the TestisČlánek The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I ofČlánek GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and AnnotationČlánek Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant BiomassČlánek Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant GeneČlánek p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide SecretionČlánek The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of MitophagyČlánek Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of SenescenceČlánek ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 11- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Establishing a Multidisciplinary Context for Modeling 3D Facial Shape from DNA
- RNA Processing Factors Swd2.2 and Sen1 Antagonize RNA Pol III-Dependent Transcription and the Localization of Condensin at Pol III Genes
- Inversion of the Chromosomal Region between Two Mating Type Loci Switches the Mating Type in
- A Thermolabile Aldolase A Mutant Causes Fever-Induced Recurrent Rhabdomyolysis without Hemolytic Anemia
- The Role of Regulatory Evolution in Maize Domestication
- Stress Granule-Defective Mutants Deregulate Stress Responsive Transcripts
- 24-Hour Rhythms of DNA Methylation and Their Relation with Rhythms of RNA Expression in the Human Dorsolateral Prefrontal Cortex
- Pseudoautosomal Region 1 Length Polymorphism in the Human Population
- Fungal Communication Requires the MAK-2 Pathway Elements STE-20 and RAS-2, the NRC-1 Adapter STE-50 and the MAP Kinase Scaffold HAM-5
- The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural Competence
- The Protein -glucosyltransferase Rumi Modifies Eyes Shut to Promote Rhabdomere Separation in
- The Talin Head Domain Reinforces Integrin-Mediated Adhesion by Promoting Adhesion Complex Stability and Clustering
- Quantitative Genetics of CTCF Binding Reveal Local Sequence Effects and Different Modes of X-Chromosome Association
- Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the Testis
- Genetic Analysis of a Novel Tubulin Mutation That Redirects Synaptic Vesicle Targeting and Causes Neurite Degeneration in
- A Systems Genetics Approach Identifies , , and as Novel Aggressive Prostate Cancer Susceptibility Genes
- Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in
- Approximation to the Distribution of Fitness Effects across Functional Categories in Human Segregating Polymorphisms
- The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I of
- SAS-1 Is a C2 Domain Protein Critical for Centriole Integrity in
- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and Annotation
- Let's Face It—Complex Traits Are Just Not That Simple
- Glutamate Receptor Gene , Coffee, and Parkinson Disease
- The Red Queen Model of Recombination Hotspots Evolution in the Light of Archaic and Modern Human Genomes
- The Ethics of Our Inquiry: An Interview with Hank Greely
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
- Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-response
- Lack of Replication of the -by-Coffee Interaction in Parkinson Disease
- Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity
- A Germline Polymorphism of Thymine DNA Glycosylase Induces Genomic Instability and Cellular Transformation
- Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene
- ATPase-Independent Type-III Protein Secretion in
- p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide Secretion
- The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of Mitophagy
- Evolution of DNA Methylation Patterns in the Brassicaceae is Driven by Differences in Genome Organization
- Regulation of mRNA Abundance by Polypyrimidine Tract-Binding Protein-Controlled Alternate 5′ Splice Site Choice
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of Senescence
- A Functional Portrait of Med7 and the Mediator Complex in
- Systematic Analysis of the Role of RNA-Binding Proteins in the Regulation of RNA Stability
- ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
- Genomic Evidence of Rapid and Stable Adaptive Oscillations over Seasonal Time Scales in Drosophila
- Genome-Wide Associations between Genetic and Epigenetic Variation Influence mRNA Expression and Insulin Secretion in Human Pancreatic Islets
- HAM-5 Functions As a MAP Kinase Scaffold during Cell Fusion in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



