-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Morphogenesis and Cell Fate Determination within the Adaxial Cell Equivalence Group of the Zebrafish Myotome
One of the central questions of developmental biology is how cells of equivalent potential—an equivalence group—come to adopt specific cellular fates. In this study we have used a combination of live imaging, single cell lineage analyses, and perturbation of specific signaling pathways to dissect the specification of the adaxial cells of the zebrafish embryo. We show that the adaxial cells are myogenic precursors that form a cell fate equivalence group of approximately 20 cells that consequently give rise to two distinct sub-types of muscle fibers: the superficial slow muscle fibers (SSFs) and muscle pioneer cells (MPs), distinguished by specific gene expression and cell behaviors. Using a combination of live imaging, retrospective and indicative fate mapping, and genetic studies, we show that MP and SSF precursors segregate at the beginning of segmentation and that they arise from distinct regions along the anterior-posterior (AP) and dorsal-ventral (DV) axes of the adaxial cell compartment. FGF signaling restricts MP cell fate in the anterior-most adaxial cells in each somite, while BMP signaling restricts this fate to the middle of the DV axis. Thus our results reveal that the synergistic actions of HH, FGF, and BMP signaling independently create a three-dimensional (3D) signaling milieu that coordinates cell fate within the adaxial cell equivalence group.
Published in the journal: . PLoS Genet 8(10): e32767. doi:10.1371/journal.pgen.1003014
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003014Summary
One of the central questions of developmental biology is how cells of equivalent potential—an equivalence group—come to adopt specific cellular fates. In this study we have used a combination of live imaging, single cell lineage analyses, and perturbation of specific signaling pathways to dissect the specification of the adaxial cells of the zebrafish embryo. We show that the adaxial cells are myogenic precursors that form a cell fate equivalence group of approximately 20 cells that consequently give rise to two distinct sub-types of muscle fibers: the superficial slow muscle fibers (SSFs) and muscle pioneer cells (MPs), distinguished by specific gene expression and cell behaviors. Using a combination of live imaging, retrospective and indicative fate mapping, and genetic studies, we show that MP and SSF precursors segregate at the beginning of segmentation and that they arise from distinct regions along the anterior-posterior (AP) and dorsal-ventral (DV) axes of the adaxial cell compartment. FGF signaling restricts MP cell fate in the anterior-most adaxial cells in each somite, while BMP signaling restricts this fate to the middle of the DV axis. Thus our results reveal that the synergistic actions of HH, FGF, and BMP signaling independently create a three-dimensional (3D) signaling milieu that coordinates cell fate within the adaxial cell equivalence group.
Introduction
The mechanisms that are utilised to generate individual cell types from a set of equivalently fated set of precursors remains a central experimental focus of developmental biology. Studies from invertebrate systems have defined the concept of an equivalence group, where small clusters of lineage related cells are determined by a combination of inductive and intrinsic signals to adopt individual fates [1]–[6]. This concept faces many difficulties when applied to complex three dimensional tissues such as those that typify vertebrate development, where the direct lineage relationships of many cells remain ill defined and the complicated morphogenesis of many tissues precludes definition of models of equivalence.
Zebrafish provides perhaps one of the most tractable contexts in which to examine concepts of cell fate determination in a vertebrate embryo, as a variety of lineage tracing techniques can be deployed in different genetic contexts in real time within an optically accessible embryo. One zebrafish lineage that has been examined in some detail is the embryonic myotome of zebrafish. As in all vertebrates, the majority of skeletal muscle in zebrafish forms from precursor cells present in the somites, which arise by segmentation of paraxial mesoderm in a rostral to caudal progression on either side of neural tube and notochord along the main body axis of the embryo. This process, referred to as myogenesis, gives rise to distinct slow and fast twitch muscle populations that differ in contraction speeds, metabolic activities and motoneuron innervation. In zebrafish, the location and origin of these two different cell populations are topographically separable [7], [8]. The early differentiating slow-muscle cells arise from a particular subset of presomitic mesodermal cells, termed the adaxial cells, which at the end of gastrulation align medially against the notochord [8]. These precursors initially adopt a pseudo epithelial morphology but shortly after their incorporation within the formed somite, undergo stereotypic morphogenetic cell shape changes, moving from their columnar shape to flatten and interleave, adopting a triangular shape, that upon further differentiation results in single adaxial cells extending from one somite boundary to the other. These cells collectively flatten medio-laterally to form a set of elongated myocytes that span the somite, positioned against the notochord [9].
Ultimately, adaxial cells give rise to two distinct sub-types of slow muscle fibers: the superficial slow-twitch muscle fibers (SSFs) and the muscle pioneer cells (MPs). SSFs and MPs possess distinct morphological, molecular and functional properties. After undergoing the initial morphogenetic cell shape changes described above, SSFs migrate from their notochord-associated midline position to traverse the entire extent of the forming myotome and come to lie at its most lateral surface. There, the SSF precursors complete their differentiation to form a monolayer of approximately 20 slow twitch muscle fibers. By contrast, MPs (2 to 6 per somite) do not migrate from the midline and are the first cells of the zebrafish myotome to differentiate, forming slow twitch muscle fibers immediately adjacent to the notochord [10]. All slow fibers express slow isoforms of myosin heavy-chain (SMyHC) as well as the homeodomain protein Prox1 and are mono nucleated cells [11]. MPs, in addition, express high levels of homeodomain-containing Engrailed proteins [12], [13]. By contrast to slow precursors, differentiating fast precursors originate from the lateral somite and fuse to form multinucleated fibers, subsequently to SSF migration, and are distinguished by their expression of fast MyHC. A subset of these fibers, known as medial fast fibers (MFFs) also expresses Engrailed at lower levels than MPs [14]. The timing of the fate determination of these distinct cell types has been examined by a rigorous in vivo transplantation assays. By interchanging slow and fast muscle precursors at specific points in their development it has been demonstrated that at the time of gastrulation, although slow and fast muscle precursors are already spatially segregated, they remain uncommitted to their individual fates until they have entered into the segmental plate. Furthermore, the subdivision of adaxial compartment in to MP and non MP cell fates occurs at a similar period of development, with MP becoming irreversibly fated within the posterior part of the segmental plate during early somite formation [7].
In vertebrates, the specification and differentiation of the somite into specific cell types is under the influence of inductive signals from the somite itself or those derived from the surrounding tissues (reviewed in [15], [16]). In the case of zebrafish myogenesis, by far the most well understood inductive signals controlling myogenesis are the Hedgehog (HH) family of secreted glycoproteins, which emanate from the embryonic midline. Numerous studies in the last two decades have demonstrated that HH is necessary and sufficient for induction of the slow twitch muscle fate. Indeed, analysis of loss of function mutants in HH pathway genes and the use of the HH pathway inhibitor cyclopamine have demonstrated that the timing and the level of HH signaling are critical for the formation of different muscle identities, including the MP cells, which require the highest level of HH signaling for their formation [14], [17]–[19]. However, even though HH over-expression can induce supernumerary MP cells, this is not sufficient to convert the dorsal and ventral extremes of the myotome into MP cells [20], [21] suggesting that other signals could induce MP in the midline region or repress MP differentiation in the dorsal and ventral muscle cells [21].
A further complication of these analyses is that they fail to explain how the symmetry of the adaxial cell compartment is initially broken to generate the dichotomy of MP and SFF fates within equivalent sets of cells. As the adaxial cells flank the notochord and floorplate, the source of HH peptide secretion, all adaxial cells would initially be exposed to the same level of secreted HH peptides. Hence, it is unclear how different levels of HH could act to generate the MP cell fate within a subset of adaxial cells and suggests that additional signals must influence adaxial cell fate. Recent studies have shed some light on the nature of other secreted signals that may act to influence muscle cell formation. Several studies have shown that manipulation of BMP signaling can alter MP number [21], [22]. Furthermore, Smad5, a downstream effector of BMP signaling has been shown to be activated in the dorsal and ventral adaxial cells and is absent within the central region of the somite [22], [23]. In addition, Smad binding sites have been shown to regulate activity of the eng2a promoter, the eng gene expressed the earliest within MP precursors [12], [21]–[23]. Collectively, these studies suggest that BMP signaling can influence the number of different cell types within the embryonic zebrafish myotome, but exactly how this is achieved has yet to be determined mechanistically.
In this study, we utilize a combination of live imaging, retrospective and indicative fate mapping, molecular and genetic studies to demonstrate that MP and SSF precursors arise from distinct regions along the anterior-posterior (AP) and dorsal-ventral (DV) axes of the adaxial cell compartment. Uniquely, this regionalization is controlled by the action of different signal transduction pathways that act specifically to direct specification in distinct axial dimensions. We demonstrate that the sprouty4-mediated inhibition of FGF signaling induces MP cell fate in the anterior-most adaxial cells in each somite and that radar-mediated BMP signaling restricts this fate to the middle of the DV axis. Our results indicate that HH, FGF and BMP signaling synergize to determine cell fate within the adaxial cell equivalence group.
Results
Superficial slow twitch muscle and muscle pioneer precursors arise from distinct locations within the adaxial compartment
In order to understand the origins of SSF and MP precursors from within the adaxial cell compartment (Figure 1A–1B), we examined adaxial cell behaviors during the first phase of their differentiation via continuous 4D time-lapse analysis and retrospective fate map analysis of the entire forming myotome. The position and shape of the adaxial cells were followed using a membrane-bound GFP and a nuclear localized mCherry whose expression in all cells was achieved after mRNA injection at 1-cell-stage. This analysis identified that the first adaxial cells to initiate differentiation and elongation arise adjacent to the anterior border of each somite at its DV mid-point (Figure 1C–1M and Video S1). These cells are most likely MPs, which previously have been shown to differentiate precociously [10]. To confirm this, we analyzed the expression of the MP marker gene engrailed2a (eng2a) during early somitogenesis by in situ hybridization. At the 10-somite stage eng2a transcripts were detected within newly formed somites exclusively within a subset of adaxial cells, adjacent to the anterior somitic border, located precisely at the mid-point of the DV axis of the somite (Figure 1N). To more precisely localize eng2a expression within the somite, we undertook dual in situ hybridization with myod, which marks the adaxial cells and the posterior aspect of the lateral somite, which contains the differentiating fast muscle progenitors (Figure 1O). This analysis confirmed that the expression of eng2a initiates specifically in the anterior-most cells of the newly formed somites. The positioning of cells initiating eng2a expression to the dorsal ventral midline of the forming myotome was confirmed in transverse sections of similarly staged embryos individually stained for eng2a and slow myosin heavy chain 1 (smyhc1) gene expression (Figure 1P, 1Q).
Fig. 1. Distinct muscle precursor populations within the adaxial compartment. 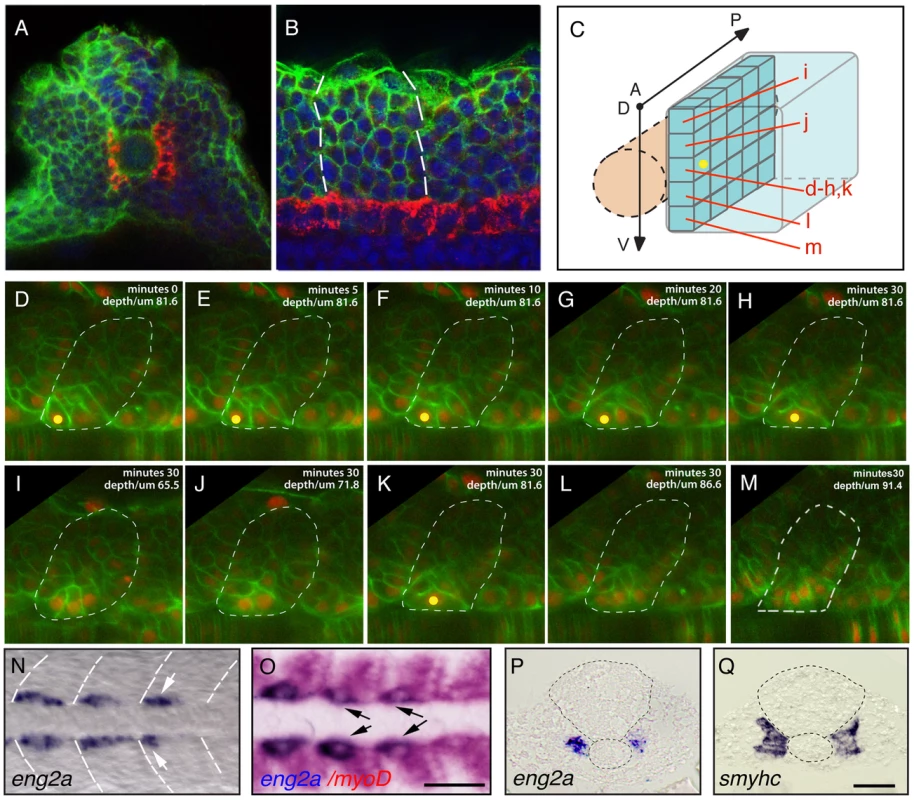
(A–C) The adaxial cell compartment. (A) Cross-section and (B) single confocal scan, dorsal view of a newly formed somite where the adaxial cells express the sMyHC (red). Nuclei and membranes are also marked with DAPI (blue) and membrane-bound GFP (green). (C–M) Restrospective fate map of the adaxial cell compartment. Adaxial cell behaviors occurring during the first phase of differentiation were analyzed in time lapse using a membrane-bound GFP (green) and a nuclear localized mCherry (red). The anterior-most adaxial cells in the dorso-ventral midline (yellow dot) are the first to differentiate and elongate. Adaxial cells above and below remain undifferentiated. The positions of individual confocal planes on the dorso-ventral axis are represented in (C). (N–Q) eng2a expression initiates in anterior adaxial cells at the dorso ventral midline of the myotome. 10-somite stage embryos on which in situ hybridization (ISH) for eng2a was performed alone (Blue, N, P) or in combination with myoD (Red, O,) that marks the posterior somitic region and the adaxial cells. Arrows indicate eng2a expression in the anterior adaxial cells. (Q) In situ hybridisation for smyhc demonstrates the location of the adaxial cells. (N, O) dorsal view, anterior toward the left, (P, Q) cross sections. Scale bar 50 µM. Collectively, these results suggest that SSF and MP precursors arise from distinct positions within the adaxial equivalence group. To test this hypothesis, we fate mapped the entire adaxial compartment by systematic iontophoretic injection of tetra-methyl rhodamine dextran (TMRD) lineage tracer dye into individual adaxial cells located at various AP and DV positions. Adaxial cells were labeled within the three most newly formed somites at the 10–15-somite stage and the fates of individually labeled cells were analyzed after the muscle fibers had terminally differentiated at 30 hpf. Individual injected embryos were sequentially incubated and imaged, first with an anti-Eng antibody and secondly with an anti-SMyHC antibody to unambiguously determine the fate of marked cells. This analysis confirmed that MP cells arise from the anterior-most adaxial cells at the dorso ventral midline of the somite (n = 8/8, Figure 2A, 2C, 2B, 2H) while posterior adaxial cells at this DV level make SSFs (n = 32/32, Figure 2A, 2D, 2E). Furthermore, we found that based on the initial position of a SSF precursor within the adaxial cell pool, we could predict its final location with the post migratory slow muscle palisade such that the dorsal - and ventral-most adaxial cells generate the dorsal and ventral-most post-migratory differentiated slow fibers respectively (n = 83, Figure 2B, 2F–2G, 2I–2J). This analysis not only demonstrates that MP and SSF precursors segregate at the beginning of somitogenesis but also determines the exact position of the precursors of every slow fiber. To further validate the fate of the adaxial cells located in the anterior somite at the DV mid-point, we examined their behaviour during the migration period. We thus performed a time-lapse analysis during a 20 hour period on embryos that were injected with a DNA construct containing the GFP gene under the control of the slow-twitch muscle-specific, slow myosin heavy chain 1 (smyhc1) promoter. When located in the anterior margin of the somite, the transgenically labeled adaxial cell elongates in an anterior to posterior movement but remains adjacent to the notochord identifying the labelled cell as a MP (Video S2).
Fig. 2. The fate map of the adaxial compartment. 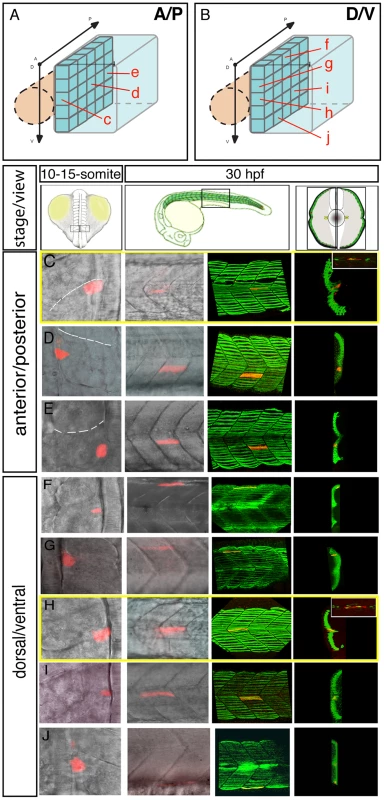
The fate of individual adaxial cells were identified using iontophoretic injections of TMRD (red) into individual adaxial cells located at various AP (C–E) and DV (F–J) positions within the three most newly formed somites of 10–15-somite stage embryos (C–J, left panels, dorsal views). The AP (A) and DV (B) position of labeled cells is represented schematically. The fate of cells was then analyzed at 30 hpf (C–J, other panels). (C–J) At 30 hpf, the panels (from the left to the right) represent the position of the labeled fiber in the myotome (lateral view), the expression of sMyHC (green, 3D reconstruction of multiple confocal scans) in lateral view and in cross section, and the expression of Engrailed (green, small upper panels for C and H, lateral view). MPs (C, H) were identified due to their position in the midline and simultaneous expression of Slow MyHC and Eng. FGF signaling controls the AP positioning of muscle pioneer precursors
We next turned our attention to the molecular basis of the adaxial cell fate specification events that we had defined by our fate mapping strategies. A candidate approach, examining AP restricted inductive signals within the myotome, highlighted the FGF pathway as a putative regulator of AP patterning in the adaxial progenitors. Indeed, in zebrafish, at least two of the genes encoding fgf ligands, fgf8 and fgf17b have been shown to be restricted in expression to the anterior somite [24], [25], [26]. However an analysis of the expression of the downstream targets of the FGF cascade, erm and pea3 surprisingly revealed that asymmetric FGF responses occur specifically within the adaxial cells such that the anterior-most cells lose expression of FGF target genes during somite formation (Figure 3A, 3B–3B″ and data not shown). The temporal and spatial regulation of FGF signal activation during zebrafish myogenesis suggests a simple hypothesis. Distinct levels of FGF activation along the AP axis of the somite inform the adaxial cells of their position within this axis and consequently control their fate. In order to test this hypothesis we disrupted FGF signaling by the addition of the pharmacological inhibitor SU5402, a drug that blocks the phosphorylation of FGF receptors (FGFRs) and so prevents downstream signaling, as revealed by the downregulation of the target genes erm, pea3 and spry4 in SU5402 treated embryos (Figure 4A–4C, [25], [27]). SU5402 treatments at the 6-somite stage did not affect the number of slow muscle fibers (Table S1) but instead increased the number of MPs at the expense of SSFs, as revealed by a failure in slow-muscle fiber migration to the surface of the myotome and a corresponding increase in Engrailed positive MP cells evident at the midline (Figure 4D, 4E). Furthermore FGF inhibition does not alter the number of En positive medial fast fibers (Figure S1). The increase in MP number is foreshadowed by an expansion of the eng2a expression domain throughout the AP dimension of the adaxial cell compartment at the 10-somite stage (Figure 4F). Furthermore, the heat shock induced expression of a dominant negative form of FGFR1 that blocks the FGF/ERK signaling cascade also causes a similar increase of eng2a expression at the expense of SSF migration at 1 dpf (Figure 4G–4J). Collectively, these results show that FGF inhibition promotes the specification of the MP fate. Importantly, delayed addition of SU5402 until the 10-somite stage revealed that the more rostral 5–6 somites, which had already formed at the time of treatment, remained unaffected revealing a discrete temporal window of action for FGF signaling in MP specification within the newly formed somite (Figure 4K). This correlates specifically with the period of development when cuboidal cells are arrayed along the AP axis, prior to their differentiation (Figure 1A–1B). These data shows that FGF signaling inhibition specifies anterior identity and consequently MP fate within the adaxial cell equivalence group.
Fig. 3. FGF signaling is down regulated in muscle pioneer precursor domain. 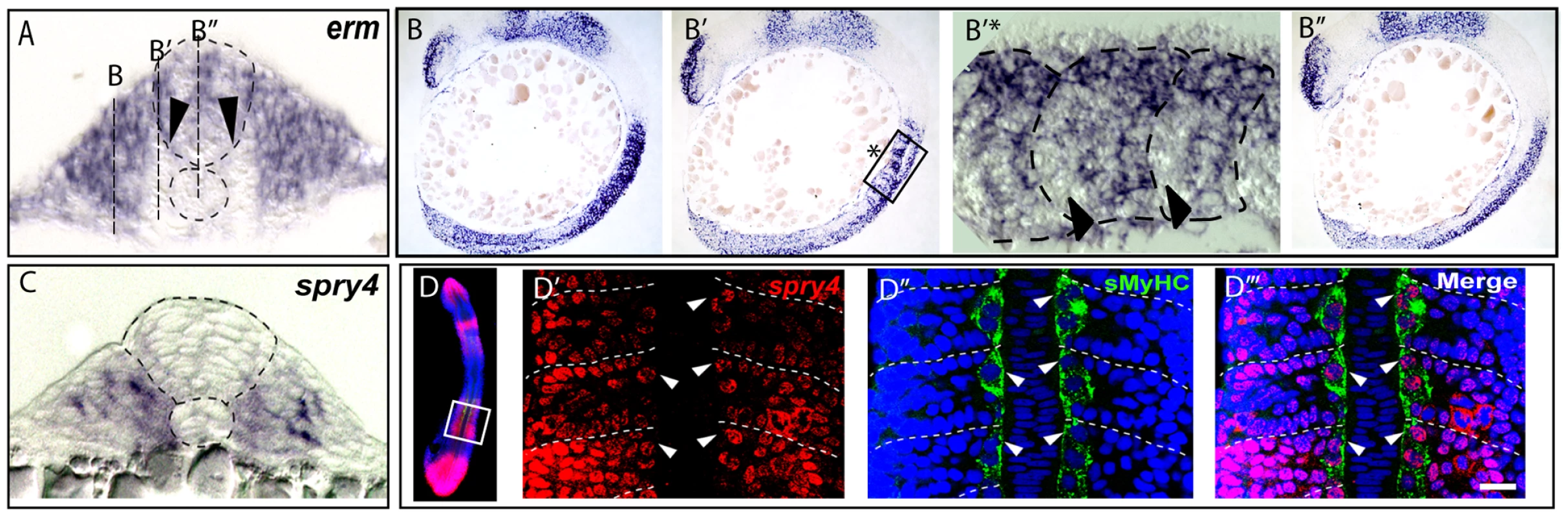
(A–B) Asymmetric expression of erm at 9-somites as determined by ISH on (B–B″) serial sagittal sections and (A) cross-section. Panel (B′*) represents a high magnification of the region in box in (B′) panel. The different positions of sagittal sections are represented in (A). Expression of erm is absent in anterior adaxial cells (arrow heads). (C) spry4 expression in a 9-somite embryo determined by ISH on a cross section of the anterior somitic region. (D–D‴) Expression of spry4 mRNA (D′, red), sMyHC (D″, green) and DAPI (D–D‴, blue) in 9-somite embryos, dorsal view. spry4 expression in the anterior adaxial cells is indicated (arrow heads). D′–D‴ high magnification view of the area boxed in D. Fig. 4. sprouty4-mediated FGF inhibition promotes muscle pioneer formation. 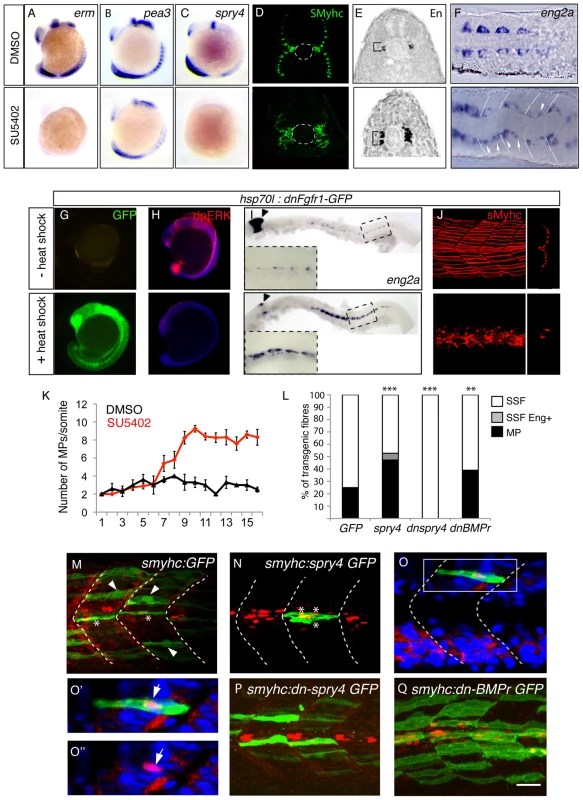
Erm (A), pea3 (B) and spry4 (C) expressions in 10–13-somite embryos after DMSO or SU5402 treatments as determined by in situ hybridization. (D) sMyHC (green) and (E) Eng (black) expression in transverse sections of 1 dpf embryos, after treatment with DMSO or SU5402 applied at the 6-somite stage. (F) eng2a expression at 10-somite after treatment with DMSO or SU5402, as determined by ISH (Dorsal view, anterior towards the left). (G–J) Expression of a dominant negative form of fgfr1 fused with GFP (green) (G) is induced after heat shock in hsp70l:dnfgfr1-GFP embryos at 13–15-somite stage. (H) Expression of the target gene of FGF signaling cascade, diphosphorylated ERK (dpERK, red), which is down regulated in 13–15 somite embryos after heat shock compared to no heat shocked control. By contrast (I) eng2a is up-regulated at 1 dph after heat shock as revealed by ISH and (J) slow fibre migration is impaired, as revealed by 3D reconstructed sMyHC expression (lateral view and cross section). (K) Graphic representation of the number of MPs per somite in 1 dpf embryos after treatment with DMSO or SU5402 applied at the 10-somite stage. The horizontal axis represents the position of the somite along the body axis, 1 being the most rostral somite, values = means, error bars = standard error of the mean (s.e.m.). (L–Q) Ectopic expression of spry4, dn-spry4 or dn-BMPr in slow precursors was obtained after injection of (L, N, O, O′, O″) smyhc:spry4-ires-GFP, (L, P) smyhc:dn-spry4-ires-GFP and (L, Q) smyhc:dn-BMPr-GFP DNA constructs into one cell embryo, respectively. (L, M) smyhc:GFP construct was used as the injection control. (L) Graphic representation of the different fates of transgenic slow fiber (SSF, MP and SSF en+) upon ectopic expression of GFP, spry4, dnspry4 or dnBMPr. Values = means of percentages of transgenic muscle fibers per injected embryo (nembryo = 6–9 per condition). We performed analysis of variance (ANOVA) to determine statistical difference within a 95% confidence interval: ** p<0.005, ***p<0.001. (M–Q) Images reveal the expression of GFP (green) and Eng (red) in the somites of 1 dpf embryos by maximum projections of multiple confocal scans (M, N, P, Q) or single confocal scan (O–O″). SSFs (arrow heads) and MPs (*) are indicated. (O–O″) In 4.19% of cases, the spry4 transgene triggers expression of Engrailed in post-migratory slow fibers (arrows, p<0.05). (O′, O″) high magnification of the region boxed in O. Scale bar = 25 µm. FGF signaling patterns the adaxial cells independently from HH signaling
As described above the adaxial cells, and thus the slow twitch muscle lineage are highly dependent on Hedgehog (HH) signaling with the MP fate requiring higher levels and longer exposure to HH for proper specification than SSFs [14]. To test a possible cross talk between FGF and HH signaling, we analyzed the expression of HH target gene patched1 (ptc1). However ptc1 expression remains unaffected by SU5402 treatment (Figure S2A). FGF signaling was also recently shown to control the length of motile cilia within Kupffer's vesicle [28]. Although non-motile cilia are a distinct class of cell organelle, one possible mechanism for FGF action could be to regulate HH signal reception through the length or number of primary cilia on adaxial cells, as reception and activation of the HH pathway is controlled within the primary cilia in vertebrate cells [29]. However, our analysis suggests that SU5402 treatment doesn't affect the length or the number of primary cilia within the adaxial cells (Figure S2B). Therefore, the effect of FGF signaling on MP specification cannot be explained by modulation of HH transduction within adaxial cells.
Sprouty4 controls muscle pioneer fate specification through FGF signal inhibition
To understand how the precise spatial activity of FGF is regulated to control the dichotomy of the cell fate decision evident with the adaxial cells, we systematically examined known inhibitors of the FGF pathway for their expression within the adaxial cells. This analysis revealed that sprouty4 (spry4), which encodes a known intracellular inhibitor of receptor tyrosine kinases (RTKs), including the Fgfrs [30]–[33], becomes specifically activated in the anterior adaxial cells. Furthermore, the loss of expression of FGF target gene erm in the anterior adaxial cells correlates spatially and temporally with the induction of expression of the spry4 gene in the identical cells (Figure 3C, 3D–3D‴). To test whether spry4 expression influences MP and SSF fate specification, we ectopically expressed it within the adaxial cell compartment. Mosaic overexpression of spry4 from the promoter of the smyhc1 gene (smyhc1:spry4-IRES-GFP), which drives expression throughout the adaxial cell compartment [34], [35] doubled the number of MP cells (47.29% of transgenic fibres, nfibres = 143) within the embryo compared to control embryos expressing GFP alone (24.9%, nfibres = 521) (Figure 4L–4N). Furthermore, over-expression of spry4 induces a third population of transgenic fibers that possess attributes of both MPs and SSFs. These rare fibers (4.19%, nfibres = 143) are able to migrate to the surface of the myotome and express Engrailed, a unique behavior never observed in control embryos (untreated or smyhc:GFP injected) (Figure 4L, 4O, 4O′ and 4O″). Reciprocally when we express a dominant negative form of spry4, using the identical smyhc1 promoter (smyhc1:dn-spry4-IRES-GFP) cell autonomous loss of spry4 leads to a loss of MP identity and adaxial cells that express dnspry4 are incapable of making MPs (0% of transgenic fibres, nfibres = 48, Figure 4L, 4P, [36]).
We next analyzed muscle development in mutants that have had the spry4 gene inactivated. spry4fl117 mutants carry a single A-to-T transversion, which introduces a stop codon early in the ORF of the gene (Figure 5A). The mutant allele encodes for a truncated protein, which lacks the putative activation domain involved in FGF signaling inhibition and is consequently predicted to engender a full loss of function in spry4 (Figure 5B). Maternal zygotic (MZ) spry4fh117 homozygous mutant embryos, but not heterozygous or zygotic (Z) mutants, exhibit a marked increase in FGF target gene expression (erm, n = 9/9; dpERK, n = 13/13; and spry4, n = 8/8, Figure 5C–5F and 5G–5H and data not shown), showing that FGF activity is increased in MZ spry4fh117 mutants. Furthermore, while we could show that both the number of slow fibers and their position was unaffected in MZ spry4fh117 mutant embryos (Figure 5I–5J, Table S1 and Figure S3) the number of MPs was less than half that of controls (n = 31, Figure 5K, 5L and 5N) a deficit that was rescued by SU5402 treatment (n = 32, Figure 5K, 5M and 5O), indicating that the deficiency of MPs associated with the loss of spry4 is directly due to FGF over-activation and not modulation of other RTKs.
Fig. 5. sprouty4 loss-of-function leads to the reduction of MP formation. 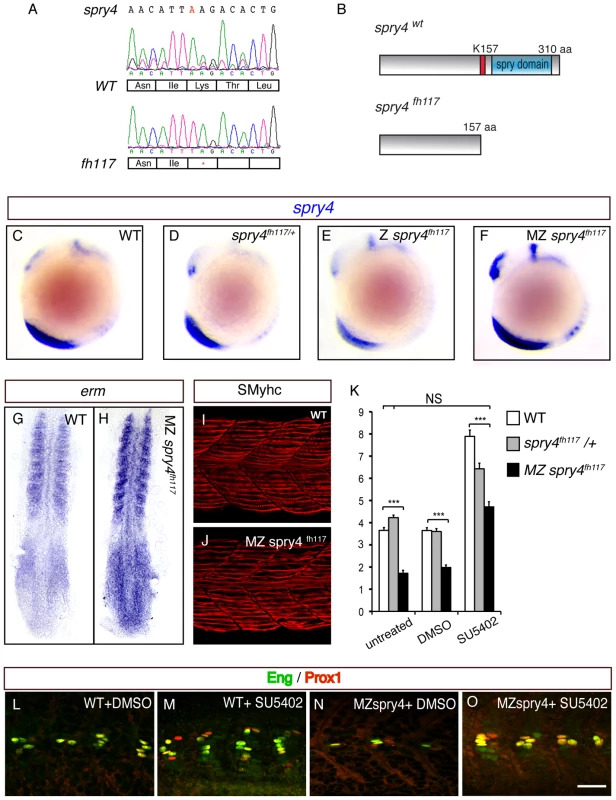
(A) Sequencing of spry4WT and spry4fh117 confirms the single A to T mutation in position 469 of the spry4 ORF, resulting in the premature stop codon. (B) Predicted peptide arising from spry4WT and spry4fh117. The mutation results in a truncated protein of 157 amino acids (K157 in red is replaced by a stop codon) lacking the conserved cysteine rich domain [31] also called the spry domain (blue). This domain has been involved in RTK inhibition in other systems. (C–F) spry4 expression at 10-somite in WT, spry4fh117/+, Z spry4fh117 and MZ spry4fh117 embryos as determined by in situ hybridization. Whole mounted embryos, lateral view. (G–H) Erm expression in 9-somite WT and MZ spry4fh117 embryos. Flat mounted embryos, anterior toward the top. (I–J) sMyhc expression (red) in the somites of (I) WT and (J) MZ spry4fh117 embryos at 1 dpf. Maximal projection of multiple confocal scans, lateral view. (K) Graphic representation of the number of MPs per somite of 1 dpf embryos in different conditions: WT, spry4fh117/+, and MZ spry4fh117 untreated embryos or after DMSO or SU5402 treatments, values = means, error bars = s.e.m, ***p<0.001. (L–O) Engrailed (green) and Prox1 (red) expression in the somites of 1 dpf WT or MZ spry4fh117 embryos, after DMSO or SU5402 treatments, scale bar = 50 µm. Radar-mediated Bmp signaling coordinates MP and SSF fate specification synergistically with FGF signaling
Although the regional inhibition of FGF signaling can explain the localization of the MP precursors to anterior adaxial cells, it cannot explain the positioning of these progenitors to the DV midline of the somite. Several recent studies have shown that manipulation of BMP signaling can alter MP number [21], [22] and these studies also show that Smad5, a downstream effector of BMP signaling is activated in the dorsal and ventral adaxial cells and but not within cells of the central region of the compartment [22], [23]. Furthermore, Smad binding sites have been shown to regulate activity of the eng2a promoter [22], [23]. This has led to the suggestion that BMP activity could influence the fate of the myotome along the DV axis, although direct evidence for this assertion is lacking. Furthermore, Smad5 is also known to be activated by the TGF-ß signaling pathway in many biological systems and a number of tgf-ß genes are expressed during zebrafish during myogenesis complicating interpretation of these data [37]. To visualize BMP signaling more specifically we generated a transgenic line that expresses GFP under the control of a BMP Responsive Element which contains 5 tandem BRE elements derived from the Xenopus vent2 gene coupled to a minimal Xenopus id3 promoter, promoter elements known to specifically respond to BMP signal transduction. The activation of this transgene (Tg(5XBRE[vent2]:-20lid3:GFP) has been shown to occur specifically via the BMP signaling pathway, and not by other TGF-ß-related ligands [38], [39], [40] Figure 6A–6D). The expression of GFP in Tg(5XBRE[vent2]:-20lid3:GFP) embryos correlates with the distribution of phospho-Smad5 (Figure 6F). By early somitogenesis, BMP signaling is activated in the adaxial cells specifically in cells of the dorsal and ventral edges of the myotome, and reporter expression decreases in the midline (n = 14/14, Figure 6A, 6B, 6D, 6H) where MP precursor formation occurs (Figure 7C). Subsequent activity of the transgene is restricted to migrating adaxial cells but not to MPs (n = 12/12, Figure 6C and 6G). These data suggest that the different levels of BMP activation along the DV axis could control the dichotomy of the MP/SSFs cell fate choice.
Fig. 6. BMP signaling forms a dorsal and ventral gradient within the myotome. 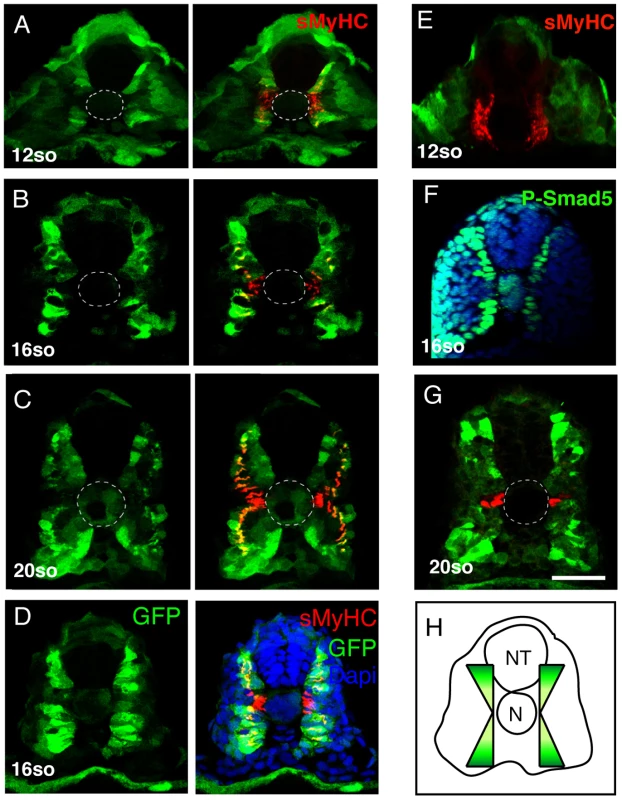
(A, B, C, D, E) Expression of GFP (green) and sMyHC (red) at indicated stages in Tg(5XBre[vent2]:-201id3:gfp) embryos uninjected or (E) after radar morpholino (rdrMO) injection. (F) Expression of phosphorylated-Smad5 (green) and nuclei (DAPI, blue) in 15-somite WT embryos. (G) Engrailed (red) and GFP (green) expression in Tg(5XBre[vent2]:-201id3:gfp) embryos at 20-somites. Cross-sections, maximum projections of multiple confocal scans. Scale bar 50 µm. Fig. 7. Radar or BMP signaling knock-down impairs MP formation. 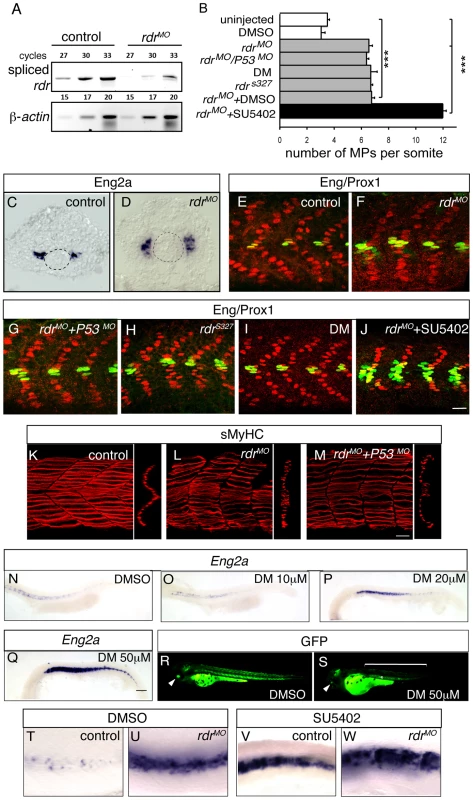
(A) The level of expression of spliced radar and β-actin mRNAs was analysed by semi-quantitative RT-PCR. Total RNA extracts were prepared at 1 dpf from uninjected embryos and from embryos that were injected with morpholinos blocking the splicing of radar (rdrMO). The number of cycles is shown. (B) Graphic representation of the number of MPs per somite at 1 dpf in the indicated conditions, values = mean, error bars = s.e.m., ***p<0.001. (C–D) eng2a expression at the 12-somite stage in (C) uninjected, or (D) rdrMO-injected embryos, as determined by ISH. (E–J) MPs staining with anti-Eng (green) and anti-Prox1 (red) antibodies at 1 dpf embryos in (E) uninjected, (F) rdrMO-injected embryos, (G) p53MO- and rdrMO-injected embryos, (H) radar mutant (rdrs327), (I) Dorsomorphin (DM) treated embryos and (J) SU5402-treated embryos that were injected with rdrMO, scale bar = 20 µm. (K–M) sMyhc (red) expression in the somites of (K) WT, (L) radar morphans and (M) P53/radar double morphans (rdrMO/P53MO) at 1 dpf, 3D reconstructions of multiple confocal scans, lateral and cross section views, scale bar = 20 µm. (N–Q) Eng2a expression in the somites of 1 dpf embryos after treatment with increasing doses of Dorsomorphin (10, 20 and 50 µM) or with DMSO (1%), scale bar 100 µm. (R–S) GFP expression in Tg(5XBRE[vent2]:-20lid3:GFP) embryos at 60 hpf after (S) DM (50 µM) or (R) DMSO (1%) treatments, showing that DM treatment inhibits BMP signaling. (T–W) Eng2a expression in 1 dpf embryos that were (U–W) injected or (T–V) not with rdrMO and treated with either (T–U) DMSO or (V–W) SU5402. As mentioned above, several BMP-like ligands are present in the tissues surrounding the myotome. gdf6a/radar exhibits polarized expression in the DV axis, with expression evident in the dorsal neural tube, hypochord, and the primitive gut endothelium [41], [42]. The specific temporal and spatial aspects of its expression suggest radar/gdf6a is the most likely BMP ligand to influence the DV patterning of the zebrafish myotome [41], [42]. To examine this question, we genetically down-regulated radar/gdf6a by the injection of antisense morpholinos specifically targeted to the zygotic radar/gdf6a mRNA (rdrMO, Figure 7A). Loss of zygotic radar/gdf6a function in Tg(5XBre[vent2]:-201id3:gfp) embryos causes a reduction of BMP activation evident within this line (n = 5, Figure 6E versus 6A), and a concomitant medio-ventral expansion of both the MP precursor domain (n = 6/6, Figure 7D versus 7C, Figure S5) and the number of differentiated MP cells at 24 hpf (nsomite = 21, Figure 7B and 7F versus 7E, Figure S5) consistent with previously reported results [43]. To confirm the specific effect of the rdrMO we generated a p53 and radar double morphant in which the number of MPs was similarly increased (Figure 7B and 7G) but non-Eng-positive SSFs now migrated properly at 24 hpf compared to the single radar morphant, (Figure 7M versus 7K, 7L). Furthermore, the phenotype of the p53/rdrMO injected embryos was identical to homozygous rdrs327 mutant embryos [44] (nsomite = 17, Figure 7B and 7H), an phenotype that could be reversed by careful titration with WT rdr mRNA injection (Figure S4). Embryos treated with Dorsomorphin (DM) (nsomite = 17), a specific pharmacological inhibitor of BMP signaling [45], exhibited a dose dependent increase in MP number (Figure 7B and 7I, 7N–7Q) and a concomitant reduction of GFP expression in Tg(5XBre[vent2]:-201id3:gfp) embryos (Figure 7R, 7S and [22]). A similar increase in MP number is also seen when adaxial cells are cell autnomously inhibited from responding to BMP like ligands through use of a dominant negative form of the BMP receptor (dnbmpr) expressed from the adaxial specific smyhc promoter (smyhc:dn-BMPr GFP) (39.01% of transgenic fibres, nfibres = 326, Figure 4L and 4Q).
To elucidate whether FGF and BMP signaling co-operate to control adaxial cell fate, we examined the formation of MPs and SSFs when both pathways were simultaneously knocked down. rdr morpholino injections into SU5402-treated embryos caused an increase in MPs and eng2a expression compared to controls (DMSO, SU5402 treatment or rdrMO alone) that was essentially additive (nsomite = 17, Figure 7B, 7J and 7T–7W), demonstrating that FGF and BMP cooperate to control the MP/SSF decision, and do so independently of one another.
FGF and BMP signaling independently coordinate specification of adaxial cells in the AP and DV planes
While the experiments outlined above, together with those of previously published studies, clearly show that BMP and FGF signaling can influence MP formation, they do not provide direct evidence for a role in DV or AP axis specification. It is possible that these signals could influence proliferation of MP precursors or recruitment to the adaxial cell compartment. In order to examine these issues more directly, we fate mapped the adaxial cell compartment using iontophoresis of TMRD into embryos where FGF signaling (SU5402 treatment) or FGF and BMP (SU5402+DM treatment) signaling had been inhibited (Figure 8A). According to our model, the MP domain should expand in the AP axis without FGF signaling and along both the AP and DV axes in the absence of either signal. Consistent with these predictions we found that MPs in SU5402-treated embryos could be derived from posterior adaxial cells (n = 8/12), a situation never observed in untreated embryos, but remained restricted to the mid-point of the DV axis (Figure 8B–8D). MPs in SU5402+DM-treated embryos arose from a pool of progenitors expanded in both the DV and AP axes of the adaxial cell equivalence group (n = 7/11, Figure 8E). Collectively, these results demonstrate that FGF and BMP signaling synergize to control specification of adaxial cells in the AP and DV axis, respectively.
Fig. 8. FGF and BMP signaling coordinates the fate of adaxial cells. 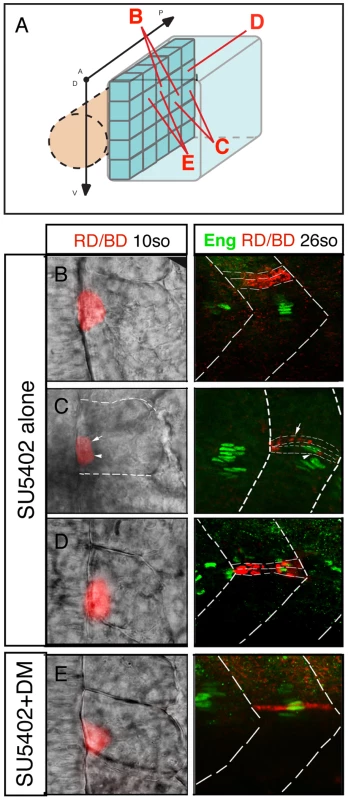
(A) Individual adaxial cells located in the posterior region of the adaxial cells at various D/V positions were labeled according to the diagram. (B–C–D) The fate of the adaxial cells after treatment of SU5402 alone or (E) in combination with Dorsomorphin (DM) was followed using iontophoretic injections of TMRD (red) within the newly formed somites of 10- to 15-somite embryos (left panels, dorsal views). Their fate was analyzed using anti-Engrailed antibody staining (green) at 30 hpf (right panels, lateral views). Discussion
A fate map for the HH-dependent adaxial cell compartment
At the beginning of segmentation all adaxial cells are columnar shaped epithelial-like precursors that align medially along the notochord, and display no morphological asymmetry. By initially undertaking fate map analyses of the entire forming myotome we have defined the adaxial cell compartment as a cell fate equivalence group that gives rise to these two specific slow muscle cell fates, the MPs and the SSFs. We have further defined mechanistically how these precursors are induced to give rise to these two distinct populations. The adaxial cells differentiate asynchronously within newly formed somites, with the cells adjacent to the anterior somitic border and located at the mid-point of the DV axis of the somite being the first to initiate the morphogenetic and differentiation movements we have previously describe [9]. This morphogenetic asymmetry is mirrored at the molecular level where the same cells that undergo precocious differentiation simultaneously initiate expression of the MP specific marker gene eng2a. This analysis suggests that these cells are the progenitors of the MP cells. In order to examine this question directly we generated a fate map of the adaxial compartment and found that each slow muscle fiber type (SSF and MP) arose from a specific region of the adaxial cell array. While the anterior adaxial cells at the DV mid-point of the somite give rise to MP within the midline, the non-MP precursor adaxial cells go on to form the SSF palisade at the lateral surface of the myotome in direct topographical reflection of their position in the pre-migratory adaxial compartment. These data indicate that both dorso-ventral and the anterior-posterior identities need to be determined coordinately within the adaxial cell equivalence group for cell fate determination to occur correctly.
Previous analyses have indicated that HH signaling is required to specify the adaxial cells prior to the onset of segmentation and that levels of HH influence the fate of these cells [7], [14], [18]. However, in the absence of HH signaling, cells with a distinct morphology still form adjacent to the notochord, indicating that not all aspects of adaxial cell morphogenesis are controlled by HH signal transduction [7]. In the absence of HH signal activation, a fast twitch muscle gene expression profile is activated within these cells instead of genes indicative of the slow muscle lineage. Consequently, these cells differentiate as fast MyHC expressing, cells stochastically dispersed throughout the myotome [7]. Despite the ability of HH signaling to control the determination of the slow muscle fate, the three HH ligands expressed in the embryonic midline (ehh/ihhb, shh/shha, twhh/shhb, [12], [20], [46], [47] are not restricted in the anterior-posterior direction, nor is there any indication that HH target genes are asymmetrically activated within the nascent adaxial cell compartment in either the anterior-posterior or dorso-ventral planes. Furthermore, we could also find no variation in the length of the primary cilia in adaxial cells, in line with the lack of modulation of HH target gene expression within adaxial cells. Thus, a model involving distinct regulators of cell fate needed to be invoked in order to conceptually generate the MP fate from the anterior-most cells of the dorso-ventral midline of the adaxial cell equivalence group.
A lack of FGF signaling in anterior adaxial cells induces MP fate
Many studies have examined the role of FGF signaling during myogenesis in vitro, where it has been shown to promote cell proliferation and represses myoblast differentiation. It has also been shown that early myoblast precursors require FGF in order to subsequently express their myogenic phenotype [48], [49]. However, despite these extensive in vitro studies the exact function of Fgf in the activation or the repression of muscle differentiation in vivo is controversial and appears to often to contradict this simple repressive role defined in vitro [50]. For example, zebrafish Fgf8-mediated signaling has been shown to drive the terminal differentiation of fast-twitch but not slow-twitch muscle fibers, and simultaneously also controls proliferation of the external cell progenitor layer, the equivalent of the amniote dermyotome [25], [51]. In amniote embryos, FGF signaling has been implicated in myogenesis in vivo, both in promoting progenitor cell proliferation [52] and in promoting their differentiation [53], [54]. In chick embryos most, if not all, replicating myoblasts present within the skeletal muscle masses of the limb express high levels of the FGF receptor FREK/FGFR4 and the inhibition of FgfR4 leads to a dramatic loss of limb muscle [54], [55]. Conversely, over expression of FGF in the chick somite leads to muscle differentiation suggesting that, as in the zebrafish lateral myotome, myogenic differentiation is positively controlled by FGF signaling [25], [54]. This is consistent with observations in mouse where ectopic expression of the cell autonomous negative regulator of FGF signaling sprouty2 in myogenic progenitors inhibits their differentiation [53].
Here we show that the FGF pathway does play a role in muscle formation but it is downstream of the HH dependent process of slow-twitch fiber specification. FGF signaling is asymmetrically activated in the adaxial cells. Specifically, within anterior adaxial cells it is strongly reduced, to the point of complete inhibition of specific FGF target genes. We have shown, using a combination of genetic and pharmacological approaches that down regulation of the FGF pathway promotes MP formation at the expense of SSFs within the adaxial cell compartment. This does not appear to be driven by the restriction of the expression of FGF ligands, since the FGF encoding genes, Fgf8a and Fgf17, are both localized to the anterior somite [25]. Rather, FGF signaling in anterior adaxial cells is inhibited by a cell autonomous negative regulator of the FGF signaling cascade, spry4 [30]. spry4 expression is induced by FGF signaling and has been shown to act in a negative feed back loop on the FGF pathway in a number of contexts (this present study and [27], [31]). The direct role of spry4 in MP formation is demonstrated by data that shows that the ectopic expression of spry4 in the adaxial cells induces MPs while its inactivation in spry4 mutant embryos inhibits this fate. Therefore, our results suggest a model where spry4 is activated within the anterior adaxial cell compartment in response to high levels of adjacent FGF ligands that ultimately suppress FGF signaling within these cells, thereby breaking equivalence in the anterior posterior dimension. This role appears to be more analogous to that played by FGF signaling during organogenesis rather than those outlined above for myogenesis, where the fate of various stem and progenitor cells are partitioned by activation or inhibition of FGF signaling in organs as diverse as the liver and pancreas [56], ear [57], [58] and teeth [59] often in conjunction with opposing cell fate determining signals, including BMP signaling.
BMP signaling determines dorso-ventral identity of the adaxial cell equivalence group
While FGF signaling restricts the fate of the adaxial cells to the anterior most cells of the myotome, a second signal is needed to restrict the positioning of these cells in the dorso-ventral dimension. Recently, studies have demonstrated that the downstream effector of BMP signaling, p-Smad5 is specifically restricted to the dorsal and ventral adaxial cells, and is absent from cells of the dorso-ventral midline of the myotome [22], [23]. Furthermore, several previous studies have shown that manipulation of BMP signaling can influence the number of engrailed positive MPs [21], [43]. Indeed, the ectopic expression of chick Dorsalin-1, a BMP-like family member, in the zebrafish notochord inhibits MP development [21]. More recent studies have shown that inhibition of BMP via use of the small molecule inhibitor Dorsomorphin, or morpholinos against the BMP receptor bmpr1ba, results in an increase of MPs [22], [23]. However, exactly how BMP influences the formation of these muscle subtypes has remained unclear. Here we show that the fate of the adaxial cells is specified in the DV axis by a radar-mediated BMP signaling. This statement is supported by several lines of evidence. Firstly, a transgenic reporter line specific for BMP signaling reveals that at the onset of segmentation, BMP signaling is active in the dorsal and ventral most adaxial cells, but absent from in the DV mid-point of the forming myotome. This region of low BMP activity of correlates with the location of MP precursor specification, as specifically determined via our fate map analysis. Secondly, BMP signaling is mediated by gdf6a/radar in the adaxial cells and knockdown of BMP activity modifies the fate muscle precursors in the adaxial compartment and promotes MP formation in a dose dependant manner.
Previous analysis of the activity of BMP signaling during muscle formation in amniotes has provided evidence that it negative regulates the myogenic program [60], [61] a role it appears to also play in controlling the proliferation and the onset of myogenesis within the external progenitor cell layer of the zebrafish myotome [62]. However, in the context of the adaxial cells it does not appear to influence the proliferation of these progenitors, the timing of entry of these cells into myogenesis or the differentiation of the adaxial cells themselves. Our lineage analysis specifically illustrates that it alters the fate of this progenitor compartment.
The activities of the HH, FGF, and BMP signaling pathways specify MP identity
In this study we show that in contrast to HH signal transduction, FGF and BMP signaling has no effect on the slow muscle fate but instead regulates the decision of adaxial cell progenitors to become either SSF or MP cells. Indeed, as discussed above, the activation of these signaling pathways promotes SSF formation while their decrease or absence promotes MP formation. Modulation of FGF or BMP signals does not affect HH signaling and the consequences of their knockdown on the adaxial fate are additive (this study and [22]). Similarly, manipulation of the level of HH signaling (mutants within the HH pathway or cyclopamine treatment) does not affect the expression pattern of phospho-Smad 5, suggesting that HH signaling does not influence cell fate indirectly through BMP signaling [22], [23]. Thus the FGF and BMP signals act independently of, and synergistically with, each other to control the SSF/MP cell fate dichotomy. Intriguingly, the application of both FGF and BMP is required for the induction of a specific muscle cell fate, the Pax7-positive satellite cell progenitors, in Xenopus animal caps [63]. This suggests that the synergistic action of BMP and FGF may operate to specify other muscle cell types.
While HH and BMP signalling have been demonstrated to coordinate cell fate determination in the chick neural tube [64] and HH, BMP and FGF signalling collectively control the specification of numerous cell types in vertebrate and invertebrate systems, the majority of these studies do not examine the fate of individual cells in real time. The developmental paradigm of the adaxial cells allows single cells to be labelled and tracked and their fate determined within a genetically defined cellular equivalence group in the living animal a set of attributes that is to our knowledge unique in vertebrate developmental systems. We therefore believe that our study suggests that the adaxial of zebrafish could emerge as a paradigmatic example of a vertebrate cell fate equivalence group, in the same manner as the, Drosophila neuroectoderm, parasegment and imaginal disc and the C. elegans vulva [1]–[6] which have provided exquisite cellular and genetic resolution to generate a detailed understanding of cell specification mechanisms within invertebrate systems.
Our results also demonstrate an integrated signaling milieu that coordinates the specification of muscle cell fates within the adaxial cell compartment. The adaxial cell pool is initially specified in the somitic region adjacent to the notochord by HH signal transduction from the embryonic midline. This, together with regional inhibition of FGF in the anterior-most adaxial cells and a lack of BMP signaling at the DV midpoint of the somite, creates a 3-Dimensional network of signals that restricts the MP fate to the most anterior cells within a specific cellular equivalence group in the developing myotome (Figure 9). These signals act independently from each other to determine fate and uniquely MP specification is controlled by the action of different signal transduction pathways that act specifically to direct specification in distinct axial dimensions. This essentially Cartesian system of cell fate determination is somewhat reminiscent of that deployed during the development of the ventral nerve chord of Drosophila where a complex series of patterning genes are deployed in gradients along the DV and AP axes to induced specific fate determining genes within individual neuroblasts within the neuroectodermal sheet [3], [4], [5]. However, in the case of the adaxial cells there is no evidence for a role of lateral inhibition, which in the Drosophila ventral neuroectoderm is required for the expression of individual proneural genes and adoption of specific fates [4], [5]. Furthermore, our results reveal that individual secreted signals act in specific dimensions within this Cartesian system, rather than in a cooperative or mutually exclusive manner to specify cell fate, the prevalent ways by which cells are determined in vertebrate systems.
Fig. 9. Model of the synergic action of FGF and BMP signaling on adaxial cell specification. 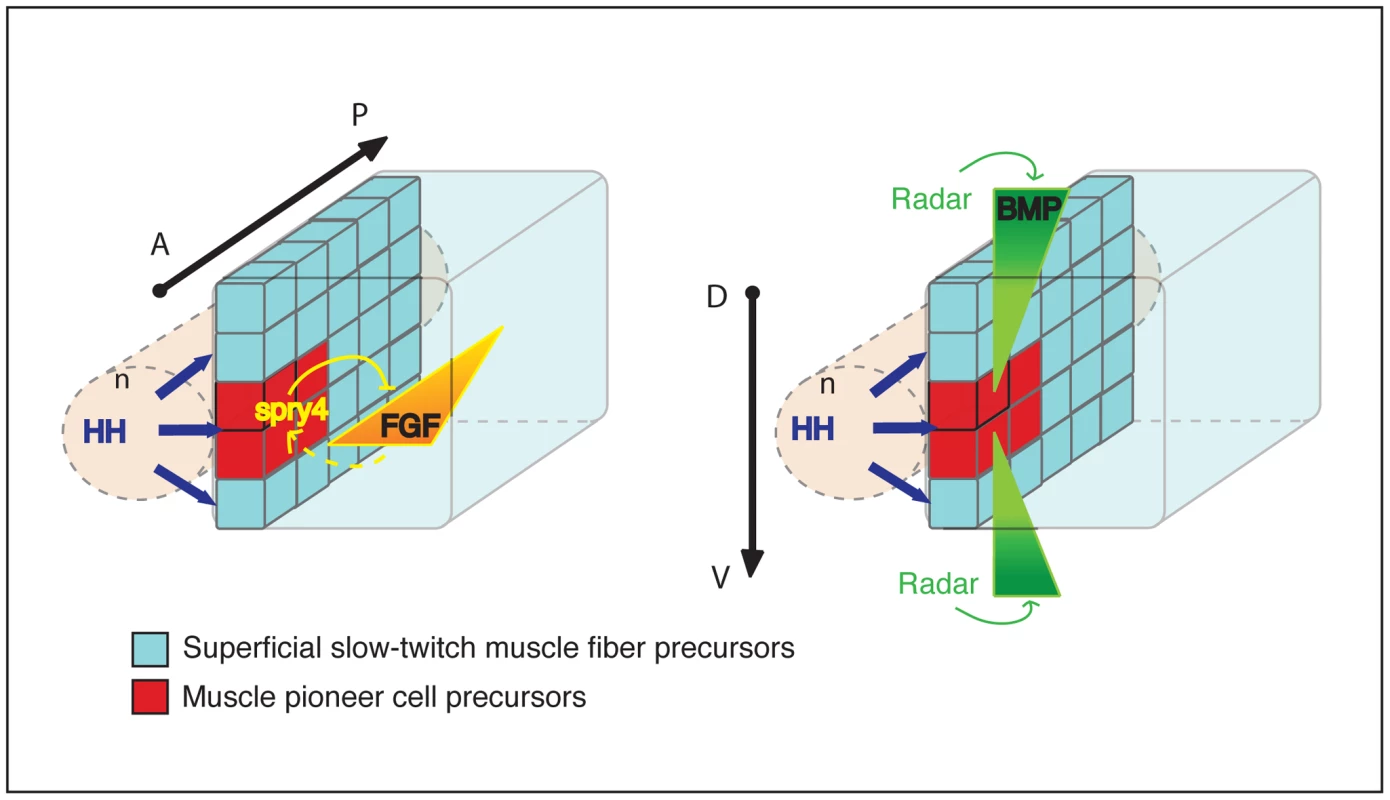
The diagram represents the adaxial compartment in the zebrafish somite, adjacent to the notochord (n), at the beginning of segmentation. The anterior-posterior (A/P) and dorsal-dentral (D/V) axes are shown. The adaxial cell pool is initially specified by HH signal transduction from the embryonic axial structure. Spry4-mediated regional inhibition of FGF restricts the MP fate to the most anterior adaxial cells. By contrast, the absence of Radar-mediated-BMP signaling at the dorso-ventral mid-point of the somite restricts MP fate at the dorso-ventral mid-point of the myotome. The combination of the three signals forms a 3-D patterning system that coordinates the specification of adaxial cells into muscle pioneer cells and superficial slow fibers. Materials and Methods
Zebrafish line and maintenance
Fish maintenance, staging and husbandry were as described previously [65]. Wild-type embryos of the TE strain were used in all staining and manipulation. Mutant alleles used were spry4fh117 (ZIRC, direct submission from the laboratory of Cecila Moens), radar/gdf6as327 (kind gift of Herwig Baier). Transgenic lines used were Tg (hsp70:dnFgfr1-EGFP)pd1 and Tg(5XBre[vent2]:-201id3:gfp).
In situ hybridization, antibody staining, sectioning, microscopy, and statistical analysis
In situ hybridization, antibody staining, and microtome sectioning were performed as previously described [65]. Probes were obtained by PCR amplification or from existing clones: sprouty4, erm (cb805), pea3 (IRBOp991G0430D) and eng2a [65]. In situ hybridizations on whole mount embryos were performed using digoxigenin (DIG)-labeled (Roche) antisense RNA probes and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) or fast red (Sigma). Microtome sectioning was performed on ISH stained embryos. Antibodies used were: anti-sMyHC (1/10, F59, DSHB Iowa, USA), anti-GFP (1/500, Rockland), anti-Engrailed (1/10, 4D9, DSHB Iowa, USA), anti-Prox1 (1/150, Fitzgerald), anti-Phospho-Smad5 (1/100, Cell signaling technology), anti-diphospho-ERK (1/10000, Sigma) streptavidin-alexa546 (1/1000, Molecular Probe). Vibratome sectioning was performed before antibody staining when necessary. 3D reconstructions were performed using Nikon C1 and Leica SP5 Confocal microscopes and Imaris software.
Statistical analysis
Counts of the number of differentiated MPs or SSFs were performed in the yolk extension region of 6 to 15 embryos. Analysis of variance (ANOVA) determined statistical significance of differences within a 95% confidence interval. In specific figures the following statistics were applied: Figure 4L: ANOVA analysis, Figure 5K: ANOVA analysis, Figure 7B: ANOVA analysis, Figures S1, S3, S5: Student test, 2 tails, unpaired, Table S1: ANOVA analysis.
Assembly of DNA constructs and RNA for live imaging
All constructs were assembled from entry clones using the Tol2kit (Kwan et al 2007). For transcription of RNA for whole-somite imaging, we assembled CMV/SP6-EGFPcaax and CMV/SP6-H2/afz-mCherry. Plasmids were linearized with NotI before transcription of capped RNA using an mMessage-mMachine kit (Ambion). Vectors used for mosaic analysis of single cells were smyhc1:spry4-IRES-EGFP, smyhc1:EGFP, smyhc1:dnBMPr GFP and smyhc1:dnspry4-IRES-EGFP. The new entry clone p5E-smyhc1 was made by subcloning the smyhc1 promoter from the plasmid p9.7kbsmyhc1:GFP-I-SceI (Elworthy et al 2008) into p5E-MCS (Kwan et al 2007). The pME-spry4 clone was made by cloning the full-length spry4 ORF into pDONR221. Similarly, the ORF of Xenopus type Ia BMPr truncated in C terminal (BMPrΔC) from BMPR22 construct ([66] or of dominant negative form of spry4 (spry4Y52A) from the pCS2-spry4Y52A were also cloned into pDONR221.
Injections, drug treatments, and heat shock inductions
Injections were performed as described previously [65]. 40 ng/µl of DNA encoding smyhc:spry4 ires GFP or smyhc:GFP were injected in one cell stage. Adaxial cells were imaged in embryos where 25 ng/µl of both CAAX-GFP and NLS-mCherry encoding mRNAs were injected at the one cell stage. 3 ng/µl of radar morpholino alone (5′-GCAATACAAACCTTTTCCCTTGTCC-3′) or in combination with 3 ng/µl of p53 morpholinos (5′-GCGCCATTGCTTTGCAAGAATTG-3′) were injected at the once cell stage. SU5402 (calbiochem) was added to the embryo medium at gastrulation or between 6 - to 10-somites at a final concentration of 80 µM and maintained until the appropriate stage. 10 to 50 µM Dorsomorphin (Sigma) was applied to similarly staged embryos. Heat shock induction of dn-fgfr1 expression was carried out at 6-somite stage. (hsp70:dnFgfr1-EGFP)pd1 transgenic embryos in there plate were placed at 38° during 2 hours. GFP expression was visualized immediately after heat shock to confirm the expression of the transgenic protein.
Iontophoresis injections
Iontophoresis injections as described in [67] with the following modifications: rhodamine dextran (10,000 MW, Molecular Probes, 5 mg/ml) combined with Biotin dextran (10,000 MW, Molecular Probes, 1.5 mg/ml) were injected into cells of agarose-imbedded, 10 - to 15-somite stage embryos. Adaxial cell labelings were positioned on the dorso-ventral axis via references to adjacent tissue landmarks within injected embryos and were imaged as previously described. The labeled embryo was dissected free of agarose and was allowed to develop until 30 hpf; it was then remounted in a 3% solution of methylcellulose (Sigma) and imaged. Subsequently, the embryo was fixed 2H in 4% paraformaldehyde and sequentially stained for Engrailed and sMyhc as described above.
Supporting Information
Zdroje
1. BakerNE, YuSY (1998) The R8-photoreceptor equivalence group in Drosophila: fate choice precedes regulated Delta transcription and is independent of Notch gene dose. Mech Dev 74 : 3–14.
2. HuangFZ, WeisblatDA (1996) Cell fate determination in an annelid equivalence group. Development 122 : 1839–1847.
3. SimpsonP, CarteretC (1990) Proneural clusters: equivalence groups in the epithelium of Drosophila. Development 110 : 927–932.
4. SkeathJB (1999) At the nexus between pattern formation and cell-type specification: the generation of individual neuroblast fates in the Drosophila embryonic central nervous system. Bioessays 21 : 922–931.
5. SkeathJB, ThorS (2003) Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol 13 : 8–15.
6. WangM, SternbergPW (2001) Pattern formation during C. elegans vulval induction. Curr Top Dev Biol 51 : 189–220.
7. HirsingerE, StellabotteF, DevotoSH, WesterfieldM (2004) Hedgehog signaling is required for commitment but not initial induction of slow muscle precursors. Dev Biol 275 : 143–157.
8. DevotoSH, MelanconE, EisenJS, WesterfieldM (1996) Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122 : 3371–3380.
9. DaggettDF, DomingoCR, CurriePD, AmacherSL (2007) Control of morphogenetic cell movements in the early zebrafish myotome. Dev Biol 309 : 169–179.
10. FelsenfeldAL, CurryM, KimmelCB (1991) The fub-1 mutation blocks initial myofibril formation in zebrafish muscle pioneer cells. Dev Biol 148 : 23–30.
11. RoyS, WolffC, InghamPW (2001) The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes Dev 15 : 1563–1576.
12. EkkerM, WegnerJ, AkimenkoMA, WesterfieldM (1992) Coordinate embryonic expression of three zebrafish engrailed genes. Development 116 : 1001–1010.
13. HattaK, BremillerR, WesterfieldM, KimmelCB (1991) Diversity of expression of engrailed-like antigens in zebrafish. Development 112 : 821–832.
14. WolffC, RoyS, InghamPW (2003) Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol 13 : 1169–1181.
15. Bryson-RichardsonRJ, CurriePD (2008) The genetics of vertebrate myogenesis. Nat Rev Genet 9 : 632–646.
16. BuckinghamM (2001) Skeletal muscle formation in vertebrates. Curr Opin Genet Dev 11 : 440–448.
17. BarresiMJ, StickneyHL, DevotoSH (2000) The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development 127 : 2189–2199.
18. InghamPW, KimHR (2005) Hedgehog signalling and the specification of muscle cell identity in the zebrafish embryo. Exp Cell Res 306 : 336–342.
19. LewisKE, CurriePD, RoyS, SchauerteH, HaffterP, et al. (1999) Control of muscle cell-type specification in the zebrafish embryo by Hedgehog signalling. Dev Biol 216 : 469–480.
20. CurriePD, InghamPW (1996) Induction of a specific muscle cell type by a hedgehog-like protein in zebrafish. Nature 382 : 452–455.
21. DuSJ, DevotoSH, WesterfieldM, MoonRT (1997) Positive and negative regulation of muscle cell identity by members of the hedgehog and TGF-beta gene families. J Cell Biol 139 : 145–156.
22. DolezM, NicolasJF, HirsingerE (2011) Laminins, via heparan sulfate proteoglycans, participate in zebrafish myotome morphogenesis by modulating the pattern of Bmp responsiveness. Development 138 : 97–106.
23. MauryaAK, TanH, SourenM, WangX, WittbrodtJ, et al. (2011) Integration of Hedgehog and BMP signalling by the engrailed2a gene in the zebrafish myotome. Development 138 : 755–765.
24. CaoY, ZhaoJ, SunZ, ZhaoZ, PostlethwaitJ, et al. (2004) fgf17b, a novel member of Fgf family, helps patterning zebrafish embryos. Dev Biol 271 : 130–143.
25. GrovesJA, HammondCL, HughesSM (2005) Fgf8 drives myogenic progression of a novel lateral fast muscle fibre population in zebrafish. Development 132 : 4211–4222.
26. HamadeA, DeriesM, BegemannG, Bally-CuifL, GenetC, et al. (2006) Retinoic acid activates myogenesis in vivo through Fgf8 signalling. Dev Biol 289 : 127–140.
27. RoehlH, Nusslein-VolhardC (2001) Zebrafish pea3 and erm are general targets of FGF8 signaling. Curr Biol 11 : 503–507.
28. NeugebauerJM, AmackJD, PetersonAG, BisgroveBW, YostHJ (2009) FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature 458 : 651–654.
29. HuangP, SchierAF (2009) Dampened Hedgehog signaling but normal Wnt signaling in zebrafish without cilia. Development 136 : 3089–3098.
30. CasciT, VinosJ, FreemanM (1999) Sprouty, an intracellular inhibitor of Ras signaling. Cell 96 : 655–665.
31. FurthauerM, ReifersF, BrandM, ThisseB, ThisseC (2001) sprouty4 acts in vivo as a feedback-induced antagonist of FGF signaling in zebrafish. Development 128 : 2175–2186.
32. HacohenN, KramerS, SutherlandD, HiromiY, KrasnowMA (1998) sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 92 : 253–263.
33. ReichA, SapirA, ShiloB (1999) Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development 126 : 4139–4147.
34. Bryson-RichardsonRJ, DaggettDF, CortesF, NeytC, KeenanDG, et al. (2005) Myosin heavy chain expression in zebrafish and slow muscle composition. Dev Dyn 233 : 1018–1022.
35. ElworthyS, HargraveM, KnightR, MebusK, InghamPW (2008) Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135 : 2115–2126.
36. LabaletteC, BouchouchaYX, WassefMA, GongalPA, Le MenJ, et al. (2011) Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4. Development 138 : 317–326.
37. DalyAC, RandallRA, HillCS (2008) Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol Cell Biol 28 : 6889–6902.
38. AlexanderC, ZunigaE, BlitzIL, WadaN, LePabicP, et al. (2011) Combinatorial roles for Bmps and Endothelin1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development 138 : 5135–46.
39. ColleryRF, LinkBA Dynamic smad-mediated BMP signaling revealed through transgenic zebrafish. Dev Dyn 240 : 712–722.
40. LauxDW, FebboJA, RomanBL (2011) Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Dev Dyn 240 : 682–694.
41. HallCJ, FloresMV, DavidsonAJ, CrosierKE, CrosierPS (2002) Radar is required for the establishment of vascular integrity in the zebrafish. Dev Biol 251 : 105–117.
42. SidiS, GoutelC, PeyrierasN, RosaFM (2003) Maternal induction of ventral fate by zebrafish radar. Proc Natl Acad Sci U S A 100 : 3315–3320.
43. KawakamiA, NojimaY, ToyodaA, TakahokoM, SatohM, et al. (2005) The zebrafish-secreted matrix protein you/scube2 is implicated in long-range regulation of hedgehog signaling. Curr Biol 15 : 480–488.
44. GosseNJ, BaierH (2009) An essential role for Radar (Gdf6a) in inducing dorsal fate in the zebrafish retina. Proc Natl Acad Sci U S A 106 : 2236–2241.
45. YuPB, HongCC, SachidanandanC, BabittJL, DengDY, et al. (2008) Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol 4 : 33–41.
46. KraussS, ConcordetJP, InghamPW (1993) A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75 : 1431–1444.
47. SchauerteHE, van EedenFJ, FrickeC, OdenthalJ, StrahleU, et al. (1998) Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development 125 : 2983–2993.
48. CleggCH, LinkhartTA, OlwinBB, HauschkaSD (1987) Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J Cell Biol 105 : 949–956.
49. OlwinBB, HauschkaSD (1986) Identification of the fibroblast growth factor receptor of Swiss 3T3 cells and mouse skeletal muscle myoblasts. Biochemistry 25 : 3487–3492.
50. SeedJ, HauschkaSD (1988) Clonal analysis of vertebrate myogenesis. VIII. Fibroblasts growth factor (FGF)-dependent and FGF-independent muscle colony types during chick wing development. Dev Biol 128 : 40–49.
51. HammondCL, HinitsY, OsbornDP, MinchinJE, TettamantiG, et al. (2007) Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev Biol 302 : 504–521.
52. von SchevenG, AlvaresLE, MootoosamyRC, DietrichS (2006) Neural tube derived signals and Fgf8 act antagonistically to specify eye versus mandibular arch muscles. Development 133 : 2731–2745.
53. LaghaM, KormishJD, RocancourtD, ManceauM, EpsteinJA, et al. (2008) Pax3 regulation of FGF signaling affects the progression of embryonic progenitor cells into the myogenic program. Genes Dev 22 : 1828–1837.
54. MaricsI, PadillaF, GuillemotJF, ScaalM, MarcelleC (2002) FGFR4 signaling is a necessary step in limb muscle differentiation. Development 129 : 4559–4569.
55. MarcelleC, WolfJ, Bronner-FraserM (1995) The in vivo expression of the FGF receptor FREK mRNA in avian myoblasts suggests a role in muscle growth and differentiation. Dev Biol 172 : 100–114.
56. CalmontA, WandziochE, TremblayKD, MinowadaG, KaestnerKH, et al. (2006) An FGF response pathway that mediates hepatic gene induction in embryonic endoderm cells. Dev Cell 11 : 339–348.
57. HammondKL, WhitfieldTT (2011) Fgf and Hh signalling act on a symmetrical pre-pattern to specify anterior and posterior identity in the zebrafish otic placode and vesicle. Development
58. ShimK, MinowadaG, ColingDE, MartinGR (2005) Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell 8 : 553–564.
59. KleinOD, LyonsDB, BaloochG, MarshallGW, BassonMA, et al. (2008) An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development 135 : 377–385.
60. DuprezDM, ColteyM, AmthorH, BrickellPM, TickleC (1996) Bone morphogenetic protein-2 (BMP-2) inhibits muscle development and promotes cartilage formation in chick limb bud cultures. Dev Biol 174 : 448–452.
61. PourquieO, FanCM, ColteyM, HirsingerE, WatanabeY, et al. (1996) Lateral and axial signals involved in avian somite patterning: a role for BMP4. Cell 84 : 461–471.
62. PattersonSE, BirdNC, DevotoSH (2010) BMP regulation of myogenesis in zebrafish. Dev Dyn 239 : 806–817.
63. DaughtersRS, ChenY, SlackJM (2011) Origin of muscle satellite cells in the Xenopus embryo. Development 138 : 821–830.
64. PriceSR, BriscoeJ (2004) The generation and diversification of spinal motor neurons: signals and responses. Mech Dev 121 : 1103–1115.
65. HollwayGE, MauleJ, GautierP, EvansTM, KeenanDG, et al. (2006) Scube2 mediates Hedgehog signalling in the zebrafish embryo. Dev Biol 294 : 104–118.
66. PyatiUJ, WebbAE, KimelmanD (2005) Transgenic zebrafish reveal stage-specific roles for Bmp signaling in ventral and posterior mesoderm development. Development 132 : 2333–2343.
67. HollwayGE, Bryson-RichardsonRJ, BergerS, ColeNJ, HallTE, et al. (2007) Whole-somite rotation generates muscle progenitor cell compartments in the developing zebrafish embryo. Dev Cell 12 : 207–219.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 10- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- The Germline Genome Provides a Niche for Intragenic Parasitic DNA: Evolutionary Dynamics of Internal Eliminated Sequences
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Calpain-5 Mutations Cause Autoimmune Uveitis, Retinal Neovascularization, and Photoreceptor Degeneration
- Cofilin-1: A Modulator of Anxiety in Mice
- The Date of Interbreeding between Neandertals and Modern Humans
- Embryos of Robertsonian Translocation Carriers Exhibit a Mitotic Interchromosomal Effect That Enhances Genetic Instability during Early Development
- Viral Evasion of a Bacterial Suicide System by RNA–Based Molecular Mimicry Enables Infectious Altruism
- Phosphatase-Dead Myotubularin Ameliorates X-Linked Centronuclear Myopathy Phenotypes in Mice
- Full-Length Synaptonemal Complex Grows Continuously during Meiotic Prophase in Budding Yeast
- MOV10 RNA Helicase Is a Potent Inhibitor of Retrotransposition in Cells
- A Likelihood-Based Framework for Variant Calling and Mutation Detection in Families
- The Contribution of RNA Decay Quantitative Trait Loci to Inter-Individual Variation in Steady-State Gene Expression Levels
- New Partners in Regulation of Gene Expression: The Enhancer of Trithorax and Polycomb Corto Interacts with Methylated Ribosomal Protein L12 Its Chromodomain
- Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- The Many Landscapes of Recombination in
- Faster-X Evolution of Gene Expression in
- Loss of Slc4a1b Chloride/Bicarbonate Exchanger Function Protects Mechanosensory Hair Cells from Aminoglycoside Damage in the Zebrafish Mutant
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
- and the BTB Adaptor Are Key Regulators of Sleep Homeostasis and a Dopamine Arousal Pathway in Drosophila
- Mutation and Fetal Ethanol Exposure Synergize to Produce Midline Signaling Defects and Holoprosencephaly Spectrum Disorders in Mice
- Specific Missense Alleles of the Arabidopsis Jasmonic Acid Co-Receptor COI1 Regulate Innate Immune Receptor Accumulation and Function
- Deep Genome-Wide Measurement of Meiotic Gene Conversion Using Tetrad Analysis in
- Mismatch Repair Balances Leading and Lagging Strand DNA Replication Fidelity
- Distinguishing between Selective Sweeps from Standing Variation and from a Mutation
- Cytokinesis-Based Constraints on Polarized Cell Growth in Fission Yeast
- Deposition of Histone Variant H2A.Z within Gene Bodies Regulates Responsive Genes
- Functional Antagonism between Sas3 and Gcn5 Acetyltransferases and ISWI Chromatin Remodelers
- The SET-Domain Protein SUVR5 Mediates H3K9me2 Deposition and Silencing at Stimulus Response Genes in a DNA Methylation–Independent Manner
- Morphogenesis and Cell Fate Determination within the Adaxial Cell Equivalence Group of the Zebrafish Myotome
- Muscle-Specific Splicing Factors ASD-2 and SUP-12 Cooperatively Switch Alternative Pre-mRNA Processing Patterns of the ADF/Cofilin Gene in
- Maize Is Required for Maintaining Silencing Associated with Paramutation at the and Loci
- Increasing Signal Specificity of the TOL Network of mt-2 by Rewiring the Connectivity of the Master Regulator XylR
- Use of Pleiotropy to Model Genetic Interactions in a Population
- RAB-Like 2 Has an Essential Role in Male Fertility, Sperm Intra-Flagellar Transport, and Tail Assembly
- Variants Affecting Exon Skipping Contribute to Complex Traits
- Topoisomerase II– and Condensin-Dependent Breakage of -Sensitive Fragile Sites Occurs Independently of Spindle Tension, Anaphase, or Cytokinesis
- Comparison of Family History and SNPs for Predicting Risk of Complex Disease
- Recovery of Arrested Replication Forks by Homologous Recombination Is Error-Prone
- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Comparative Genomics Suggests an Independent Origin of Cytoplasmic Incompatibility in
- It Was Heaven: An Interview with Evelyn Witkin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Mutation in the Gene Causes Alternative Splicing Defects and Deafness in the Bronx Waltzer Mouse
- Mutations in (Hhat) Perturb Hedgehog Signaling, Resulting in Severe Acrania-Holoprosencephaly-Agnathia Craniofacial Defects
- Classical Genetics Meets Next-Generation Sequencing: Uncovering a Genome-Wide Recombination Map in
- Regulation of ATG4B Stability by RNF5 Limits Basal Levels of Autophagy and Influences Susceptibility to Bacterial Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



