-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
The highly conserved, Nxf/Nxt (TAP/p15) RNA nuclear export pathway is important for export of most mRNAs from the nucleus, by interacting with mRNAs and promoting their passage through nuclear pores. Nxt1 is essential for viability; using a partial loss of function allele, we reveal a role for this gene in tissue specific transcription. We show that many Drosophila melanogaster testis-specific mRNAs require Nxt1 for their accumulation. The transcripts that require Nxt1 also depend on a testis-specific transcription complex, tMAC. We show that loss of Nxt1 leads to reduced transcription of tMAC targets. A reporter transcript from a tMAC-dependent promoter is under-expressed in Nxt1 mutants, however the same transcript accumulates in mutants if driven by a tMAC-independent promoter. Thus, in Drosophila primary spermatocytes, the transcription factor used to activate expression of a transcript, rather than the RNA sequence itself or the core transcription machinery, determines whether this expression requires Nxt1. We additionally find that transcripts from intron-less genes are more sensitive to loss of Nxt1 function than those from intron-containing genes and propose a mechanism in which transcript processing feeds back to increase activity of a tissue specific transcription complex.
Published in the journal: . PLoS Genet 9(6): e32767. doi:10.1371/journal.pgen.1003526
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003526Summary
The highly conserved, Nxf/Nxt (TAP/p15) RNA nuclear export pathway is important for export of most mRNAs from the nucleus, by interacting with mRNAs and promoting their passage through nuclear pores. Nxt1 is essential for viability; using a partial loss of function allele, we reveal a role for this gene in tissue specific transcription. We show that many Drosophila melanogaster testis-specific mRNAs require Nxt1 for their accumulation. The transcripts that require Nxt1 also depend on a testis-specific transcription complex, tMAC. We show that loss of Nxt1 leads to reduced transcription of tMAC targets. A reporter transcript from a tMAC-dependent promoter is under-expressed in Nxt1 mutants, however the same transcript accumulates in mutants if driven by a tMAC-independent promoter. Thus, in Drosophila primary spermatocytes, the transcription factor used to activate expression of a transcript, rather than the RNA sequence itself or the core transcription machinery, determines whether this expression requires Nxt1. We additionally find that transcripts from intron-less genes are more sensitive to loss of Nxt1 function than those from intron-containing genes and propose a mechanism in which transcript processing feeds back to increase activity of a tissue specific transcription complex.
Introduction
Export of mRNAs from the nucleus is a fundamental aspect of gene expression regulation. This involves RNA processing and packaging followed by translocation via nuclear pores (reviewed in [1], [2]). During transcription, nascent RNA associates with hnRNPs and a succession of export factors. The THO complex binds transcripts as they emerge from the RNA polymerase, and promotes transcription elongation and 3′ end processing [3]. Associated with THO are UAP56 (an RNA helicase) and REF (an adaptor protein), to make a super-complex, TREX [4]. Splicing occurs co-transcriptionally, and intron excision leads to deposition of the exon junction complex (EJC) 5′ of the splice site [5]. The EJC is also implicated in recruitment of TREX to the nascent transcript [6]. After this initial processing, a dimer comprising Nxf1 (TAP) and Nxt1 (p15) is recruited via interaction of Nxf1 with REF [7]. Recruitment of these export factors renders the mature poly adenylated mRNA competent for export via nuclear pores. mRNAs derived from intronless genes also interact with REF and UAP56, and this again recruits Nxf1/Nxt1 [8]. The Nxf1/Nxt1 dimer is released from the mRNA in the cytoplasm, and recycled to the nucleus. Most RNA polymerase II transcripts are exported via this Nxf1/Nxt1-dependent route, however some endogenous mRNAs and viral RNAs use a parallel route, dependent on the nuclear protein export factor Crm1, acting via unknown RNA-binding adapters [9]. The components of the canonical pathway are well conserved throughout eukaryotes. REF (RNA export factor) is also known as ALY or Aly (ally of AML1 and LEF1), however the protein product of the Drosophila melanogaster meiotic arrest transcriptional regulator aly (always early) is also abbreviated Aly. To avoid confusion, throughout this paper we will use REF to denote the RNA export factor, encoded in Drosophila by Ref1 and ref2, and aly (Aly) to refer to the always early gene (and protein).
Differential gene expression underlies the morphological diversity of differentiated cell types, and depends on regulation at both transcriptional and post-transcriptional levels. The production of mature sperm demonstrates the extreme morphological specialisation achievable, and relies on testis-specific expression of many genes. About 8% of annotated Drosophila protein-coding genes are expressed exclusively in testes [10], [11]. Transcription of these genes occurs predominantly in primary spermatocytes [12], [13]. The mRNAs are exported from the nucleus and either translated immediately, or, more typically, are stored in translationally inactive RNPs for translation during spermiogenesis (reviewed in [14]). Most of the “meiotic arrest” genes promote the spermatocyte-specific transcriptional programme (reviewed in [15]). Meiotic arrest testes contain only stages up to and including mature primary spermatocytes and the spermatocytes of almost all meiotic arrest mutants fail to express a very large subset of the testis-specific transcripts, as well as some ubiquitous transcripts [16], [17]. Most meiotic arrest genes encode proteins predicted to form two distinct complexes, tMAC and a testis-specific version of TFIID (reviewed in [15]). These complexes are believed to primarily regulate transcription within primary spermatocytes, rather than acting post-transcriptionally. tMAC comprises at least 6 subunits that co-purified from testis extract, encoded by aly, comr, tomb, topi, mip40 and Caf1 [18]. Genetic and protein interaction data indicate that the complex also probably contains Wuc and Achi/Vis [16], [19]. At least four tMAC subunits (Comr, Tomb, Topi and Achi+Vis) contain predicted DNA-binding motifs, and the complex localises to chromatin in primary spermatocytes, consistent with a transcriptional transactivator role [20], [21], [22], [23], [24]. The testis-specific TATA-binding proteins (tTAFs), subunits of a putative testis form of TFIID, and encoded by can, mia, nht, rye and sa, localise to promoters of target genes and the nucleolus in primary spermatocytes [25], [26], [27]. The meiotic arrest mutants are classified according to phenotype. aly-class mutants (aly, comr, tomb, topi and achi+vis) have virtually undetectable levels of a very large number of transcripts in mutant testes. All aly-class mutants encode tMAC subunits. can-class mutants (encoded by tTAF genes) have dramatically reduced expression of a large subset of the genes affected in aly-class mutants, although they express normal levels of some aly-class dependent transcripts [15]. Interestingly, wuc;aly double mutant testes, despite being mutant for two tMAC subunits actually has a can-class phenotype with respect to defects in gene expression [16]. Recently a new meiotic arrest mutant, thoc5, which encodes a THO complex subunit was reported [28]. No transcriptional defects were reported in this mutant, although transcription of only a few genes was assayed.
Here we identify Nxt1 as a meiotic arrest gene in Drosophila, and show that it is a founding member of a new meiotic arrest gene class. Partial loss of function of Nxt1 results in failure of primary spermatocytes to accumulate many transcripts. Transcripts that were sensitive to loss of Nxt1 were also dependent on tMAC for their transcription, indicating a mechanistic link between the testis-specific transcription machinery and the core mRNA export pathway. We demonstrate that the link depends on the promoter used to generate the mRNA, and not on the mRNA sequence itself. We found that spliced transcripts are expressed more efficiently in the Nxt1 loss of function background than unspliced transcripts, and propose a model in which RNA processing feeds back to increase the activity of the tissue-specific transcription complex.
Results
Nxt1 is a novel meiotic arrest gene
The line z2-0488 was identified by screening a collection of EMS-induced male sterile mutants using phase-contrast microscopy of live testis squash preparations [29]. z2-0488 homozygote and hemizygote males showed a typical meiotic arrest phenotype; the mutant testes had stages up to and including mature primary spermatocytes, however the meiotic divisions and spermatid differentiation were absent (Figure 1C). Positional cloning (see Materials and Methods and Figure S1) revealed that z2-0488 causes an amino acid substitution (D126N) in the RNA export protein Nxt1. An independent allele. Nxt1DG05102, which has a P-element insertion in the coding region, and is expected to be a null, was obtained from Bloomington Drosophila stock centre. Nxt1z2-0488 homozygotes, Nxt1z2-0488 hemizygotes and Nxt1z2-0488/Nxt1DG05102 transheterozygotes were semi-lethal and male and female sterile. Most of these pupae had head eversion defects during the pupal stages (Figure 1 H–K), accounting for the semi-lethality. Testes from Nxt1z2-0488/Nxt1DG05102 displayed a meiotic arrest phenotype (Figure 1B). Nxt1DG05102 homozygotes and hemizygotes were embryonic lethal. Depletion of Nxt1 from spermatocytes by RNAi also generated a meiotic arrest phenotype (Figure 2A), indicating that Nxt1 functions autonomously in these cells.
Fig. 1. Mutation of Nxt1 leads to meiotic arrest in testes and failure of head eversion in pupae. 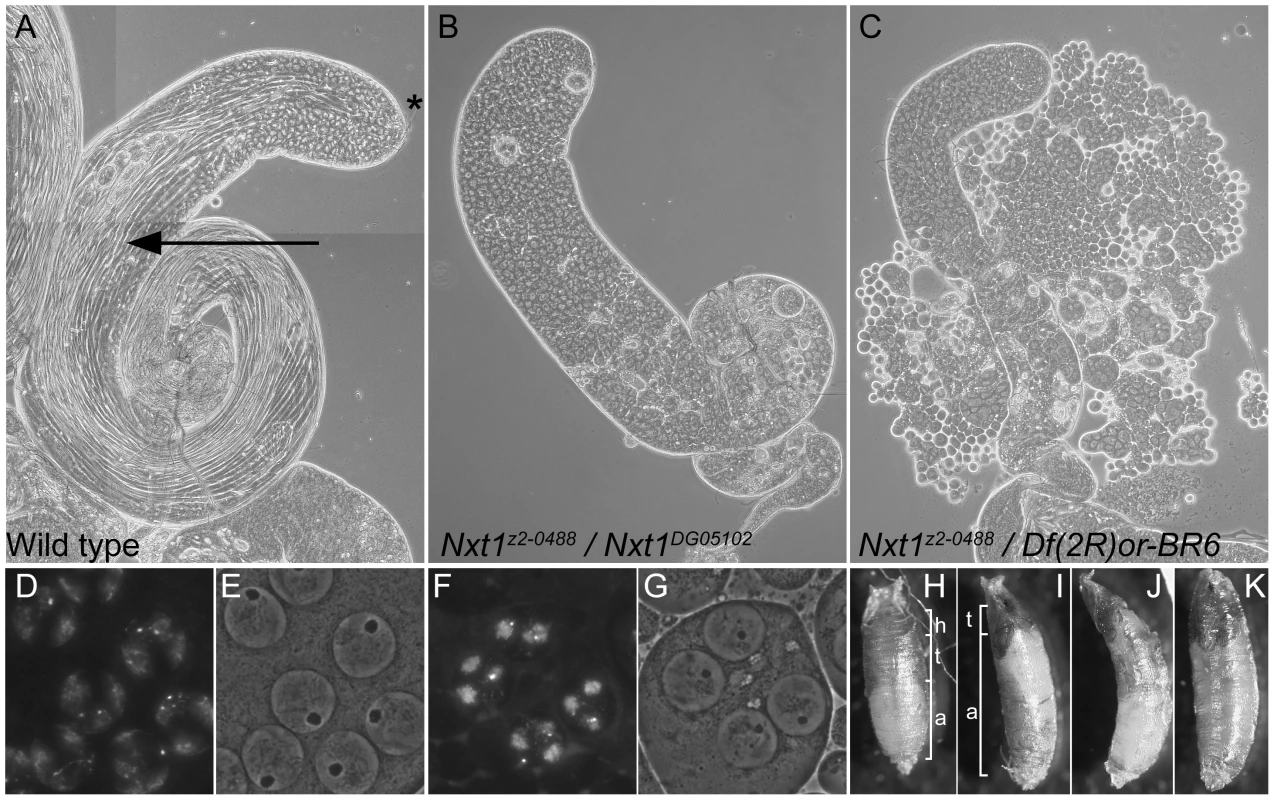
(A) All stages of spermatogenesis are seen in wild type testes by phase contrast microscopy. Large round cells near the apical region (*) are primary spermatocytes, while spermatids elongated up the length of the testis (arrow). In Nxt1z2-0488/Nxt1DG05102 mutant (B) or Nxt1z2-0488/Deficiency (C) testes, only stages up to mature primary spermatocytes are present. (D) Hoechst fluorescence and (E) phase contrast images of wild type mature primary spermatocytes reveals a prominent nucleolus and distinct chromosome territories in each nucleus. (F) Hoechst fluorescence and (G) phase contrast imaging of Nxt1 mutant spermatocytes reveals that the cells arrest with partially condensed chromatin and a prominent nucleolus. (H) Nxt1/+ pupae had everted spiracles, and distinct head, thorax and abdomen (h, t, a), while Nxt1 mutant pupae (I–K) often had only partially everted spiracles, and the thorax was at the extreme anterior of the pupal case. Many mutant pupae were also curved (J). Fig. 2. RNAi of Nxt1 in spermatocytes phenocopies the Nxt1z2-0488 defect. 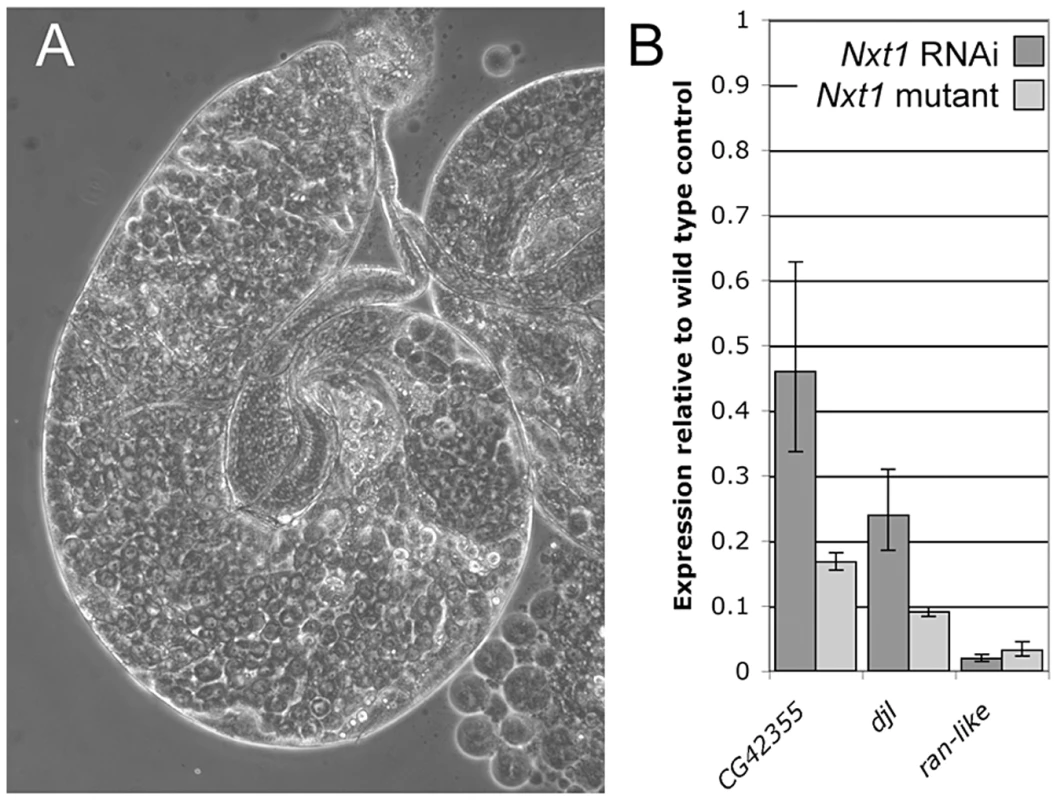
(A) Expression of an Nxt1 RNAi hairpin construct in the male germline causes a highly penetrant meiotic arrest phenotype. (B) Q-RT-PCR reveals that expression of Nxt1 target genes is reduced in Nxt1 RNAi testes (dark bars), and that the effect is similar to that seen in Nxt1z2-0488/Nxt1DG05102 mutant testes (light bars). We conducted a detailed phenotypic analysis to compare the meiotic arrest caused by Nxt1 mutation with previously described meiotic arrest loci. RNA in situ hybridisation revealed that Nxt1 mutant testes had defects in accumulation of some tMAC target mRNAs, while other tMAC target transcripts were detected in the cytoplasm of the mutant primary spermatocytes (Figure 3). Nxt1 mutant testes failed to express some genes that were expressed in nht, a can-class mutant (Figure 3). The RNA in situ hybridisation profiles thus did not match those seen in any characterised class of meiotic arrest mutant (Figure 3), suggesting that Nxt1 is the founding member of a novel class. Surprisingly, given the known role of Nxt1 in mRNA export from the nucleus [30], we detected no accumulation of transcripts within the nucleoplasm. All mRNAs detected were exclusively cytoplasmic (Figure 3H–H′″).
Fig. 3. Gene expression defects in Nxt1 mutant testes places Nxt1 in a novel meiotic arrest class. 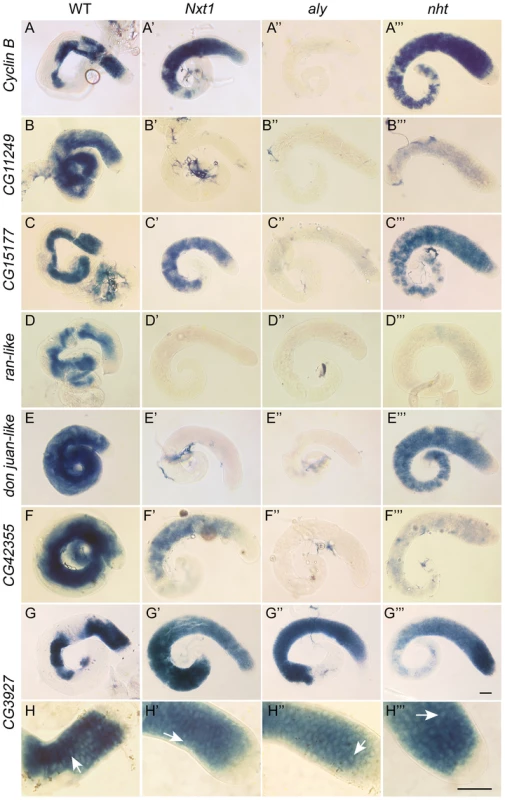
Comparison of gene expression defects in Nxt1, aly (aly-class mutant) and nht (can-class mutant) by RNA in situ hybridisation. All test genes were expressed in primary spermatocytes of control testes (A–F), with some transcripts persisting into spermatid elongation stages. No signal was detected in tMAC mutant testes (aly) (A″–F″), while all were detected at basal level (CG11249 and ran-like) or higher (CycB, CG15177, djl) in nht testes (A′″–F′″). CycB, CG15177 and CG42355 expression was detected in Nxt1z2-0488/Nxt1DG05102 mutant testes (A′″, C′″, F′″) while CG11249, ran-like and djl, were not detected in Nxt1 testes. CG3927 expression (control) is restricted to primary spermatocytes, and robust expression was detected in all four genotypes (G–G′″). Higher magnification images of the CG3927 staining (H–H′″) show a honey-comb appearance of the signal in all four genotypes, indicating mRNA accumulation predominantly in the cytoplasm rather than the nucleus (arrows). Scale bars are 50 µm. Bar in G′″ applies to A–G′″, bar in H′″ applies to H–H′″. Uniquely among meiotic arrest mutants, thoc5 primary spermatocytes showed severe fragmentation and disorganisation of the nucleolus [28]. Phase contrast microscopy revealed no overt nucleolar organisation defect in Nxt1 mutant testes, although the nucleoli in some mature primary spermatocytes appeared smaller than in wild type (Figure 1D–G). Sa-GFP, whose localisation is abnormal in both aly-class and thoc5 mutant testes, was localised to the nucleolus and indistinguishable from wild type in Nxt1 primary spermatocyte (data not shown). The chromatin in arrested spermatocytes was partially condensed, and displaced from the nuclear envelope (Figure 1F, G).
The subcellular localisation of Nxt1 is dynamic in male germline cells
We determined the sub-cellular localisation of Nxt1 protein by expressing eGFP-Nxt1 using the GAL4-UAS system. When expressed in spermatocytes (with bam-Gal4VP16), the protein was primarily nuclear, although some cytoplasmic signal was detected (Figure 4A, B). Within early primary spermatocyte nuclei the signal was weaker in the nucleolus, and enriched in a peri-nucleolar dot (Figure 4E, F). As primary spermatocytes matured this peri-nucleolar dot disappeared, and one or more cytoplasmic puncta appeared, typically apposed to the nuclear membrane (Figure 4G, H). The protein remained stable into spermatid differentiation, and was localised to one face of the nuclear envelope of early elongation stage spermatids (Figure 4I, J). The protein remained detectable in the needle-shaped nuclei of spermatids (Figure 4K, L), but was not detected in mature sperm. When eGFP-Nxt1 was expressed in ovarian somatic cells using a ubiquitous driver (tubulin-Gal4) it was predominantly nuclear; relatively uniformly distributed within the nucleoplasm, but weaker in the nucleolus. Some cytoplasmic signal was detected (Figure 4M, N).
Fig. 4. The dynamic localisation of eGFP-Nxt1. 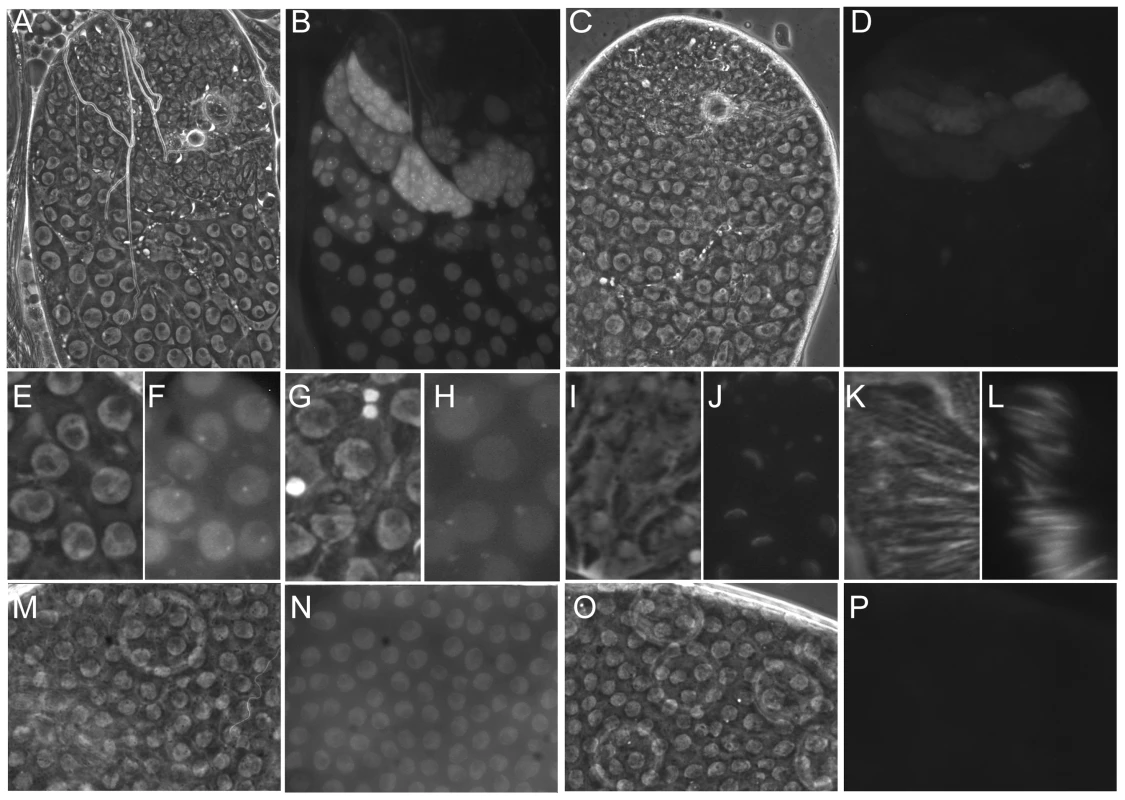
Phase contrast (A, C, E, G, I, K, M, O) and fluorescence images (B, D, F, H, J, L, N, P) of ectopically expressed eGFP-Nxt1(wild type) or eGFP-Nxt1-D126N (D, P). (A–D) Testes tips oriented with younger cells towards the top. When expressed in male germline cells the wild type protein (A, B) localised predominantly to the nucleus, and remained stable as spermatocytes matured. The signal from mutant protein was dramatically weaker, and predominantly cytoplasmic (C, D). In early primary spermatocytes the WT protein localised to an intra-nuclear dot, adjacent to the nucleolus (F) as well as throughout the nucleoplasm. As spermatocytes matured the protein relocated to one or more cytoplasmic puncta, frequently found adjacent to the uniformly labelled nucleus (H). Early elongation spermatids showed cytoplasmic puncta and eGFP-Nxt1 localisation to one face of the nuclear envelope (J). Label persisted until late elongation in spermatid nuclei (L). In the ovarian follicular epithelium the wild type eGFP-tagged protein was predominantly nuclear (N), while the D126N form of the protein was not detected (P). Nxt1 protein instability caused by D126N
The crystal structure of the human Nxf1/Nxt1 dimer has been solved, and we used this structure to predict the molecular effects of the D126N mutation [31]. The residue is conserved in metazoa (D131 in human Nxt1), and lies on a beta sheet at the Nxt1/Nxf1 dimer interface. The D131 side chain is oriented towards the core of the protein, and participates in a hydrogen bonding network with three other residues, Y24, Y39 and Q111. All three of these residues are conserved throughout metazoa, suggesting that this H-bonding network is conserved. Substitution of D131N disrupts the H-bonding network (Figure 5). This is likely to destabilise the folding around the core of the protein, particularly the interaction of the beta sheets of the dimer interface and an alpha helix on the opposite protein face. Expression of GFP-Nxt1-D126N revealed a dramatic decrease in protein stability (Figure 4). When driven with bam-Gal4VP16, fusion protein was only detected in the cells in which the driver itself was active (Figure 4C, D). The fusion protein in these cells was uniformly distributed. When expressed using tubulin-Gal4 no fusion protein could be detected under conditions in which the wild type protein was detected (Figure 4O, P). The mutation is not predicted to affect the dimer interaction with Nxf1, thus we conclude that the primary defect in the mutant is reduced protein stability, but expect that any mutant protein that does get folded is likely to be able to participate in mRNA nuclear export.
Fig. 5. Modelling the potential effect of D126N. 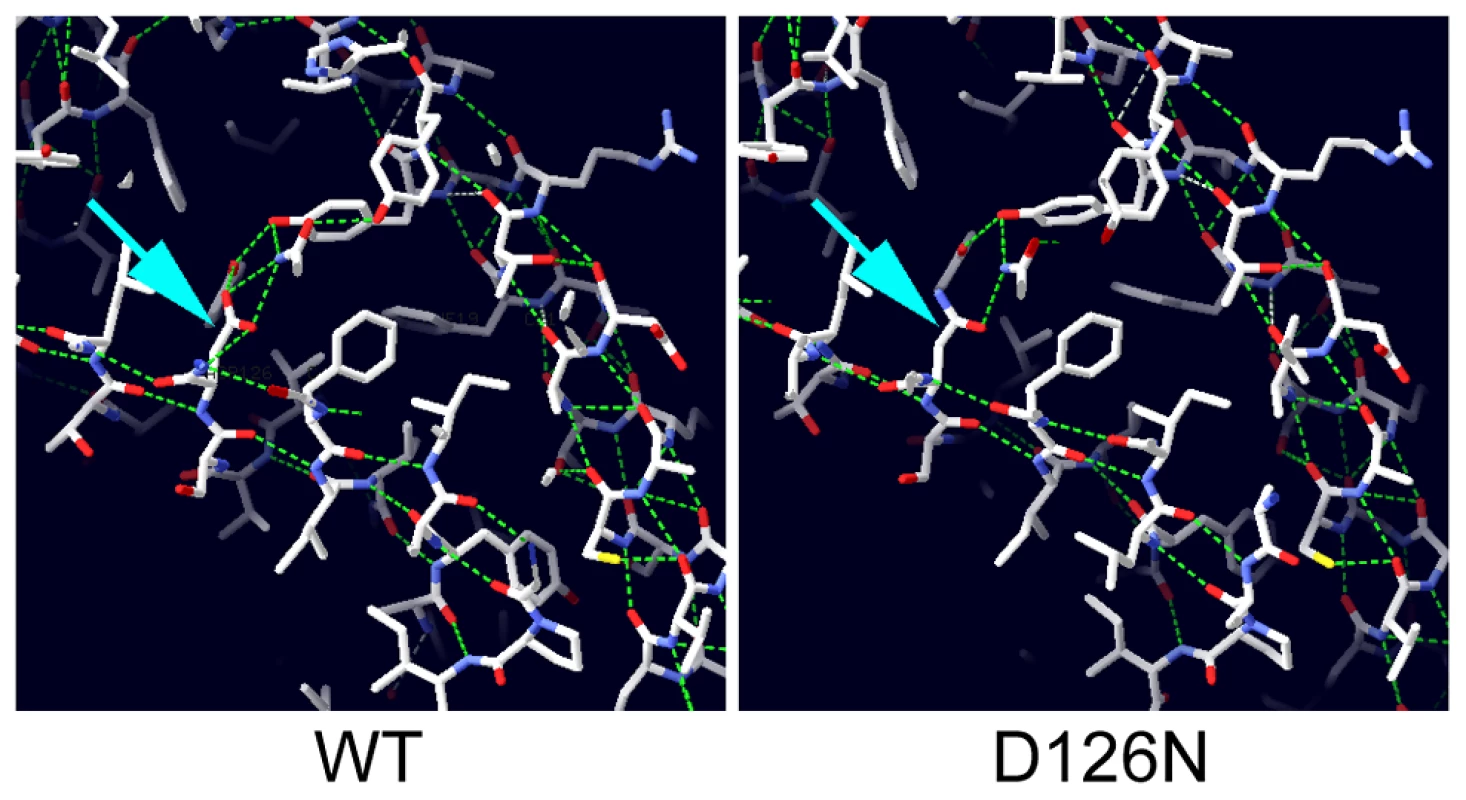
The human Nxt1 protein structure was used to model the Drosophila Nxt1 protein folding (WT) using Swisspdb viewer, and the region containing the mutated residue is shown. The side chain of D126 is indicated with an arrow. Computed H-bonds are shown in green. Running the model with the D126N substitution (arrow) revealed disruption of two computed H-bonds. Many testis-specific transcripts fail to accumulate in Nxt1 mutant testes
We used microarrays to characterise the gene expression defects in Nxt1 mutant testes. We found that Nxt1 mutant testes had dramatic defects in transcript accumulation, with many transcripts being reduced in abundance, or undetectable (Figure 6A). As with other meiotic arrest loci, there were very few transcripts whose level was elevated in mutant testes compared to controls (Figure 6A, Figure S2). 485 probes showed a 16 fold or more decrease in signal in Nxt1 mutant testes compared to normal testes. Comparison with FlyAtlas data revealed that the transcripts highly dependent on Nxt1 for expression in testes were strongly biased towards testis-specific expression (Figure 6B). 78% of all highly Nxt1-dependent transcripts were testis-specific (Figure 6D), although clearly the majority of testis-specific transcripts are not highly Nxt1-dependent (defined as 16× or more down-regulated in Nxt1 testes). To test whether the transcription defects were a peculiarity of our EMS allele, we assayed gene expression changes in the meiotic arrest testes generated by Nxt1 RNAi in spermatocytes. We found a very dramatic down-regulation of ran-like, and a mild down-regulation of expression of djl and CG42355, mirroring the results seen with Nxt1z2-0488/Nxt1DG05102 (Figure 2B).
Fig. 6. Microarray analysis of Nxt1-dependent gene expression in testes. 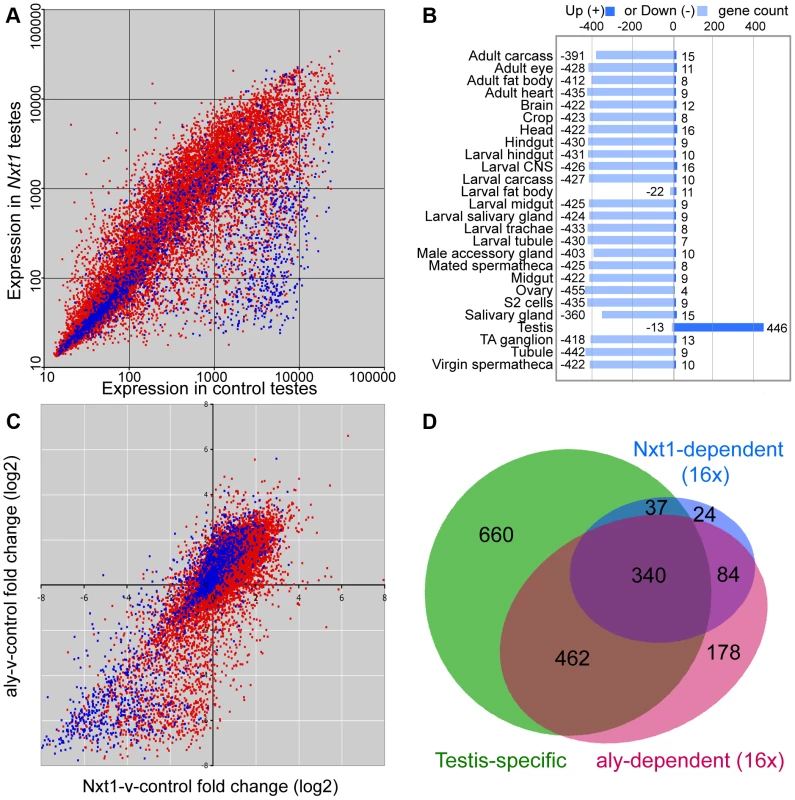
(A) Scatter plot showing normalised expression levels in control testes vs. Nxt1z2-0488/Nxt1DG05102 mutant testes of all probes on the Affymetrix expression arrays. Red dots correspond to probes for genes with introns, blue dots are probes for intron-less genes. (B) FlyAtlas expression data for 485 probes that are 16× or more down-regulated in Nxt1 testes compared to control. “Up” indicates probes whose signal is 2× or more higher in the specific tissue than whole fly, “Down” indicates 2× or more lower signal in the specific tissue than whole fly. TA ganglion = Thoracioabominal ganglion. (C) Scatter plots showing log2-transformed fold changes in Nxt1-vs-control and aly-vs.-control pairwise combination. Red indicates intron-containing genes, blue represents intron-less genes. (D) Venn diagram to show that most highly Nxt1-dependent genes (blue) have testis-specific expression (green), and that most highly Nxt1-dependent genes are also highly aly-dependent (red), but not vice versa. We compared the global effects on gene expression of Nxt1 mutation with microarray data from aly testes [16]. 1064 probes were highly (16× or more) down-regulated in aly testes. Comparison of the fold-change in Nxt1 vs. control and aly vs. control revealed that 424 (87%) of the 485 probes that were highly dependent on Nxt1 for expression were also highly dependent on aly for expression. (Figure 6C, D; Figure S2). Of the remaining 61 highly Nxt1-dependent probes, 34 were 8–16 fold down-regulated in aly, 24 were 2–8 fold down-regulated in aly and only 3 were normal or nearly-normal (less than 2× down) in aly. In contrast, the 1064 highly aly-dependent probes showed a graded response to loss of Nxt1. 424 (40%) were also highly dependent on Nxt1 (16 fold change), 186 (17.5%) showed a strong requirement for Nxt1 (8–16 fold change), 313 (29.5%) showed a mild-moderate reduction in expression (2–8 fold change) while 141 (13%) were detected at normal, or near-normal (less than 2 fold change) levels in mutant testes (Figure 6C, D; Figure S2). We did not see any reduction in the levels of transcript for any of the known tMAC subunits in Nxt1 testes (Figure S2). Immunohistochemistry indicated that Aly protein is expressed and localised to the nucleus as per wild type in Nxt1 testes (data not shown). The finding that many highly aly-dependent transcripts are expressed at near-normal levels in Nxt1 testes, coupled with the observation that no known tMAC components are under-expressed, indicate that the transcript accumulation defects seen in Nxt1 testes are very unlikely to be caused indirectly, via reduction in expression of a tMAC subunit. These data confirm that Nxt1 founds a novel meiotic arrest class of mutant, and is required for accumulation in testes of a large set of predominantly testis-specific transcripts that depend on tMAC for transcription.
The failure to accumulate target mRNAs in Nxt1 testes is primarily due to reduced transcription
In light of the known role of Nxt1 in RNA nuclear export we tested whether the gene expression defects in Nxt1 testes are the consequence of reduced transcription, or whether normal transcription occurs but the transcripts have reduced stability, or both. We compared the levels of nascent RNA to mRNA in wild type and mutant cells. If transcripts are produced in the mutant, and then degraded, the level of nascent RNA would be similar to WT, while the mRNA level is reduced. If transcription itself is reduced in the mutant then both RNA species would be lower than in WT. To distinguish between nascent and mature RNA we generated cDNA from DNase-treated total spermatocyte RNA using random primers. We amplified nascent RNA with at least one primer in an intron; for mRNA we placed one primer in an exon and the other spanning an exon-exon junction (Figure 7A). All genes selected for this analysis had a single, small, intron. Control genes, ocn and CG12699 were selected that showed no defect in expression in Nxt1 according to the microarrays. mRNAs and nascent RNAs of these control genes were expressed in mutant spermatocytes at levels similar to WT (Figure 7B). 9 genes (CG4907, CG10478, CG11249, CG14546, CG16736, CG17380, CG32487, CG33125 and pif2) were selected as being highly dependent on Nxt1 from the microarrays. For all these test genes we saw a reduction in the mature transcript level of 20–40 fold. Analysis of the nascent transcripts revealed a reduction of about 5 fold (Figure 7B). Thus the failure of Nxt1 mutant testes to accumulate testis-specific transcripts can partially be explained by reduced transcription of these genes.
Fig. 7. Expression of both nascent and mature mRNA of target genes is reduced in Nxt1 mutant spermatocytes. 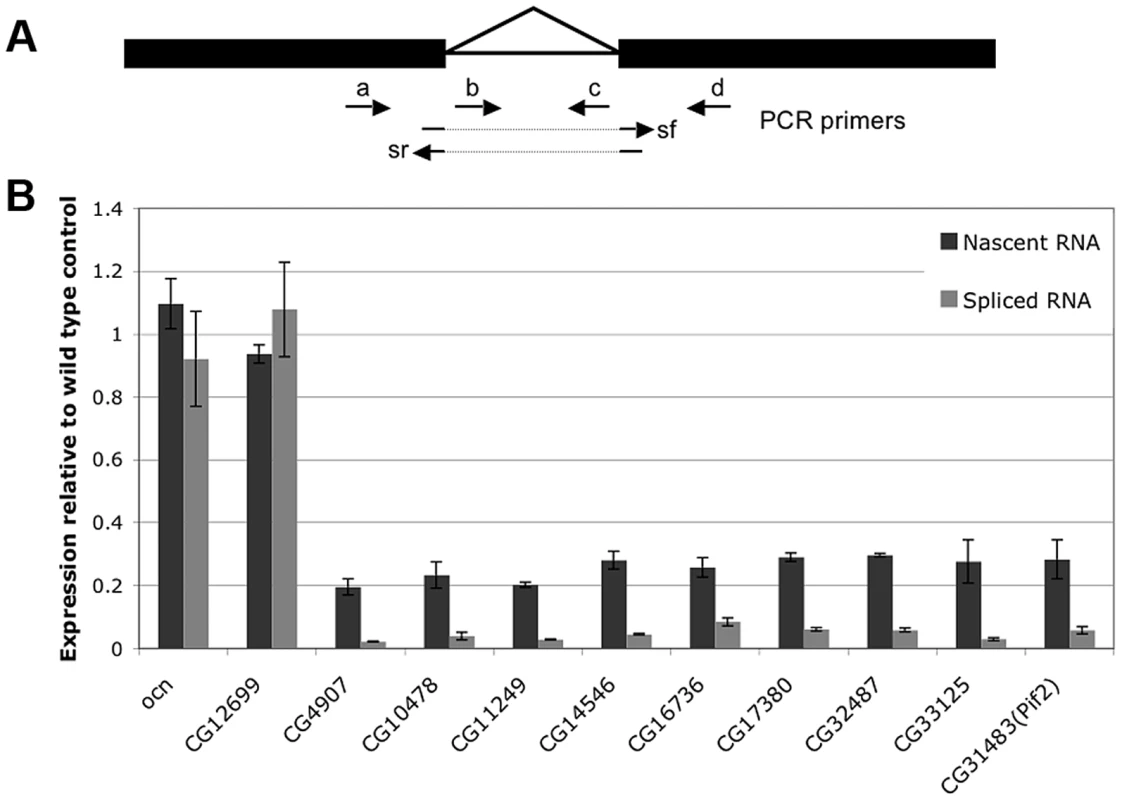
(A) Expression of nascent (unspliced) RNA was quantified using primer pairs a+c, b+c or b+d, while mRNA levels were determined with primer pairs a+sr or sf+d. (B) CG12699 and ocn control genes were expressed at broadly similar levels in WT and mutant testes, while expression of both nascent and mature RNA for the Nxt1-dependent test genes CG4907, CG10478, CG11249, CG14546, CG16736, CG17380, CG32487, CG33125 and pif2 were reduced. For each gene, the expression levels of nascent transcript (dark gray) and mature transcript (light gray) in Nxt1z2-0488/Nxt1DG05102 are presented as relative levels to those found in the wild-type (arbitrarily assigned as 1). tMAC imposes a requirement for Nxt1 on the transcripts whose expression it promotes
The finding that Nxt1-dependent transcripts all rely on tMAC for their expression suggests that the transcriptional regulation could be key to determining whether a specific transcript requires Nxt1 for accumulation. We tested this using reporter constructs in which a testis-specific transcriptional control element is used to drive expression of LacZ (Figure 8A). djl is highly transcribed in primary spermatocytes, and the transcript remains stable until late elongation, when it is translated. djl is testis-specifically expressed and depends strongly on the tMAC component aly, but less strongly on Nxt1, for its expression (Figure 3, E–E′″, Figure 8C). A genomic fragment spanning the region from −555 to +95 (relative to the transcription start site, TSS) is sufficient to drive transcription of the reporter in primary spermatocytes, and to confer translational repression on the mRNA [32]. This transcription has already been shown to be reduced in tTAF mutants [32]. RNA in situ hybridisation (data not shown) and q-RT-PCR (Figure 8C) revealed, as expected, that this djl+95-LacZ reporter construct expression is extremely low in a tMAC mutant (comr). We found a dramatic reduction in reporter expression in Nxt1 mutant primary spermatocytes compared to wild type by q-RT-PCR and RNA in situ hybridisation (Figure 8B, C). This indicates that the promoter and/or 5′UTR are critical for determining Nxt1 dependence.
Fig. 8. Nxt1 is required for full expression of tMAC dependent reporter constructs. 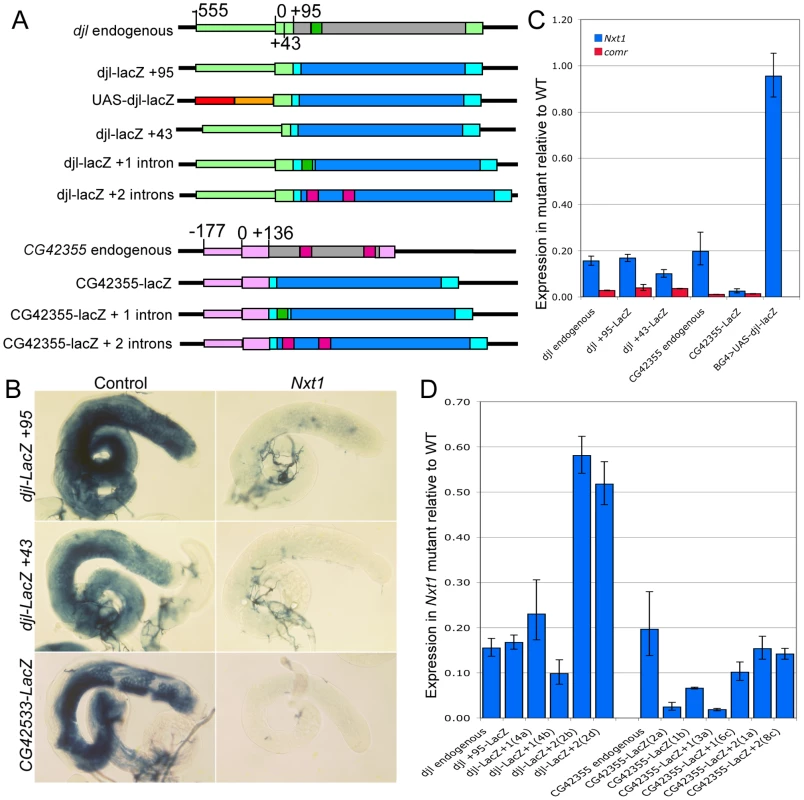
(A) Schematic diagram to illustrate reporter construct design. Promoter regions are narrow coloured boxes, transcribed regions are broad boxes. The djl promoter and UTRs are light green; ORF grey; intron dark green. CG42355 promoter and UTRs are pale pink; ORF grey; introns dark pink. UAS-djl-LacZ uses 5×UAS (red) and hsp70 minimal promoter to the TSS (orange) as the promoter. All reporter transcripts comprise 5′ UTR (from djl or CG42355) fused to Adh 5′UTR (cyan) LacZ ORF (blue) and SV40 3′UTR (cyan). (B) RNA in situ hybridisation with a LacZ probe reveals reduced expression of reporter constructs in Nxt1z2-0488/Nxt1DG05102 mutant testes compared to control. (C) q-RT-PCR to test LacZ reporter expression in mutant spermatocytes compared to controls. Expression for each transgene was normalised as 1 in the control, and the relative expression in Nxt1z2-0488 homozygotes (blue) and comr (red) mutants was calculated. (D) q-RT-PCR to test the effect of inclusion of introns on reporter expression in Nxt1z2-0488 homozygote spermatocytes compared to controls. Expression for each transgene was normalised to 1 in the control samples and relative expression in mutant was calculated. Introduction of two introns into the reporters increased expression in the mutant testes compared to the no intron version. Two separate reporter lines for transgenes are shown. To test whether the Nxt1-dependence of a specific transcript depends on the transcript sequence or on the promoter used to activate transcription, we drove expression of the djl+95-LacZ transcript independently of the djl promoter by replacing the djl promoter with 5×UAS and the Hsp70 minimal promoter (up to the TSS, Figure 8A). q-RT-PCR of spermatocyte samples revealed that reporter expression in Nxt1 mutant cells (w ; Nxt1z2-0488/Nxt1DG05102 ; UAS-djl-LacZ/bamGal4VP16) was essentially equal to that in control cells (w ; Sco/Nxt1DG05102 ; UAS-djl-LacZ/bamGal4VP16) when driven using bamGal4VP16 (Figure 8C). The transcripts from djl+95-LacZ and UAS-djl-LacZ have the same sequence; they differ only in the promoter sequence used to activate their expression. Their differential requirement for Nxt1 in accumulation of the transcript therefore reveals that the promoter sequence, rather then the RNA sequence, imposes Nxt1-dependence on the transcript.
To test whether this finding is a general property of tMAC-dependent promoters we generated a reporter using the CG42355 promoter and 5′ UTR. CG42355 is expressed specifically in testes in the same pattern as djl. CG42355 expression depends on tMAC but less strongly on Nxt1 (Figure 3 F–F′″, Figure 8C). CG42355 promoter + 5′UTR (−177/+136) - LacZ reporter lines recapitulated the native expression of the mRNA in control testes (Figure 8B), and revealed that CG42355, like djl, is significantly translationally repressed (Figure S3). Expression of this construct was barely detected in both comr and Nxt1 mutant spermatocytes (Figure 8C, Figure 9). We conclude that the tMAC-dependence of a promoter + 5′UTR is sufficient to confer Nxt1-dependence onto an exogenous transcript. The native CG42355 gene must contain features that allow the transcript to overcome this requirement for Nxt1, and be expressed at only 5× reduced levels in Nxt1 mutant cells.
Fig. 9. Addition of introns partially restores reporter expression in Nxt1 but not comr. 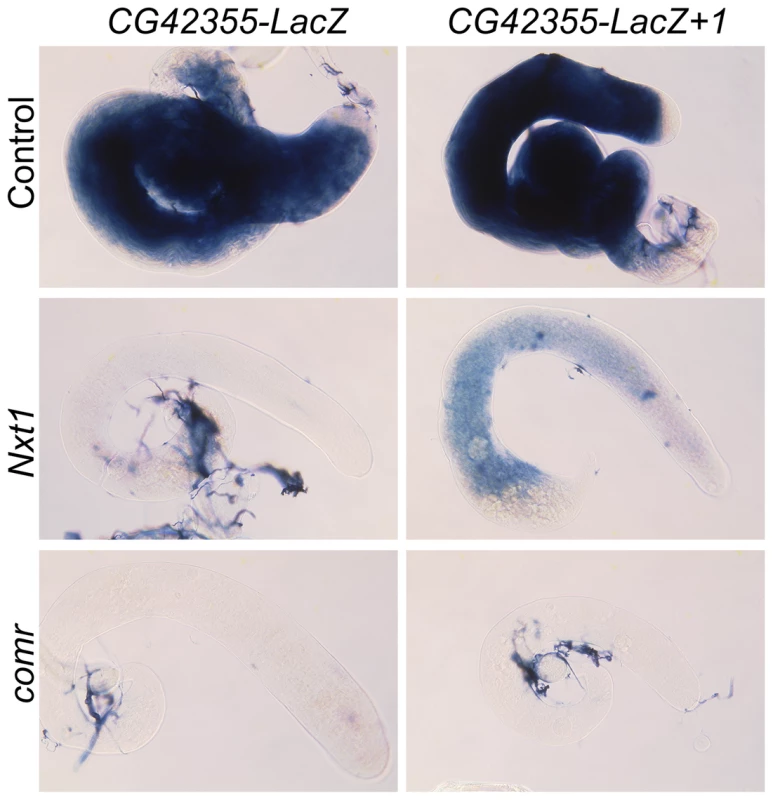
RNA in situ hybridisation reveals CG42355-LacZ reporter construct expression in control testes, and no signal in Nxt1Z2-0488/Nxt1DG05102 or comr mutant testes. Addition of one intron generates detectable, although weak, expression in Nxt1Z2-0488/Nxt1DG05102 testes, but not in comr mutant testes. Translational repression does not confer Nxt1 dependence
tMAC predominantly regulates genes whose transcripts are normally translationally repressed in the male germline; if Nxt1 is required to recognise and protect translationally repressed mRNAs then the overlap in target genes would be merely co-incidental. To test this we used a djl+43-LacZ reporter construct (Figure 8A), which is transcribed in primary spermatocytes, but not translationally repressed (Figure S3) [32]. We confirmed by both RNA in situ hybridisation (data not shown) and q-RT-PCR (Figure 8C) that transcription of this reporter depends on tMAC function. Expression of this reporter in Nxt1 mutant testes was even lower than expression of the djl+95-LacZ construct, indicating that the promoter and first 43 bp of 5′UTR, and not the translational repression sequence of the mRNA, is responsible for conferring Nxt1 dependence on this exogenous reporter transcript (Figure 8B, C). This correlates with the results obtained from the UAS-djl+95-LacZ construct, in which an mRNA that is translationally repressed accumulates normally in Nxt1 mutant testes (Figure 8B, Figure S3).
Short and intron-less genes are particularly sensitive to the lack of Nxt1 function
To understand why some tMAC-dependent transcripts require Nxt1 for their expression, while others show much lower dependence on Nxt1, we further analysed the microarrays, and focused on genes 16× or more down-regulated in Nxt1 or aly mutants. Highly aly-dependent transcripts were significantly longer (mean 1694 bp, SD 1209) than highly Nxt1-dependent transcripts (mean 1518 bp, SD 1170) (2-tailed Mann-Whitney U test, p = 0.0024). This difference was even more dramatic when the length of highly Nxt1-dependent transcripts was compared to those that depend highly on aly but are not 16× or more down-regulated in Nxt1 (mean 1800 bp, SD 1238) (2-tailed Mann-Whitney U test, p<0.0001). Thus we conclude that shorter transcripts are more likely to be dramatically under-expressed in Nxt1 testes than longer transcripts. However CG42355 encodes a short transcript (<1 kb) that is only mildly down-regulated in Nxt1 mutant testes, while the LacZ reporter driven by this promoter is much longer (>3 kb), and is dramatically under-expressed in the mutant, so other features of the transcript must also be implicated.
Approximately 22% of all annotated Drosophila genes lack introns. 1412 genes are expressed specifically in testis (based on FlyAtlas data) and 521 (37%) of these lack introns, a significantly higher proportion than expected by chance (Chi-squared, p<0.0001). Thus, intron-less genes are enriched for testis-specific expression. Of the 1019 genes that were 16× or more reduced in aly testes, 404 (40%) lack introns, indicating that the presence or absence of introns does not affect whether a testis-specific transcript is also highly aly-dependent (Chi-squared, p = 0.18). In contrast, of the 456 highly Nxt1-dependent genes, 303 (66%) lack introns, revealing that highly Nxt1-dependent genes are significantly more likely to lack introns than randomly selected testis-specific genes (Chi-squared, p<0.0001). Genes that are highly dependent on both aly and Nxt1 for their expression are predominantly intron-less while genes that are highly dependent on aly, but are only mildly dependent on Nxt1 predominantly have introns (Figure 6D). Notably, the more introns present in a strongly tMAC-dependent gene, the higher its expression in an Nxt1 background (Figure S4). We analysed the 300 most down-regulated genes in a tMAC mutant, and correlated fold change compared to control in Nxt1 mutants with intron number (0, 1, 2, 3+). Relative expression of genes lacking introns was significantly lower than that of genes containing introns (t-test, p = 1.18889E-32). Relative expression of genes with 1 intron was not significantly different than that of 2-intron genes (t-test, p = 0.126), however genes with 1 or 2 introns had significantly lower expression in Nxt1 mutants compared to wild type than genes with 3 or more introns (t-test, p = 0.00013). This indicates a dose-response relationship between gene expression in Nxt1 mutant testes and intron number, where the more introns a tMAC-dependent gene, has the lower its requirement for Nxt1 for expression.
The presence of introns partially negates the requirement for Nxt1 in reporter gene expression
The microarray results indicate that having at least one intron is beneficial for expression of tMAC-dependent genes in Nxt1 mutants. We used reporter constructs to test how the presence or absence of introns impacts on reporter gene expression in Nxt1 testes (Figure 8A). We inserted the djl intron into the 5′UTR of (the previously intron-less) djl −555/+95 reporter sequence, and found that the reporter was testis-specifically expressed, and translation was delayed until spermatid elongation stages, in a pattern indistinguishable from the original construct. Similarly, insertion of the CG42355 introns into the LacZ coding sequence of the djl construct resulted in expression mirroring the original reporter (Figure S3). Equivalent one and two intron CG42355 reporters were constructed (Figure 8A). We confirmed correct splicing of these transcripts by RT-PCR (data not shown).
When we tested expression of these intron-containing reporters in Nxt1 spermatocytes we found that insertion of the introns partially restored the reporter expression (Figure 8D, Figure 9). Strikingly, the djl - reporter with two introns was expressed at over 50% of wild type levels. Similarly, introduction of two introns into the CG42355 reporter elevated expression to 15% of that seen for the reporter in wild type spermatocytes. This represents a 10× increase in expression in mutants compared to the intron-less version of the reporter. Insertion of introns did not increase reporter expression in comr mutant testes (Figure 9). We conclude that, in spermatocytes, tMAC-dependent promoters confer dependence on Nxt1 for transcript expression, but that this dependence can be partially overcome if the transcript has one or more introns.
Discussion
Identification of Nxt1 as a novel regulator of gene expression in the male germline
To date 14 meiotic arrest loci have been described (aly, achi-vis, topi, tomb, comr, wuc, mip40, can, mia, sa, rye, nht, thoc5, Nurf301) [15], [28], [33]. Most of these encode subunits of either a testis-specific TFIID complex or tMAC. The exceptions are mutations resulting in a C-terminally truncated product from Nurf301 [33], and loss of thoc5 [28]. In both these cases the effect on gene expression is apparently much less dramatic than loss of either tTAFs or tMAC. Here we identify Nxt1 as a novel meiotic arrest locus, and show that the phenotype does not fit with any previous classification. Notably, mutation of Nxt1 has much more dramatic effects on gene expression than thoc5, Nurf301 or tTAFs, although the defect is not as pronounced as that seen in most tMAC mutants.
Nxt1 protein acts in the same biochemical pathway as the TREX subunit, Thoc5, suggesting that the entire RNA export pathway might be critical for testis gene expression. In support of this, in preliminary experiments we have observed incomplete meiotic arrest phenotypes in testes with spermatocyte specific knock down (by RNAi) of either Ref1 or sbr (data not shown). No defects in gene expression were detected in the thoc5 mutant [28], but we note that the genes assayed (bol, twe, cycB, dj, fzo, Mst87F) all contain at least one intron, and, although fzo, bol and dj have reduced expression in Nxt1 testes, none is on the list of genes 16× or more down regulated in this background. It would be very interesting to determine whether the intron-less genes most dependent on Nxt1 for expression are also dependent on thoc5 or other RNA export factors. Notably, Nxt1 and Thoc5 localise to a punctate structure adjacent to the nucleolus in early primary spermatocytes and to a structure adjacent to the nuclear envelope in late primary spematocytes. This localisation adjacent to the nuclear envelope is also seen for Nxf1, the binding partner of Nxt1, encoded in Drosophila by the sbr gene [28], [30], [34]. Both Nxt1 and Sbr also localise to one face of the nucleus in spermatids. This co-localisation suggests that the proteins are all working together in these cells.
A model to explain how tMAC and the RNA export factors together regulate testis gene expression
The direct link we describe here, between tMAC and RNA export in regulating testis gene expression, can be explained by two theoretical mechanisms. (1) tMAC could promote transcription of target genes and feed-forward to promote their stability and RNA export. In the absence of Nxt1 the mRNAs would not be protected, and instead would be degraded. (2) Initially low tMAC activity promotes production of low levels of transcript. Binding of the export factors to these transcripts during processing would feed-back to increase the activity of tMAC. This would then increase the transcription of the tMAC-dependent gene. The ability of introns to rescue expression in the mutant indicates that the mechanism must be mediated through the RNA transcript, and presumably depends on the higher affinity of spliced transcripts for the Nxt1/Nxf1 dimer. If the feed-forward mechanism predominates, the rate of nascent transcript production should be equal to wild type. If feed-back predominates, the rate of nascent transcript production would be lower than wild type. We found a reduction in both nascent and mature transcripts in the mutant, although the reduction in mRNA is greater than the reduction in nascent RNA. We propose that both feed-forward and feedback occur, to give an amplification loop. This amplification would depend on a specific interaction between the RNA export factors and tMAC.
The RNA nuclear export pathway and tissue specific transcript accumulation
The RNA nuclear export pathway has been deduced primarily using experiments in yeast, and in tissue culture cells (reviewed in [1]). Drosophila culture cells depleted of components of the pathway, including Nxt1, accumulate poly-adenylated mRNAs within the nucleus. Most transcripts in the cell are also reduced in abundance, typically a 1.5–2 fold level change for altered transcripts [35]. Very few transcripts were reduced more than 5 fold after RNAi treatment, and very few were significantly increased in abundance. The RNAi treatments used in these experiments also caused defects in cell proliferation or viability. The in vivo role of sbr, which encodes the Nxf1 partner of Nxt1, has been investigated in embryos and larvae, and again defects have been detected in bulk export of mRNAs from nuclei [36]. In none of these experiments has a link been detected between the role of the Nxt1/Nxf1 and transcription.
Factors in the mRNA export pathway, upstream of Nxt1/Nxf1 have been linked to transcription, consistent with the fact that RNA splicing and processing is co-transcriptional. Specifically the TREX complex interacts with chromatin and RNA polymerase II, and is important for facilitating transcription elongation, 3′ end formation, mRNA processing, and transfer to the nuclear pore [4]. While the interaction of TREX with RNA polymerase is well established, there has been no data to implicate Nxt1/Nxf1 in this process. TREX promotes association of REF with the transcript, and this in turn promotes Nxt1/Nxf1 association [37]. Similarly, the spliceosome can have a stimulatory effect on transcription, both at the level of initiation and elongation [38]. Our study is the first, to our knowledge, to implicate these pathways in the regulation of a programme of gene expression promoted by a specific transcription factor complex.
Recently two studies have demonstrated that the stability of specific S. cerevisiae mRNAs can depend on non-transcribed promoter sequences. RPL30 mRNA was shown to have a short half-life imposed by the transcriptional activator Rap1 and its binding site in the UAS, although how this affects the RNA is not yet determined [39]. The cell cycle regulation of the half-lives of SWI5 and CLB2 was independent of the transcript sequences, was coordinated with transcription, and was promoter dependent [40]. Dbf2, a kinase, is recruited by specific promoter sequences and co-transcriptionally deposited on the SWI5 and CLB2 mRNAs. Dbf2 is a component of the CCR4-NOT cytoplasmic deadenylase complex, and the activity of this complex could then normally de-stabilise these transcripts. In both these examples the promoter acts to confer a short half-life on the transcripts it regulates. We report a similar phenomenon in an animal system, however the outcome is increased mRNA expression, with the RNA processing pathway feeding back to increase transcription.
Why don't transcripts accumulate in Nxt1 mutant nuclei?
The canonical role for Nxt1 is export of mRNAs from the nucleus, however, paradoxically, and in contrast to the results from tissue culture studies [41], we do not detect accumulation of mRNAs in Nxt1 mutant nuclei. The majority of mRNAs expressed in the mutant spermatocytes are present at normal levels, and are detected in the cytoplasm. This mRNA export capability could be provided by residual Nxt1 function, or by a parallel pathway. We suggest instead that residual Nxt1 function provided by the hypomorphic allele is sufficient for mRNA export. This interpretation is supported by the observation that the null allele is embryonic lethal while the z2-0488 allele is viable, thus sufficient activity remains in this allele to support normal function of most cells. The fact that reduction in Nxt1 levels by RNAi in spermatocytes phenocopies the z2-0488 allele indicates that the effect is due to reduced activity, rather than an allele-specific effect. A well characterised alternative mRNA export pathway involves the Crm1 protein, binding RNA via an unidentified adaptor protein. Crm1 is implicated in export of a subset of endogenous RNAs, as well as HIV mRNA in humans [42]. The Drosophila Crm1 protein is encoded by the gene embargoed. This gene is expressed at much lower levels in testes than in other adult tissues, and thus is unlikely to represent the major RNA export pathway in spermatocytes, given their high level of transcriptional activity.
How does the presence of introns promote transcript accumulation in Nxt1 mutant spermatocytes
Nxt1/Nxf1 heterodimers are already known to be implicated in export of some mRNAs with retained introns, and have been demonstrated to interact with constitutive transport elements present in some cellular and viral mRNA [43]. Similarly, in human cells naturally intronless transcripts tested were transported from the nucleus in a TREX and Nxf1 dependent manner [44]. In the absence of Nxf1 the transcripts remained nuclear, but apparently were not destabilised. When nascent transcripts are being produced they are bound by hnRNPs, and those with introns are processed by the spliceosome. During this processing the EJC associates with the transcript. This is then responsible for recruitment of Ref1, which recruits Nxt1/Nxf1. Transcripts that lack introns also associate with Ref1 and thus Nxt1/Nxf1, but without the help of the EJC. Thus the EJC increases the affinity of Nxt1/Nxf1 to the transcript. For a transcript being synthesised in an Nxt1-depleted background this could be sufficient to ensure association of the export factor, and thus to ensure proper processing and export. We detect a dose response for intron number, specifically, the more introns a tMAC dependent gene has the higher its expression in an Nxt1 mutant background. If, as we propose, there is feedback from the export pathway to increase tMAC activity, then the presence of the EJC and the increased affinity for Nxt1/Nxf1 would result in higher transcript levels from intron-containing genes than intron-less genes in an Nxt1-depleted background, consistent with our findings.
Introns and evolution of testis-enriched genes
It is well demonstrated that the presence of introns in primary transcripts correlates with higher gene expression levels [45]; this is true for almost all Drosophila adult tissues. Indeed, most ectopic expression systems include at least one intron to facilitate higher transcription levels, higher transcript stability and higher translation efficiency. However the testis is unique among Drosophila adult tissues in that, amongst genes whose expression is detected, the expression level of intron-containing genes is significantly lower than that of intron-less genes (Mann-Whitney U test, p<0.001) (deduced from FlyAtlas data; expression is in arbitrary units). Specifically, the median expression signal for intron-containing genes in brain (a typical somatic tissue) is 139, while that for intron-less genes is 78.1. In contrast, the equivalent figures for testes are 84 and 168.5. In testis, and in somatic tissues, approximately 60–65% of all annotated intron-containing protein coding genes are expressed. In somatic cells only 30–35% of all annotated intron-less genes are expressed. In contrast, in testes, 63% of all annotated intron-containing genes are expressed (Figure S5). Thus spermatocytes must have evolved a mechanism to support high level expression of intron-less genes. Many of the intron-less genes expressed in testes are retroposed copies of intron-containing genes. Typically the parent gene will have a broad or ubiquitous expression pattern while the retroposed gene's expression is highly restricted [46]. The new gene is often subject to rapid evolution under positive selection, and contributes to sperm-specific cell biology. Most of these retroposed gene copies are regulated by the tMAC transcriptional regulation complex, and, like other tMAC-dependent genes, they typically have short promoters. Surprisingly, the promoter regions, and even transcription start sites, are not highly conserved between species [47], although the insertions are biased towards genomic regions already containing testis-specific transcription units [48]. This could be explained by low-affinity tMAC binding occurring by virtue of the chromosomal domain, promoting transcription of the newly inserted sequence, and feedback from RNA export factors serving to increase tMAC activity, and thus expression of the new gene.
Nxt1 function is not limited to the male germline
In this study we have demonstrated a link between the core RNA export pathway and testis-specific transcription. Null alleles of Nxt1 are recessive lethal, and the hypomorph (homozygous or hemizygous has low viability. The animals often pupate with a distinctive elongated curved shape, uneverted spiracles, and then fail in head eversion, consistent with defects in air bubble movement. These processes are controlled by transcriptional responses downstream of ecdysone signalling, and the mutant phenotype is highly reminiscent of mutants in Eip74EF, an ets family transcription factor [49]. Nxt1z2-0488 homozygous or hemizygous adult females are also sterile. These highly reproducible defects potentially also arise as a result of defects in gene expression in the relevant tissues, rather than being caused by a general defect in export of all mRNAs from the nuclei of somatic or female germline cells. In testes we have demonstrated that the effect is via a specific transcription complex, tMAC. The soma and female germline defects cannot be caused by an interaction between Nxt1 and tMAC since tMAC expression is restricted to testes. We postulate that Nxt1 might be important for regulating the transcriptional response to ecdysone during pupariation, and expression of specific genes in the female germline. Thus we suggest that Nxt1 could work with, as yet unidentified, specific transcription factors to control gene expression in other tissues.
Materials and Methods
Cloning of z2-0488
We mapped the male and female sterility, and semi-lethality, of z2-0488 using deficiency chromosomes to a region of approximately 100 kb containing 22 known or predicted genes at the distal end of chromosome 2R (Figure S1). 9 candidate genes were sequenced, and a single non-synonymous change was found (D126N, codon GAT-AAT) in the Nxt1 gene. A P-element insertion allele of Nxt1, P{wHy}DG05102, failed to complement the semi-lethality and sterility of the z2-0488 mutation. This P-element insertion, which is in the first exon, just downstream of the ATG, and predicted to be a null allele, results in homozygous and hemizygous lethality. We conclude that z2-0488 is a hypomorphic allele of Nxt1, and designate the allele Nxt1z2-0488.
Microscopy and in situ hybridisation
Testes were dissected from young adults, prepared for phase contrast and fluorescence microscopy as in [50] and imaged using a Hamamatsu Orca-05G camera driven by HCImage software on an Olympus BX50 microscope. RNA in situ hybridisation using dig-labelled RNA probes was as in [51]. Primer sequences used to generate templates are in Text S1. CycB probe was made from a cDNA clone as in [17]. Templates for CG42355 and djl were subcloned into pGEM-T-Easy, while other probes were generated directly from PCR products. Beta-galactosidase activity assays on whole mount testes were as in [17]. Colour images were taken with a JVC-F75U camera run by KY-LINK software mounted on the Olympus BX50 microscope. Image composites were assembled using Adobe Photoshop.
Plasmid construction
djl-lacZ −555/+95 and djl-lacZ −555/+43 [32] were provided by Renate Renkawitz-Pohl. PCR from genomic DNA yielded a fragment 5′-EcoRI-CG42355-177/+136-BamHI-3′ which was cloned into pCaSpeR4-AUG-betaGAL [52] to yield CG42355-LacZ −177/+136. The plasmids djl-lacZ -555/+95 +1 intron, djl-lacZ −555/+95 +2 introns, djl-lacZ −555/+43 +1 intron, djl-lacZ −555/+43 +2 introns, CG42355-lacZ −177/+136 +1 intron and CG42355-lacZ +2 introns were derived from these basic promoter-reporter constructs by incorporation of a synthetic DNA fragment containing the intron(s) (see Text S1 for sequences and construction details). pUAST-eGFP-Nxt1 and pUAST-eGFP-Nxt1-D126N were generated by PCR and subcloning of the Nxt1 ORF from Nxt1z2-0488/CyO flies [53]. pCaSpeR4-UAS-djl - AUG-betaGAL was made by cloning a synthetic fragment comprising 5× UAS; Hsp70 minimal promoter (up to the TSS); +1–+95 of djl into the MCS of pCaSpeR4-AUG-betaGAL using EcoRI and BamH1 (Text S1).
Drosophila methods
Drosophila were maintained on cornmeal, sucrose, yeast, agar medium, at 25°C (29°C for RNAi crosses). Mutant alleles were aly5, comrZ1340, nhtZ2-5946, Nxt1DG05102, Nxt1z2-0488. w1118 was used as a control. For RNAi against Nxt1 we used UAS-hairpin lines from the Vienna Drosophila Resource Centre [54] driven by bam-Gal4VP16, with both constructs heterozygous. bam-Gal4VP16 was also used to drive fusion protein constructs in spermatocytes. tub-Gal4 was from the Bloomington Drosophila stock centre and was used to drive ubiquitous fusion protein expression. Transgenic lines were selected after injection of P-element constructs into w1118 using standard methods. Insertions on the third chromosome were selected, balanced, and crossed into Nxt1z2-0488, Nxt1DG05102 and comrZ1340 mutant backgrounds.
Purification of RNA from spermatocytes
For q-RT-PCR, total RNA was extracted from purified spermatocyte samples using the RNAqueous micro kit (Invitrogen). Testes were dissected from 1–3 adult males of the appropriate genotype in TB (183 mM KCl, 47 mM NaCl, 10 mM Tris pH7.4), transferred to a small drop of TB on a hydrophobic plate, and the testis tip was cut open using a tungsten needle. Spermatocytes were pushed out of the testis sheath into the buffer with the needle and the remainder of the testis discarded. Most of the buffer was removed, leaving approximately 1–2 µl, and 10 µl lysis solution was added. The lysate was added to 90 µl lysis solution in a microfuge tube; immediately frozen in liquid nitrogen, and stored at −80°C. Preparation of each sample took no more than 10 minutes. RNA was purified according the manufacturer's instructions. RNA was eluted from the spin column with 2×6.5 µl of elution buffer. The total eluate volume was approximately 11 µl.
RT-PCR and q-RT-PCR
For conventional q-RT-PCR, cDNA was synthesised using Superscript III (Invitrogen) and oligo-dT primers. For analysis of nascent RNA and spliced products the purified RNA was treated with DNase I, half of the RNA sample was reserved as a control to assay DNA contamination and the remainder was reverse transcribed using Superscript III and random hexamer primers. PCR primers (sequences in Text S1) were used that recognised exon, intron or exon-exon junction sequences. The cDNA reaction was diluted to 60 µl with water, and 1 µl of this was used as a template in PCR reactions, using PowerSybr reagent (ABI) in a Chromo4 instrument (MJR). For q-RT-PCR of reporter gene expression and CG42355 and djl levels, the entire RNA eluate was used for cDNA synthesis with oligo dT primers. CG3927 was used as a control gene for relative quantitation using the ΔΔCt method. Expression of the test gene relative to CG3927 gene was normalised to 1 in the control sample. Control testes had the relevant transgene present in the same copy number as the test sample, but had normal testis morphology. CG3927 expression is restricted to primary spermatocytes (Figure 3). CG3927 expression in Nxt1 and comr mutant testes is similar to wild type, as judged by both RNA in situ hybridisation and microarray analysis. All reactions were performed in triplicate. We performed biological replicates for the djl+95 and djl+43 lines, for which only one insertion was available. For other reporters we used two different third-chromosome insertion lines, and show both results.
Microarray and bioinformatics
Testes from Nxt1z2-0488/Nxt1DG05102 transheterozygote males, raised at 25°C were used for Affymetrix microarray analysis, and were compared to our existing microarray data sets. Sample preparation, processing and data analysis were as described in [16]. The FlyMine interface was used to extract lists of probes associated with intron-containing and intron-less genes, as well as to extract data from FlyAtlas on gene expression profiles in adult and larval tissues. A list of 1412 genes with testis-specific expression (1523 probes) was created by filtering of FlyAtlas data [11]. We selected those probes where the present call in testis was 4/4, and sum of present calls in all other adult tissues was <4.
FlyMine (v27.0, Feb 2011) was used to generate lists of genes with and without introns. First a list of all annotated Drosophila melanogaster genes was created. A list of genes with introns were selected using the “Gene → Introns” template, and selecting the column “genes” as output. A list of all genes without introns was generated by subtracting the genes with introns list from the all genes list. The “Gene → Affymetrix probe” template was used to generate a list of all probes for intron-less genes. This was then combined with our Affymetrix data in Microsoft Excel to allow us to analyse intron-containing and intron-less genes separately. The number of introns present in each of the 300 genes most down regulated in tomb was determined by first ranking Affymetrix fold change data for tomb-vs.-control, then manually checking annotations in FlyBase. Annotated transcripts were cross-referenced with the RNA-seq data for adult males to ensure accuracy. All genes with valid male - (probably testis-) expressed spliced transcripts were scored as having an intron, even if they also encoded alternative intron-less transcripts.
Supporting Information
Zdroje
1. CarmodySR, WenteSR (2009) mRNA nuclear export at a glance. J Cell Sci 122 : 1933–1937.
2. ErkmannJA, KutayU (2004) Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp Cell Res 296 : 12–20.
3. JimenoW, RondónA, LunaR, AfuileraA (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21 : 3526–3535.
4. SträsserK, MasudaS, MasonP, PfannstielJ, OppizziM, et al. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417 : 304–308.
5. Le HirH, GatfieldD, IzaurraldeE, MooreM (2001) The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20 : 4987–4997.
6. ZhouZ, LuoM, StraesserK, KatahiraJ, HurtE, et al. (2000) The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407 : 401–405.
7. SträsserK, HurtE (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 19 : 410–420.
8. AbruzziK, LacadieS, RosbashM (2004) Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J 23 : 2620–2631.
9. WatanabeM, FukudaM, YoshidaM, YanagidaM, NishidaE (1999) Involvement of CRM1, a nuclear export receptor, in mRNA export in mammalian cells and fission yeast. Genes Cells 4 : 291–297.
10. AndrewsJ, BouffardGG, CheadleC, LuJN, BeckerKG, et al. (2000) Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Research 10 : 2030–2043.
11. ChintapalliV, WangJ, DowJ (2007) Using FlyAtlas to identify better Drosophila models of human disease. Nat Genet 39 : 715–720.
12. OlivieriG, OlivieriA (1965) Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. Mutat Res 2 : 366–380.
13. ZhaoJ, KlyneG, BensonE, GudmannsdottirE, White-CooperH, et al. (2010) FlyTED: the Drosophila testis gene expression database. Nucleic Acids Research 38: D710–715.
14. SchäferM, NayerniaK, EngelW, SchäferU (1995) Translational control in spermatogenesis. Developmental Biology 172 : 344–352.
15. White-CooperH (2010) Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 139 : 11–21.
16. DoggettK, JiangJ, AletiG, White-CooperH (2011) Wake-up-call, a lin-52 paralogue, and Always early, a lin-9 homologue physically interact, but have opposing functions in regulating testis-specific gene expression. Dev Biol 355 : 381–393.
17. White-CooperH, SchaferMA, AlpheyLS, FullerMT (1998) Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 125 : 125–134.
18. BeallEL, LewisPW, BellM, RochaM, JonesDL, et al. (2007) Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-MuvB. Genes & Development 21 : 904–919.
19. WangZ, MannRS (2003) Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogensis. Development 130 : 2853–2865.
20. AyyarS, JiangJ, ColluA, White-CooperH, WhiteR (2003) Drosophila TGIF is essential for developmentally regulated transcription in spermatogenesis. Development 130 : 2841–2852.
21. JiangJ, BensonE, BausekN, DoggettK, White-CooperH (2007) Tombola, a tesmin/TSO1 family protein, regulates transcriptional activation in the Drosophila male germline and physically interacts with Always early. Development 134 : 1549–1559.
22. JiangJ, White-CooperH (2003) Transcriptional activation in Drosophila spermatogenesis involves the mutually dependent function of aly and a novel meiotic arrest gene cookie monster. Development 130 : 563–573.
23. PerezgazgaL, JiangJ, BolivalB, HillerMA, BensonE, et al. (2004) Regulation of transcription of meiotic cell cycle and terminal differentiation genes by the testis-specific Zn finger protein matotopetli. Development 131 : 1691–1702.
24. White-CooperH, LeroyD, MacQueenA, FullerMT (2000) Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development 127 : 5463–5473.
25. ChenX, HillerMA, SancakY, FullerMT (2005) Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310 : 869–872.
26. HillerMA, ChenX, PringleMJ, SuchorolskiM, SancakY, et al. (2004) Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131 : 5297–5308.
27. HillerMA, LinT-Y, WoodC, FullerMT (2001) Developmental regulation of transcription by a tissue-specific TAF homolog. Genes and Development 15 : 1021–1030.
28. MoonS, ChoB, MinS-H, LeeD, ChungYD (2011) The THO complex is required for nucleolar integrity in Drosophila spermatocytes. Development 138 : 3835–3845.
29. KoundakjianE, CowanD, HardyR, BeckerA (2004) The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167 : 203–206.
30. HeroldA, KlymenkoT, IzaurraldeE (2001) NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7 : 1768–1780.
31. FribourgS, BraunIC, IzaurraldeE, ContiE (2001) Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Molecular Cell 8 : 645–656.
32. HempelL, RathkeC, RajaS, Renkawitz-PohlR (2006) In Drosophila, don juan and don juan like encode proteins of the spermatid nucleus and the flagellum and both are regulated at the transriptional level by the TAFII80 cannonball while translational repressio is achieved by distinct elements. Developmental Dynamics 235 : 1053–1064.
33. KwonS, XiaoH, WuC, BadenhorstP (2009) Alternative splicing of NURF301 generates distinct NURF chromatin remodelling complexes with altered modified histone binding specificities. PLoS Genet 5: e1000574.
34. AtsapkinaA, GolubkovaE, KasatkinaV, AvanesyanE, IvankovaN, et al. (2010) Peculiarities of spermatogenesis in Drosophila melanogaster: role of main transport receptor of mRNA (Dm NXF1). Cell and Tissue Research 4 : 1–7.
35. HeroldA, TeixeiraL, IzaurraldeE (2003) Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J 22 : 2472–2483.
36. WilkieGS, ZimyaninV, KirbyR, KoreyC, Francis-LangH, et al. (2001) Small bristles, the Drosophila ortholog of NXF1, is essential for mRNA export throughout development. RNA 7 : 1781–1792.
37. KatahiraJ (2012) mRNA export and the TREX complex. Biochimica et Biophysica Acta (BBA) 18119 : 507–513.
38. ManleyJ (2002) Nuclear coupling: RNA processing reaches back to transcription. Nature Structural Biology 9 : 790–791.
39. BregmanA, Avraham-KelbertM, BarkaiO, DuekL, GutermanA, et al. (2011) Promoter elements regulate cytoplasmic mRNA decay. Cell 147 : 1473–1483.
40. TrcekT, LarsonD, MoldónA, QueryC, SingerR (2011) Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147 : 1484–1497.
41. HeroldA, KlymenkoT, IzaurraldeE (2001) NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7 : 1768–1780.
42. CullenB (2003) Nuclear mRNA export: insights from virology. Trends Biochem Sci 28 : 419–424.
43. GrüterP, TaberneroC, von KobbeC, SchmittC, SaavedraC, et al. (1998) TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cel 1 : 649–659.
44. LeiH, DiasA, ReedR (2011) Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc Natl Acad Sci U S A 108 : 17985–17990.
45. Le HirH, NottA, MooreM (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28 : 215–220.
46. DaiH, YoshimatsuT, LongM (2006) Retrogene movement within - and between-chromosomes in the evolution of Drosophila genomes. Gene 385 : 96–102.
47. SorourianM, BetranE (2010) Turnover and lineage-specific broadening of the transcription start site in a testis-specific retrogene. Fly 4 : 3–11.
48. BaiY, CasolaC, BetranE (2008) Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics 9 : 241.
49. FletcherJ, BurtisK, HognessD, ThummelC (1995) The Drosophila E74 gene is required for metamorphosis and plays a role in the polytene chromosome puffing response to ecdysone. Development 121 : 1455–1465.
50. White-Cooper H (2004) Spermatogenesis: analysis of meiosis and morphogenesis. In: Henderson D, editor. Drosophila Cytogenetics Protocols. Totowa, New Jersey: Humana Press. pp. 45–75.
51. MorrisC, BensonE, White-CooperH (2009) Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nature Protocols 4 : 1807–1819.
52. ThummelC, BouletA, LipshitzHD (1988) Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene 74 : 445–456.
53. ParkerL, GrossS, AlpheyL (2001) Vectors for the expression of tagged proteins in Drosophila. Biotechniques 31 : 1280–1286.
54. DietzlG, ChenD, SchnorrerF, SuK, BarinovaY, et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 : 151–156.
55. TweedieS, AshburnerM, FallsK, LeylandP, McQuiltonP, et al. (2009) FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Research 37: D555–D559.
Štítky
Genetika Reprodukční medicína
Článek PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant TransformationČlánek The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene ClusterČlánek Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish PathogenČlánek USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and TranscriptionČlánek Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA EditingČlánek Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 6- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- BMS1 Is Mutated in Aplasia Cutis Congenita
- High Trans-ethnic Replicability of GWAS Results Implies Common Causal Variants
- How Cool Is That: An Interview with Caroline Dean
- Genetic Architecture of Vitamin B and Folate Levels Uncovered Applying Deeply Sequenced Large Datasets
- Juvenile Hormone and Insulin Regulate Trehalose Homeostasis in the Red Flour Beetle,
- Meiosis-Specific Stable Binding of Augmin to Acentrosomal Spindle Poles Promotes Biased Microtubule Assembly in Oocytes
- Environmental Dependence of Genetic Constraint
- H3.3-H4 Tetramer Splitting Events Feature Cell-Type Specific Enhancers
- Network Topologies and Convergent Aetiologies Arising from Deletions and Duplications Observed in Individuals with Autism
- Effectively Identifying eQTLs from Multiple Tissues by Combining Mixed Model and Meta-analytic Approaches
- Altered Splicing of the BIN1 Muscle-Specific Exon in Humans and Dogs with Highly Progressive Centronuclear Myopathy
- The NADPH Metabolic Network Regulates Human Cardiomyopathy and Reductive Stress in
- Negative Regulation of Notch Signaling by Xylose
- A Genome-Wide, Fine-Scale Map of Natural Pigmentation Variation in
- Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals mC within Archaeal mRNAs
- Multiplexin Promotes Heart but Not Aorta Morphogenesis by Polarized Enhancement of Slit/Robo Activity at the Heart Lumen
- Latent Effects of Hsp90 Mutants Revealed at Reduced Expression Levels
- Impact of Natural Genetic Variation on Gene Expression Dynamics
- DeepSAGE Reveals Genetic Variants Associated with Alternative Polyadenylation and Expression of Coding and Non-coding Transcripts
- The Identification of -acting Factors That Regulate the Expression of via the Osteoarthritis Susceptibility SNP rs143383
- Pervasive Transcription of the Human Genome Produces Thousands of Previously Unidentified Long Intergenic Noncoding RNAs
- The RNA Export Factor, Nxt1, Is Required for Tissue Specific Transcriptional Regulation
- Inferring Demographic History from a Spectrum of Shared Haplotype Lengths
- Histone Acetyl Transferase 1 Is Essential for Mammalian Development, Genome Stability, and the Processing of Newly Synthesized Histones H3 and H4
- PARP-1 Regulates Metastatic Melanoma through Modulation of Vimentin-induced Malignant Transformation
- DNA Methylation Restricts Lineage-specific Functions of Transcription Factor Gata4 during Embryonic Stem Cell Differentiation
- The Genome of : Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Deregulation of the Protocadherin Gene Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy
- Evidence for Two Different Regulatory Mechanisms Linking Replication and Segregation of Chromosome II
- USF1 and hSET1A Mediated Epigenetic Modifications Regulate Lineage Differentiation and Transcription
- Methylation of Histone H3 on Lysine 79 Associates with a Group of Replication Origins and Helps Limit DNA Replication Once per Cell Cycle
- A Six Months Exercise Intervention Influences the Genome-wide DNA Methylation Pattern in Human Adipose Tissue
- The Gene Desert Mammary Carcinoma Susceptibility Locus Regulates Modifying Mammary Epithelial Cell Differentiation and Proliferation
- Hooked and Cooked: A Fish Killer Genome Exposed
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
- Mutations in Cause Autosomal Recessive Congenital Ichthyosis in Humans
- Integrated Transcriptomic and Epigenomic Analysis of Primary Human Lung Epithelial Cell Differentiation
- RSR-2, the Ortholog of Human Spliceosomal Component SRm300/SRRM2, Regulates Development by Influencing the Transcriptional Machinery
- Comparative Polygenic Analysis of Maximal Ethanol Accumulation Capacity and Tolerance to High Ethanol Levels of Cell Proliferation in Yeast
- SPO11-Independent DNA Repair Foci and Their Role in Meiotic Silencing
- Budding Yeast ATM/ATR Control Meiotic Double-Strand Break (DSB) Levels by Down-Regulating Rec114, an Essential Component of the DSB-machinery
- Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing
- Functional Analysis of Neuronal MicroRNAs in Dauer Formation by Combinational Genetics and Neuronal miRISC Immunoprecipitation
- DNA Ligase IV Supports Imprecise End Joining Independently of Its Catalytic Activity
- Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene
- Heritable Change Caused by Transient Transcription Errors
- From Many, One: Genetic Control of Prolificacy during Maize Domestication
- Neuronal Target Identification Requires AHA-1-Mediated Fine-Tuning of Wnt Signaling in
- Loss of Catalytically Inactive Lipid Phosphatase Myotubularin-related Protein 12 Impairs Myotubularin Stability and Promotes Centronuclear Myopathy in Zebrafish
- H-NS Can Facilitate Specific DNA-binding by RNA Polymerase in AT-rich Gene Regulatory Regions
- Prophage Dynamics and Contributions to Pathogenic Traits
- Global DNA Hypermethylation in Down Syndrome Placenta
- Fragile DNA Motifs Trigger Mutagenesis at Distant Chromosomal Loci in
- Disturbed Local Auxin Homeostasis Enhances Cellular Anisotropy and Reveals Alternative Wiring of Auxin-ethylene Crosstalk in Seminal Roots
- Causes and Consequences of Chromatin Variation between Inbred Mice
- Genome-scale Analysis of FNR Reveals Complex Features of Transcription Factor Binding
- Distinct and Atypical Intrinsic and Extrinsic Cell Death Pathways between Photoreceptor Cell Types upon Specific Ablation of in Cone Photoreceptors
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- BMS1 Is Mutated in Aplasia Cutis Congenita
- Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits
- Distinctive Expansion of Potential Virulence Genes in the Genome of the Oomycete Fish Pathogen
- Distinct Neuroblastoma-associated Alterations of Impair Sympathetic Neuronal Differentiation in Zebrafish Models
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



