-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Operon Essential for Biofilm and Rugose Colony Development in
Biofilms are specialized and highly differentiated three-dimensional communities of bacteria encased in an extracellular polymeric matrix (EPM), and the bacteria’s mechanisms to form biofilm are closely linked to their virulence. The EPM often consists of polysaccharides, proteins, nucleic acids, and lipids. Compared to extracellular polysaccharides, little is known about the protein components in the biofilm matrix of Vibrio vulnificus, a foodborne pathogen. In this study, we identified and characterized cabABC genes which were preferentially expressed in biofilms. CabA is a calcium-binding protein and is secreted through functional CabB and CabC. Our results indicated that CabA contributes to the development of biofilm and rugose colony morphology under elevated c-di-GMP conditions. CabA is an extracellular matrix protein crucial for the structural integrity of robust biofilm in flow cells and on oyster shells. Calcium binding induces conformational changes and multimerization of CabA that may render the protein functional to build a well-structured matrix. CabA can assemble a functional matrix extracellularly only when exopolysaccharides (EPS) coexist, indicating that both CabA and EPS are required for the scaffold of V. vulnificus biofilm matrix. This is the first report on a non-polysaccharide matrix component that is essential for the development of the V. vulnificus biofilm structure.
Published in the journal: . PLoS Pathog 11(9): e32767. doi:10.1371/journal.ppat.1005192
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1005192Summary
Biofilms are specialized and highly differentiated three-dimensional communities of bacteria encased in an extracellular polymeric matrix (EPM), and the bacteria’s mechanisms to form biofilm are closely linked to their virulence. The EPM often consists of polysaccharides, proteins, nucleic acids, and lipids. Compared to extracellular polysaccharides, little is known about the protein components in the biofilm matrix of Vibrio vulnificus, a foodborne pathogen. In this study, we identified and characterized cabABC genes which were preferentially expressed in biofilms. CabA is a calcium-binding protein and is secreted through functional CabB and CabC. Our results indicated that CabA contributes to the development of biofilm and rugose colony morphology under elevated c-di-GMP conditions. CabA is an extracellular matrix protein crucial for the structural integrity of robust biofilm in flow cells and on oyster shells. Calcium binding induces conformational changes and multimerization of CabA that may render the protein functional to build a well-structured matrix. CabA can assemble a functional matrix extracellularly only when exopolysaccharides (EPS) coexist, indicating that both CabA and EPS are required for the scaffold of V. vulnificus biofilm matrix. This is the first report on a non-polysaccharide matrix component that is essential for the development of the V. vulnificus biofilm structure.
Introduction
Biofilm formation provides bacteria with protection from antimicrobial agents and host immune defense systems during infection as well as from a variety of stresses in the environment [1,2]. Therefore, biofilms of pathogenic bacteria are considered as one of the most important causes for new outbreaks and account for 65% of bacterial infections in humans [3]. Biofilm formation can be divided into sequential developmental stages beginning from initial surface attachment of planktonic bacteria continuing to multicellular structures (microcolony), subsequent maturation to biofilms, and detachment (dispersal) of bacterial cells from mature biofilms [1,2,4]. Mature biofilms are specialized and highly differentiated three-dimensional communities of bacteria encased in an extracellular polymeric matrix (EPM), the framework contributing to the organization and maintenance of biofilm structure and stability [2]. The major components of the EPM are polysaccharides, proteins, nucleic acids, and lipids, which are distributed between the cells in a non-homogeneous pattern [5]. Changes in colony morphology frequently reflect variations in biofilm matrix production levels, as smooth colony morphology of bacterial cells can switch to rugose (or wrinkled) by producing increased levels of EPM [4].
Exopolysaccharides (EPS) are the most prevalent component of Vibrio EPM and the structure, composition, and function of the Vibrio cholerae exopolysaccharide (VPS) and regulatory networks involved in its production have been studied as a model to delineate the molecular basis of bacterial biofilm formation [4,6]. Like many other pathogenic bacteria, Vibrio vulnificus, a pathogenic marine bacterium, forms biofilms in natural ecosystems [7,8,9]. There are several lines of evidence supporting that V. vulnificus embed themselves in oyster tissues and form biofilms to colonize and to persist in oyster as the primary infection route of the pathogen [10,11]. Diverse extracellular polysaccharides are produced by V. vulnificus and their functions in the development of biofilms are well characterized [8,12,13,14]. Among them, lipopolysaccharide (LPS) is known to play a role in the initial adhesion of planktonic cells to the surfaces of assay tubes [12]. A capsular polysaccharide (CPS), which is more tightly associated with the cell envelope than EPS, is also produced after maturation of biofilms [14,15,16]. CPS contributes to colony opacity and is required for determining biofilm size by limiting continual growth of mature biofilms [14,17]. Many, at least three, types of EPS are thought to play essential roles in the development of robust structures of EPM and mature biofilms of V. vulnificus [7,8,13,18,19].
The structural and regulatory genes involved in the V. vulnificus EPS production and, in turn, biofilm formation have been extensively studied [7,8,13,18,19,20]. The structural genes required for EPS production are organized into three clusters, EPS-I, II, and III on chromosome [13]. The cluster EPS-I consists of the rbd genes which are highly homologous to the syp genes involved in synthesis of the symbiotic-related polysaccharides of Vibrio fischeri [4,8]. The EPS-I genes were preferentially expressed in the estuarine-like conditions as determined by an RNA sequencing analysis [21]. The cluster EPS-II contains the brp genes (also wcr genes) which are organized into brpT encoding a putative transcription regulator and the brpABCDFHIJK operon required for the synthesis and transport of EPS [7,8,18,19]. The brpA mutant lost the ability to produce EPS, the parent biofilm phenotype, and rugose colony morphology [19]. Mutational analyses demonstrated that the third EPS cluster, EPS-III, is also involved in the production of EPS and formation of biofilms, but the functions of the EPS-III genes have not yet been addressed in details [13]. The rbd genes are regulated by RbdG [8], and all three clusters are positively regulated by NtrC at the transcription level [13]. Expression of the brp genes and production of EPS are regulated by 3′,5′-cyclic diguanylic acid (c-di-GMP) [7,22]. The secondary messenger c-di-GMP is synthesized by diguanylate cyclases (DGCs) containing the GGDEF domain and degraded by c-di-GMP-specific phosphodiesterases (PDEs) containing the EAL (or HD-GYP) domain which are ubiquitous in bacteria (for a recent review, [23]).
However, compared to EPS, little is known about the non-EPS components of the V. vulnificus EPM. Although a type IV pilin required for the initial adherence to HEp-2 cells and biofilm formation has been reported [24], no matrix proteins essential for the mature biofilm structures have been previously identified. In this study, a transcriptomic comparison of planktonic and biofilm cells of V. vulnificus led us to identify three genes, named cabABC, which are among the genes that are preferentially expressed in biofilms. Genetic, biochemical, and microscopic analyses were performed to demonstrate that CabA is a calcium-binding protein, produced as the biofilm matures, and secreted into the medium of the extracellular matrix through functional CabB and CabC. Construction of the cabA mutant and evaluation of its phenotypes provided evidences that CabA is a matrix protein that plays a crucial role in the c-di-GMP-induced development of robust biofilm and rugose colony in a calcium-dependent manner. Finally, a possible mechanism by which CabA can assemble a functional matrix was also proposed by examining the extracellular complementation of the cabA mutant and the brpA mutant which lacks EPS, and by determining the influence of calcium on the conformational changes and multimerization of CabA.
Results
Identification of the cabABC genes
A whole-genome microarray analysis was used to compare the transcriptional profiles of the biofilm and planktonic cells of V. vulnificus. The microarray analysis predicted 321 genes differentially regulated in biofilm cells, of which 177 genes were up-regulated and 144 were down-regulated. A complete list of the 321 genes is shown at Supporting Information (S1 Table). Among the genes that were up-regulated in biofilms, three consecutive genes VV2_1571, VV2_1572, and VV2_1573 were identified and annotated to encode a protein with putative calcium-binding motifs, an ATP-binding cassette (ABC)-type transporter, and a membrane-fusion protein (MFP), respectively (Fig 1A). Since the up-regulation of the three genes in biofilms was not previously reported, their preferential expression was experimentally verified. Quantitative real-time PCR (qRT-PCR) revealed that expression of the three genes increased 2.7–6.8 fold in biofilms (S2 Table). The three genes were named cabABC (for calcium-binding protein in biofilms A to C), since the three genes encode proteins involved in the furnishing of a calcium-binding and matrix-associated protein essential for biofilm development as demonstrated in the later parts of this study. It was noteworthy that the cabABC genes were 9–69 fold upregulated by elevated intracellular c-di-GMP levels, indicating that cabABC genes are regulated by c-di-GMP in cells [25].
Fig. 1. Genetic organization of the cabABC operon and amino acid sequence of CabA. 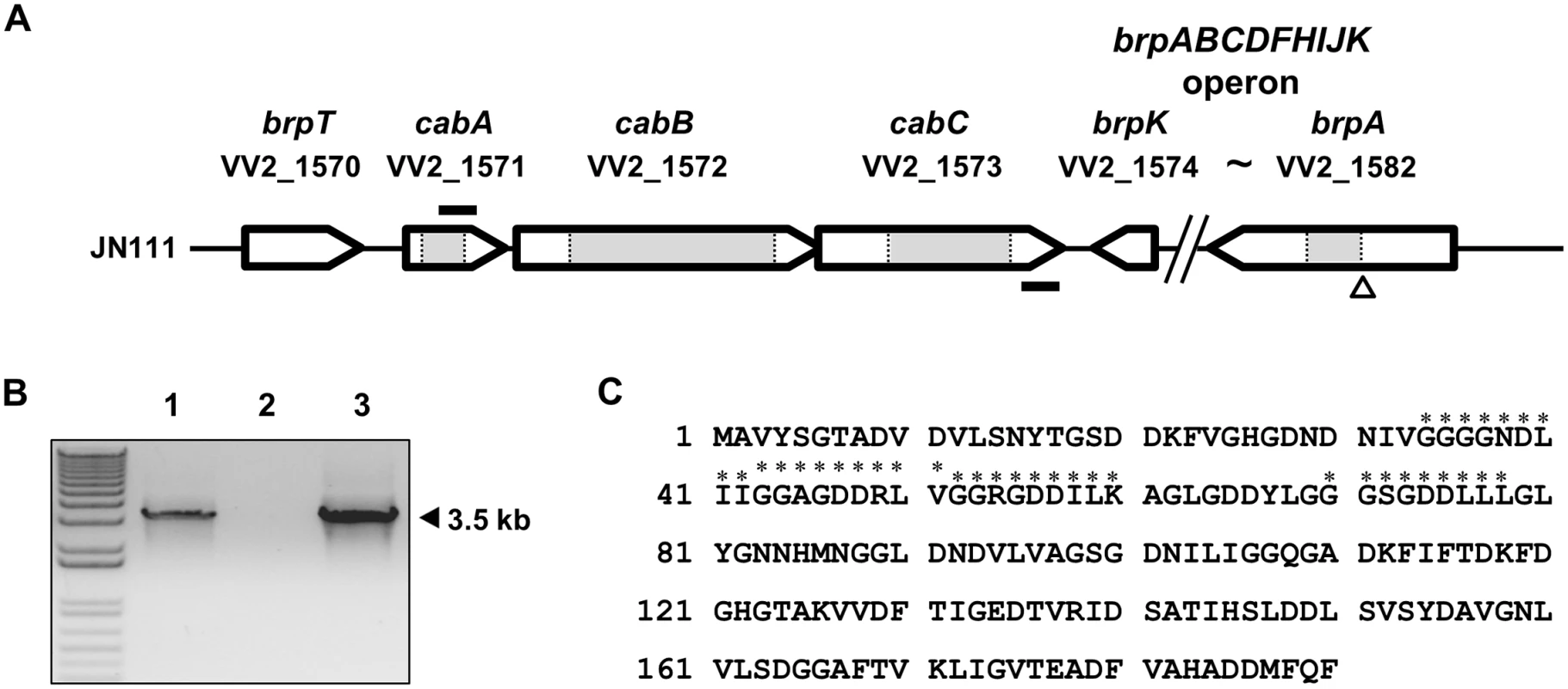
(A) The physical map of the cabABC operon located between brpT and brpABCDFHIJK gene cluster. The open arrows represent the coding regions and transcriptional directions of the genes. The figure was derived using the V. vulnificus CMCP6 genome sequences. The gene identifications are shown above each coding region. The size of the brpABCDFHIJK operon is reduced as indicated. The grey boxes represent the deleted regions of the cabA, cabB, cabC, and brpA mutants, respectively. The nptI insertion site is indicated as a triangle. The primers RTcabA and RTcabC2, used for the RT-PCR analysis, are depicted by solid bars. (B) Analysis of the cabABC transcript by RT-PCR. Total RNA was isolated from JN111 grown with LBS containing 0.01% arabinose and used for the synthesis of cDNA by reverse transcription. The cDNA (lane 1), DNase I-treated RNA (negative control; lane 2), and genomic DNA (positive control; lane 3) were used as templates for PCR. Molecular size markers (1 kb plus ladder; Invitrogen) and a PCR product are indicated. (C) The amino acid sequence was deduced from the nucleotide sequence of the cabA coding region (VV2_1571) and the four putative calcium-binding motifs (GGXG(N/D)DX(L/I/F)X) [28] are indicated with asterisks. Genetic organization of cabABC
Many coding regions that participate in biofilm formation were identified immediately upstream and downstream of the cabABC genes from the V. vulnificus genome sequence (Fig 1A). The brpABCDFHIJK operon is located downstream of the cabC gene and brpT is located upstream of the cabA gene. Consequently, the cabABC genes are positioned in the intergenic region separating the brpT and brpABCDFHIJK genes, and are organized in the same orientation as that of brpT (Fig 1A). To analyze the expression pattern of the cabABC genes at the transcriptional level, cDNA was synthesized from the RNA isolated from JN111 (hereafter referred as a parent strain, constructed to manipulate intracellular c-di-GMP levels as demonstrated in the later parts of this study and S1 Fig) grown with LBS containing 0.01% of arabinose and amplified by PCR using primers RTcabA and RTcabC2 (S3 Table and Fig 1B). As shown in Fig 1B, a single band of, approximately 3.5-kb DNA was amplified. Based on the DNA sequence of cabABC, the 3.5-kb PCR product corresponded to the expected size of the amplification product between the RTcabA and RTcabC2 primers. This revealed that cabA, cabB, and cabC are transcribed as a single transcriptional unit. A positive control reaction with V. vulnificus genomic DNA as a template produced the same-sized 3.5-kb PCR product, and no product was observed when total RNA was directly used as a PCR template (a negative control). Therefore, it appeared that the cabABC genes are transcribed as a single operon rather than as three independent genes.
Sequence analysis of CabA, CabB, and CabC
The amino acid sequence deduced from the cabA revealed a protein, CabA, composed of 190 amino acids with a theoretical molecular mass of 19,424 Da and a pI of 3.95 (Fig 1C). CabA revealed a high level of similarity (63% identical in amino acid sequence) with MfpA (http://blast.ncbi.nlm.nih.gov/Blast.cgi), which was previously predicted as a putative calcium-binding protein of Vibrio parahaemolyticus [26,27]. The predicted profile of the hydrophobicity (http://web.expasy.org/protscale/) suggested that CabA is a soluble protein rather than a membrane-associated protein. The in silico analysis of the amino acid sequences revealed that CabA harbors neither motif nor sequences frequently conserved in the known catalytic enzymes and predicted that CabA is a non-enzymatic structural protein (http://www.ncbi.nlm.nih.gov/cdd). Amino acid sequence analysis of CabA revealed the presence of at least four repeats of the calcium-binding motifs, the acidic glycine-rich nonapeptides, GGXG(N/D)DX(L/I/F)X [28], as depicted in Fig 1C.
The amino acid sequences of V. vulnificus CabB and CabC were 67 and 65% identical to those of V. parahaemolyticus MfpB and MfpC, respectively (http://blast.ncbi.nlm.nih.gov/Blast.cgi). MfpB and MfpC have been predicted as an ABC-type transporter and a MFP, respectively, which most likely constitute a type 1 secretion system (T1SS) [26,27]. The predicted profiles of the hydrophobicity (http://web.expasy.org/protscale/) of CabB and CabC indicated that both proteins are membrane-associated proteins. These sequence analyses suggested that CabA is a calcium-binding and non-enzymatic structural protein, and CabB and CabC are membrane-associated proteins homologous to the members of T1SS.
Calcium binding to CabA
To examine whether calcium binds specifically to CabA, the metal content of the purified CabA was analyzed using inductively coupled plasma-atomic emission spectrometry (ICP-AES). Only calcium was detected in significant amounts (39 μM) in the protein while other metal elements including magnesium, manganese, iron and zinc were not identified (Fig 2A). The concentration of calcium in the CabA solution (210 μM) indicated that the molar ratio of calcium ion bound to CabA was about 0.19. However, it is noteworthy that the recombinant CabA protein was synthesized in the Escherichia coli cytosol which contains limited concentration of calcium ion around 90 nM [29]. Thus, the molar ratio of calcium to CabA determined by the ICP-AES does not necessarily reflect actual stoichiometry of calcium bound to CabA. To evaluate calcium-binding parameters for CabA, isothermal titration calorimetry (ITC) experiments were carried out. The calcium-free CabA protein was titrated with calcium, and the ITC data were integrated and fit into a binding model consisting of two sets of binding sites which yields three very tight (Kd = 3 nM) and one weaker (Kd = 11 μM) sites (Fig 2B). The thermodynamic parameters of calcium binding to CabA determined by ITC are summarized in Fig 2C.
Fig. 2. Calcium binding to CabA. 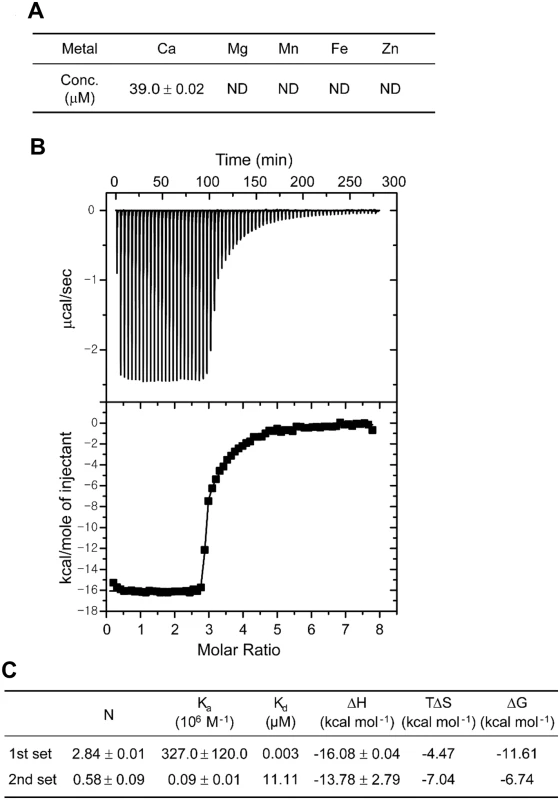
(A) The concentrations of metal ions in CabA analyzed by ICP-AES. ND, non-detection. (B) Typical isothermal titration calorimetric measurement of the calcium binding to CabA. The raw data displayed in the upper panel were integrated in the lower panel. The data points represent the experimental injection heat and the solid line corresponds to the calculated fit of the data using a model with two sets of binding sites of the ORIGIN software package (MicroCal Inc.). The heat data of calcium into the reaction buffer was subtracted from the reaction heat data between CabA and calcium. The experimental parameters were as follows: 70 injections of 3 μl of calcium solution, stirring speed of 307 rpm, duration 12 s, and spacing 240 s. (C) Thermodynamic parameters of the calcium binding to CabA. Construction of JN111 and effects of c-di-GMP on the expression of cabA
Since expression of the cabABC genes is regulated by c-di-GMP [25], V. vulnificus JN111, in which dcpA encoding a DGC is controlled under PBAD, was constructed to manipulate the intracellular c-di-GMP levels (Materials and Methods and S1 Fig). The intracellular c-di-GMP levels of JN111, determined using liquid chromatography–mass spectrometry (LC-MS) [30], were gradually elevated as concentrations of arabinose increased in the growth media (Fig 3A). The intracellular c-di-GMP level of the wild type CMCP6, in which the expression of dcpA was not manipulated, was approximately equivalent to the uninduced (or induced with 0.005% arabinose) JN111 level, indicating that the over-expression of dcpA leads to the increase of c-di-GMP well above the wild type level (Fig 3A). The results indicated that JN111 was an appropriate strain to manipulate intracellular c-di-GMP levels. The cellular levels of the cabA transcript and CabA protein in JN111 grown with VFMG-CF (the modified V. fischeri minimal medium as described in Materials and Methods) containing different concentrations of arabinose were also determined. In the absence of arabinose, the qRT-PCR and Western blot analyses revealed little, if any, expression of cabA. However, cabA expression increased along with the increased concentrations of arabinose in the growth media (Fig 3B and 3C). The combined results confirmed that cabA expression is induced by c-di-GMP and the increased expression of cabA in biofilms, as observed in microarray analysis, was probably attributed to elevated c-di-GMP. Strikingly, the presence of more than 0.02% arabinose was detrimental to the biofilm formation of the V. vulnificus strains (S2 Fig). Therefore, 0.01 and 0.02% arabinose were used in the hereafter experiments for biofilm and colony morphology analyses, respectively.
Fig. 3. Intracellular levels of c-di-GMP and cabA expression. 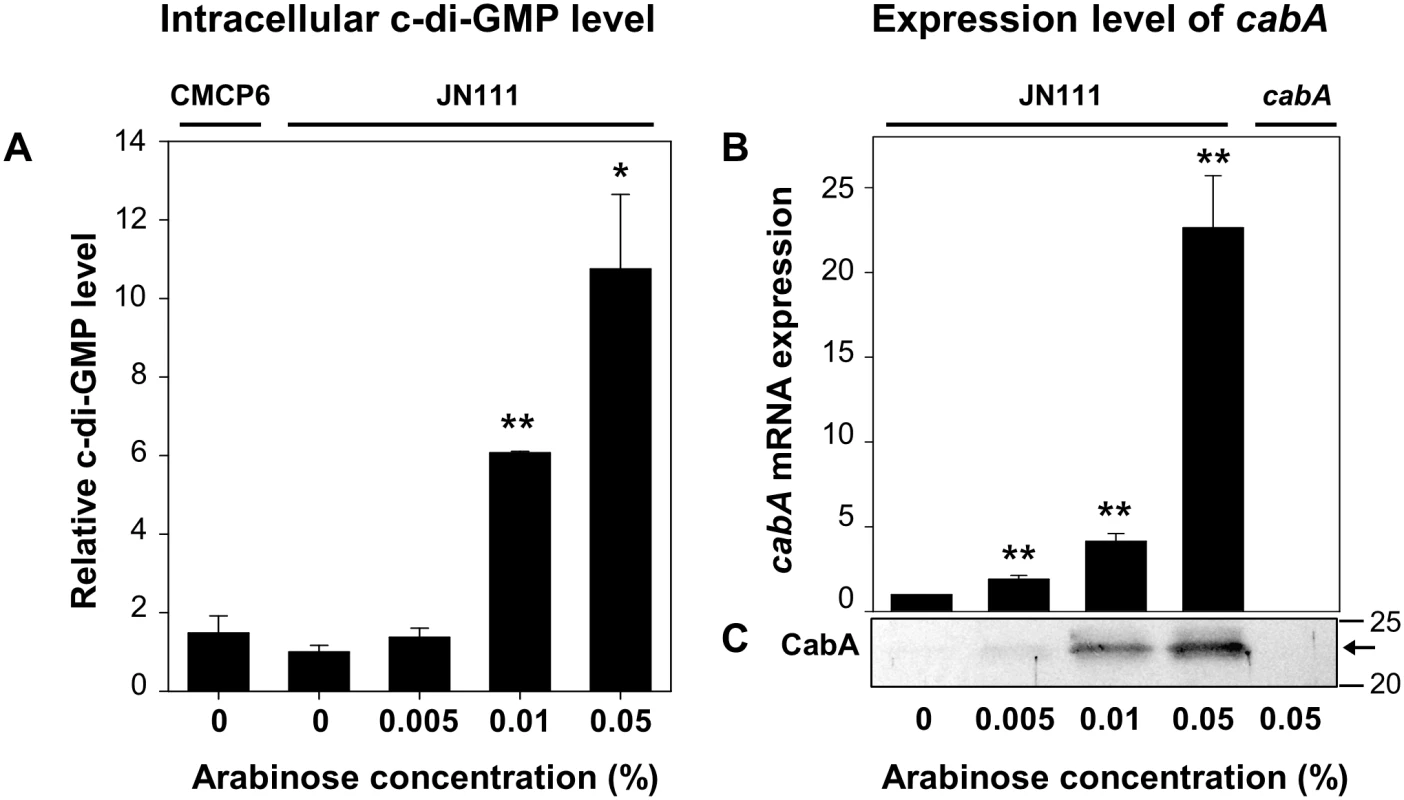
The strains were grown to A600 of 0.15 with VFMG-CF containing 10 mM of CaCl2, and different concentrations of arabinose as indicated. (A) Intracellular levels of c-di-GMP were measured using LC-MS. (B) Transcripts of cabA were determined using qRT-PCR. Relative levels of the c-di-GMP and cabA mRNA were presented as those levels in JN111 grown without arabinose as 1, respectively. *, P<0.05; **, P<0.005 relative to the c-di-GMP or cabA mRNA levels in JN111 grown without arabinose. Error bars represent the SD. (C) Protein samples were resolved on SDS-PAGE and immunoblotted using the rabbit anti-CabA antibody. The protein size markers (Precision Plus Protein Standards; Bio-Rad Laboratories) and CabA (arrows) are shown in kDa. CMCP6, wild type; JN111, parent strain; cabA, cabA mutant. Effects of the cabA mutation on biofilm formation
Biofilms of JN111 and the isogenic cabA mutant were formed in microtiter plates, and quantified using crystal violet (CV) staining assays (Fig 4A,4B and 4C). CV staining revealed that the level of biofilm formation is consistently reduced in the cabA mutant under conditions of elevated c-di-GMP (0.01% arabinose) (Fig 4A). This difference in the biofilm-forming ability of JN111 and the cabA mutant was apparent when increasing concentrations of calcium were present in the growth media (Fig 4A). These results indicated that CabA induced by c-di-GMP plays a role in biofilm formation in a calcium-dependent manner. Complementation with a functional cabA gene (pYM1109) restored the biofilm-forming ability of the cabA mutant to the level comparable to that of JN111 (Fig 4B). Therefore, the reduced biofilm-forming ability of the cabA mutant resulted from the inactivation of functional cabA rather than any polar effects on genes downstream of cabA. Noticeably, when grown in the absence of arabinose, JN111 as well as the cabA mutant produced very low levels of biofilm (Fig 4C). Furthermore, no obvious differences in the amounts of biofilm formed by JN111 and the cabA mutant in the absence of arabinose were observed. This indicated that the trace amounts of CabA expressed in the cells grown in the absence of arabinose would be not sufficient to contribute to biofilm formation (Fig 3B). Biofilm-forming abilities of the strains, in the same conditions used for Fig 4A and 4C but in a larger scale in test tubes, were similar to those observed in microtiter plates as determined by the pictures of the resulting CV staining (S3 Fig). These results demonstrated that CabA contributes to the c-di-GMP-mediated biofilm formation and requires calcium for its optimum activity.
Fig. 4. Effects of the cabA mutation on biofilm formation. 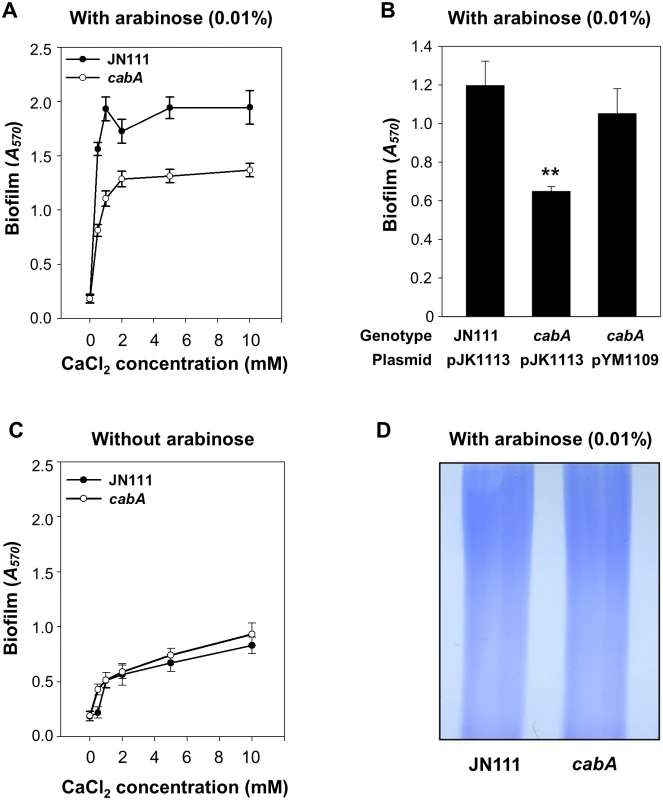
(A and C) Biofilms of the strains were grown for 24 h on 96-well microtiter plate wells containing VFMG-CF supplemented with or without 0.01% arabinose and with various levels of CaCl2, and quantified using CV staining. (B) Biofilms of the strains were grown with VFMG-CF containing 0.01% arabinose and 10 mM CaCl2, and quantified as described above. Ampicillin and kanamycin (100 μg/ml for each) were used to maintain plasmids in the strains. JN111 (pJK1113), parent strain; cabA (pJK1113), cabA mutant; cabA (pYM1109), complemented strain. **, P<0.005 relative to the JN111 biofilm. Error bars represent the SD. (D) EPS extracts were prepared from the strains grown on the LBS agar plates supplemented with 0.02% arabinose and resolved on a 4% stacking SDS-PAGE. JN111, parent strain; cabA, cabA mutant. The decreased formation of biofilm by the cabA mutant could be indicative of defects in the production of EPS which is known to be a major component of the Vibrio spp. biofilm matrix [4]. However, the amount of total EPS isolated from JN111 was indistinguishable from that isolated from the cabA mutant when determined based on the intensities of EPS bands resolved on the SDS-PAGE gel (Fig 4D). The result implied that CabA did not significantly influence the levels of EPS produced.
CabB - and CabC-dependent secretion of CabA into extracellular matrix
To get a better understanding on the role of CabA in biofilm formation, the localization of the protein in the biofilms was determined. Biofilms were fractionated to separate cells from the extracellular matrix and subjected to Western blot analyses. As evident from the result shown in Fig 5A, CabA was detected in the matrix fraction as well as the cell fraction of the JN111 biofilms, indicating that CabA is a protein secreted to the cell exterior in the matrix of biofilms. In contrast, CabA was detected neither in the cell nor the matrix fractions of the cabA mutant biofilms. This lack of CabA in the cabA mutant biofilms was restored by the introduction of pYM1109. To verify that the fractionation was performed without lysis of cells, a cytosolic regulatory protein IscR [31] was determined in each fraction by Western blot analysis. The result that IscR was found exclusively in the cell fraction ruled out the possibility that CabA in the matrix fraction resulted from cell lysis (Fig 5A).
Fig. 5. CabA in the cell and matrix fraction of biofilms. 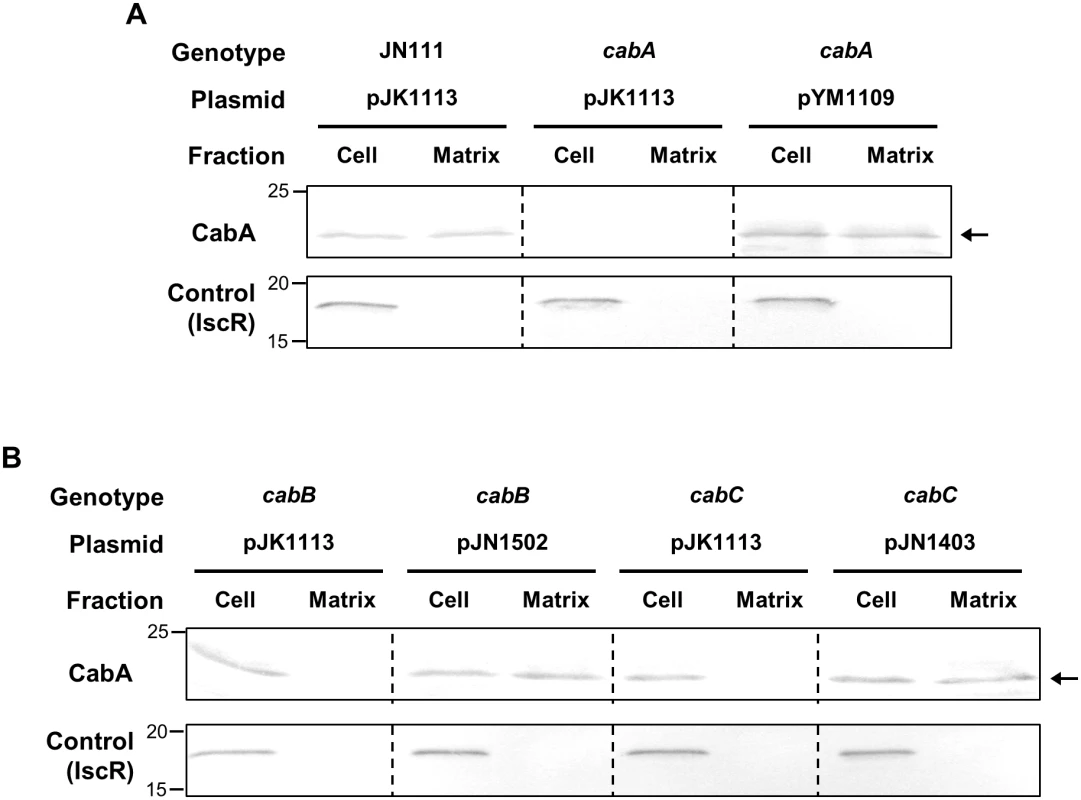
Biofilms of the strains were grown in the flask containing VFMG-CF supplemented with 0.01% of arabinose and 10 mM CaCl2. (A and B, upper panels) Cell and matrix fractions of the biofilms were prepared and the resulting fractions, equivalent to 10 μg of total proteins, were resolved on SDS-PAGE and immunoblotted using the rabbit anti-CabA antibody. (A and B, lower panels) In order to demonstrate that cells were not lysed during the fractionation procedures, an intracellular regulator IscR was detected in each fraction using the rabbit anti-IscR antibody prepared previously [31]. The protein size markers (Precision Plus Protein Standards; Bio-Rad Laboratories) and CabA (arrows) are shown in kDa. JN111 (pJK1113), parent strain; cabA (pJK1113), cabA mutant; cabA (pYM1109), complemented strain; cabB (pJK1113), cabB mutant; cabB (pJN1502), complemented strain; cabC (pJK1113), cabC mutant; cabC (pJN1403), complemented strain; Cell, cell fraction; Matrix, matrix fraction. As mentioned above, the deduced amino acid sequences of CabB and CabC share homology with those of MfpB and MfpC which are predicted to constitute a T1SS [26,27]. This information prompted us to examine whether CabA secretion to the cell exterior indeed depends on the presence of functional CabB and CabC. For this purpose, the isogenic cabB and cabC mutants were constructed (Table 1) and the CabA localization in biofilms was determined (Fig 5B). Western blot analyses revealed that CabA was detected exclusively in the cell fraction, but not in the matrix fraction, of the cabB and cabC mutant biofilms. The failure to detect CabA in the matrix fraction is indicative of defects in the secretion of CabA. The defect of CabA secretion in the cabB and cabC mutant was restored by the introduction of pJN1502 or pJN1403 carrying either recombinant cabB or cabC, respectively. IscR was detected once again exclusively in the cell fraction (Fig 5B). The results indicated that CabB and CabC play a critical role in the secretion of CabA, possibly by composing a T1SS. Importantly, the cabB and cabC mutant failed to accumulate additional CabA in the cell fraction as determined based on band intensity (Fig 5B). This was not surprising since previous studies have shown that the T1SS-impaired strains prevent accumulation of the protein substrates (effectors) in cells by degrading them by an unknown mechanism [32,33]. This observation was not pursued further. These combined results suggested that CabA is a secreted protein and localized at the extracellular matrix.
Tab. 1. Bacterial strains and plasmids used in this study. 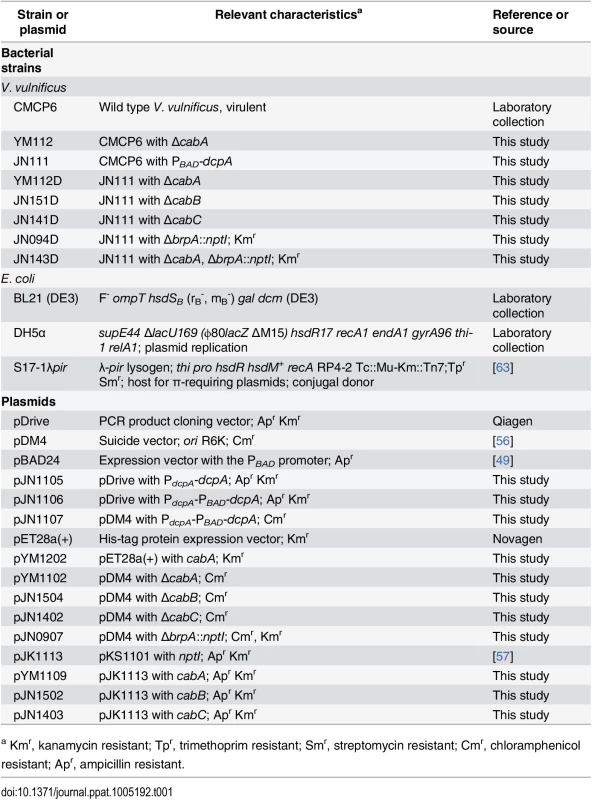
a Kmr, kanamycin resistant; Tpr, trimethoprim resistant; Smr, streptomycin resistant; Cmr, chloramphenicol resistant; Apr, ampicillin resistant. In situ determination of the CabA localization in biofilms
The proteins secreted to the cell exterior might perform diverse functions such as initial attachment to surfaces and subsequent formation of matrix. It has been reported that proper localization of the proteins is crucial for their functions in biofilm formation [34]. Many, if not all, proteins involved in the initial surface attachment, namely adhesins, are somehow retained on the cell envelope as observed with Pseudomonas fluorescens LapA and Salmonella enteritidis SiiE [35,36]. Therefore, to examine whether CabA is another adhesin, biofilms of the strains were formed on nickel grids and the localization of the functional form of CabA was determined in situ with a rabbit anti-CabA antibody and labeled with gold particles. When visualized using transmission electron microscopy (TEM), secreted CabA proteins were not concentrated or localized along the cell boundaries, but rather distributed distally from the cells throughout the extracellular medium of the biofilm matrix (Fig 6). In contrast, CabA was not detected at all in the cabA mutant biofilm, and this absence of CabA was restored when the cabA mutant was complemented with pYM1109. The throughout, albeit not uniform, distribution of CabA in the extracellular matrix indicated that CabA is not a cell envelope-localized adhesin. We therefore hypothesize that CabA is a structural component of the extracellular matrix.
Fig. 6. Distribution of CabA in biofilms. 
Biofilms of the strains were grown on the nickel grids containing VFMG-CF supplemented with 0.01% of arabinose and 10 mM of CaCl2. CabA in the biofilms was detected with the rabbit anti-CabA antibody, labeled with the secondary antibody conjugated to 10-nm gold particles, and then visualized using TEM (JEM1010, JEOL) at a 40000× magnification. Black dots represent the CabA protein labeled with the antibody conjugated to gold particles. Bars, 200 nm. JN111 (pJK1113), parent strain; cabA (pJK1113), cabA mutant; cabA (pYM1109), complemented strain. Kinetics of biofilm formation and cabA expression
Biofilm development of JN111 and cabA mutant was assessed at different time points using CV staining. Biofilm formation of both JN111 and the cabA mutant initiated at 2 h, reached maximum levels at 44 h, and then steadily decreased during extended periods of incubation (Fig 7A). During the first 10 h of biofilm development, the amounts of both biofilms of JN111 and the cabA mutant were not significantly different. In contrast, the biofilm formation of the cabA mutant was consistently reduced than that of JN111 after 10 h. The results indicated that CabA is not crucial for the initial surface attachment but plays an important role in the subsequent growth and maturation of biofilms. Consistent with this, cabA expression increased greatly at both transcript and protein levels after 12 h (Fig 7B). Interestingly, the CabA protein increased up to 44 h whereas the cabA transcript arrived at the maximum at 12 h and then decreased (Fig 7C). The prolonged increase of CabA may result from the accumulation of the protein in the extracellular matrix during biofilm development as observed in Pseudomonas putida biofilms [37]. These results indicated that CabA is a matrix-associated protein and plays a crucial role as the biofilm matures.
Fig. 7. Expression of CabA at the different development stages of biofilm. 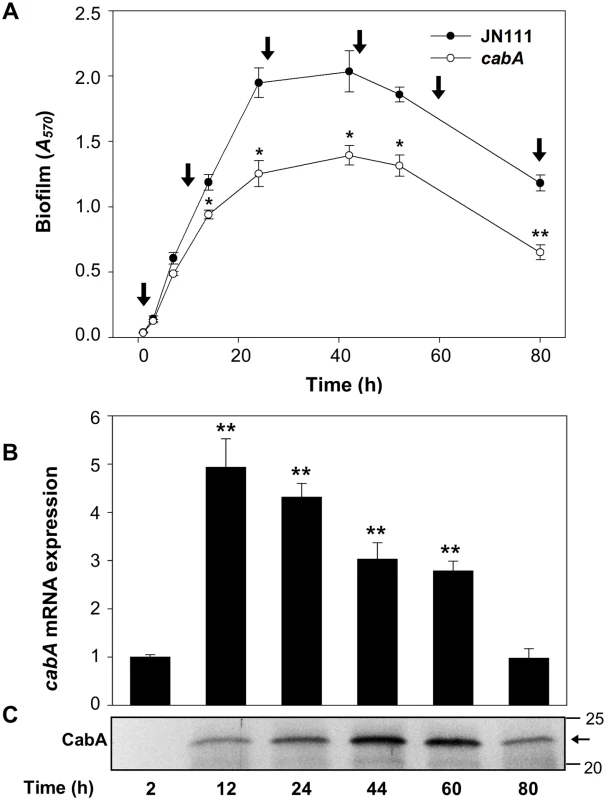
(A) Biofilm development of JN111 and the cabA mutant with VFMG-CF containing 10 mM of CaCl2 and 0.01% of arabinose was monitored using CV staining. Total RNAs and proteins were prepared from the JN111 biofilms (that is, from the cells and matrices) harvested at different stages as indicated by vertical arrows. *, P<0.05; **, P<0.005 relative to the JN111 biofilm. JN111, parent strain; cabA, cabA mutant. (B) The cabA mRNA levels were determined by qRT-PCR analyses, and the cabA mRNA level in the biofilms harvested after 2 h incubation was set as 1. **, P<0.005 relative to the cabA mRNA level in the biofilms harvested after 2 h incubation. Error bars represent the SD. (C) Protein samples were resolved on SDS-PAGE and immunoblotted using the rabbit anti-CabA antibody. The protein size markers (Precision Plus Protein Standards; Bio-Rad Laboratories) and CabA (horizontal arrow) are shown in kDa. Effects of the cabA mutation on colony morphology
In order to determine the role of CabA on colony morphology, JN111 and the cabA mutant were grown on the VFMG-CF agar plate supplemented with different concentrations of arabinose and 10 mM of CaCl2 (Fig 8). When grown in the absence of arabinose, JN111 revealed smooth colony morphology. However, the smooth colony morphology of JN111 switched to rugose (wrinkled) in the presence of 0.02% of arabinose, indicating that elevating c-di-GMP induced the development of the rugose colony morphology. In contrast to JN111, the cabA mutant formed smooth colony morphology regardless of the presence of arabinose and the smooth colony morphology of the cabA mutant did not switch to rugose under conditions of elevated c-di-GMP (Fig 8). These results indicated that the rugose colony formation induced by the elevated c-di-GMP is CabA-dependent. Complementation of the cabA mutant by the introduction of pYM1109 carrying a recombinant cabA shifted the smooth colony morphology of the cabA mutant to the rugose comparable to that of JN111 (Fig 8). Since rugose colony morphology frequently reflects increased levels of EPM [4], these results reinforce the view that CabA is an important constituent of the V. vulnificus biofilm matrix.
Fig. 8. Effects of the cabA mutation on colony morphology. 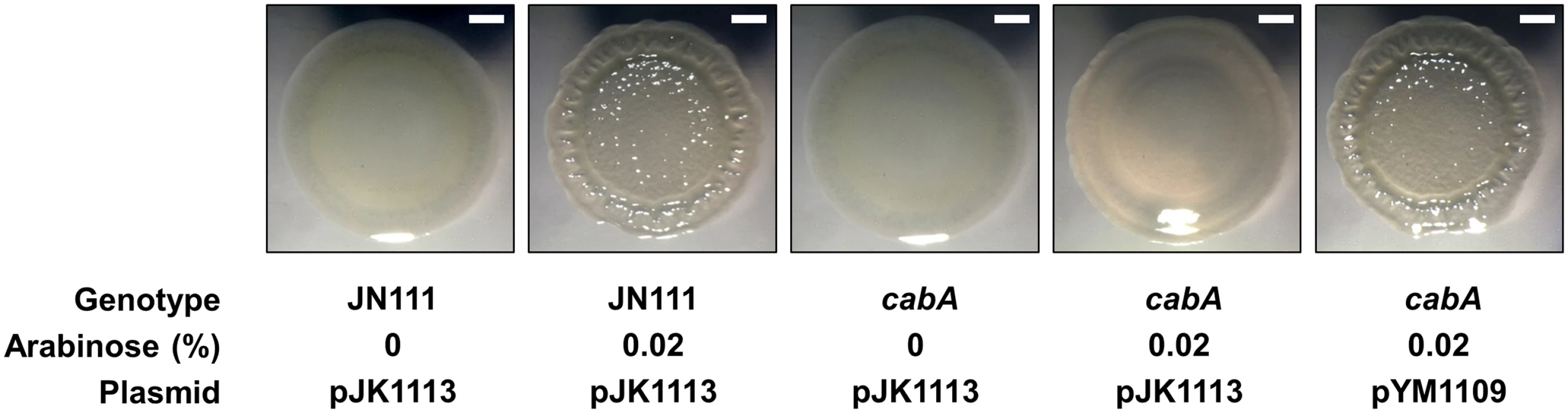
(A) The strains were spotted onto VFMG-CF agar plates supplemented with or without 0.02% of arabinose and 10 mM of CaCl2. Then, the plates were incubated at 30°C for 3 d. Ampicillin and kanamycin (200 μg/ml for each) were used to maintain plasmids in the strains. Each colony representing the mean rugosity of colony morphology from at least three independent experiments was visualized using a stereomicroscope (Stemi DV4, Zeiss) at an 8× magnification. Bars, 1 mm; JN111 (pJK1113), parent strain; cabA (pJK1113), cabA mutant; cabA (pYM1109), complemented strain. Effects of the cabA mutation on biofilm structure
The biofilm biomass of JN111 was less than 2-fold greater than that of the cabA mutant as determined by CV staining (Fig 4A and 4B). This limited CabA-dependent increase of biofilm formation led us to examine whether the cabA mutation has effects on the structure of biofilm. The biofilms produced on the coverslips in flow cells were stained using LIVE/DEAD BacLight Viability Kit (Invitrogen, Carlsbad, CA) and visualized by confocal scanning laser microscopy (CSLM). As shown in Fig 9A, JN111 displayed the structure of robust, large, uniform, and mushroom-shaped biofilms, whereas the cabA mutant produced weak, sparse, inconsistent, and markedly unstructured biofilms. JN111 formed much thicker biofilms than the cabA mutant in flow cells, and Z-stack measurements indicated that the depths of the JN111 and the cabA mutant biofilms were about 112 and 66 μm, respectively. When determined by the flow cell images, the JN111 biofilms were stable and evenly covered the glass coverslip, whereas the cabA mutant biofilms seemed to be easily washed away and disintegrated by medium flow, which resulted in uncovered surfaces of the glass coverslip (Fig 9A, upper-right corner). The results indicated that the structure of the CabA-deficient matrix is not robust enough to withstand medium flow in flow cells, and CabA is essential more for the structure rather than the quantity of the V. vulnificus biofilm.
Fig. 9. Effects of the cabA mutation on biofilm structure. 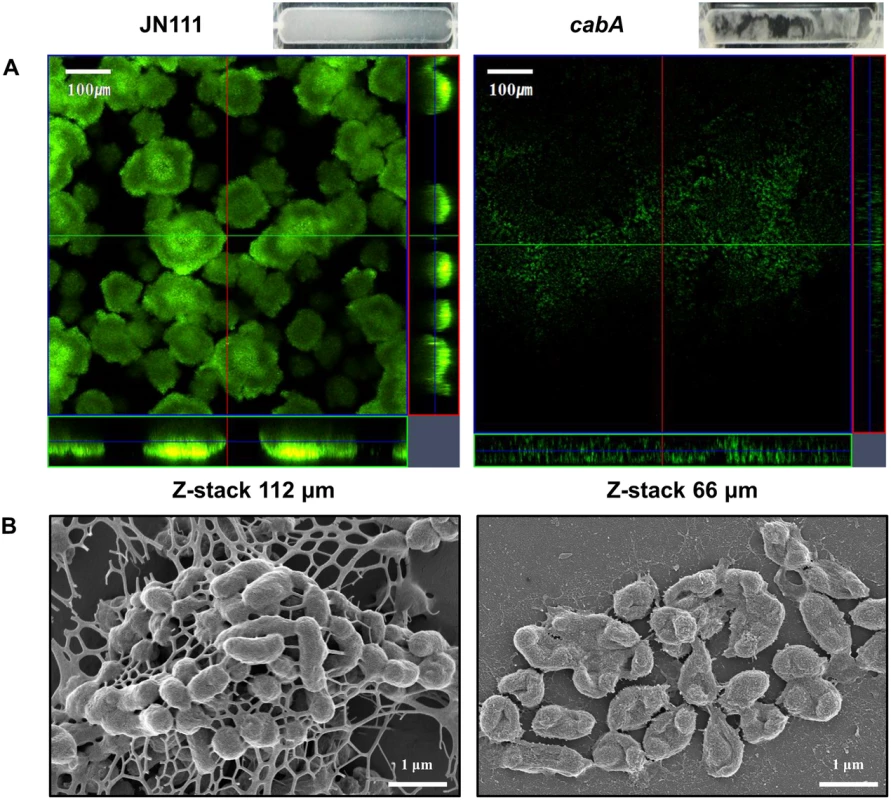
Biofilms of the strains were grown for 3 d in the flow cells containing VFMG-CF supplemented with 0.01% arabinose and 10 mM CaCl2. (A) Biofilms were stained by LIVE/DEAD BacLight Viability Kit (Invitrogen), and CSLM (LSM710, Zeiss) images were acquired at a 100× magnification. The depth of the Z-stack is indicated below the images in μm. Images of flow cell chambers were presented on the upper-right corner of each CSLM image. (B) Biofilms of the strains were fixed, dehydrated, coated with platinum, and visualized using SEM (Supra 55VP, Zeiss) at a 30000× magnification. Bars, 100 μm (A) and 1 μm (B); JN111, parent strain; cabA, cabA mutant. To further investigate the role of CabA in biofilm formation, biofilm structures of the strains were examined in more detail using scanning electron microscopy (SEM) (Fig 9B). JN111 produced compact and thick biofilms in which bacterial cells were bound together and encased within extracellular matrices. Filamentous materials that connect the bacterial cells to each other were visible within the extracellular matrix of JN111. In contrast, the cabA mutant cells were scattered without any extracellular filaments binding the cells together. The cabA mutant formed only few bacterial cell clusters that were left nearly bare, albeit the occasional appearance of thread-like strands. The observation that the cabA mutant made no matrices at all emphasizes the importance of CabA to the extracellular matrix formation. Since the amounts of EPS produced by JN111 and the cabA mutant grown on the LBS agar plates were not significantly different as described above (Fig 4D), it is reasonable to conclude that CabA appears to be important specifically for the structure of the extracellular matrix.
Biofilms of the strains were grown on oyster shells and their structures were examined using SEM (Fig 10). As observed in flow cells, JN111 formed robust biofilms on the oyster shells with filamentous materials connecting bacterial cells in the extracellular matrix whereas its isogenic cabA mutant (YM112D) formed only few bacterial cell clusters with the significantly smaller amount of extracellular filaments (Fig 10A). Similarly, the wild type CMCP6, in which the intracellular c-di-GMP levels were not manipulated, produced markedly greater amounts of extracellular matrix compared to its isogenic cabA mutant (YM112) (Fig 10B). Both cabA mutants, YM112D and YM112, were not able to develop considerable amounts of multicellular layers of bacterial cells on the surfaces of oyster shells. These results, combined with the results of Fig 9A and 9B, suggested that CabA is essential for the structural integrity of the V. vulnificus biofilms in the natural environments as well as in vitro.
Fig. 10. Structure of biofilms formed on oyster shells. 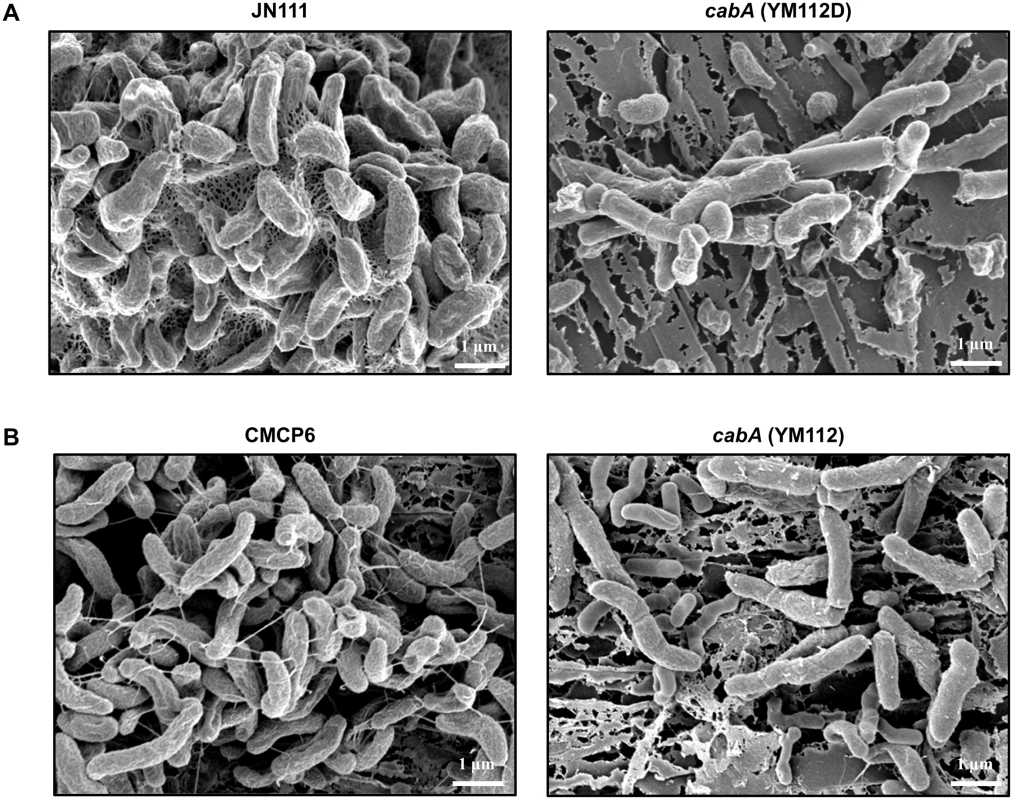
Biofilms of JN111 and its isogenic mutant (A) or CMCP6 and its isogenic mutant (B) were grown on oyster shells for 24 h in 24-well microtiter plates containing VFMG-CF supplemented with or without 0.01% arabinose and with 10 mM CaCl2. The biofilms were fixed, dehydrated, coated with platinum, and visualized using SEM (Supra 55VP, Zeiss) at a 20000× magnification. Bars, 1 μm; JN111, parent strain; YM112D, cabA mutant (of JN111); CMCP6, wild type; YM112, cabA mutant (of CMCP6). Exogenous addition of purified CabA rescues the cabA mutant
We examined if exogenous CabA can rescue the biofilm defect of the cabA mutant. The cabA mutant in microtiter plates was allowed to form static biofilm in the presence of increasing amounts of purified CabA. Purified CabA was able to restore the biofilm - and rugose colony forming ability of the cabA mutant (Fig 11). The cabA mutant incubated in the presence of purified CabA of more than 0.8 μM formed biofilms that were comparable to that of JN111, in terms of their biomass (Fig 11A). This CabA complementation of the cabA mutant to form biofilm exhibited dependence on calcium and was not apparent unless calcium was present in the growth media (Fig 11A). Similarly, the smooth colony morphology of the cabA mutant on VFMG-CF agar plate was shifted to rugose in the presence of increasing amounts of CabA (Fig 11B). However, this CabA complementation of the cabA mutant to develop the rugose colony morphology was not obtainable when calcium was not added to the growth media (Fig 11B). The rugose colony morphology developed by JN111 exhibited a similar dependence on the calcium concentration. We interpret these results as an indication that CabA can function properly outside the cell to yield a functional matrix, supporting the previous hypothesis that CabA is a protein released into the extracellular matrix. Taken together, the results led us to reconfirm that CabA is an extracellular matrix protein and requires calcium for its optimum activity.
Fig. 11. Effects of purified CabA protein on biofilm formation and colony morphology. 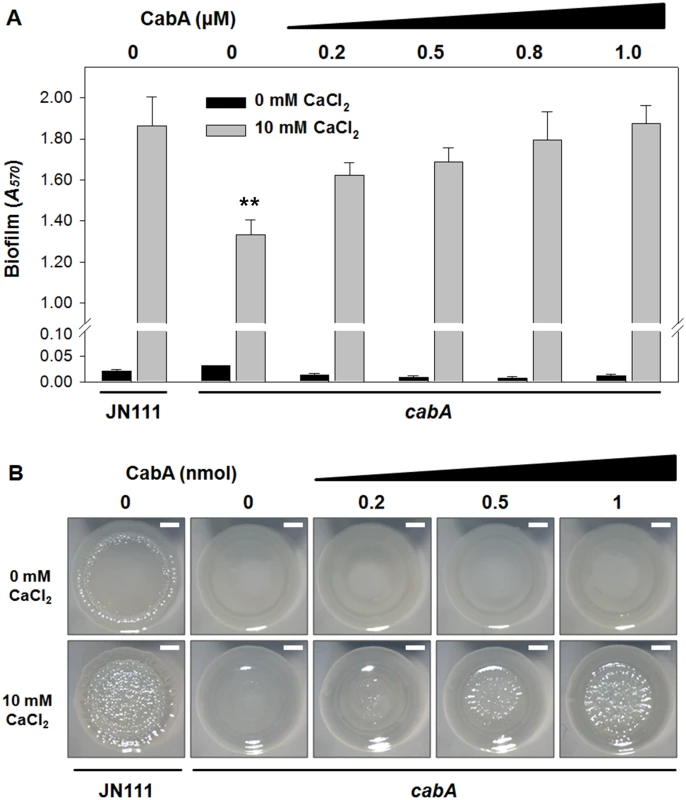
(A) Biofilms of the strains were grown with VFMG-CF supplemented with 0.01% arabinose, and various amounts of exogenously-provided calcium-free CabA in the absence or presence of 10 mM CaCl2. After 24 h incubation, biofilms were quantified using CV staining. Error bars represent the SD. **, P<0.005 relative to the JN111 biofilm grown with 10 mM CaCl2. (B) The strains were spotted onto VFMG-CF agar plates supplemented with 0.02% of arabinose and with or without 10 mM CaCl2. After being grown for 12 h at 30°C, calcium-free CabA were added exogenously onto the growing colonies as indicated and then further incubated for 2.5 d. Each colony representing the mean rugosity of colony morphology from at least three independent experiments was visualized using a stereomicroscope (Stemi DV4, Zeiss) at an 8× magnification. Bars, 1 mm; JN111, parent strain; cabA, cabA mutant. Calcium-induced conformational change and multimerization of CabA
The inability of CabA to rescue the cabA defects in the absence of calcium (Fig 11A and 11B) postulated that calcium is probably required for the shift of CabA to a conformation functional as a biofilm matrix protein. The analysis of the far-UV circular dichroism (CD) spectrum showed that the calcium-free CabA is predominantly unfolded and lacks secondary structures (Fig 12A). However, the secondary structures of CabA were obviously formed upon addition of 10 mM calcium (Fig 12A) [38]. The properties of the CabA protein were further assessed by performing size-exclusion chromatography. The calcium-free CabA showed a broader elution pattern than the calcium-binding monomeric CabA, indicating that calcium has an important role in CabA folding (Fig 12B). A monomeric form of CabA showed a tendency to form a multimer (dimer or tetramer) in the presence of high concentration of calcium (above 20 mM) (Fig 12B). By contrast, the multimeric CabA was converted to a monomeric form by reducing calcium concentration (Fig 12B). Collectively, these results led us to hypothesize that CabA undergoes calcium-dependent conformational change and multimerization, which may be linked to the development of biofilm matrix and rugose colony. Supporting the hypothesis, the calcium in the matrix of biofilm, which was grown with 10 mM CaCl2 added externally, was concentrated to reach 43 mM, which is enough for induction of CabA multimerization (Fig 13).
Fig. 12. Calcium-induced conformational change and multimerization of CabA. 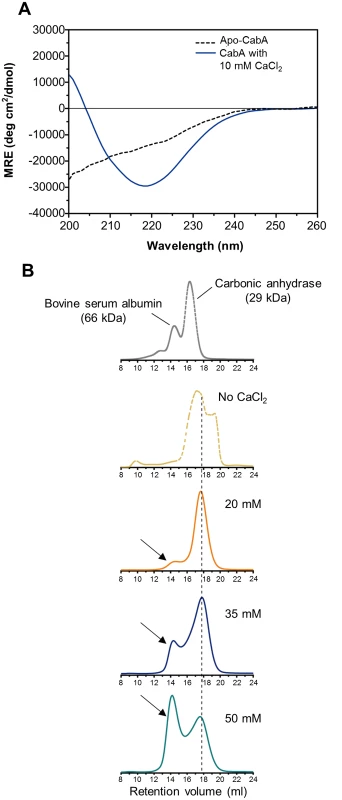
(A) Typical far-UV CD spectra of CabA in the absence (black dotted line) and presence (blue line) of calcium. MRE, mean residue ellipticity. (B) Calcium-dependent multimerization of CabA was assessed using size-exclusion chromatography. CabA was eluted in the absence (yellow dotted line) and presence of calcium (20 mM, orange line; 35 mM, blue line; 50 mM, green line). The molecular weight standards (gray dotted line), bovine serum albumin (66 kDa) and carbonic anhydrase (29 kDa) are indicated. Elution peaks corresponding to monomeric and multimeric CabA are indicated by a black dashed line and arrows, respectively. Fig. 13. Calcium accumulation in the biofilm matrix. 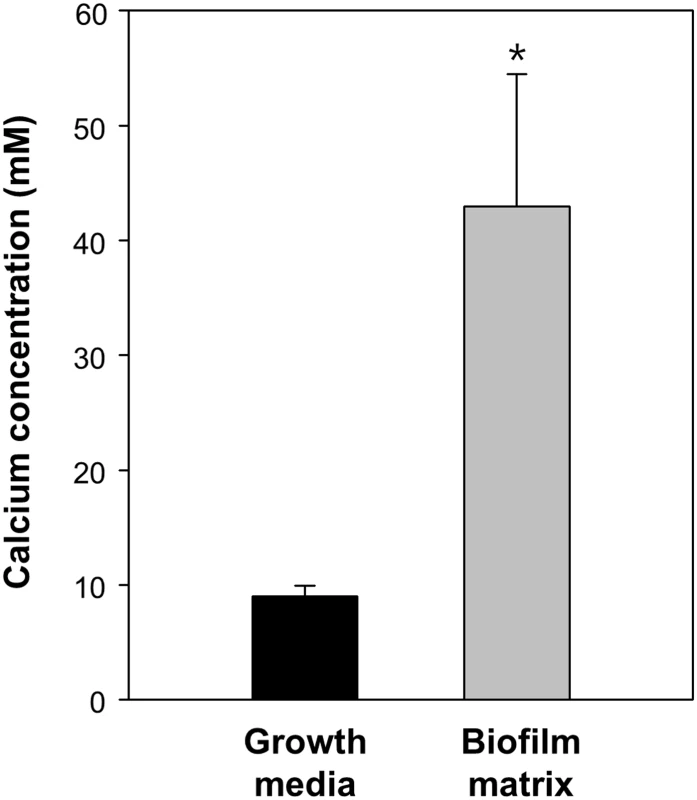
The JN111 Biofilm was grown in the flask containing VFMG-CF supplemented with 0.01% of arabinose and 10 mM CaCl2. Growth media and matrix fraction of the biofilms were prepared and the concentrations of calcium ion were measured using ICP-AES. The calcium concentrations were normalized to each volume of the matrix and growth media, and presented in molarity (mM). Error bars represent the SD. *, P<0.05 relative to the calcium in the biofilm growth media. Extracellular complementation of the cabA and brpA mutants
Since the brp genes (EPS-II) are known as the most important genes for EPS production and thereby biofilm formation of V. vulnificus [7,8,13,18,19], a brpA mutant was constructed and used to assess the EPS effects on the CabA function. Both the brpA and cabA mutants were defective in the biofilm formation when co-cultured with the cabA brpA double mutant which lack production of both EPS and CabA (Fig 14A). The residual biofilm levels of the brpA and cabA mutants were similar to each other and significantly lower than that of JN111 (Fig 14A). The biofilm defect of the brpA mutant supported the previous observation that EPS synthesized by EPS-II is an essential constituent of the V. vulnificus biofilm matrix [13] and further implied that CabA is unable to develop a functional matrix unless EPS is also present. When the brpA mutant was co-cultured with the cabA mutant, however, the resulting biofilm was similar to that of the JN111 (Fig 14A). This result indicated that CabA can extracellularly assemble an apparently functional matrix with EPS that is produced by different cells. Similarly, the smooth colony morphologies of the brpA and cabA mutants became rugose when the mutants were co-cultured (Fig 14B). The results suggested that CabA requires EPS in the matrix to perform its function for the development of mature biofilm structures and rugose colony morphology.
Fig. 14. Extracellular complementation by co-culturing the mutants which lack either EPS or CabA. 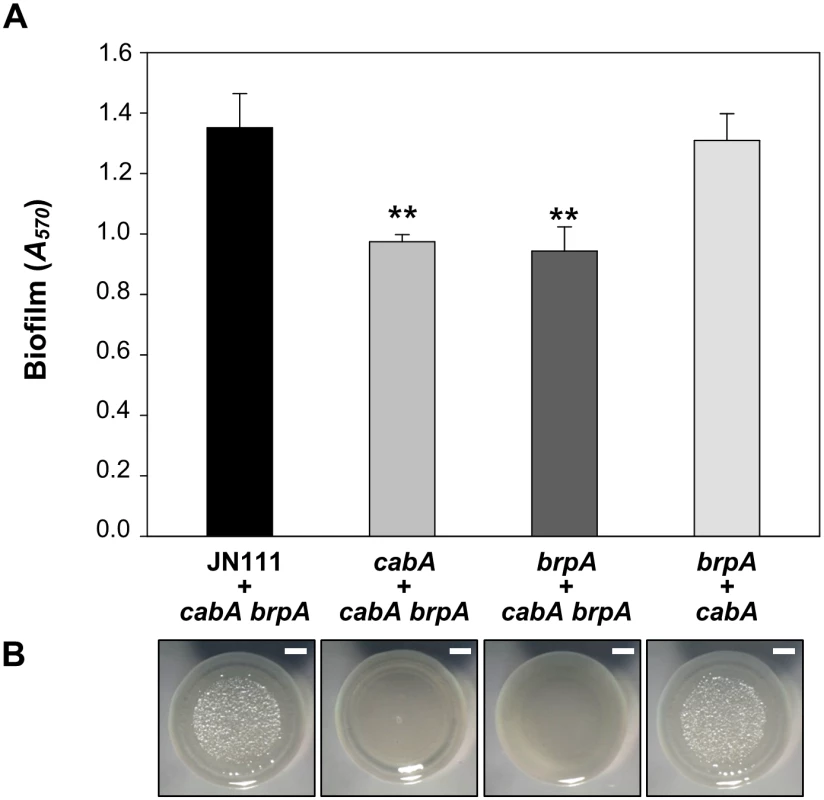
(A) Biofilms of the co-cultures prepared by mixing JN111, the cabA mutant, or the brpA mutant with either the cabA brpA double mutant or the brpA mutant at a 1:1 ratio were grown with VFMG-CF supplemented with 0.01% of arabinose and 10 mM of CaCl2 for 24 h, and quantified using CV staining. Error bars represent the SD. **, P<0.005 relative to the biofilm of co-culture of JN111 and the cabA brpA double mutant. (B) The co-cultures prepared as described above were spotted onto VFMG-CF agar plates supplemented with 0.02% of arabinose and 10 mM CaCl2, incubated at 30°C for 3 d, and then visualized using a stereomicroscope (Stemi DV4, Zeiss) at an 8× magnification. Bars, 1 mm; JN111, parent strain; cabA, cabA mutant; brpA, brpA mutant; cabA brpA, cabA brpA double mutant. Discussion
The bacterial secondary messenger, c-di-GMP, is a global regulatory molecule that controls multiple phenotype changes in the bacterial world. Among the phenotype changes affected by c-di-GMP, life style switches from free-living planktonic cells to sessile biofilms have been most extensively studied [23]. Although little is known about the c-di-GMP regulation of the phenotype changes of V. vulnificus, it has been reported that increased intracellular c-di-GMP levels induced production of EPS, a component of the Vibrio spp. biofilm matrix, and subsequent development of biofilm and rugose colonies [7,22]. In this study, elevated c-di-GMP induced the expression of cabA encoding a calcium-binding matrix protein CabA (Fig 3B and 3C). A mutation in cabA significantly impaired biofilm development even in the presence of elevated c-di-GMP without reducing the amount of EPS produced (Figs 4, 7, 9, and 10). The results indicated that EPS alone is not sufficient to develop the well-structured robust biofilm and that CabA is another key component of the V. vulnificus biofilm matrix.
The data presented here and bioinformatic analysis of CabA suggested that CabA is a structural protein rather than an enzymatic protein modifying or catalyzing other matrix components such as EPS to facilitate biofilm development (http://www.ncbi.nlm.nih.gov/cdd). Exogenous addition of purified CabA was able to rescue the biofilm defect of the cabA mutant only when calcium was available at the same time (Fig 11A). The ability of exogenously provided CabA to switch the smooth colony morphology of the cabA mutant to rugose disappeared when calcium was not added to the growth media (Fig 11B). These observations suggested that apo-CabA without calcium is not functional as a biofilm matrix component. Calcium-dependent multimerization of CabA, as shown in Fig 12B, indicated that the multimeric form of CabA could be a functional conformation which is competent as a biofilm matrix component. This calcium dependency of CabA is especially intriguing since calcium is a relatively abundant element in the environment of oyster, the natural niche and primary route of infection for V. vulnificus [10,39]. Therefore, it is a reasonable hypothesis that calcium is an environmental cue that multimerizes CabA to the functional conformation, rendering V. vulnificus to switch from a planktonic to a sessile life style, survive in hostile environments, and reach a concentrated infective dose when consumed by humans. Supporting the hypothesis, CabA is essential for the robust structures of the V. vulnificus biofilms on oyster shells (Fig 10).
It was noteworthy that calcium was still able to promote biofilm formation under the conditions in which cabA expression is limited or deficient (Figs 4C and 11A). These results suggested that calcium regulates cellular functions involved in biofilm formation besides mediating the multimerization of CabA. The previous observation that calcium alters the extracellular polysaccharides to a more biofilm-proficient phase in V. vulnificus could be a possible explanation for the calcium-dependent, yet CabA-independent, biofilm formation [40].
The majority of CabA present as a monomer under conditions of low calcium shifted to multimeric form in the presence of increasing concentrations of calcium (Fig 12B). However, the concentrations of calcium required for multimerization of CabA in vitro (20–50 mM) were significantly higher than those added externally to induce biofilm formation (10 mM). One possible explanation for this discrepancy is that calcium could be concentrated in the matrix by yet unknown mechanisms and the concentrations of calcium in the microenvironment where CabA multimerization occurs could be higher than those added externally for the biofilm formation experiments (Figs 4, 7, 9, 10, 11, and 14). To test this possibility, the matrix of biofilms was fractionated and the calcium concentration in the matrix was determined using ICP-AES. As shown in Fig 13, the calcium concentration in the matrix was 4.8-fold (P<0.05) higher than that in the biofilm growth media, indicating that calcium could be concentrated in the matrix. Consistent with this, it has been reported that excess of calcium is accumulated and immobilized within the biofilm matrices of Pseudomonas aeruginosa and Enterococcus faecalis [41,42].
There are several different mechanisms for structural proteins in the matrix to contribute to biofilm maturation [5]. Recent works demonstrated that an extracellular protein TasA forms amyloid fibers that glue cells together and provide structure to the extracellular matrix in Bacillus subtilis biofilms [43]. P. putida large surface-associated protein LapF, and probably S. enteritidis BapA, accumulate in certain cell surface regions and interact with each other to promote aggregations of adjacent cells, which lead to microcolony formation and subsequent biofilm maturation [44,45]. However, the amino acid sequence of CabA is not homologous to that of LapF and BapA (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Furthermore, in situ TEM analysis revealed that functional CabA irregularly diffused into the matrix rather than retained on or concentrated along the cell envelopes (Fig 6). Another mechanism adopted by P. aeruginosa CdrA, and V. cholerae RbmC harboring carbohydrate binding domains is that the proteins bind to extracellular polysaccharides and cross-link them to polymers. Polymerization of the polysaccharides promote aggregation of cells and thus increase structural integrity of the biofilm [46,47]. Successful formation of biofilms by co-culturing the mutants defective in the synthesis of either EPS or CabA indicates that CabA requires EPS to assemble an apparently functional matrix (Fig 14). However, CabA does not harbor carbohydrate binding domains and share homology with any of the proteins in amino acid sequences (http://www.ncbi.nlm.nih.gov/cdd). Additional work is needed to clarify precise mechanisms by which CabA enable cells to build more structured biofilms.
In summary, CabA is a calcium-binding protein contributing to biofilm formation in a calcium-dependent manner and secreted via functional CabB and CabC. CabA is distributed distally from the cells throughout the biofilm matrix and produced as the V. vulnificus biofilm matures under conditions of elevated c-di-GMP. The cabA mutant was significantly defective in the development of rugose colony morphology and robust biofilms in flow cells and on oyster shells. Purified CabA, added exogenously along with calcium, was able to restore the cabA mutant abilities of forming biofilm and rugose colony morphology. Calcium binding induces conformational changes of CabA to multimeric forms and CabA can assemble a functional matrix when EPS coexist. Consequently, CabA is a calcium-binding protein functional in the extracellular matrix and essential for the development of well-structured mature biofilms and rugose colony of V. vulnificus.
Materials and Methods
Culture conditions, construction of JN111, and measurement of intracellular c-di-GMP
The strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, the V. vulnificus strains were grown in Luria-Bertani (LB) medium supplemented with 2.0% (w/v) NaCl (LBS) at 30°C. The V. fischeri minimal medium [48] modified to omit calcium and contain glycerol (50 mM Tris-HCl, pH 7.2, 50 mM MgSO4, 300 mM NaCl, 10 mM KCl, 0.33 mM K2HPO4, 18.5 mM NH4Cl, and 32.6 mM glycerol) (VFMG-CF) was used for biofilm formation. To manipulate c-di-GMP levels in cells, V. vulnificus JN111, which carries dcpA encoding a DGC [22] on the chromosome under the control of arabinose-inducible promoter PBAD [49], was constructed as depicted in S1 Fig. JN111 was used as a parent strain in this study (Table 1), and intracellular c-di-GMP levels of the V. vulnificus strains were manipulated by adding different concentrations of arabinose in the growth media. Levels of c-di-GMP in cells were measured using LC-MS as previously described [30]. Briefly, the strains grown with VFMG-CF containing 10 mM CaCl2 and different concentrations of arabinose was lysed with 0.6 M HClO4, and spin down to obtain supernatants. Measurements of c-di-GMP in the supernatants were performed using an Acuity UPLC with a Synergi 4μ Hydro RP 80A column and a C18 Guard Cartridge (Phenomenex, Torrance, CA) on a Premier XL triple-quadrupole electrospray mass spectrometer (Waters, Milford, MA).
Microarray analysis
A Center for Disease Control (CDC) reactor (Biosurface Technologies, Bozeman, MT) [50] containing 300 ml AB medium [300 mM NaCl, 50 mM MgSO4, 0.2% (w/v) vitamin-free casamino acids, 10 mM potassium phosphate, 1 mM L-arginine, pH 7.5] [51] was inoculated using 3 ml of overnight culture of V. vulnificus CMCP6 and biofilms were formed for 4 d at 30°C using the procedures developed by Park and colleagues [52]. Total cellular RNAs were isolated from the biofilm cells attached on the surfaces of the coupons and the planktonic cells suspended in the culture broth using an RNeasy Mini Kit (Qiagen, Valencia, CA). Aminoallyl-cDNAs were synthesized using an Amino Allyl cDNA Labeling Kit (Ambion, Austin, TX) and were respectively labeled with Cy3 or Cy5 (Amersham Pharmacia, Uppsala, Sweden). Equal amounts of the labeled cDNAs were combined to hybridize the V. vulnificus Whole-Genome Twin-Chip [53] at 42°C for 16 h and the arrays were washed, dried, scanned, and analyzed by GenePix Pro 3.0 software (Axon Instruments, Union City, CA). The ORF spots that showed a 2.5 (that is, 1.322 in log2 scale)-fold or greater difference in expression with a P value of < 0.05 were considered to be differentially expressed in biofilms.
RNA purification and transcript analysis
Total RNAs were isolated from JN111 grown to A600 of 0.6 with LBS containing 0.01% (w/v) of arabinose using an RNeasy Mini Kit (Qiagen) and used for reverse-transcription PCR (RT-PCR) analyses of the cabABC transcription pattern. For RT-PCR, a series of reactions was performed with Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) according to the manufacturer’s procedures to synthesize cDNA. PCR amplification of the cDNA was performed using standard protocols with a pair of primers, RTcabA and RTcabC2, which are designed to hybridize to the 3′ ends of cabA and cabC, respectively (S2 Table and Fig 1A). For quantitative real-time PCR (qRT-PCR) analyses of cabA expression, total RNAs were isolated from JN111 grown for various periods with VFMG-CF containing 10 mM CaCl2 and different concentrations of arabinose. cDNA was synthesized from the RNAs by using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) and real-time PCR amplification of the cDNA was performed by using the Chromo 4 real-time PCR detection system (Bio-Rad Laboratories) with a pair of primers, qRTcabA_F and qRTcabA_R (S2 Table). Relative expression levels of the cabA transcripts were calculated by using the 16S rRNA expression level as the internal reference for normalization.
Purification of CabA, ICP-AES, and ITC
The coding region of cabA was amplified by PCR using the V. vulnificus CMCP6 chromosomal DNA and a pair of primers, cabAexp_F and cabAexp_R (S2 Table), and cloned into a His6 tag expression vector, pET28a(+) (Novagen, Madison, WI) to result in pYM1202 (Table 1). The His-tagged CabA was then expressed in E. coli BL21 (DE3), and purified by affinity chromatography according to manufacturer’s procedure (Qiagen). The purified CabA was dialyzed for 12 h against the storage buffer [20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT and 50% (v/v) glycerol] and kept frozen until use. The calcium-free CabA was prepared by extensively dialyzing the purified CabA against the storage buffer with EGTA (10 mM) and then subsequently against the storage buffer without EGTA. No remaining calcium in the calcium-free CabA protein was confirmed by the analysis of ICP-AES.
The purified CabA (210 μM) was digested with concentrated HNO3 and diluted with distilled water. The diluted CabA solution was nebulized into the plasma using MiraMist nebulizer (Burgener Research Inc., Mississauga, Canada) and the concentrations of five metal ions (Ca, Mg, Mn, Fe, and Zn) were measured using ICP-AES (Optima-4300 DV, PerkinElmer, Waltham, MA). The operating conditions developed by Cubadda and Raggi [54] were adopted and calibration was performed using the reference metal solutions as external standards.
ITC experiments were performed to determine calcium-binding parameters for CabA. For this purpose, the calcium-free CabA was reconstituted in 300 mM NaCl and 50 mM Tris-HCl, pH 8.0, and CaCl2 solution was prepared in the same buffer. The protein and CaCl2 solution were degassed by vacuum aspiration for 20 min prior to loading and titration at 25°C. The calorimetric assays were performed using a VP-ITC (MicroCal Inc., Northampton, MA). The calcium (1.2 mM) in the syringe was titrated against the 0.024 mM CabA in the reaction cell at 25°C. The stirring speed was 270 rpm, and the thermal power was recorded every 15 s. Raw data were processed and plotted using the Origin program (version 7) supplied with the instrument. The ITC experiments were repeated three times.
Generation of the cabA, cabB, cabC, brpA, and cabA brpA mutants and complementation of the cabA, cabB, and cabC mutants
The cabA gene was inactivated in vitro by deletion (530-bp of 573-bp) of the cabA coding region using the PCR-mediated linker-scanning mutation method as described previously [31]. Briefly, pairs of primers cabA_F1 and cabA_R1 (for amplification of the 5’ amplicon) or cabA_F2 and cabA_R2 (for amplification of the 3’ amplicon) were designed and used as listed in S2 Table. The ΔcabA was amplified by PCR using the mixture of both amplicons as the template and cabA_F1 and cabA_R2 as primers. Similar experimental procedures were adopted for construction of the ΔcabB, ΔcabC and ΔbrpA in vitro, except that primers cabB_F1, cabB_R1, cabB_F2 and cabB_R2 (for 1191-bp deleted ΔcabB), cabC_F1, cabC_R1, cabC_F2 and cabC_R2 (for 849-bp deleted ΔcabC) and brpA_F1, brpA_R1, brpA_F2, and brpA_R2 (for 448-bp deleted ΔbrpA) were used (S2 Table). An 1.2-kb nptI DNA conferring resistance to kanamycin [55] was also inserted into a unique BamHI site present within the ΔbrpA to result in ΔbrpA::nptI. The resulting ΔcabA, ΔcabB, ΔcabC, and ΔbrpA::nptI were ligated with SpeI-SphI-digested pDM4 [56] to form pYM1102, pJN1504, pJN1402, and pJN0907, respectively (Table 1). The E. coli S17-1 λ pir, tra containing either pYM1102, pJN1504, pJN1402, or pJN0907 was used as a conjugal donor to CMCP6 or JN111. The conjugation and isolation of the transconjugants were conducted as previously described [31], and resulted in the cabA mutants (YM112, YM112D), the cabB mutant (JN151D), the cabC mutant (JN141D), or the brpA mutant (JN094D), respectively (Table 1). Similarly, the E. coli S17-1 λ pir, tra containing pJN0907 was used as a conjugal donor to YM112D to construct the cabA brpA double mutant JN143D.
For complementation, pairs of primers, cabAcom_F and cabAcom_R, cabBcom_F and cabBcom_R or cabCcom_F and cabCcom_R, were designed and used to amplify coding regions of cabA, cabB or cabC, respectively (S3 Table). The amplified ORFs of cabA, cabB, and cabC were cloned into pJK1113 [57] under an arabinose-inducible promoter PBAD to create pYM1109. pJN1502, and pJN1403, respectively (Table 1). The plasmids pYM1109, pJN1502, and pJN1403 were transferred into YM112D, JN151D, and JN141D, respectively, by conjugation as described above.
Biofilm formation
Biofilms were formed using the procedure developed by O’Toole and Kolter [58] with minor modifications. Briefly, each well of the 96-well polystyrene microtiter plates (Nunc, Roskilde, Denmark) was inoculated with 200 μl of each culture diluted to an A600 0.05 with VFMG-CF and then incubated for 24 h at 30°C without shaking. Once the planktonic cells were removed, the biofilm cells on the wall were washed with phosphate-buffered saline (PBS, pH 7.4), and then stained with 220 μl of 1% (w/v) CV solution for 15 min at room temperature. Biofilms were quantified by elution of CV with 220 μl 100% ethanol and measurement of absorbance at 570 nm (A570). When required, VFMG-CF supplemented with various amounts of CaCl2, arabinose, and/or calcium-free CabA was used. Similarly, inoculums of the co-cultures were prepared by mixing JN111, cabA mutant, or brpA mutant with either cabA brpA mutant or brpA mutant at a 1 : 1 ratio and used to form biofilms. For isolation of total RNAs and proteins from biofilms at different development stages, biofilm of JN111 was formed as described above but in a larger scale (800 μl) using 24-well polystyrene microtiter plates (SPL, Seoul, Korea).
EPS analysis
EPS was prepared following the procedures previously described by Kim et al. [12]. Briefly, JN111 cells grown on LBS agar plates containing 0.02% (w/v) arabinose were suspended in PBS, and were vigorously shaken to elute EPS from cells. Cells and debris were removed by centrifugation and EPS in the supernatants was treated with RNase A (50 μg/ml), DNase I (50 μg/ ml with 10 mM MgCl2), and proteinase K (200 μg/ml). Subsequently, the remained polysaccharide fractions were extracted twice with phenol-chloroform, and precipitated with 2.5× volumes of 100% ethanol and resuspended in distilled water. EPS resuspensions were resolved using a 4% stacking SDS-PAGE, stained with Stains-All (Sigma-Aldrich, St. Louis, MO). The gel was subsequently destained as described previously [59], and photographed using a digital camera (PowerShot SX220 HS, Canon, Tokyo, Japan).
Biofilm fractionation, ICP-AES, and Western blot analysis
Biofilms were formed by gentle shaking of the glass flask (Duran, Mainz, Germany) containing 100 ml VFMG-CF supplemented with 0.01% (w/v) of arabinose and 10 mM of CaCl2 for 8 h at 30°C. Planktonic cells were removed by rinsing gently with PBS, after which the biofilms were collected with a cell scraper (SPL) and resuspended in PBS. Biofilms were disrupted by sonication (VC130 Ultrasonic Processor, Sonics & Materials, Inc., Newtown, CT) and then fractionated into cell fractions (pellets) and matrix fractions (supernatants) by centrifugation as previously described [60]. The calcium concentrations in the biofilm matrix (equivalent to 30 μl) and growth media were determined using ICP-AES (Optima-4300 DV, PerkinElmer) as described above. For Western blot analysis, proteins (10 μg) from the lysates of cell fractions and concentrates of matrix fractions were resolved by SDS-PAGE [61]. Immunoblotting of CabA was performed using the rabbit anti-CabA antibody (AbFrontier, Seoul, Korea) as previously described [31]. Similar Western blot analyses were adopted to quantify the levels of CabA in the JN111 biofilms (that is, in the cells and matrices) grown with different concentrations of arabinose or at different biofilm development stages.
Immunogold labeling and TEM
Immunogold labeling of CabA was performed following the procedures developed by Martinez-Gil et al. [45] with minor modifications. Biofilms were grown on nickel grids in 24-well polystyrene microtiter plate wells (SPL) containing VFMG-CF supplemented with 0.01% (w/v) of arabinose and 10 mM CaCl2 for 12 h at 30°C. The grids were treated with blocking buffer [1% (w/v) skim milk in PBS with 0.1% (v/v) Tween20], incubated for 3 h with the rabbit anti-CabA antibody (AbFrontier), and exposed to a goat anti-rabbit secondary antibody conjugated to 10-nm gold particles (Sigma-Aldrich) for 80 min. The biofilms on the grids were negatively stained with 2% (v/v) solution of uranyl acetate and visualized using TEM (JEM1010, JEOL, Tokyo, Japan) at 80 kV accelerating voltage.
Flow cells, CSLM, and SEM
For structural analyses, biofilms of the strains were formed in flow cell chambers as described elsewhere [46]. Glass coverslips were attached on polycarbonate flow cells with individual channel dimensions of 1 × 4 × 40 mm. Each flow cell was inoculated with 100 μl of the culture diluted to A600 of 0.1, and inverted to allow bacteria to attach to the coverslip for 1 h without flow. Then VFMG-CF supplemented with 0.01% (w/v) arabinose and 10 mM CaCl2 was flowed at a constant rate of 8 ml/h using a Minipuls Evolution peristaltic pump (Gilson, Villiers, France) to grow biofilm for 3 d. The flow cell chambers with grown biofilms were photographed using a digital camera (PowerShot SX220 HS, Canon).
For CSLM analysis, biofilms on the coverslips were stained with LIVE/DEAD BacLight Viability Kit containing SYTO9 and propidium iodide (Invitrogen) for 15 min in the dark and visualized by CSLM (LSM710, Zeiss, Jena, Germany). The biofilm images were processed using Zeiss Zen software (Zeiss). Biofilms on the coverslips were fixed, and dehydrated for SEM analysis by the procedures developed by Lim et al. [62]. Briefly, the biofilms were fixed using a buffer [2% (v/v) paraformaldehyde, 0.2% (v/v) glutaraldehyde, 0.1 M sodium cacodylate, pH 7.3] for 1 h at 4°C, washed with 0.05 M sodium cacodylate for 20 min at 4°C, and then postfixed with 1% (v/v) osmium tetroxide in 0.05 M sodium cacodylate (pH 7.3) for 45 min at 4°C. Fixed biofilms were washed with distilled water, dehydrated in a series of increasing concentrations of ethanol (50, 70, 80, 90, and 100%), and then dried in hexamethyldisilazane. Dried biofilms were mounted on an aluminum stub, coated with platinum using a sputter coater (BAL-TEC SCD 005, BAL-TEC AG, Balzers, Liechtenstein), and visualized using SEM (Supra 55VP, Zeiss). To determine the structure of biofilms on oyster shells, biofilms of the strains were grown for 24 h on fragmented oyster shells (1 × 1 cm) in 24-well polystyrene microtiter plates (SPL) containing VFMG-CF supplemented with 10 mM CaCl2 and with or without 0.01% arabinose. Biofilms on oyster shells were fixed, washed, dehydrated, dried, mounted, coated and visualized using SEM as described above.
Colony morphology assay
For the analysis of colony morphology, 2 μl of cultures grown to A600 of 0.8 was spotted onto VFMG-CF agar plates supplemented with or without 0.02% (w/v) of arabinose and 10 mM CaCl2. When required, various amounts of calcium-free CabA was exogenously provided to the spotted colonies. Similarly, 2 μl of co-cultures prepared by mixing JN111, the cabA mutant, or the brpA mutant with either the cabA brpA double mutant or the brpA mutant at a 1 : 1 ratio were spotted as described above. The resulting colonies grown at 30°C for 3 d were visualized using a stereomicroscope (Stemi DV4, Zeiss).
CD and size-exclusion chromatography analysis
CD spectroscopy was used to evaluate the effect of calcium on the CabA conformation. The calcium-free CabA (0.05 mg/ml) in 300 mM NaCl and 50 mM Tris-HCl, pH 8.0 was subjected to far-UV CD measurements at 25°C using a 1-mm path length quartz cuvette in a Jasco J-815 CD spectrometer (Jasco, Tokyo, Japan). The CabA protein was incubated with 10 mM CaCl2 for 1 h at room temperature, and its CD spectra were measured under the same conditions. CD Spectra were acquired over the wavelength range of 200–260 nm and converted into mean residue ellipticity (MRE, degree cm2/dmol). Blank spectra of the buffer without the protein were subtracted.
Calcium-induced conformational change of CabA was further assessed by size-exclusion chromatography using a Superdex 75 10/300 GL gel filtration column (GE Healthcare, Waukesha, WI) installed on an AKTA FPLC system (GE Healthcare). The multimeric CabA obtained with 100 mM CaCl2 was serially diluted to various calcium concentrations. The diluted samples were then injected onto the pre-equilibrated column with 300 mM NaCl, 20 mM Tris-HCl, pH 8.0 and corresponding concentrations of calcium and eluted at a flow rate of 0.4 ml/min at room temperature. The effluent was monitored by measuring absorbance at 280 nm. Bovine serum albumin (66 kDa) and carbonic anhydrase (29 kDa) were used as the molecular weight standards.
Data analyses
Averages and standard deviation (SD) were calculated from at least three independent experiments. All other data were analyzed by Student's t tests with the SAS program (SAS software; SAS Institute Inc.). Significance of differences between experimental groups was accepted at a P value of < 0.05.
Accession numbers
GenBank accession numbers for the proteins and genome sequences are as follows: DcpA, AAO08284.1; CabA, AAO08434.1; CabB, AAO08435.2; CabC, AAO08436.1; BrpA, AAO08445.1; V. vulnificus CMCP6 genome, AE016795 and AE016796. All primary microarray data were deposited in the Gene Expression Omnibus [GEO (http://www.ncbi.nlm.nih.gov/projects/geo/)] database under accession number (GSE59753).
Supporting Information
Zdroje
1. Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13 : 7–10. 15639625
2. Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73 : 310–347. doi: 10.1128/MMBR.00041-08 19487730
3. Costerton JW. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 2001;9 : 50–52. 11173226
4. Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17 : 109–118. doi: 10.1016/j.tim.2008.12.004 19231189
5. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8 : 623–633. doi: 10.1038/nrmicro2415 20676145
6. Teschler JK, Zamorano-Sánchez D, Utada AS, Warner CJ, Wong GC, Linington RG, Yildiz FH. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol. 2015;13 : 255–268. doi: 10.1038/nrmicro3433 25895940
7. Guo Y, Rowe-Magnus DA. Identification of a c-di-GMP-regulated polysaccharide locus governing stress resistance and biofilm and rugose colony formation in Vibrio vulnificus. Infect Immun. 2010;78 : 1390–1402. doi: 10.1128/IAI.01188-09 20065022
8. Guo Y, Rowe-Magnus DA. Overlapping and unique contributions of two conserved polysaccharide loci in governing distinct survival phenotypes in Vibrio vulnificus. Environ Microbiol. 2011;13 : 2888–2900. doi: 10.1111/j.1462-2920.2011.02564.x 21895917
9. Marco-Noales E, Milán M, Fouz B, Sanjuán E, Amaro C. Transmission to eels, portals of entry, and putative reservoirs of Vibrio vulnificus serovar E (biotype 2). Appl Environ Microbiol. 2011;67 : 4717–4725.
10. Paranjpye RN, Johnson AB, Baxter AE, Strom MS. Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters. Appl Environ Microbiol. 2007;73 : 5041–5044. 17557854
11. Froelich B, Oliver JD. The interactions of Vibrio vulnificus and the oyster Crassostrea virginica. Microb Ecol. 2013;65 : 807–816. doi: 10.1007/s00248-012-0162-3 23280497
12. Kim HS, Lee MA, Chun SJ, Park SJ, Lee KH. Role of NtrC in biofilm formation via controlling expression of the gene encoding an ADP-glycero-manno-heptose-6-epimerase in the pathogenic bacterium, Vibrio vulnificus. Mol Microbiol. 2007;63 : 559–574. 17241201
13. Kim HS, Park SJ, Lee KH. Role of NtrC-regulated exopolysaccharides in the biofilm formation and pathogenic interaction of Vibrio vulnificus. Mol Microbiol. 2009;74 : 436–453. doi: 10.1111/j.1365-2958.2009.06875.x 19737353
14. Lee KJ, Kim JA, Hwang W, Park SJ, Lee KH. Role of capsular polysaccharide (CPS) in biofilm formation and regulation of CPS production by quorum-sensing in Vibrio vulnificus. Mol Microbiol. 2013;90 : 841–857. doi: 10.1111/mmi.12401 24102883
15. Chatzidaki-Livanis M, Jones MK, Wright AC. Genetic variation in the Vibrio vulnificus group 1 capsular polysaccharide operon. J Bacteriol. 2006;188 : 1987–1998. 16484211
16. Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75 : 39–68. 16756484
17. Joseph LA, Wright AC. Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J Bacteriol. 2004;186 : 889–893. 14729720
18. Grau BL, Henk MC, Garrison KL, Olivier BJ, Schulz RM, O'Reilly KL, et al. Further characterization of Vibrio vulnificus rugose variants and identification of a capsular and rugose exopolysaccharide gene cluster. Infect Immun. 2008;76 : 1485–1497. doi: 10.1128/IAI.01289-07 18212074
19. Garrison-Schilling KL, Kaluskar ZM, Lambert B, Pettis GS. Genetic analysis and prevalence studies of the brp exopolysaccharide locus of Vibrio vulnificus. PLoS One. 2014;9: e100890. doi: 10.1371/journal.pone.0100890 25013926
20. Kim M, Park JM, Um HJ, Lee KH, Kim H, Min J, et al. The antifouling potentiality of galactosamine characterized from Vibrio vulnificus exopolysaccharide. Biofouling. 2011;27 : 851–857. doi: 10.1080/08927014.2011.605521 21827336
21. Williams TC, Blackman ER, Morrison SS, Gibas CJ, Oliver JD. Transcriptome sequencing reveals the virulence and environmental genetic programs of Vibrio vulnificus exposed to host and estuarine conditions. PloS One. 2014; 9: e114376. doi: 10.1371/journal.pone.0114376 25489854
22. Nakhamchik A, Wilde C, Rowe-Magnus DA. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl Environ Microbiol. 2008;74 : 4199–4209. doi: 10.1128/AEM.00176-08 18487410
23. Krasteva PV, Giglio KM, Sondermann H. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci. 2012;21 : 929–948. doi: 10.1002/pro.2093 22593024
24. Paranjpye RN, Strom MS. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect Immun. 2005;73 : 1411–1422. 15731039
25. Park JH, Lim JG, Choi SH. Effects of elevated intracellular cyclic di-GMP levels on biofilm formation and transcription profiles of Vibrio vulnificus. Food Sci Biotech. 2015;24 : 771–776.
26. Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol. 2005;55 : 1160–1182. 15686562
27. Ferreira RB, Chodur DM, Antunes LC, Trimble MJ, McCarter LL. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J Bacteriol. 2012;194 : 914–924. doi: 10.1128/JB.05807-11 22194449
28. Satchell KJ. Structure and function of MARTX toxins and other large repetitive RTX proteins. Annu Rev Microbiol. 2011;65 : 71–90. doi: 10.1146/annurev-micro-090110-102943 21639783
29. Gangola P, Rosen BP. Maintenance of intracellular calcium in Escherichia coli. J Biol Chem. 1987;262 : 12570–12574. 2442165
30. Irie Y, Parsek MR. LC/MS/MS-based quantitative assay for the secondary messenger molecule, c-di-GMP. Methods Mol Biol. 2014;1149 : 271–279. doi: 10.1007/978-1-4939-0473-0_22 24818912
31. Lim JG, Choi SH. IscR is a global regulator essential for pathogenesis of Vibrio vulnificus and induced by host cells. Infect Immun. 2014;82 : 569–578. doi: 10.1128/IAI.01141-13 24478072
32. Cosme AM, Becker A, Santos MR, Sharypova LA, Santos PM, Moreira LM. The outer membrane protein TolC from Sinorhizobium meliloti affects protein secretion, polysaccharide biosynthesis, antimicrobial resistance, and symbiosis. Mol Plant Microbe Interact. 2008;21 : 947–957. doi: 10.1094/MPMI-21-7-0947 18533835
33. Hahn A, Stevanovic M, Brouwer E, Bublak D, Tripp J, Schorge T, et al. Secretome analysis of Anabaena sp. PCC 7120 and the involvement of the TolC-homologue HgdD in protein secretion. Environ Microbiol. 2014.
34. Chagnot C, Zorgani MA, Astruc T. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol. 2013;4 : 303. doi: 10.3389/fmicb.2013.00303 24133488
35. Griessl MH, Schmid B, Kassler K, Braunsmann C, Ritter R, Barlag B, et al. Structural insight into the giant Ca2+-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells. Structure. 2013;21 : 741–752. doi: 10.1016/j.str.2013.02.020 23562396
36. Boyd CD, Smith TJ, El-Kirat-Chatel S, Newell PD, Dufrêne YF, O'Toole GA. Structural features of the Pseudomonas fluorescens biofilm adhesin LapA required for LapG-dependent cleavage, biofilm formation, and cell surface localization. J Bacteriol. 2014;196 : 2775–2788. doi: 10.1128/JB.01629-14 24837291
37. Jahn A, Griebe T, Nielsen PH. Composition of Pseudomonas putida biofilms: accumulation of protein in the biofilm matrix. Biofouling. 1999;14 : 49–57.
38. Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751 : 119–139. 16027053
39. Yoon GL, Kim BT, Kim BO, Han SH. Chemical–mechanical characteristics of crushed oyster-shell. Waste Manag. 2003;23 : 825–834. 14583245
40. Garrison-Schilling KL, Grau BL, McCarter KS, Olivier BJ, Comeaux NE, Pettis GS. Calcium promotes exopolysaccharide phase variation and biofilm formation of the resulting phase variants in the human pathogen Vibrio vulnificus. Environ Microbiol. 2010;13 : 643–654. doi: 10.1111/j.1462-2920.2010.02369.x 21059165
41. Turakhia MH, Characklis WG. Activity of Pseudomonas aeruginosa in biofilms: effect of calcium. Biotechnol Bioeng. 1989;20 : 406–414.
42. Kishen A, George S, Kumar R. Enterococcus faecalis-mediated biomineralized biofilm formation on root canal dentine in vitro. J Biomed Mater Res A. 2006;77 : 406–415. 16444682
43. Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11 : 157–168. doi: 10.1038/nrmicro2960 23353768
44. Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penadés JR, et al. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol. 2005;58 : 1322–1339. 16313619
45. Martínez-Gil M, Romero D, Kolter R, Espinosa-Urgel M. Calcium causes multimerization of the large adhesin LapF and modulates biofilm formation by Pseudomonas putida. J Bacteriol. 2012;194 : 6782–6789. doi: 10.1128/JB.01094-12 23042991
46. Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2009;75 : 827–842.
47. Berk V, Fong JC, Dempsey GT, Develioglu ON, Zhuang X, Liphardt J, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337 : 236–239. doi: 10.1126/science.1222981 22798614
48. Cao X, Studer SV, Wassarman K, Zhang Y, Ruby EG, Miyashiro T. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. mBio. 2012;3: e00285–11.
49. Guzman L, Dominique B, Michael JC, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177 : 4121–4130. 7608087
50. Donlan RM, Piede JA, Heyes CD, Sanii L, Murga R, Edmonds P, et al. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl Environ Microbiol. 2004;8 : 4980–4988.
51. Greenberg EP, Hastings JW, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120 : 87–91.
52. Park H, Park HJ, Kim JA, Lee SH, Kim JH, Yoon J, et al. Inactivation of Pseudomonas aeruginosa PA01 biofilms by hyperthermia using superparamagnetic nanoparticles. J Microbiol Methods. 2011;84 : 41–45. doi: 10.1016/j.mimet.2010.10.010 20971135
53. Jeong HG, Choi SH. Evidence that AphB, essential for the virulence of Vibrio vulnificus, is a global regulator. J Bacteriol. 2008;90 : 3768–3773.
54. Cubadda F, Raggi A. Determination of cadmium, lead, iron, nickel and chromium in selected food matrices by plasma spectrometric techniques. Microchem J. 2005;79 : 91–96.
55. Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147 : 217–226. 6270337
56. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178 : 1310–1319. 8631707
57. Lim JG, Bang YJ, Choi SH. Characterization of the Vibrio vulnificus 1-Cys peroxiredoxin Prx3 and regulation of its expression by the Fe-S cluster regulator IscR in response to oxidative stress and iron starvation. J Biol Chem. 2014;289 : 36263–36274. doi: 10.1074/jbc.M114.611020 25398878
58. O’Toole GA, Kolter R. The initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;30 : 449–461.
59. Enos-Berlage JL, McCarter LL. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J Bacteriol. 2000;182 : 5513–5520. 10986256
60. Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59 : 1229–1238. 16430696
61. Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001.
62. Lim JY, May JM, Cegelski L. Dimethyl sulfoxide and ethanol elicit increased amyloid biogenesis and amyloid-integrated biofilm formation in Escherichia coli. Appl Environ Microbiol. 2012;778 : 3369–3378.
63. Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1 : 784–791.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Ross River Virus: Many Vectors and Unusual Hosts Make for an Unpredictable Pathogen
- Distinct but Spatially Overlapping Intestinal Niches for Vancomycin-Resistant and Carbapenem-Resistant
- Intracellular Survival of Depends on Uptake and Degradation of Extracellular Matrix Glycosaminoglycans by Macrophages
- Type IX Secretion Substrates Are Cleaved and Modified by a Sortase-Like Mechanism
- Structural and Functional Characterization of Anti-A33 Antibodies Reveal a Potent Cross-Species Orthopoxviruses Neutralizer
- Suppression of a Natural Killer Cell Response by Simian Immunodeficiency Virus Peptides
- Inhibition of Translation Initiation by Protein 169: A Vaccinia Virus Strategy to Suppress Innate and Adaptive Immunity and Alter Virus Virulence
- Enteropathogenic Uses NleA to Inhibit NLRP3 Inflammasome Activation
- Flavodoxin-Like Proteins Protect from Oxidative Stress and Promote Virulence
- Cullin4 Is Pro-Viral during West Nile Virus Infection of Mosquitoes
- The NLRP3 Inflammasome and IL-1β Accelerate Immunologically Mediated Pathology in Experimental Viral Fulminant Hepatitis
- DYRK2 Negatively Regulates Type I Interferon Induction by Promoting TBK1 Degradation via Ser527 Phosphorylation
- A KSHV microRNA Directly Targets G Protein-Coupled Receptor Kinase 2 to Promote the Migration and Invasion of Endothelial Cells by Inducing CXCR2 and Activating AKT Signaling
- The Operon Essential for Biofilm and Rugose Colony Development in
- ADAP2 Is an Interferon Stimulated Gene That Restricts RNA Virus Entry
- The Role of the Antiviral APOBEC3 Gene Family in Protecting Chimpanzees against Lentiviruses from Monkeys
- The Deacetylase Sirtuin 1 Regulates Human Papillomavirus Replication by Modulating Histone Acetylation and Recruitment of DNA Damage Factors NBS1 and Rad51 to Viral Genomes
- Experimental Malaria in Pregnancy Induces Neurocognitive Injury in Uninfected Offspring via a C5a-C5a Receptor Dependent Pathway
- Intrahepatic Transcriptional Signature Associated with Response to Interferon-α Treatment in the Woodchuck Model of Chronic Hepatitis B
- Adipose Tissue Is a Neglected Viral Reservoir and an Inflammatory Site during Chronic HIV and SIV Infection
- Infection Is Associated with Impaired Hepatic Dimethylarginine Dimethylaminohydrolase Activity and Disruption of Nitric Oxide Synthase Inhibitor/Substrate Homeostasis
- Conserved Motifs within Hepatitis C Virus Envelope (E2) RNA and Protein Independently Inhibit T Cell Activation
- The RelA/SpoT Homolog and Stringent Response Regulate Survival in the Tick Vector and Global Gene Expression during Starvation
- Hybridization in Parasites: Consequences for Adaptive Evolution, Pathogenesis, and Public Health in a Changing World
- KSHV Latency Locus Cooperates with Myc to Drive Lymphoma in Mice
- Immunostimulatory Defective Viral Genomes from Respiratory Syncytial Virus Promote a Strong Innate Antiviral Response during Infection in Mice and Humans
- Retraction: Extreme Resistance as a Host Counter-counter Defense against Viral Suppression of RNA Silencing
- Appetite for a Foodborne Infection
- Here I Am, Despite Myself
- Microbial Regulation of p53 Tumor Suppressor
- Fiat Luc: Bioluminescence Imaging Reveals In Vivo Viral Replication Dynamics
- Knocking on Closed Doors: Host Interferons Dynamically Regulate Blood-Brain Barrier Function during Viral Infections of the Central Nervous System
- Rapid Lymphatic Dissemination of Encapsulated Group A Streptococci Lymphatic Vessel Endothelial Receptor-1 Interaction
- Simian Immunodeficiency Virus Infection of Chimpanzees () Shares Features of Both Pathogenic and Non-pathogenic Lentiviral Infections
- Epicellular Apicomplexans: Parasites “On the Way In”
- The Depsipeptide Romidepsin Reverses HIV-1 Latency
- Skin-Derived C-Terminal Filaggrin-2 Fragments Are -Directed Antimicrobials Targeting Bacterial Replication
- Type IV Pili Composed of Sequence Invariable Pilins Are Masked by Multisite Glycosylation
- Heterosexual Transmission of Subtype C HIV-1 Selects Consensus-Like Variants without Increased Replicative Capacity or Interferon-α Resistance
- Prevention of Influenza Virus-Induced Immunopathology by TGF-β Produced during Allergic Asthma
- Global Analysis of Mouse Polyomavirus Infection Reveals Dynamic Regulation of Viral and Host Gene Expression and Promiscuous Viral RNA Editing
- Modulation of the Host Lipid Landscape to Promote RNA Virus Replication: The Picornavirus Encephalomyocarditis Virus Converges on the Pathway Used by Hepatitis C Virus
- Intrinsic MyD88-Akt1-mTOR Signaling Coordinates Disparate Tc17 and Tc1 Responses during Vaccine Immunity against Fungal Pneumonia
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Epicellular Apicomplexans: Parasites “On the Way In”
- Fiat Luc: Bioluminescence Imaging Reveals In Vivo Viral Replication Dynamics
- Knocking on Closed Doors: Host Interferons Dynamically Regulate Blood-Brain Barrier Function during Viral Infections of the Central Nervous System
- A KSHV microRNA Directly Targets G Protein-Coupled Receptor Kinase 2 to Promote the Migration and Invasion of Endothelial Cells by Inducing CXCR2 and Activating AKT Signaling
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



