-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Calpain-5 Mutations Cause Autoimmune Uveitis, Retinal Neovascularization, and Photoreceptor Degeneration
Autosomal dominant neovascular inflammatory vitreoretinopathy (ADNIV) is an autoimmune condition of the eye that sequentially mimics uveitis, retinitis pigmentosa, and proliferative diabetic retinopathy as it progresses to complete blindness. We identified two different missense mutations in the CAPN5 gene in three ADNIV kindreds. CAPN5 encodes calpain-5, a calcium-activated cysteine protease that is expressed in retinal photoreceptor cells. Both mutations cause mislocalization from the cell membrane to the cytosol, and structural modeling reveals that both mutations lie within a calcium-sensitive domain near the active site. CAPN5 is only the second member of the large calpain gene family to cause a human Mendelian disorder, and this is the first report of a specific molecular cause for autoimmune eye disease. Further investigation of these mutations is likely to provide insight into the pathophysiologic mechanisms of common diseases ranging from autoimmune disorders to diabetic retinopathy.
Published in the journal: . PLoS Genet 8(10): e32767. doi:10.1371/journal.pgen.1003001
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003001Summary
Autosomal dominant neovascular inflammatory vitreoretinopathy (ADNIV) is an autoimmune condition of the eye that sequentially mimics uveitis, retinitis pigmentosa, and proliferative diabetic retinopathy as it progresses to complete blindness. We identified two different missense mutations in the CAPN5 gene in three ADNIV kindreds. CAPN5 encodes calpain-5, a calcium-activated cysteine protease that is expressed in retinal photoreceptor cells. Both mutations cause mislocalization from the cell membrane to the cytosol, and structural modeling reveals that both mutations lie within a calcium-sensitive domain near the active site. CAPN5 is only the second member of the large calpain gene family to cause a human Mendelian disorder, and this is the first report of a specific molecular cause for autoimmune eye disease. Further investigation of these mutations is likely to provide insight into the pathophysiologic mechanisms of common diseases ranging from autoimmune disorders to diabetic retinopathy.
Introduction
Autosomal dominant neovascular inflammatory vitreoretinopathy (ADNIV) is a heritable autoimmune condition. It is characterized by various stages that mimic several much more common eye diseases, including: uveitis, retinitis pigmentosa, proliferative diabetic retinopathy and proliferative vitreoretinopathy [1], [2]. Together, these diseases account for a significant fraction of visual morbidity and human blindness [3], [4]. Identification of a gene that generates the varied pathological features of these common conditions could have a significant impact on the understanding and treatment of blindness [5]. Although there are numerous causative genes for retinitis pigmentosa, only a handful of genes have previously been associated with intraocular inflammation, neovascularization and fibrotic disease [6].
Because of its similarity to other common eye diseases, ADNIV patients are often misdiagnosed, unless the familial nature of their disease is recognized. Bennett and co-workers described the original ADNIV family, ADNIV-1 in this study [1], in which the characteristic clinical findings were transmitted in an autosomal dominant fashion through eight-generations. The disease onset in this family varies between 10 and 30 years of age and the disease course can be divided into five stages, each lasting approximately ten years (Figure 1, Figure S1) [2]. In the first stage, ADNIV is clinically indistinguishable from an autoimmune, non-infectious uveitis [7]. Although the retina appears normal, an abnormality is detectable with electroretinography very early in the course of the disease. In the second stage, retinitis pigmentosa-like photoreceptor degeneration is apparent. In the third stage, retinal neovascularization develops, which is very similar to the pathologic angiogenesis of proliferative diabetic retinopathy [8]. In the fourth stage, intraocular fibrosis leads to retinal detachment, similar to that seen in proliferative vitreoretinopathy [9]. In the fifth stage, continued inflammation, intraocular hemorrhage, neovascular glaucoma, fibrosis and retinal detachment eventually lead to phthisis and complete blindness. There are no systemic features in this condition. This combination of overlapping clinical conditions is unusual and suggests that the disease-causing mutations may act through multiple pathways.
Fig. 1. ADNIV phenotype. 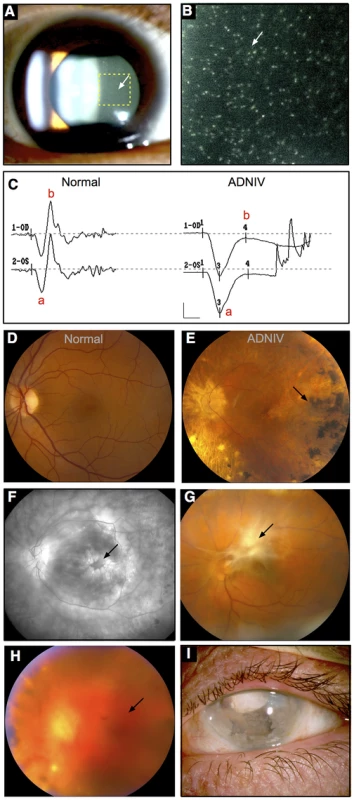
A–B. Clusters of autoimmune reactive leukocytes are visible in the vitreous cavity (inset, arrows). C. Electroretinography shows loss of the b-wave. D. Fundus image of the normal retina. E. Fundus image of the ADNIV retina shows pigmentary degeneration (arrow) similar to retinitis pigmentosa. F. Fluorescein angiography reveals cystoid macular edema at the fovea (arrow), a consequence of autoimmune intraocular inflammation. G. Preretinal fibrosis leads to tractional retinal detachments. H. Vitreous hemorrhage (arrow) from retinal neovascularization. I. Phthisis bulbi and involution of eye tissues in end-stage ADNIV. Stone and co-workers previously mapped the genetic locus for ADNIV to chromosome 11q13 [10]. In this study, we identified two new ADNIV families, and these additional subjects provided an opportunity to refine the genetic interval and identify the causative gene. Two different mutations were identified among three ADNIV families in the gene encoding calpain-5, an intracellular calcium-activated cysteine protease with an unknown physiological function.
Results
Clinical Findings
The new ADNIV families displayed a phenotype very similar to the original ADNIV-1 pedigree. Specifically, affected members showed all of the previously reported clinical signs of the disease (Figure 1, Figure S1) including: non-infectious uveitis (Figure 1A, 1B), early loss of the b-wave on electroretinography (Figure 1C), pigmentary retinal degeneration (Figure 1D, 1E), cystoid macular edema, (Figure 1F), retinal and iris neovascularization, vitreous hemorrhage (Figure 1H), epiretinal membrane formation, proliferative vitreoretinopathy (Figure 1G), retinal detachment, cataract, neovascular glaucoma and ultimately phthisis and complete blindness (Figure 1I) [1]. Each pedigree was consistent with autosomal dominant inheritance with complete penetrance (Figure 2). There were sixty-one affected subjects in ADNIV-1, seven in ADNIV-2, and thirty-one in ADNIV-3 (Figure 2). Forty-two of these 99 affected individuals (42%) were male. The clinical severity of the disease was indistinguishable between affected males and females. With the exception of psoriasis in one individual, there were no other systemic, autoimmune or inflammatory conditions present in any of the affected family members.
Fig. 2. ADNIV pedigrees. 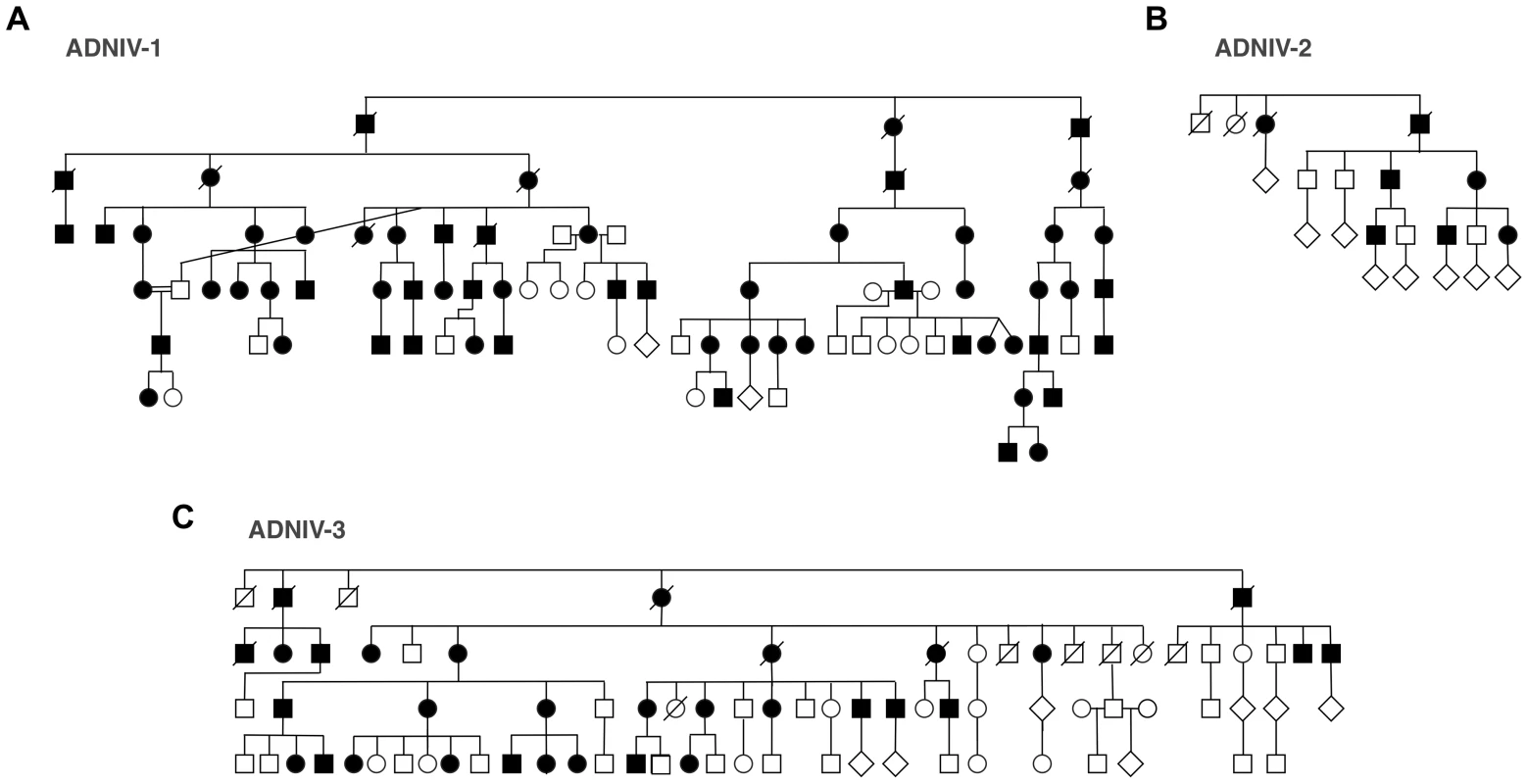
A–C. Three families with clinical features of ADNIV exhibit a dominant pattern of inheritance. Black symbols represent clinically affected subjects. Open symbols represent unaffected subjects. Deceased individuals are marked by a slash. Linkage Analysis
Prior linkage analysis of the ADNIV-1 family mapped the disease-causing mutation to a 22-megabase (chr11 : 91,760,018–69,339,635) interval on chromosome 11q13 (Figure 3A) [10]. Genotyping of the ADNIV-2 and ADNIV-3 families with short tandem repeat polymorphisms was consistent with linkage to the same locus. Haplotype analysis was suggestive of an ancestral relationship between ADNIV-1 and ADNIV-2. In addition, two affected individuals in the ADNIV-3 family were found to be recombinant within the disease interval, narrowing it to 6.5 megabases between D11S4139 and D11S1789. High resolution SNP genotyping of ADNIV-1 and ADNIV-3 further reduced the interval to the 6 megabases between rs879380 and D11S1789, a region harboring 86 known genes (Figure 3A).
Fig. 3. The CAPN5 gene harbors mutations in exon 6 of ADNIV subjects. 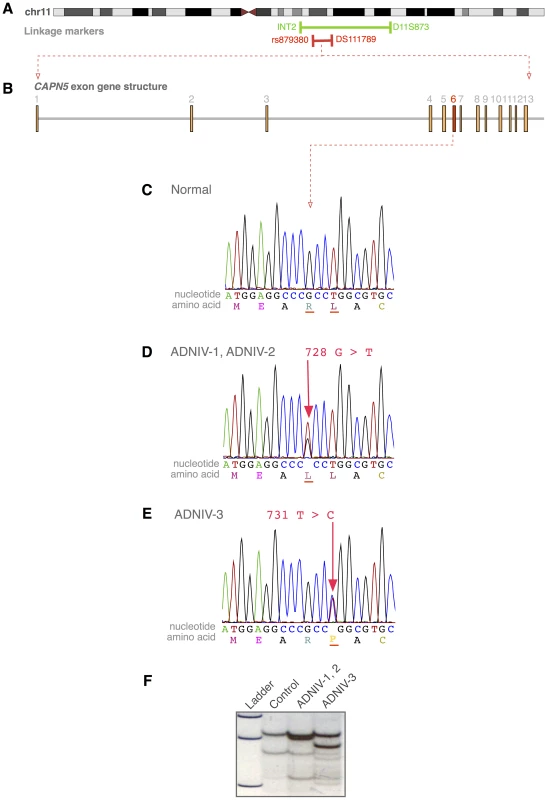
A. The ADNIV locus was previously mapped to chromosome 11q13 (green bar). STRP and SNP array mapping narrowed the interval (red bar). B. CAPN5 gene structure is composed of 13 exons. C. Chromatogram of normal CAPN5 DNA sequence in exon 6. D–E. Chromatograms of ADNIV subjects shows mutations in CAPN5 exon 6. F. SSCP distinguishes between normal sequence and ADNIV mutations. Exome Sequencing
Whole-exome sequencing was performed using DNA from two affected family members from the ADNIV-1 pedigree who were separated by seven meioses. Only one of the resultant sequence variations met the following four criteria: located within the 6 Mb ADNIV interval, shared by the two affected members of ADNIV-1, not previously reported as a polymorphism, and nonsynonymous. This variant is a guanine to thymine nucleotide change (c.728G>T, p.Arg243Leu) in exon 6 of the CAPN5 gene (NM_004055) (Figure 3B–3D). A combination of SSCP and Sanger sequencing of CAPN5 exon 6 verified that this mutation was present in all the affected members and none of the unaffected members of ADNIV-1.
The coding sequence of CAPN5 was then sequenced in affected members of the two other ADNIV pedigrees. All affected members of ADNIV-2 were found to harbor the same heterozygous variant (c.728G>T, p.Arg243Leu) found in ADNIV-1, supporting the suspected ancestral relationship between these two families (Figure 3D). All affected members of the ADNIV-3 family, were found to harbor a heterozygous variant in the adjacent codon, a thymine to cytosine change (c.731T>C, p.Leu244Pro) (Figure 3E). Both of these putative disease-causing variants in exon 6 of CAPN5 were easily detectable by SSCP in ADNIV patients, but were absent from all unaffected adult members of the ADNIV families (no asymptomatic minors were tested) as well as 272 ethnically similar control individuals (Figure 3F). None of the three variants were listed in the dbSNP or 1000 Genome databases. In addition, none of the variant alleles were found in the over 10,700 CAPN5 alleles sequenced in the NHLBI Exome Sequencing Project (http://evs.gs.washington.edu/EVS/)
Structural Analysis of Calpain-5 ADNIV Mutations
Calpain-5 is an intracellular calcium-activated cysteine protease (NP_004046) with evolutionarily conserved domains required for protease activity. Both ADNIV-causing mutations were found in exon 6, which encodes a major part of the catalytic domain and contains two of the three catalytic residues that compose the active site (Figure 4). Modeling of secondary structure suggests that both the ADNIV-1/2 (p.Arg243Leu) and ADNIV-3 (p.Leu244Pro) mutations lie within a nearby alpha helical domain. The first of these mutations removes a charged residue while the second disrupts the putative helical structure (Figure 4A).
Fig. 4. Protein structure modeling of calpain-5 and ADNIV mutants. 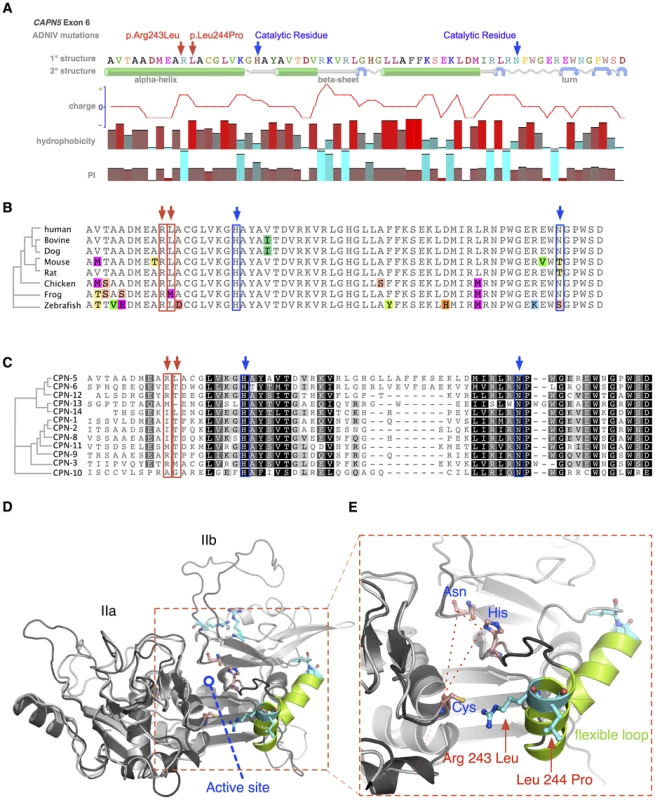
A. Both ADNIV mutations (red arrows) are located in exon 6, which encodes a portion of the catalytic domain and two of three catalytic residues (blue arrows). Primary protein sequence analysis shows the ADNIV-1/2 and ADNIV-3 mutations to be 8–9 amino acids upstream of the catalytic histidine residue. Secondary structure modeling shows that the two mutations are within a putative alpha helical domain. One mutated codon results in the loss of a basic residue, while the other introduces a proline into the putative alpha helix. B. Alignment of calpain-5 orthologs shows very high evolutionary conservation of the mutated residues (red arrows). Amino acid mismatches are color-highlighted. C. Twelve human calpain paralogs show significant differences in exon 6 (Black, 100% similarity; Dark grey, 80–100% similarity; Light grey, 60–80% similarity; White, less than 60% similarity). D. Three-dimensional modeling of the catalytic domain shows the location of the active site cleft (red outline). E. Magnified view of the active site cleft shows the catalytic triad (dashed line – blue text) and location of the two mutations (red arrows and text). Both mutations are located within a peptide loop that is homologous to a flexible loop of calpain-1 that undergoes a calcium-induced conformational change in association with regulation of the active site cleft (see text). The amino acid sequence of CAPN5 exon 6 is highly conserved across vertebrate species (Figure 4B). The catalytic residues show 100% conservation among CAPN5 orthologs. Interestingly, there is also 100% conservation of the Arg243 residue mutated in ADNIV-1/2 and 88% conservation of the Leu244 mutated in ADNIV-3. This small evolutionary divergence at the latter codon is also quite conservative: methionine for leucine in the frog. In contrast, the disease-causing mutation at this codon introduces a new proline bend within a putative alpha helix.
Previous comparisons of calpain-5 to its human paralogs demonstrated that it has diverged significantly and now belongs to its own subfamily with calpain-6 [11]. This divergence is also evident within exon 6 alone, where the calpain-5 catalytic domain shows relatively low homology to other human calpains (Figure 4C). Each of the ADNIV mutant residues is conserved in four or fewer of 12 calpain paralogs, suggesting that the residues mutated in ADNIV are specifically important to calpain-5 function and may physiologically distinguish it from the other calpains. The PolyPhen2 sequence analysis program predicted both ADNIV mutations to have damaging effects on protein function (0.999 for Arg243Leu and 0.998 for Leu244Pro) comparable to an active site Cys81Ser mutation (1.0). The SIFT program predicted the Leu244Pro mutation to be comparably pathogenic (0.04) to Cys81Ser (0.03) but predicted the Arg243Leu mutation to be better tolerated (0.1).
To better examine the relationship of ADNIV mutations within the calpain-5 catalytic domain, homology modeling to calpain-2 (m-calpain) was used to generate a three-dimensional structure for calpain-5 (Figure 4D) [12], [13]. Both mutations were outside the active site cleft and relatively far removed from the calcium-binding domains and the binding site of the endogenous inhibitor calpastatin. Interestingly, both the ADNIV-1/2, and ADNIV-3 mutations fell into a region of low electron density, suggesting the presence of a flexible loop (Figure 4E). In calpain-1 (μ-calpain) models, the homologous loop undergoes calcium-induced conformational changes that regulate the proximity of catalytic residues within the active site cleft [14]. This putative loop contains both ADNIV mutants and is highly conserved among all calpain-5 orthologs (Figure 4B).
Calpain-5 Expression in the Retina
We evaluated the CAPN5 transcript in human retinal tissue using RNA sequencing. The transcript was observed at a level of 4.63 fragments per kilobase of exon per million, which places it between the first quartile and the median level of expression for all transcripts observed in the retina. No significant splice variants were detected. Two antibodies against calpain-5 were used to determine whether calpain-5 protein could also be detected in human retinal tissue sections. Both antibodies showed strong calpain-5 expression in the photoreceptor cells (Figure 5A, 5B). There was a punctate pattern of labeling over the nuclei and inner segments with less expression along the outer segments and outer plexiform layer. There was no significant expression in the nerve fiber layer, ganglion cell layer, inner nuclear layer, inner plexiform layer, or retinal pigment epithelium. The localization to the photoreceptor cells is consistent with both the early electrophysiologic abnormalities and the later photoreceptor degeneration seen in ADNIV patients.
Fig. 5. Calpain-5 expression in human retinal photoreceptor cells and cultured cells. 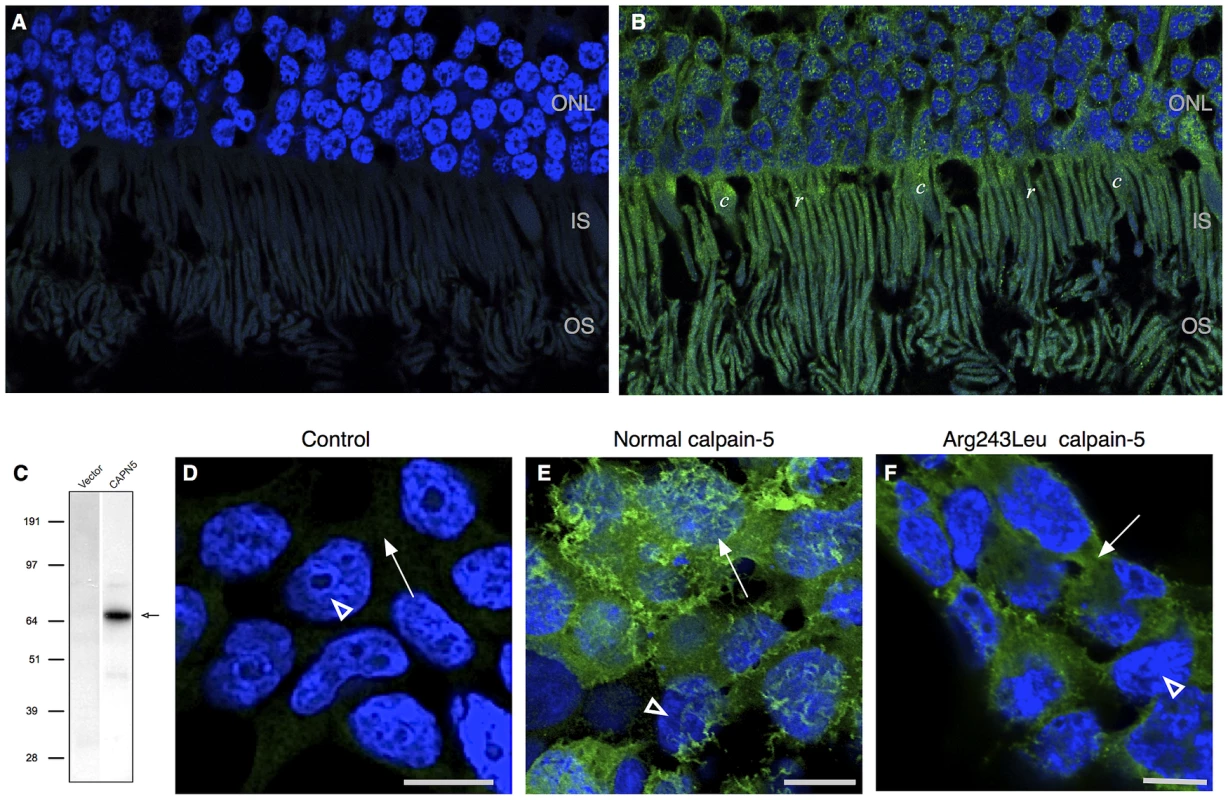
A. No significant signal was detected in control retinal sections probed with secondary antibody or primary antibody blocked with recombinant calpain-5. DAPI highlights the cell nuclei (blue) B. Calpain-5 was detected in photoreceptor cells (green). A punctate expression pattern was most prominent overlying the photoreceptor nuclei in the outer nuclear layer (ONL) and inner and outer segments (IS, OS). C. Anti-myc antibody western blot detects a single species (black arrow) of the appropriate size in HEK293T cells transfected with a vector bearing normal, myc-tagged CAPN5. D. No significant anti-myc signal was detected in the nuclei (white arrowhead) or cytoplasm (white arrow) of control cells that were treated with transfection reagent alone, vector alone, or secondary antibody alone. E. Anti-myc antibody shows that calpain-5 (green) is expressed in a ruffled pattern (white arrow) that obscures the underlying nuclei (white arrowhead) suggesting a location near the cell surface in these very thin cultured cells. F. Transfection with a mutant CAPN5 (Arg243Leu) exhibits a more uniform anti-myc signal that does not obscure the nuclei (white arrowhead) and is thus more compatible with localization to the cytoplasm (white arrow). Effect of Calpain-5 Mutations on Intracellular Location
Intracellular compartmentalization is a key regulatory mechanism for calpains [15], [16], [17]. For example, mutations that disturb localized protein interactions with calpain-3 cause limb-girdle muscular dystrophy type 2A [17], [18]. To determine the effect of the ADNIV-causing mutations on the intracellular compartmentalization of calpain-5, HEK293T cells were transfected with normal and mutant CAPN5 constructs. A western blot with anti-myc antibody revealed a single protein species of the expected size for myc-tagged calpain-5 (Figure 5C). Immunocytochemistry of HEK293T cells showed normal calpain-5 to be localized near the cell surface (Figure 5E). In contrast, both ADNIV mutants were found largely within the cytoplasm (Figure 5F and Figure S2). This suggests that the ADNIV-causing mutations may alter a membrane binding property of the protein.
Discussion
The calpains are an evolutionarily ancient family of calcium dependent intracellular proteases that utilize a cysteine residue in the active site to mediate limited proteolysis. The multifunctional calpains require careful regulation, since they target multiple intracellular proteins and pathways [15], [16], [17], [19], [20]. Their activity is regulated by intracellular calcium, lipid and protein interactions, subcellular localization, autocatalysis and inhibition by the endogenous peptide calpastatin [18], [20], [21]. There is no consensus amino acid sequence or structural motif that is targeted for cleavage by calpains, and as a result it is often difficult to identify the physiologic substrates of these enzymes, including calpain-5 [22], [23]. Capn5 is expressed during nematode and mouse embryogenesis [24], [25]. In adults, CAPN5 is highly expressed in the colon, kidney, liver, trachea, uterus, eye and brain [11], [25]. Calpains have been implicated in the pathogenesis of a wide range of human diseases including cancer, multiple sclerosis, Alzheimer's disease, cataract, diabetes and muscular dystrophy [17], [26]. Some polymorphisms in CAPN10 and CAPN5 have been shown to be risk factors for type II diabetes [27], [28]. However, prior to this report, limb girdle muscular dystrophy (LGMD) type 2A was the only disease shown to be caused in a monogenic fashion by variations in a calpain's protein sequence [29].
The evidence that the two missense mutations we observed in CAPN5 are responsible for ADNIV is compelling. The gene lies within the critical region previously linked to the disease, and all living subjects in the study who are clinically affected were found to harbor a CAPN5 mutation in exon 6. Each of these two mutations alters an amino acid in the catalytic domain that has been highly conserved throughout evolution. Neither of these mutations was found among any of the clinically unaffected members of the three kindreds we studied, or among thousands of normal individuals.
There are interesting similarities and differences between calpain-associated LGMD and ADNIV. In both disorders, the affected cells (skeletal muscle fibers and photoreceptor cells) experience large changes in membrane potential and intracellular calcium concentration as part of their normal behavior. In both disorders, the non-mutant calpain molecules display functionally critical subcellular localization [17], [18], [20], [30]. A subset of LGMD is associated with leukocyte infiltration into the tissue [31], and all cases of ADNIV are marked by severe intraocular inflammation. The differences in these diseases are also noteworthy. Calpain-associated LGMD is inherited in a recessive fashion and appears to result from loss of calpain-3 function [29]. In contrast, ADNIV is inherited in an autosomal dominant fashion and is caused by missense mutations near the active site. Although these mutations could cause disease through haploinsufficiency, it seems more likely that they result in a gain of function of calpain-5 that causes harm to the photoreceptor cells. Capn5 knockout mice have no observable phenotype [32], and several human neurological disorders have been associated with excess calpain activity [17], [33], including photoreceptor degeneration [34].
A gain of function mechanism for ADNIV is also supported by the unusual inflammation and neovascularization associated with the disease. There are dozens of monogenic disorders that cause the apoptotic death of photoreceptor cells without causing severe intraocular inflammation or neovascularization of the retina. The latter is much more typical of proliferative diabetic retinopathy than it is heritable photoreceptor disease [8]. It is easier to imagine an unregulated or mislocalized calpain promiscuously activating different signaling pathways, or being released into the extracellular space after photoreceptor death and causing inappropriate angiogenesis and leukocyte recruitment, than it is to imagine a 50% reduction of such a protein causing these dramatic complications.
Whether caused by a gain or loss of function of CAPN5, it is likely that the further elucidation of the pathogenic mechanism of ADNIV will provide important new insight into some of the most important causes of irreversible human blindness: autoimmune uveitis, retinitis pigmentosa, proliferative vitreoretinopathy and diabetic retinopathy. The latter condition alone is responsible for as much as 17% of blindness in some regions of the world [4]. The fact that an amino acid change in a single protein can lead to such a phenotype raises the possibility that a common, therapeutically accessible pathway may be shared among these conditions that could be targeted with drugs, antibodies or gene transfer approaches. It is possible that variations in the structure or expression of CAPN5 cause or modify some of these common disorders and this hypothesis will be important to test in future studies. However, given the extreme phenotypic heterogeneity of these disorders, it will be important to study a large number of subjects in such an experiment, to subdivide the patient cohorts into clinically well-characterized groups, and to screen an equal number of ethnically matched controls for each of these groups.
Materials and Methods
The study was approved by the University of Iowa's Institutional Review Board and adheres to the tenets set forth in the Declaration of Helsinki. Informed consent was obtained from all study participants. Informed consent was obtained from all study participants.
Genetic Analysis
Phenotypic ascertainment of the pedigrees included complete ocular examination as previously described [1]. Following genotyping of all adult members of the three ADNIV pedigrees with short tandem repeat polymorphisms within the original disease interval, three members of ADNIV-1 and three members of ADNIV-3 were also genotyped with an Affymetrix GeneChip Human Mapping 50K Array. Exome sequencing was performed using NimbleGen's SeqCap EZ Human Exome v2.0 capture and paired-end (2×100) sequencing on an Illumina HiSeq 2000 instrument at Otogenetics (Norcross, GA). The putative disease-causing mutations in CAPN5 were evaluated in the ADNIV kindreds and controls using Sanger sequencing and single-strand conformational polymorphism analysis (SSCP) [35]. Sequence analysis of human retinal cDNA was performed using the Illumina HiSeq 2000 instrument.
Structural Analysis
Primary and secondary structure protein alignments and trees were created with Geneious Pro 5.4.6 (http://www.geneiouspro.com). Yasara Structure (version 11.3.2) was used to generate a homology model of human calpain-5 using calpain-2 (m-calpain) structures (pdb id: 3BOW, 1DF0, 1U5I, 1KFU) as templates [12], [13]. Sequence alignment with the templates was first used to build a backbone model for aligned residues followed by loop modeling and side chain optimization using a combination of steepest descent and simulated annealing minimization. The top ranking of the 20 models generated was used as the homology model of calpain-5. The above steps were automated using Yasara's hm_build macro (http://www.yasara.org). Another homology model was generated using the Phyre server (version 0.2) (http://www.sbg.bio.ic.ac.uk/~phyre), which showed good agreement with the Yasara model for the domain folds and the active site region. The putative alpha-helix region in the Yasara model also formed a helix in the best Phyre model although it was positioned further away from the calpastatin binding site region than the Yasara model. PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT (http://sift.jcvi.org/) sequence analysis software were used to predict the functional effect of mutations.
RNA Sequencing
RNA sequence analysis was performed by extracting RNA from the retina of a human eye donor using an RNeasy kit from Qiagen (Valencia, CA) according to the manufacturer's instructions, preparing the sequencing library using the Illumina (San Diego, CA) RNA TruSeq sample preparation kit, and sequencing the latter on the Illumina HiSeq 2000 instrument at the Hudson Alpha Institute in Huntsville Alabama. The resulting sequence data were mapped using TopHat [36] and analyzed using Cufflinks [37]. The retinal expression of CAPN5 was compared to the expression of all other genes expressed in the retina.
Immunohistochemistry
Donor eyes were received from Iowa Lions Eye Bank (Iowa City, IA). Tissue was fixed in 4% paraformaldehyde solution diluted in 10 mM phosphate buffered saline (PBS), pH 7.4, and 7 µm sections underwent immunohistochemistry (IHC) using a polyclonal anti-calpain 5 primary antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), AlexaFluor 488 donkey anti-rabbit secondary antibody (Invitrogen, Carlsbad, CA) and 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Images were captured using an Olympus BX41 microscope equipped with fluorescent filters and the SPOT Advanced software package.
HEK293T cells (ATCC, Manassas, VA) were transfected with normal and mutant CAPN5 pCMV6-Entry vector plasmids using Turbofectin 8.0 (Origene) transfection reagent according to the manufacturer's instructions. Cells were incubated for 48 hours post transfection. For immunocytochemistry, cells were blocked using 5% bovine serum albumin (Amresco, Solon, OH) diluted in PBS with 0.1% Triton X-100. The polyclonal primary antibody, anti-Myc-tag, was diluted in PBS at 1∶500 and applied to the cells. Alexa Fluor 488 donkey anti-rabbit secondary, at concentration 10 µg/mL, and 0.0001 µg/mL of the counterstain, 4′, 6-diamidino-2-phenylindole (DAPI) (both from Molecular Probes, Eugene, OR), were applied to the cells, Images were captured using a Zeiss LSM 710 equipped with Zen2009 software (Zeiss, New York, NY).
Supporting Information
Zdroje
1. BennettSR, FolkJC, KimuraAE, RussellSR, StoneEM, et al. (1990) Autosomal dominant neovascular inflammatory vitreoretinopathy. Ophthalmology 97 : 1125–1135; discussion 1135–1126.
2. MahajanVB, FolkJC, FingertJH, SkeieJM, KinnickTR, ScheetzTC, BassukAG, BJRM, SheffieldVC, StoneEM (2011) Genetic Analysis and Phenotypic Staging of Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy. ARVO May 01 : 62/A175.
3. PascoliniD, MariottiSP (2012) Global estimates of visual impairment: 2010. Br J Ophthalmol 96 : 614–618.
4. ResnikoffS, PascoliniD, Etya'aleD, KocurI, PararajasegaramR, et al. (2004) Global data on visual impairment in the year 2002. Bull World Health Organ 82 : 844–851.
5. SheffieldVC, StoneEM (2011) Genomics and the eye. N Engl J Med 364 : 1932–1942.
6. BergerW, Kloeckener-GruissemB, NeidhardtJ (2010) The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res 29 : 335–375.
7. CaspiRR (2010) A look at autoimmunity and inflammation in the eye. J Clin Invest 120 : 3073–3083.
8. FrankRN (2004) Diabetic retinopathy. N Engl J Med 350 : 48–58.
9. PastorJC, de la RuaER, MartinF (2002) Proliferative vitreoretinopathy: risk factors and pathobiology. Prog Retin Eye Res 21 : 127–144.
10. StoneEM, KimuraAE, FolkJC, BennettSR, NicholsBE, et al. (1992) Genetic linkage of autosomal dominant neovascular inflammatory vitreoretinopathy to chromosome 11q13. Hum Mol Genet 1 : 685–689.
11. DearN, MatenaK, VingronM, BoehmT (1997) A new subfamily of vertebrate calpains lacking a calmodulin-like domain: implications for calpain regulation and evolution. Genomics 45 : 175–184.
12. HannaRA, CampbellRL, DaviesPL (2008) Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature 456 : 409–412.
13. MoldoveanuT, GehringK, GreenDR (2008) Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature 456 : 404–408.
14. MoldoveanuT, HosfieldCM, LimD, ElceJS, JiaZ, et al. (2002) A Ca(2+) switch aligns the active site of calpain. Cell 108 : 649–660.
15. LeloupL, ShaoH, BaeYH, DeasyB, StolzD, et al. (2010) m-Calpain activation is regulated by its membrane localization and by its binding to phosphatidylinositol 4,5-bisphosphate. J Biol Chem 285 : 33549–33566.
16. MichettiM, SalaminoF, TedescoI, AvernaM, MinafraR, et al. (1996) Autolysis of human erythrocyte calpain produces two active enzyme forms with different cell localization. FEBS Lett 392 : 11–15.
17. ZatzM, StarlingA (2005) Calpains and disease. N Engl J Med 352 : 2413–2423.
18. ErmolovaN, KudryashovaE, DiFrancoM, VergaraJ, KramerovaI, et al. (2011) Pathogenity of some limb girdle muscular dystrophy mutations can result from reduced anchorage to myofibrils and altered stability of calpain 3. Hum Mol Genet 20 : 3331–3345.
19. CroallDE, DeMartinoGN (1991) Calcium-activated neutral protease (calpain) system: structure, function, and regulation. Physiol Rev 71 : 813–847.
20. GollDE, ThompsonVF, LiH, WeiW, CongJ (2003) The calpain system. Physiol Rev 83 : 731–801.
21. KawasakiH, KawashimaS (1996) Regulation of the calpain-calpastatin system by membranes (review). Mol Membr Biol 13 : 217–224.
22. DuVerleDA, OnoY, SorimachiH, MamitsukaH (2011) Calpain cleavage prediction using multiple kernel learning. PLoS ONE 6: e19035 doi:10.1371/journal.pone.0019035
23. WaghrayA, WangDS, McKinseyD, HayesRL, WangKK (2004) Molecular cloning and characterization of rat and human calpain-5. Biochem Biophys Res Commun 324 : 46–51.
24. BarnesTM, HodgkinJ (1996) The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J 15 : 4477–4484.
25. DearTN, BoehmT (1999) Diverse mRNA expression patterns of the mouse calpain genes Capn5, Capn6 and Capn11 during development. Mech Dev 89 : 201–209.
26. HuangY, WangKK (2001) The calpain family and human disease. Trends Mol Med 7 : 355–362.
27. HorikawaY, OdaN, CoxNJ, LiX, Orho-MelanderM, et al. (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26 : 163–175.
28. SaezME, Martinez-LarradMT, Ramirez-LorcaR, Gonzalez-SanchezJL, ZabenaC, et al. (2007) Calpain-5 gene variants are associated with diastolic blood pressure and cholesterol levels. BMC Med Genet 8 : 1.
29. RichardI, BrouxO, AllamandV, FougerousseF, ChiannilkulchaiN, et al. (1995) Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 81 : 27–40.
30. MarcilhacA, RaynaudF, ClercI, BenyaminY (2006) Detection and localization of calpain 3-like protease in a neuronal cell line: possible regulation of apoptotic cell death through degradation of nuclear IkappaBalpha. Int J Biochem Cell Biol 38 : 2128–2140.
31. KrahnM, Lopez de MunainA, StreichenbergerN, BernardR, PecheuxC, et al. (2006) CAPN3 mutations in patients with idiopathic eosinophilic myositis. Ann Neurol 59 : 905–911.
32. FranzT, WincklerL, BoehmT, DearTN (2004) Capn5 is expressed in a subset of T cells and is dispensable for development. Mol Cell Biol 24 : 1649–1654.
33. VanderklishPW, BahrBA (2000) The pathogenic activation of calpain: a marker and mediator of cellular toxicity and disease states. Int J Exp Pathol 81 : 323–339.
34. AzumaM, ShearerTR (2008) The role of calcium-activated protease calpain in experimental retinal pathology. Surv Ophthalmol 53 : 150–163.
35. TuckerBA, ScheetzTE, MullinsRF, DeLucaAP, HoffmannJM, et al. (2011) Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci U S A 108: E569–576.
36. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
37. TrapnellC, WilliamsBA, PerteaG, MortazaviA, KwanG, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28 : 511–515.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 10- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- The Germline Genome Provides a Niche for Intragenic Parasitic DNA: Evolutionary Dynamics of Internal Eliminated Sequences
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Calpain-5 Mutations Cause Autoimmune Uveitis, Retinal Neovascularization, and Photoreceptor Degeneration
- Cofilin-1: A Modulator of Anxiety in Mice
- The Date of Interbreeding between Neandertals and Modern Humans
- Embryos of Robertsonian Translocation Carriers Exhibit a Mitotic Interchromosomal Effect That Enhances Genetic Instability during Early Development
- Viral Evasion of a Bacterial Suicide System by RNA–Based Molecular Mimicry Enables Infectious Altruism
- Phosphatase-Dead Myotubularin Ameliorates X-Linked Centronuclear Myopathy Phenotypes in Mice
- Full-Length Synaptonemal Complex Grows Continuously during Meiotic Prophase in Budding Yeast
- MOV10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells
- A Likelihood-Based Framework for Variant Calling and Mutation Detection in Families
- The Contribution of RNA Decay Quantitative Trait Loci to Inter-Individual Variation in Steady-State Gene Expression Levels
- New Partners in Regulation of Gene Expression: The Enhancer of Trithorax and Polycomb Corto Interacts with Methylated Ribosomal Protein L12 Its Chromodomain
- Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- The Many Landscapes of Recombination in
- Faster-X Evolution of Gene Expression in
- Loss of Slc4a1b Chloride/Bicarbonate Exchanger Function Protects Mechanosensory Hair Cells from Aminoglycoside Damage in the Zebrafish Mutant
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
- and the BTB Adaptor Are Key Regulators of Sleep Homeostasis and a Dopamine Arousal Pathway in Drosophila
- Mutation and Fetal Ethanol Exposure Synergize to Produce Midline Signaling Defects and Holoprosencephaly Spectrum Disorders in Mice
- Specific Missense Alleles of the Arabidopsis Jasmonic Acid Co-Receptor COI1 Regulate Innate Immune Receptor Accumulation and Function
- Deep Genome-Wide Measurement of Meiotic Gene Conversion Using Tetrad Analysis in
- Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity
- Distinguishing between Selective Sweeps from Standing Variation and from a Mutation
- Cytokinesis-Based Constraints on Polarized Cell Growth in Fission Yeast
- Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes
- Functional Antagonism between Sas3 and Gcn5 Acetyltransferases and ISWI Chromatin Remodelers
- The SET-Domain Protein SUVR5 Mediates H3K9me2 Deposition and Silencing at Stimulus Response Genes in a DNA Methylation–Independent Manner
- Morphogenesis and Cell Fate Determination within the Adaxial Cell Equivalence Group of the Zebrafish Myotome
- Muscle-Specific Splicing Factors ASD-2 and SUP-12 Cooperatively Switch Alternative Pre-mRNA Processing Patterns of the ADF/Cofilin Gene in
- Maize Is Required for Maintaining Silencing Associated with Paramutation at the and Loci
- Increasing Signal Specificity of the TOL Network of mt-2 by Rewiring the Connectivity of the Master Regulator XylR
- Use of Pleiotropy to Model Genetic Interactions in a Population
- RAB-Like 2 Has an Essential Role in Male Fertility, Sperm Intra-Flagellar Transport, and Tail Assembly
- Variants Affecting Exon Skipping Contribute to Complex Traits
- Topoisomerase II– and Condensin-Dependent Breakage of -Sensitive Fragile Sites Occurs Independently of Spindle Tension, Anaphase, or Cytokinesis
- Comparison of Family History and SNPs for Predicting Risk of Complex Disease
- Recovery of Arrested Replication Forks by Homologous Recombination Is Error-Prone
- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Comparative Genomics Suggests an Independent Origin of Cytoplasmic Incompatibility in
- It Was Heaven: An Interview with Evelyn Witkin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



