-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Molecularly Cloned, Live-Attenuated Japanese Encephalitis Vaccine SA-14-2 Virus: A Conserved Single Amino Acid in the Hairpin of the Viral E Glycoprotein Determines Neurovirulence in Mice
A group of mosquito-borne flaviviruses that cause fatal encephalitis in humans is among the most important of all emerging human pathogens of global significance. This group includes Japanese encephalitis (JE), West Nile, St. Louis encephalitis, and Murray Valley encephalitis viruses. In this work, we have developed a reverse genetics system for SA14-14-2, a live JE vaccine that is most commonly used in JE-endemic areas, by constructing an infectious bacterial artificial chromosome that contains the full-length SA14-14-2 cDNA. Using this infectious SA14-14-2 cDNA, combined with a mouse model for JEV infection, we have identified a key viral neurovirulence factor, a conserved single amino acid in the ij hairpin adjacent to the fusion loop of the viral E glycoprotein, which regulates viral infectivity into neurons within the central nervous system in vivo and neuronal cells of mouse and human in vitro. Thus, our findings elucidate the molecular basis of the neurovirulence caused by JEV and other closely related encephalitic flaviviruses, a major step in understanding their neuropathogenesis. From a clinical perspective, the discovery of the viral neurovirulence factor and its role will have direct application to the design of a novel class of broad-spectrum antivirals to treat and prevent infection of JEV and other taxonomically related neurotropic flaviviruses.
Published in the journal: . PLoS Pathog 10(7): e32767. doi:10.1371/journal.ppat.1004290
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004290Summary
A group of mosquito-borne flaviviruses that cause fatal encephalitis in humans is among the most important of all emerging human pathogens of global significance. This group includes Japanese encephalitis (JE), West Nile, St. Louis encephalitis, and Murray Valley encephalitis viruses. In this work, we have developed a reverse genetics system for SA14-14-2, a live JE vaccine that is most commonly used in JE-endemic areas, by constructing an infectious bacterial artificial chromosome that contains the full-length SA14-14-2 cDNA. Using this infectious SA14-14-2 cDNA, combined with a mouse model for JEV infection, we have identified a key viral neurovirulence factor, a conserved single amino acid in the ij hairpin adjacent to the fusion loop of the viral E glycoprotein, which regulates viral infectivity into neurons within the central nervous system in vivo and neuronal cells of mouse and human in vitro. Thus, our findings elucidate the molecular basis of the neurovirulence caused by JEV and other closely related encephalitic flaviviruses, a major step in understanding their neuropathogenesis. From a clinical perspective, the discovery of the viral neurovirulence factor and its role will have direct application to the design of a novel class of broad-spectrum antivirals to treat and prevent infection of JEV and other taxonomically related neurotropic flaviviruses.
Introduction
Japanese encephalitis virus (JEV) is the most common cause of viral encephalitis in Asia and parts of the Western Pacific, with ∼60% of the world's population at risk of infection [1]. Within the family Flaviviridae (genus Flavivirus), JEV belongs to the JE serological group, which also includes medically important human pathogens found on every continent except Antarctica [2], [3]: West Nile virus (WNV), St. Louis encephalitis virus (SLEV), and Murray Valley encephalitis virus (MVEV). Historically, the JE serological group members have clustered in geographically distinct locations, but the recent emergence and spread of JEV in Australia [4] and WNV in North America [5], [6] have caused growing concern that these viruses can spread into new territory, posing a significant challenge for global public health [3], [7]. In the US, where WNV and SLEV are endemic, the situation is particularly problematic because the likelihood of JEV being introduced is considerable [8], [9]. Worldwide, ∼50,000–175,000 clinical cases of JE are estimated to occur annually [10]; however, this incidence is undoubtedly a considerable underestimate because surveillance and reporting are inadequate in most endemic areas, and only ∼0.1–4% of JEV-infected people develop clinical disease [11], [12]. On average, ∼20–30% of patients die, and ∼30–50% of survivors suffer from irreversible neurological and/or psychiatric sequelae [13]. Most clinical cases occur in children under age 15 in endemic areas, but in newly invaded areas, all age groups are affected because protective immunity is absent [14]. Thus, given the current disease burden and significant threat of the JEV emergence, resurgence, and spread among much larger groups of susceptible populations, control of JEV remains a high public health priority.
JEV contains a nucleocapsid composed of an ∼11-kb plus-strand genomic RNA, complexed with multiple copies of the highly-basic α-helical C proteins [15], [16]. The nucleocapsid is surrounded by a host-derived lipid bilayer containing the membrane-anchored M and E proteins [17]–[19]. The initial step in the flavivirus replication cycle involves attachment of the virions to the surface of susceptible cells [20]–[24]. The viral E protein is then assumed to bind with high affinity and specificity to an as-yet unidentified cellular receptor(s), which triggers receptor-mediated, clathrin-dependent endocytosis [25]–[27]. The acidic conditions in the endosome lead to a conformational change in the E protein [28]–[32], which triggers fusion of the viral membrane with host endosomal membrane [33]. Once the genome is released into the cytoplasm, the genomic RNA is translated into a single polyprotein, which is processed co - and post-translationally by host and viral proteases to yield at least 10 functional proteins [34]: three structural (C, prM, and E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The nonstructural proteins actively replicate the viral genomic RNA in the replication complex [35]–[38] that is associated with the virus-induced, ER-derived membranes [39]–[41]. Newly synthesized genomic RNA and C proteins are initially enveloped by the prM and E proteins to generate immature virions [42], [43] that bud into the lumen of the ER [44]. These immature virions are then transported via the secretory pathway to the Golgi apparatus. In the low-pH environment of the trans-Golgi network, the furin-mediated cleavage of prM to M induces the maturation of the viral particles [45], which is also accompanied by significant structural rearrangements of the M and E proteins [42], [46], [47]. Finally, mature virions are released into the extracellular space by exocytosis.
JEV is maintained in an enzootic cycle involving multiple species of mosquito vectors (primarily Culex species) and vertebrate hosts/reservoirs (mainly domestic pigs/wading birds). Humans become infected incidentally when bitten by an infected mosquito [48]. In the absence of antiviral therapy, active immunization is the only strategy for sustainable long-term protection. Four types of JE vaccines are used in different parts of the world [49], [50]: (i) the mouse brain-derived inactivated vaccine based on the Nakayama or Beijing-1 strain, (ii) the cell culture-derived inactivated vaccine based on the Beijing-3 or SA14-14-2 strain, (iii) the cell culture-derived live-attenuated vaccine based on the SA14-14-2 strain, and (iv) the live chimeric vaccine developed on a yellow fever virus (YFV) 17D genetic background that carries two surface proteins of JEV SA14-14-2. Of the four vaccines, the only one that is available internationally is the mouse brain-derived inactivated Nakayama [11]. Unfortunately, the production of this vaccine was discontinued in 2006 [51] because of vaccine-related adverse events, short-term immunity, and high production cost [13], [52]. To date, the most commonly used vaccine in Asia is the live-attenuated SA14-14-2 [53], but this vaccine is not recommended by the WHO for global immunization [13], [54]. In addition to the dependence of the duration of immunity on the number of doses received, there is at least a theoretical risk of virus mutation and reversion of the vaccine virus to high virulence. Recently, the SA14-14-2 vaccine virus has been utilized to produce a new Vero cell-derived inactivated vaccine that has been approved in the US, Europe, Canada, and Australia since 2009 [51], [55], [56]. In the US, this vaccine is recommended for adults aged ≥17 years travelling to JEV-endemic countries and at risk of JEV exposure [51], [57], but no vaccine is currently available for children under 17 [58]. More recently, the prM and E genes of JEV SA14-14-2 have been used to replace the corresponding genes of YFV 17D [59], creating a live chimeric vaccine [60] that is now licensed in Australia and Thailand [61], [62]. Thus, the application of JEV SA14-14-2 to vaccine development and production is continuously expanding, but the viral factors and fundamental mechanisms responsible for its loss of virulence are still elusive.
The virulence of JEV is defined by two properties: (i) neuroinvasiveness, the ability of the virus to enter the central nervous system (CNS) when inoculated by a peripheral route; and (ii) neurovirulence, the ability of the virus to replicate and cause damage within the CNS when inoculated directly into the brain of a host. Over the past 20 years, many investigators have sought to understand the molecular basis of JEV virulence, by using cell and animal infection model systems to compare the nucleotide sequences of the genomes of several JEV strains that differ in virological properties [63]–[74]. These studies have identified a large number of mutations scattered essentially throughout the entire viral genome. Because of the complexity of the mutations, however, the major genetic determinant(s) critical for either JEV neurovirulence or neuroinvasiveness remains unclear. In particular, the situation is more complicated for the live-attenuated SA14-14-2 virus, which has been reported to have a number of different mutations, i.e., 47–64 nucleotide changes (17–27 amino acid substitutions), when compared to its virulent parental strain SA14; the exact number depends on both the passage history of the viruses and the type of cell substrate used for virus cultivation [63]–[65]. A more comprehensive sequence comparison with another SA14-derived attenuated vaccine strain, SA14-2-8, together with two other virulent strains, has suggested seven common amino acid substitutions that may be involved in virus attenuation: 4 in E, 1 in NS2B, 1 in NS3, and 1 in NS4B [64]. However, the genetic component directly responsible for the attenuation of SA14-14-2 is still unknown. Given that SA14-14-2 has been administered to >300 million children for >20 years in China and recently in other Asian countries [53], it is striking that there is a fundamental gap in our knowledge at the molecular level about how SA14-14-2 is attenuated.
Here we report the development of an infectious cDNA-based reverse genetics system for JEV SA14-14-2 that has enabled the analysis of molecular aspects of its attenuation in neurovirulence. By in vivo passage of a molecularly defined, cDNA-derived SA14-14-2 virus, we generated three isogenic variants, each displaying lethal neurovirulence in mice, with a common single G1708→A substitution that corresponds to a Gly→Glu change at position 244 of the viral E glycoprotein. By in vitro site-directed mutagenesis of the infectious SA14-14-2 cDNA, coupled with conventional virologic and experimental pathologic methods and homology-based structure modeling, we have demonstrated a novel regulatory role in JEV neurovirulence of a conserved single amino acid at position E-244 in the ij hairpin adjacent to the fusion loop of the E dimerization domain. These findings offer new insights into the molecular mechanism of JEV neurovirulence and will directly aid the development of new approaches to treating and preventing JEV infection.
Results
Development of a full-length infectious cDNA of SA14-14-2, a live JE vaccine virus
As an initial step in investigating the molecular basis for the virulence attenuation of SA14-14-2, we generated a full-length infectious SA14-14-2 cDNA to serve as a template for genetic manipulation of the viral genome (Fig. 1A). The 10,977-nucleotide genome of SA14-14-2 (GenBank accession number JN604986) was first cloned as four contiguous cDNAs into the bacterial artificial chromosome (BAC) designated pBAC/Frag-I to IV (Fig. 1B). pBAC/Frag-I was modified to have an SP6 promoter immediately upstream of the viral 5′-end, and pBAC/Frag-IV was engineered to contain an artificial XbaI run-off site just downstream of the viral 3′-end, allowing in vitro run-off transcription of capped, genome-length RNAs bearing authentic 5′ and 3′ ends of the genomic RNA. Since the viral genome already had an internal XbaI site at nucleotide 9131 in the NS5 protein-coding region, this pre-existing site was eliminated in pBAC/Frag-III by introducing a silent point mutation, A9134→T (Fig. 1B, asterisk), which in turn served as a genetic marker to identify the cDNA-derived virus. In the last cloning step, a panel of the four overlapping SA14-14-2 cDNAs was sequentially assembled by joining at three natural restriction sites (BsrGI, BamHI, and AvaI) to create the full-length SA14-14-2 cDNA, pBAC/SA14-14-2 (Fig. 1C).
Fig. 1. Construction of the full-length infectious SA14-14-2 cDNA as a BAC. 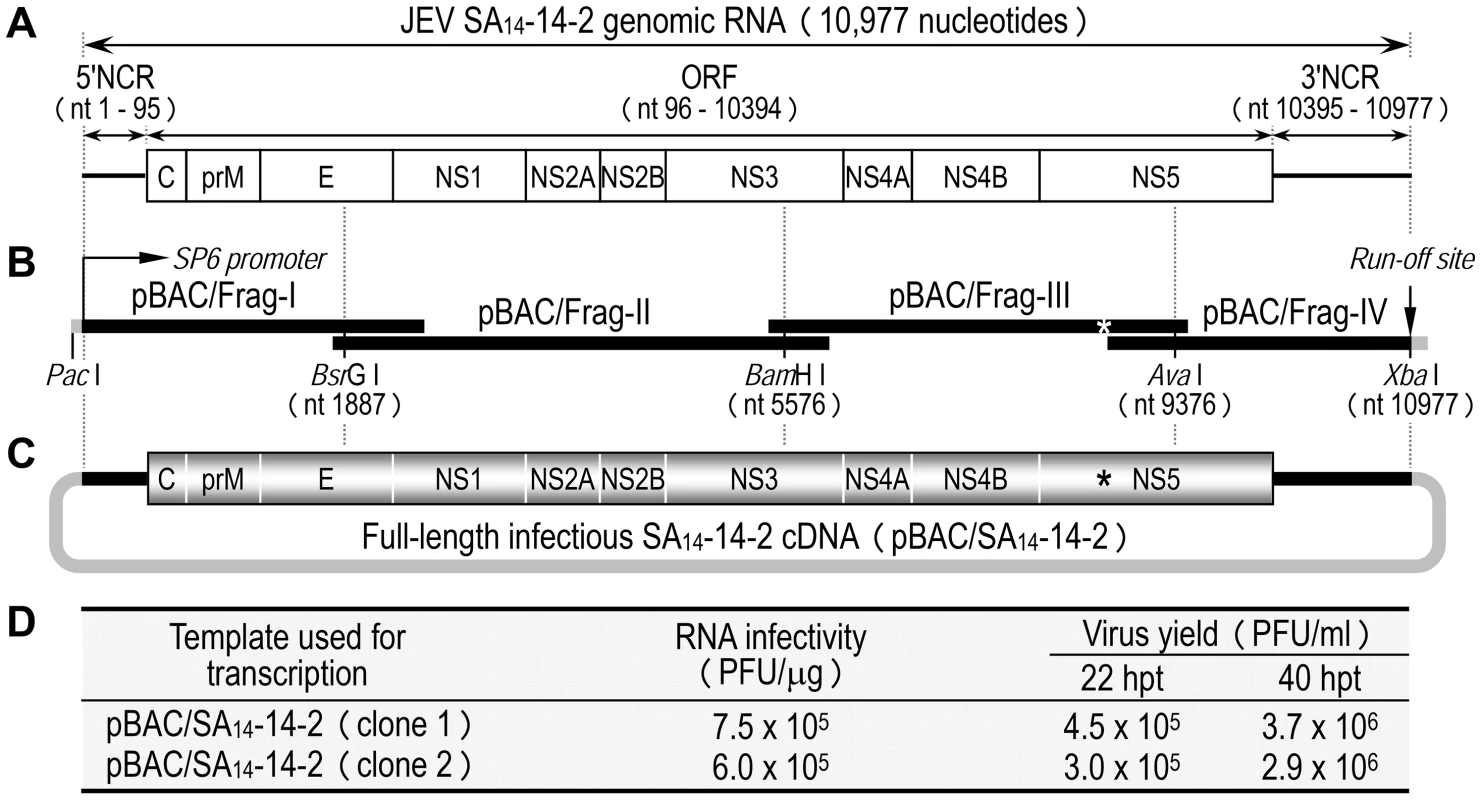
(A) Structure of the SA14-14-2 genomic RNA (GenBank accession no. JN604986). NCR, non-coding region; ORF, open reading frame. (B) Diagram of a panel of four overlapping SA14-14-2 cDNAs contained in pBAC/Frag-I to IV. SP6 promoter and an artificial run-off site are shown. An asterisk indicates a pre-existing XbaI site at nucleotide 9131 that was inactivated by introducing a silent point mutation, A9134→T. (C) Structure of the full-length SA14-14-2 cDNA (pBAC/SA14-14-2). (D) Functionality of pBAC/SA14-14-2. After in vitro transcription with SP6 RNA polymerase, RNA transcripts were electroporated into BHK-21 cells, and infectious plaque centers were determined (RNA infectivity). At 22 and 40 hpt, supernatants from RNA-transfected cells were harvested for virus titration on BHK-21 cells (Virus yield). The functionality of pBAC/SA14-14-2 was analyzed by determining the specific infectivity of the synthetic RNAs transcribed in vitro from the cDNA after RNA transfection into susceptible BHK-21 cells (Fig. 1D). Two independent clones of pBAC/SA14-14-2 were linearized by XbaI, followed by mung bean nuclease treatment to remove the 5′ overhang left by the XbaI digestion. Each was then used as a template for SP6 polymerase run-off transcription in the presence of the m7G(5′)ppp(5′)A cap structure analog. Transfection of the synthetic RNAs into BHK-21 cells gave specific infectivities of 6.0–7.5×105 PFU/µg; the virus titers recovered from the RNA-transfected cells were 3.0–4.5×105 PFU/ml at 22 h post-transfection (hpt) and increased ∼10-fold to 2.9–3.7×106 PFU/ml at 40 hpt (Fig. 1D). Unequivocally, the recovered virus contained the marker mutation (A9134→T) that had been introduced in pBAC/SA14-14-2 (data not shown). Our results show that the synthetic RNAs generated from the full-length SA14-14-2 cDNA are highly infectious in BHK-21 cells, producing a high titer of molecularly defined, infectious virus.
In cell cultures [75], [76], we assessed the in vitro growth properties of the molecularly cloned virus (SA14-14-2MCV) rescued from the infectious cDNA, as compared to those of the uncloned parental virus (SA14-14-2) used for cDNA construction. In hamster kidney BHK-21 cells, which are used most frequently for JEV propagation in laboratories, SA14-14-2MCV replicated as efficiently as SA14-14-2, with no noticeable difference in the accumulation of viral genomic RNA (Fig. 2A) and proteins (Fig. 2B) over the first 24 h after infection at a multiplicity of infection (MOI) of 1 plaque-forming unit (PFU) per cell. These observations were consistent with their growth kinetics, which were essentially identical for 4 days following infection at three different MOIs: 0.1, 1, and 10 PFU/cell (Fig. 2C and data not shown). Similarly, there was no difference in focus/plaque morphology between SA14-14-2MCV and SA14-14-2 at 4 days post-infection (dpi) (Fig. 2D); as expected, their foci/plaques were ∼30% smaller than those produced by CNU/LP2, a virulent JEV strain used as a reference (Fig. S1). Also, their growth properties were equivalent in two other cell lines, human neuroblastoma SH-SY5Y and mosquito C6/36 cells, which are potentially relevant to JEV pathogenesis and transmission, respectively (Fig. S2). These data suggest that the uncloned parental and molecularly cloned viruses are indistinguishable in viral replication and spread in both mammalian and insect cells.
Fig. 2. Characterization of biological properties of the molecularly cloned virus SA14-14-2MCV in vitro and in vivo. 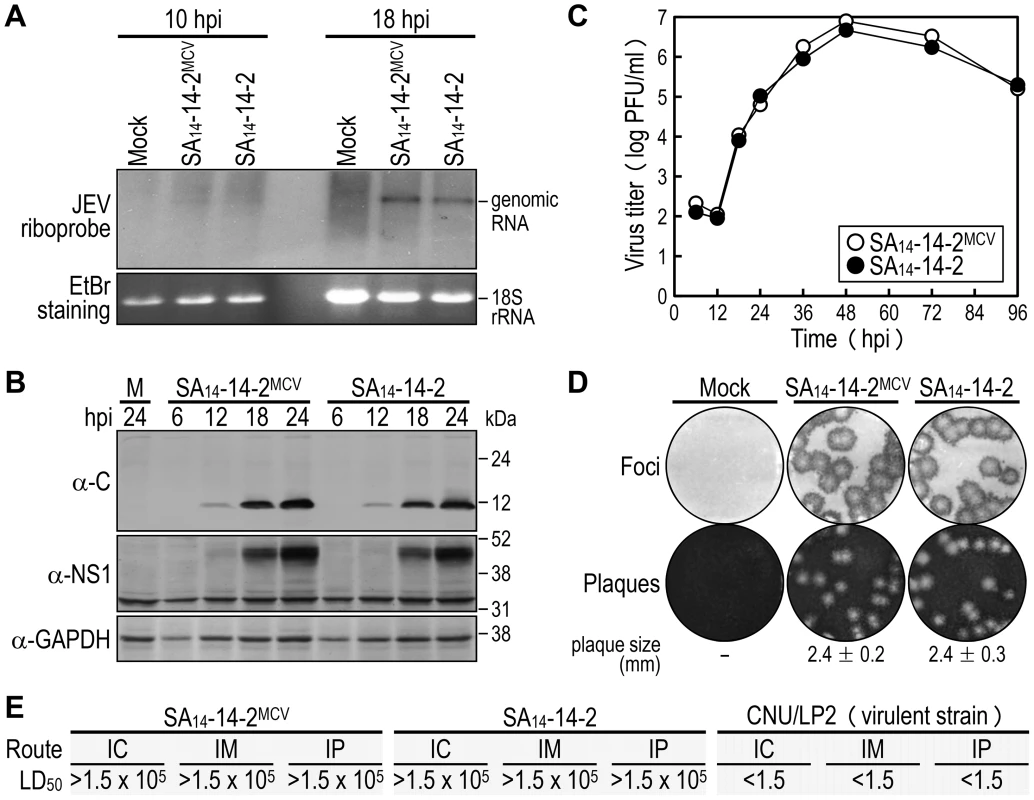
(A-D) BHK-21 cells were mock-infected or infected at an MOI of 1 with the molecularly cloned virus (SA14-14-2MCV) or the original parental virus (SA14-14-2). At the time points indicated, cells were lysed to analyze the accumulation levels of viral genomic RNA by Northern blotting (A) and viral proteins by immunoblotting (B), and culture supernatants were harvested to examine the production levels of progeny virions by plaque titration on BHK-21 cells (C). At 4 dpi, cell monolayers were first immunostained with a mouse α-JEV antiserum to visualize the infectious foci, and the same monolayers were then restained with crystal violet to observe the infectious plaques (D). The average plaque sizes (mean ± SD) were estimated by counting 10 representative plaques. (E) Groups of 3-week-old ICR mice (n = 20 per group) were infected IC, IM, or IP with serial 10-fold dilutions of each virus as indicated. The LD50 values (in PFU) were calculated by the Reed and Muench method [119]. CNU/LP2, a virulent JEV strain used as a reference. In mice [75], [77], we evaluated in vivo the attenuation phenotypes of SA14-14-2MCV and SA14-14-2, with a virulent JEV CNU/LP2 [78] in parallel. Groups of 3-week-old ICR mice (n = 20) were infected with various doses (1.5 to 1.5×105 PFU/mouse) of each virus, via three different inoculation routes: intracerebral (IC) for neurovirulence, and intramuscular (IM) and intraperitoneal (IP) for neuroinvasiveness. As with SA14-14-2, the 50% lethal doses (LD50s) of SA14-14-2MCV, regardless of the route of inoculation, were all >1.5×105 PFU (Figs. 2E and S3). Specifically, all mice infected with SA14-14-2MCV or SA14-14-2 remained healthy and displayed no clinical signs of JEV infection (e.g., ruffled fur, hunched posture, tremors, or hindlimb paralysis) after IM or IP inoculation with any of the tested doses; on the other hand, a small fraction of the mice infected with SA14-14-2MCV (5–20%) or SA14-14-2 (5–10%) developed typical symptoms and death after the IC inoculation with a relatively high dose of ≥1.5×103 PFU/mouse, but not the low dose of ≤1.5×102 PFU/mouse (Fig. S3). In all dead or surviving mice, virus titration confirmed the presence (1.8–4.1×106 PFU/brain) or absence, respectively, of viral replication in their brain tissues. As expected [75], [77], the LD50 values of CNU/LP2 [78], irrespective of the inoculation route, were always <1.5 PFU (Figs. 2E and S3); the control groups of mock-infected mice all survived with no signs of disease (Fig. S3). Thus, our data indicate that SA14-14-2MCV displays a variety of biological properties identical to those of SA14-14-2, both in vitro and in vivo.
Generation of three highly neurovirulent variants derived from SA14-14-2MCV
As was true for SA14-14-2, direct inoculation of a relatively high dose of SA14-14-2MCV into mouse brains initiated a productive infection in the CNS and caused lethal encephalitis, albeit at a very low frequency (Fig. S3). Intrigued by this observation, we decided to generate isogenic neurovirulent variants from SA14-14-2MCV by serial brain-to-brain passage in mice (Fig. 3A). At passage 1 (P1), the cDNA-derived SA14-14-2MCV was directly inoculated into the brains of 3-week-old ICR mice at 1.5×105 PFU/mouse (three groups, n = 10 per group); one or two infected mice per group exhibited clinical symptoms of JEV infection. At the onset of hindlimb paralysis (6–10 dpi), virus was harvested from the brain of a moribund mouse in each group (3 total); in each case, a brain homogenate was prepared for plaque titration and used as an inoculum for the next round of passage. Serial intracerebral passage was continued for three additional rounds, with a gradually decreasing inoculum in order to ensure the stability of selected mutations and a sufficiently pure population of viruses: 1,500 (P2), to 15 (P3), to 1.5 PFU/mouse (P4). Using this approach, we obtained three independently selected variants, SA14-14-2MCV/V1 to V3 (Fig. 3A).
Fig. 3. Development of three independent, highly neurovirulent variants from SA14-14-2MCV by serial intracerebral passage in mice. 
(A) Diagram illustrating the in vivo passage of SA14-14-2MCV. (B) Virological properties of three SA14-14-2MCV variants in mice. Groups of 3-week-old ICR mice (n = 10 per group) were inoculated IC, IM, or IP with doses of the virus stock serially diluted 10-fold. The LD50 values were determined in PFU. (C) Comparison of the complete genome sequence of the SA14-14-2MCV parental virus and its three variant viruses. We first compared the biological properties of the three SA14-14-2MCV variants, both in vitro and in vivo, to those of the parental SA14-14-2MCV. In three cell cultures (BHK-21, SH-SY5Y, and C6/36), all three variants exhibited characteristics of viral replication identical to the parent, as demonstrated by (i) quantitative real-time RT-PCRs to measure the level of viral genomic RNA production, (ii) immunoblotting with a panel of JEV-specific rabbit polyclonal antisera to probe the profile and level of viral structural and nonstructural protein accumulation, and (iii) one-step growth analyses to assess the yield of progeny virions produced during a single round of infection (data not shown). In 3-week-old ICR mice, however, there was a clear difference between the parent and the three variants in both phenotype and virulence level (Fig. 3B). When peripherally inoculated (i.e., IM and IP), neither the parent nor its three variants caused any symptoms or death at a maximum dose of 1.5×105 PFU/mouse. In contrast, when inoculated IC, the three variants, unlike the parent (IC LD50, >1.5×105 PFU), were all highly neurovirulent (IC LD50s, <1.5 PFU) (Fig. 3B and Table S1). Our findings show that all three variants still lacked a detectable level of neuroinvasiveness but gained a high level of neurovirulence after serial IC passage in mice.
Next, we determined the complete nucleotide sequence of the genome of the three SA14-14-2MCV variants to identify the nucleotide(s) and/or amino acid(s) in specific viral loci/genes that is(are) potentially responsible for the drastic increase in neurovirulence. According to our protocol [77], the consensus genome sequence of each variant was generated by direct sequencing of three overlapping, uncloned cDNA amplicons covering the entire viral RNA genome except the 5′ - and 3′-termini; the remaining consensus sequences of the 5′ - and 3′-terminal regions were obtained by 5′ - and 3′-RACE reactions, each followed by cDNA cloning and sequencing of 10–15 independent clones. In all three variants, when the consensus genome sequence was compared to that of the parent, a single nucleotide G-to-A transition was always found at nucleotide 1708, changing a Gly (GGG) to Glu (GAG) codon at amino acid 244 of the viral E glycoprotein (Fig. 3C). In addition, each of the three variants also contained a small number of unique silent point mutations scattered over the genome, confirming they were indeed independent variants (Fig. 3C): one in SA14-14-2MCV/V1 (U2580C), two in SA14-14-2MCV/V2 (G317A and U8588C), and four in SA14-14-2MCV/V3 (U419C, C3215U, C5987U, and G6551A). These results suggest that the G1708A substitution, the only mutation observed in all three variants, may contribute to the viral neurovirulence in mice.
Identification of a single Gly-to-Glu change at position E-244 that is responsible for the reversion to neurovirulence
To identify a key point mutation(s) in three variants of SA14-14-2MCV that leads to the acquisition of neurovirulence, we generated eight derivatives of SA14-14-2MCV, each containing one of the eight point mutations found in our three variants, by cloning them individually into the infectious SA14-14-2 cDNA and transfecting the synthetic RNAs derived from each mutant cDNA into BHK-21 cells. In all cases, the mutant RNA was as infectious as the parent RNA, with a specific infectivity of 6.4–8.3×105 PFU/µg; the sizes of the foci/plaques produced by each mutant RNA were indistinguishable from those generated by the parent RNA, paralleling their levels of virus production, with an average yield of 2.1–4.5×105 PFU/ml at 22 hpt (Fig. 4A). In agreement with these results, no difference was observed in the profile or expression level of the viral proteins, i.e., three structural (C, prM, and E) and one nonstructural (NS1), as determined by immunoblotting of RNA-transfected cells at 18 hpt (Fig. 4B). All the mutant viruses grew as efficiently as did the parental virus over the course of 96 h after infection at an MOI of 0.1 in BHK-21 cells (Fig. 4C). Thus, there was no apparent effect of any of the eight introduced genetic changes on virus replication.
Fig. 4. Discovery of a single locus that leads to the reversion of SA14-14-2MCV to lethal neurovirulence. 
(A-C) In vitro replicability. BHK-21 cells were mock-transfected or transfected with RNAs transcribed from the parent or each mutant cDNA, as indicated. RNA infectivity (in PFU/µg) at 4 dpt was determined by infectious center assay, combined with staining of cell monolayers using an α-JEV antiserum, and virus yield (in PFU/ml) at 22 hpt was measured by plaque titration on BHK-21 cells (A); viral protein accumulation at 18 hpt was examined by immunoblotting of cell lysates with a panel of antibodies as indicated (B). For viral growth analysis, BHK-21 cells were infected at an MOI of 0.1 with the parent or each mutant virus obtained from the corresponding RNA-transfected cells. At the indicated time points, culture supernatants harvested for virus titration on BHK-21 cells (C). (D-E) In vivo neurovirulence. Groups of 3-week-old ICR mice (n = 10 per group) were inoculated IC with doses of the virus stock serially diluted 10-fold, and the LD50 values were determined (D). For immunohistochemical staining, groups of the mice (n = 15 per group) were mock-infected or infected IC with 103 PFU of each virus. At 3, 5, and 7 dpi, five mice were processed for brain section staining with an α-NS1 antiserum. Shown are representative hippocampal slides (E). In mice, we examined the neurovirulence of these eight mutant viruses. Groups of 3-week-old ICR mice (n = 10 per group) were infected by IC inoculation with various doses (1.5 to 1.5×105 PFU/mouse) of the parent or each mutant virus. One of the eight mutants containing the G1708A substitution had an IC LD50 of <1.5 PFU, making it capable of killing all mice within ∼7 dpi with a minimum dose of 1.5 PFU/mouse; the other seven mutants had IC LD50 values all >1.5×105 PFU and behaved like the parental virus, with only <20% of infected mice developing clinical symptoms and death at a maximum dose of 1.5×105 PFU/mouse (Fig. 4D and Table S2). In all dead or surviving mice, virus titration confirmed the presence (1.4–3.5×106 PFU/brain) or absence, respectively, of productive viral replication in the brain tissues; as expected, all mock-infected mice survived with no signs of disease (data not shown). Thus, our findings showed that of the eight point mutations, a single G1708A substitution, replacing a Gly with Glu at amino acid residue 244 of the viral E glycoprotein, is sufficient to confer lethal neurovirulence in mice.
To determine whether the mutant G1708A, unlike the parent SA14-14-2MCV, is able to replicate and spread in the CNS, we immunohistochemically stained for JEV NS1 antigen in mouse brains after IC inoculation (Fig. 4E shows hippocampal slides, and Fig. S4 presents slides of other brain areas, i.e., amygdala, cerebral cortex, thalamus, hypothalamus, and brainstem): (i) In brains infected with a virulent JEV CNU/LP2 (control) [75], [77], [78], a large number of NS1-positive neurons were observed at 3 dpi in all areas we stained; this number was increased significantly at 5 dpi. In the hippocampus, most infected neurons were found in the CA2/3 region at 3 dpi and had spread to the CA1 region by 5 dpi. (ii) In brains infected with the parent SA14-14-2MCV, almost no NS1-positive cells were found in any brain region during the entire 7-day course of the experiment. In a few atypical cases, a small number of NS1-positive neurons were noted at 5–7 dpi in the hippocampal CA2/3 region, but not the CA1 region (data not shown). (iii) In brains infected with the mutant G1708A, a considerable number of NS1-positive neurons were observed at 3 dpi, mainly in the hippocampal CA2/3 region, and only a few in other areas (amygdala, cerebral cortex, thalamus, and brainstem); overall, the number of infected neurons was much lower than in brains infected with JEV CNU/LP2. At 5–7 dpi, the number of NS1-positive neurons was noticeably increased in the hippocampus (now in the CA1) and amygdala, but not in other brain regions. Our findings show that, in mice, a single G1708A substitution changing a Gly with Glu at position E-244 promotes susceptibility to SA14-14-2MCV infection of neurons.
Understanding the novel regulatory role in neurovirulence of E-244, located in the ij hairpin of the viral E glycoprotein
To probe the functional importance of the amino acid side chain at position E-244 for the viral replication and neurovirulence of SA14-14-2MCV, we performed site-directed mutagenesis, replacing G244 with 14 other amino acids of six different classes: (1) aliphatic A, V, and L; (2) hydroxyl S and T; (3) cyclic P; (4) aromatic F and W; (5) basic R and K; and (6) acidic and their amides D, E, N, and Q. We first tested the viability of synthetic RNAs transcribed in vitro from the corresponding mutant cDNAs by measuring their infectivity after transfection of BHK-21 cells. In all cases, the mutant RNA was as viable as the parent RNA, with a specific infectivity of 6.5–8.2×105 PFU/µg (Fig. 5A, RNA infectivity). However, three mutants (G244K, G244F, and G244W) were noticeably different from the parent and the other 11 mutants, as demonstrated by a ∼10-fold decrease in the yield of progeny virions released into culture medium during the first 22 hpt (Fig. 5A, virus yield) and a ∼2-2.5-fold reduction in the size of foci/plaques produced at 96 hpt (Fig. 5A, foci/plaques), although no significant difference was observed in the level of viral proteins (i.e., C, prM, E, and NS1) accumulated in RNA-transfected cells at 18 hpt (Fig. S5). As compared to G244K, the mutant G244R exhibited a barely marginal decrease in focus/plaque size and no detectable change in virus production (Fig. 5A). Overall, these findings were more evident when all mutant viruses were evaluated in multistep growth assays over the course of 96 h after infection at an MOI of 0.1, assessing their ability to grow and establish a productive infection (Fig. 5B). Our findings indicate that in BHK-21 cells, the amino acid side chain at position E-244 has no effect on the viability of the mutant RNAs, although it has a negative impact on the production and spread of infectious virions in the case of the three mutants G244K, G244F, and G244W.
Fig. 5. E-244: A key neurovirulence factor located in the ij hairpin of the viral E glycoprotein. 
(A) RNA infectivity and replicability. BHK-21 cells were mock-transfected or transfected with RNAs transcribed from the parent or one of the 14 E-244 mutant cDNAs. RNA infectivity (in PFU/µg) at 4 dpt was estimated by infectious center assay, coupled with staining of cell monolayers using an α-JEV antiserum, and virus yield (in PFU/ml) at 22 hpt was determined by plaque titration on BHK-21 cells. (B) Viral growth. BHK-21 cells were infected at an MOI of 0.1 with the parent or one of the 14 E-244 mutant viruses. At the indicated time points, culture supernatants were used for virus titration on BHK-21 cells. (C) Neurovirulence. Groups of 3-week-old ICR mice (n = 10 per group) were infected IC with serial 10-fold dilutions of each virus stock, and the LD50 values were determined. (D) Homology model. The predicted model of the E ectodomain of JEV SA14-14-2 was built based on the crystal structure of the E ectodomain of WNV NY99 [79], and the model was then fitted into the cryo-EM structure of WNV NY99 [18]. Illustrated is an icosahedral asymmetric unit of the three E monomers on the viral membrane. Highlighted in the inset is the critical residue Glu at E-244 in the ij hairpin adjacent to the fusion loop of the viral E DII. In mice, we determined the neurovirulence of our 14 mutant viruses by IC inoculating groups of 3-week-old ICR mice (n = 10 per group) with various doses ranging from 1.5 to 1.5×104 or 105 PFU/mouse of the parent or each mutant virus. According to their IC LD50 values, the 14 mutant viruses are classified into three groups (Fig. 5C and Table S3): (i) group 1 (six mutants), neurovirulent, with an IC LD50 of ≤1.5 to 31 PFU, exemplified by replacing G244 with E, D, T, S, Q, and P; (ii) group 2 (six mutants), non-neurovirulent or neuroattenuated, with an IC LD50 of >1.5×104 or 105 PFU, behaving like the parent SA14-14-2MCV and exemplified by exchanging G244 with R, K, F, W, N, and L; and (iii) group 3 (two mutants), with an intermediate phenotype and an IC LD50 of 1.2–5.8×103 PFU, exemplified by substituting G244 with A and V. We confirmed the presence or absence of viral replication in the brain tissues of all dead or surviving mice, respectively; all mock-infected mice survived with no signs of disease (data not shown). Also, the mutation and phenotype relationship was corroborated by sequence analysis of recovered viruses from brain tissues of moribund or dead mice following IC inoculation. We analyzed all of the 14 mutants except for four group 2 mutants (G244R, G244F, G244W, and G244L), which failed to produce a lethal infection. In each case, the complete 2,001-nucleotide coding region of the prM and E genes was amplified from each of four randomly selected brain samples, followed by cloning and sequencing of at least seven independent clones per brain sample. In all six group 1 and two group 3 mutants, we found that the initial mutations introduced at the G244 codon were maintained with no second-site mutations, consistent with the high and intermediate levels of their neurovirulent phenotype (Table 1). In the remaining two group 2 mutants (G244K and G244N), however, a majority of the sequenced clones contained a point mutation in the same codon that led to an amino acid substitution (i.e., K→E/T and N→D, respectively), converting both mutants into neurovirulent viruses and highlighting the biological importance of the amino acid at position E-244 for neurovirulence (Table 1).
Tab. 1. Genetic stability of 14 E-244 mutants recovered from mouse brains after IC inoculation. 
A, Attenuated; I, Intermediate; N, Neurovirulent. We next performed homology modeling to gain insight into the structural basis of E-244 function. The 3D model of the E monomer of JEV SA14-14-2 was constructed using the 3.0-Å crystal structure of the E monomer of WNV NY99 [79] as a template, with 75.5% sequence identity. The model was then fitted into the outer layer of the cryo-electron microscopy (EM) structure of WNV NY99 [18], thereby visualizing three monomers placed into an icosahedral asymmetric unit on the viral membrane. In each E monomer of SA14-14-2 containing three domains (DI, DII, and DIII), we noted that E-244 lies within the ij hairpin adjacent to the fusion loop at the tip of DII, with its amino acid side chain pointing toward the viral membrane (Fig. 5D). We also confirmed the location of E-244 in the crystal structure of the E ectodomain of JEV SA14-14-2 [80] that has been described recently (Fig. S6).
Elucidation of the functional role of E-244 during JEV infection of neuronal cells in vitro
We hypothesized that E-244, located at the ij hairpin of the viral E glycoprotein, plays an important role in JEV infection of neuronal cells. To test this hypothesis, we performed multistep growth assays in two neuronal cells, NSC-34 (mouse motor neuron) and SH-SY5Y (human neuroblastoma), by infecting at an MOI of 0.1 with the non-neurovirulent parent SA14-14-2MCV and each of the four representative E-244 mutant viruses, i.e., two neurovirulent (G244E and G244D) and two non-neurovirulent (G244R and G244K). In parallel, the non-neuronal BHK-21 cells were also infected for comparison with the same set of five viruses. In NSC-34 cells, while the two neurovirulent viruses grew rapidly and reached their maximum titers of 1.8–2.4×105 PFU/ml at 72–96 hours post-infection (hpi), the three non-neurovirulent viruses, including the parent, all replicated poorly, with peak titers only approaching 1.0–2.0×103 PFU/ml, ∼100-fold lower than those of the two neurovirulent viruses (Fig. 6A). In SH-SY5Y cells, a similar defect in viral growth was also observed, with a ∼50 - to 100-fold difference in maximum virus titers between the neurovirulent and the non-neurovirulent viruses. In addition, we noted a differential growth defect in the three non-neurovirulent viruses, with G244R replicating more poorly than the parent but better than G244K (Fig. 6B). In contrast to the pattern of viral growth observed in NSC-34 and SH-SY5Y cells, we found that in BHK-21 cells, only G244K had a noticeable defect in viral growth, with ∼20-fold lower peak titers than those of the other four viruses that grew well to maximum titers of 0.8–2.5×106 PFU/ml at 48–72 hpi (Fig. 6C). These data indicate that E-244 plays a crucial role in the productive infection of JEV in neuronal cells.
Fig. 6. E-244: A major determinant of viral infectivity in mouse and human neuronal cells in vitro. 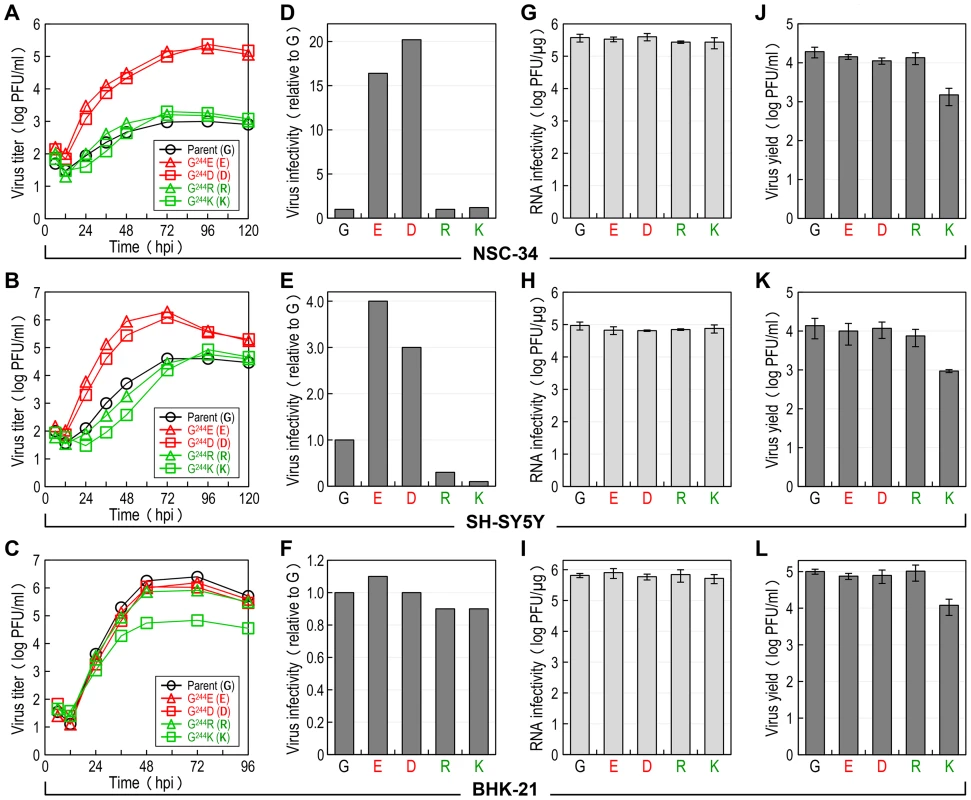
(A-C) Viral growth. NSC-34 (A), SH-SY5Y (B), or BHK-21 (C) cells were infected at an MOI of 0.1 with the parent or one of the following four E-244 mutant viruses: G244E, G244D, G244R, and G244K. At the indicated time points, culture supernatants were harvested for virus titration on BHK-21 cells. (D-F) Viral particle infectivity. NSC-34 (D), SH-SY5Y (E), or BHK-21 (F) cells were infected at an MOI of 1 with each of the five viruses, as indicated. At 12–15 hpi, the number of infected cells was measured by flow cytometry using mouse α-JEV antiserum. The results are the average of two independent experiments, presented as -fold changes in infectivity relative to the parent (infectivity value of 1). (G-I) Viral RNA infectivity/replicability. NSC-34 (G), SH-SY5Y (H), or BHK-21 (I) cells were transfected with RNAs transcribed from the parent or each mutant cDNA, as indicated. At 4 dpt, RNA infectivity was quantified by infectious center assay, coupled with staining of cell monolayers using an α-JEV antiserum. (J-L) Virus yield. At 20 h after RNA transfection, culture supernatants from RNA-transfected NSC-34 (J), SH-SY5Y (K), or BHK-21 (L) cells were harvested for virus titration on BHK-21 cells. Subsequently, we examined the infectivity/replicability of the parent and its four E-244 mutant viruses/RNAs in NSC-34 and SH-SY5Y cells, in parallel with BHK-21 cells for comparison. First, virus infectivity was quantified using flow cytometry by infecting the three cell types at an MOI of 1 with each of the five viruses and counting the number of cells stained with a mouse α-JEV antiserum at 12–15 hpi. In NSC-34 cells, the two non-neurovirulent mutants (G244R and G244K) exhibited infectivities nearly identical to that of the non-neurovirulent parent (Fig. 6D), whereas the two neurovirulent mutants (G244E and G244D) showed infectivities ∼16 - to 20-fold higher than that of the non-neurovirulent parent. Similarly, the E-244 mutation also altered virus infectivity in SH-SY5Y cells. Specifically, the two neurovirulent mutants had ∼3 - to 4-fold higher infectivities than the non-neurovirulent parent; on the other hand, the two non-neurovirulent mutants displayed even lower infectivities than the parent (∼3-fold for G244R and ∼10-fold for G244K) (Fig. 6E). In contrast, no significant difference in virus infectivity was observed among all five viruses in BHK-21 cells (Fig. 6F).
Next, the replication efficiency of the viral genomic RNA was quantified by directly transfecting the three cell types with each of the five synthetic RNAs transcribed in vitro from the respective JEV cDNAs and estimating the number of infectious foci stained with the mouse α-JEV antiserum at 4 days post-transfection (dpt). In each of the three cell types, there was no detectable difference in the specific infectivity of the five RNAs (NSC-34, Fig. 6G; SH-SY5Y, Fig. 6H; and BHK-21, Fig. 6I). Also, quantitative real-time RT-PCRs indicated that the level of the viral genomic RNAs accumulated in the RNA-transfected cells over the first 15 h of transfection was indistinguishable between the parent and the four different E-244 mutants (data not shown). Regardless of cell type, however, the G244K mutant was different from the parent and the other three E-244 mutants, as demonstrated by a ∼1-log decrease in the yield of infectious virions released into culture medium during the first 20 hpt (NSC-34, Fig. 6J; SH-SY5Y, Fig. 6K; and BHK-21, Fig. 6L). Overall, these results show that the E-244 mutation alters JEV infectivity in a neuronal cell-specific manner, in agreement with the neurovirulence phenotype observed in mice, and it also affects infectious particle production in a cell type-nonspecific manner.
Comparison of the amino acid sequences of the ij hairpin in encephalitic and non-encephalitic flaviviruses
We initially generated a multiple sequence alignment using all 154 full-length JEV genomes available from the GenBank sequence database. Of note is the fact that SA14 and SA14-14-2 have been fully sequenced by three and four independent groups, respectively; their nucleotide and deduced amino acid sequences are not identical [63]–[65], [77], [81]. The sequence alignment showed a Glu residue at position E-244 in the ij hairpin of all JEV strains isolated from infected mosquitoes, pigs, or humans, except for the Gln-encoding mosquito-derived K94P05 and three Gly-encoding SA14-derived attenuated strains (i.e., SA14-2-8, SA14-12-1-7, and all four different versions of SA14-14-2) (Fig. S7). In case of SA14, it is intriguing to note that one version has a Glu residue at position E-244, but the other two versions have a Gly residue at that position (Fig. S7); this discrepancy is likely due to variations in the cultivation history of the virus [77]. We next performed the structure-based, ij-hairpin amino acid sequence alignment with six representative flaviviruses (14 strains total), including four encephalitic (JEV, WNV, SLEV, and MVEV) and two non-encephalitic (YFV and DENV) flaviviruses. In addition to the importance of the E-244 amino acid, we noted (i) the evolutionally conserved residues in the ij hairpin and its flanking region in all six flaviviruses, i.e., W233, F242, H246, A247, V252, L255, G256, Q258, E259, and G260; (ii) the sequence similarities in the four encephalitic flaviviruses, particularly in a ∼15-aa ij-hairpin-containing region; and (iii) the sequence differences between the four encephalitic and two non-encephalitic flaviviruses, e.g., the 4-aa YFV-specific motif and the 3-aa DENV-specific motif (Fig. 7). Overall, our findings suggest that the ij hairpin of the E DII plays a key role in determining encephalitic flavivirus neurovirulence, and its function is regulated by the chemical properties of the amino acid at position E-244 in that hairpin.
Fig. 7. Structure-based, ij-hairpin amino acid sequence alignment for six representative flaviviruses (14 strains total): JEV, WNV, SLEV, MVEV, YFV, and DENV. 
All sequence information was retrieved from the GenBank database with accession numbers indicated. Multiple sequence alignments were performed using ClustalX [116]. Highlighted are the ∼15-aa ij-hairpin (yellow), the 3-aa DENV-specific motif (pink), and the 4-aa YFV-specific motif (green). The consensus sequence of the ij hairpin and its flanking region is presented on top, and only differences from that sequence are shown. Deletions are indicated by hyphens. The amino acid residue is numbered based on the JEV SA14-14-2 (GenBank accession no. JN604986). Discussion
In this work, we have developed a reverse genetics system for SA14-14-2, a live-attenuated JE vaccine [53], [82], by constructing an infectious cDNA and rescuing molecularly cloned virus from the cDNA. This reverse genetics system offers us a unique opportunity to elucidate the genetic and molecular basis of JEV neurovirulence. Using our infectious SA14-14-2 cDNA technology, we (i) generated three isogenic SA14-14-2 variants that unlike its parent, displayed lethal neurovirulence in mice; (ii) identified a single point mutation, G1708→A, causing a Gly→Glu change at position 244 of the viral E glycoprotein that is sufficient to confer a full neurovirulence by promoting viral infection into neurons in the mouse CNS in vivo and mouse/human neuronal cells in vitro; and (iii) demonstrated the structure-function relationship for neurovirulence of E-244 in the ij hairpin adjacent to the fusion loop at the tip of the viral E DII. Thus, our findings reveal fundamental insights into the neurotropism and neurovirulence of JEV and other taxonomically related encephalitic flaviviruses, including WNV, SLEV, and MVEV. Intriguingly, our results also provide a new target, the ij hairpin, for the development of novel antivirals for the prevention and treatment of infection with the encephalitic flaviviruses.
The flavivirus glycoprotein E mediates receptor-mediated endocytosis and low pH-triggered membrane fusion [33], [83], [84]. On the viral membrane, 180 E monomers are packed into 30 protein “rafts”, each composed of three E head-to-tail homodimers [17]–[19]. Each E monomer is composed of three parts: (i) an elongated ectodomain that directs receptor binding and membrane fusion; (ii) a “stem” region containing two amphipathic α-helices that lies flat on the viral membrane underneath the ectodomain; and (iii) a membrane “anchor” region containing two transmembrane antiparallel coiled-coils. The E ectodomain folds into three β-barrel domains [85]: (i) DI, a structural domain centrally located in the molecule; (ii) DII, an elongated dimerization domain containing the highly conserved fusion loop at its tip [86]; and (iii) DIII, an Ig-like domain implicated in receptor binding [20], [87], [88] and antibody neutralization [89]–[92]. Based on pre - and post-fusion crystal structures of the ectodomain [28], [30], [31], [47], [79], [80], [85] and biochemical analyses [29], [32], [93], a current, detailed model for flavivirus membrane fusion has been developed. In this model, the fusion is initiated by a low pH-induced dissociation of the antiparallel E homodimers that leads to the exposure of the fusion loops and their insertion into the host membrane, followed by a large-scale structural rearrangement into a parallel E homotrimer [33], [83], [84], [94]. In the parallel conformation, DIII folds back toward DII, presumably with the stem extended from the C-terminus of DIII along DII and toward the fusion loop (“zipping”), driving the fusion of the viral and host membranes [95]–[98]. Despite our detailed knowledge about the fusion process, there is little available structural information about how flaviviruses bind to their cellular receptors. In encephalitic flaviviruses, the presence of an RGD motif in DIII and carbohydrate moieties on the viral surface suggests a mechanism involving interaction with the RGD motif-recognizing integrins and sugar-binding lectins on the cell surface, respectively. However, blocking/alteration of either the RGD motif or glycan does not abolish viral entry [22], [99]–[101]. Thus, the viral factors and the interacting cellular counterparts required for viral entry are still elusive.
In flaviviruses, the ij hairpin is a structural motif that is closely associated with the fusion loop at the tip of the viral E DII, but its role is thus far unknown. In JEV, we now report that a single amino acid in the ij hairpin, E-244, serves as a key regulator to control the level of neurovirulence of SA14-14-2 in mice. This amino acid was also correlated with a differential ability to infect neurons, the primary target cells in the CNS. Consistent with this finding, we found that site-directed mutagenesis of the codon for E-244 in SA14-14-2 created a panel of 14 recombinant viruses of varying neurovirulence: (i) non-neurovirulent viruses, produced by substitutions of positively-charged (R, K), aromatic (F, W), polar (N), or aliphatic (L) residues; (ii) neurovirulent viruses, produced by substitutions of negatively-charged (E, D), hydroxyl (T, S), polar (Q), or cyclic (P) residues; and (iii) viruses intermediate in neurovirulence, produced by substitutions of aliphatic (A, V) residues. These results highlight the role of E-244 in neurovirulence, which was directed by a combination of three major properties of its amino acid side chain: (i) charge (R/K vs. E/D); (ii) size (N vs. Q and L vs. A/V); and (iii) functional group (N vs. D). Our data suggest that the ij hairpin acts as a viral factor that promotes JEV infection of neurons within the CNS, likely through its role in one of three major steps involved in viral entry: binding, endocytosis, or membrane fusion [33], [83], [84]. Alternatively, it is possible that the late steps in the virus life cycle in neurons, such as assembly, maturation, and release, could be affected. For JEV, WNV, and tick-borne encephalitis virus, the assembly/release of infectious virions or virus-like particles has been shown to be affected by the N-glycosylation of the viral prM and/or E protein in non-neuronal cells [44], [75], [102]–[104]. Moreover, a conserved single N-glycosylation site in the JEV prM protein has been shown to be important for viral pathogenicity in mice [75].
Over the years, the virulence of JEV has been an active area of research. Initially, comparison of the genomic sequences of several JEV strains with a different degree of pathogenicity had predicted a number of potential loci in the viral genome that are involved in virulence [63]–[74]. Due to the complexity and variation of the mutations, however, the identity of the major viral factor that is critical for JEV virulence remains unclear. In particular, SA14-14-2 has been reported to have a total of 47–64 nucleotide changes (17–27 amino acid substitutions) when compared to its virulent parental strain SA14; the number of mutations varies and depends on the cultivation history of the viruses [63]–[65]. Of the ten viral proteins, the E protein has been the primary target of genetic studies in virulence, mainly because it is involved in cell/tissue tropism and pathogenesis. Several amino acid residues in the E protein have been suggested to contribute to the neurovirulence and/or neuroinvasiveness of JEV in vivo: (i) E-123, illustrated by an S123R substitution that is capable of enhancing the neuroinvasiveness of the Mie/41/2002 strain in 3-week-old ddY mice [105]; (ii) E-279, exemplified by an M279K mutation that is able to increase the neurovirulence of ChimeriVax-JE (a chimeric virus that carries the prM and E genes of JEV SA14-14-2 on a YFV 17D genetic background) in suckling mice and rhesus monkeys [106]; and (iii) E-138, indicated by two reciprocal mutations: (1) a K138E substitution, when combined with at least two other mutations, which elevates the neurovirulence of the ChimeriVax-JE virus in 4-week-old ICR mice [107], and (2) an E138K substitution, which lowers the neurovirulence and neuroinvasiveness of three different JEVs (the JaOArS982 strain in 2 - to 5-week-old Swiss ICR mice [73] and the AT31 and NT109 strains in 3-week-old BALB/c mice [108], [109]). These data indicate that multiple amino acid residues in the E protein of JEV function in a more coordinated way to achieve the maximal level of neurovirulence and/or neuroinvasiveness [53], [107]. This notion is consistent with our finding that although a single G244E mutation in the E protein of SA14-14-2 is sufficient to confer lethal neurovirulence in 3-week-old ICR mice, the spread of the virus in the brains is still slow and limited, as compared to the highly virulent CNU/LP2 strain. These and previous findings suggest that in addition to E-244, other amino acid residues in the E protein play a role in determining the neurovirulence of SA14-14-2. In addition, JEV NS1' (a product of ribosomal frameshifting [110], [111]) is reported to be produced in cells infected with SA14 but not with SA14-14-2, and its lack of expression is shown to contribute to the attenuation phenotype of SA14-14-2 in mice [112]. Similarly, the expression of NS1' is also suggested to be associated with the neuroinvasiveness of WNV [110].
The neuroattenuation phenotype of SA14-14-2 has been tested in several laboratory animals, including mice and monkeys [53]. In 2 - to 4-week-old ICR and ddY mice, no morbidity or mortality has been observed after subcutaneous and intracerebral inoculations with 104-106 PFU of SA14-14-2 [53], [63], [113]; in a rare case, however, the virus was found to be able to cause the death of the mice following IC inoculation [113]. In line with these previous results, we also found that none of the 3-week-old ICR mice inoculated IM or IP with up to ∼105 PFU of SA14-14-2 showed clinical signs or death; on the other hand, although there was some variability among the groups of mice and the doses of virus inoculum, ∼5–30% of the mice inoculated IC with a dose of 103–105 PFU developed JEV-specific symptoms and death. This low but unexpected morbidity and mortality after the IC inoculation of SA14-14-2 is likely caused by a combination of factors and conditions imposed on our infection experiments, particularly the age and strain of mice: (i) Age-dependent susceptibility of flaviviruses, including JEV, in the murine model has been documented previously, although its molecular mechanisms remain unclear [106], [114], [115]. (ii) A noticeable variability in mortality has also been reported when two different lineages of the age-matched outbred ICR mice are inoculated IC with a mutant of ChimeriVax-JE that contains two amino acid substitutions (F107L and K138E) in the SA14-14-2 E protein-coding region, suggesting that differences in the genetic background of mice may account for the variable neurovirulence [107]. More importantly, it should be pointed out that in our study, all the revertants recovered from the mice inoculated IC with SA14-14-2 appeared to have the G244E mutation, which is sufficient to confer lethal neurovirulence to the virus, corroborating that the parental SA14-14-2 virus is highly attenuated in neurovirulence. Furthermore, it is intriguing to note that the G244E substitution has been introduced into ChimeriVax-JE, in which no mortality occurs when eight 4-week-old ICR mice are injected IC with 104 PFU of the mutant virus [107]; therefore, it appears that neurovirulence may depend on the genetic background of the pathogen. Further investigation is needed to fully elucidate the neurovirulence and neuroinvasiveness of JEV.
In summary, we show for the first time that E-244 in the ij hairpin of the viral E DII is a key regulator determining the neurovirulence of SA14-14-2, and we also provide direct evidence that viral E can contribute to the neurovirulence of JEV and possibly other closely related encephalitic flaviviruses via its role in the early or late stage of viral replication in neurons. A detailed, complete understanding of the evolutionally conserved viral ij hairpin and its function in the virus life cycle will have direct application to the design of a novel and promising class of broad-spectrum antivirals (e.g., ligands and small molecules) to expand the currently available preventive and therapeutic arsenal against infection with encephalitic flaviviruses.
Materials and Methods
Viruses and cells
An original stock of JEV SA14-14-2 was retrieved directly from a batch of commercial vaccine vials (Chengdu Institute of Biological Products, China) for viral genome sequencing and cDNA construction, to avoid any potential concern that its adaptation could occur during propagation in cell culture. This virus stock was propagated twice in BHK-21 cells to generate high-titer viral preparations for cell and mouse infection experiments. Stocks of JEV CNU/LP2 were derived from the infectious cDNA pBACSP6/JVFLx/XbaI [78]. BHK-21 cells were grown in alpha minimal essential medium (α-MEM) containing 10% fetal bovine serum (FBS), 2 mM L-glutamine, vitamins, and penicillin-streptomycin at 37°C in 5% CO2 [78]. SH-SY5Y cells were cultivated in a 1∶1 mixture of MEM and Ham's F-12 nutrient mix supplemented with 10% FBS, 0.1 mM nonessential amino acids, and penicillin-streptomycin at 37°C in 5% CO2 [75]. NSC-34 cells were maintained in Dulbecco's modified Eagle's medium containing 10% FBS and penicillin-streptomycin at 37°C in 5% CO2.
JEV reverse genetics
As a vector, we used the BAC plasmid pBeloBAC11 [78]. First, four cDNA fragments covering the entire viral genome were cloned into the vector individually, then joined sequentially at three natural restriction sites (BsrGI, BamHI, and AvaI) to generate a single BAC clone that contained the full-length SA14-14-2 cDNA, named pBAC/SA14-14-2 (Fig. 1). The SP6 promoter sequence was positioned just upstream of the viral 5′-end, and an artificial XbaI run-off site was engineered just downstream of the viral 3′-end. A pre-existing XbaI site at nucleotide 9131 was removed by introducing a silent point mutation (A9134→T); this mutation also served as a rescue marker to identify the cDNA-derived SA14-14-2. All mutations were created by overlap extension PCR. All PCR-generated fragments were sequenced. Detailed cloning procedures are described in Supporting Information.
RNA transcription and transfection
All BAC plasmids were purified by centrifugation using CsCl-ethidium bromide equilibrium density gradients. The closed circular plasmids were linearized by XbaI and mung bean nuclease digestion to produce DNA templates for in vitro run-off transcription. RNA was transcribed from a linearized plasmid with SP6 RNA polymerase as described [78]. The resulting RNA was stored at −80°C until needed. RNA yield was measured on the basis of the incorporation rate of [3H]UTP, and RNA integrity was evaluated by agarose gel electrophoresis. RNA was transfected by electroporation into cells under our optimized conditions (980 V, a 99-µs pulse length, and five pulses for BHK-21 cells; and 760 V, a 99-µs pulse length, and five pulses for NSC-34 and SH-SY5Y cells) [78]. RNA infectivity was determined by infectious center assay as reported [78]. The infectious centers of foci were detected by decorating of cells with a mouse α-JEV antibody (American Type Culture Collection [ATCC], 1∶500) and a horseradish peroxidase-conjugated goat α-mouse IgG (Jackson ImmunoResearch, 1∶1,000), followed by staining with 3,3′-diaminobenzidine (Vector).
Northern blots
Total RNA was extracted with TRIzol reagent (Invitrogen). Northern blot analysis was performed as described [78]. JEV genomic RNA was detected with an antisense riboprobe that binds to a 209-bp region (nt 9143–9351) in the NS5 protein-coding region. The probe was synthesized with [α-32P]CTP by using the T7-MEGAscript kit (Ambion). The blots were prehybridized, hybridized, and washed at 55°C. Autoradiographs were obtained by exposure to film for 24–48 h.
Immunoblots
Cells were lysed in sample buffer (80 mM Tri-HCl [pH 6.8], 2.0% SDS, 10% glycerol, 0.1 M dithiothreitol, 0.2% bromophenol blue). Equal amounts of the lysates were run on SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membranes, and subjected to immunoblotting as described [78]. The following polyclonal antisera were used as primary antibodies [75], [76]: α-JEV (mouse, 1∶1,000), α-C (rabbit, 1∶1,000), α-pr (rabbit, 1∶4,000), α-E (rabbit, 1∶500), α-NS1 (rabbit, 1∶1,000), and α-GAPDH (rabbit, 1∶10,000). An alkaline phosphatase-conjugated goat α-mouse or α-rabbit IgG (Jackson ImmunoResearch, 1∶5,000) was used for the secondary antibody, as appropriate. The specific signals were visualized by chromogenic membrane staining with a mixture of 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium (Sigma-Aldrich).
Flow cytometry
Cells (5×105) were harvested by trypsinization, washed with phosphate-buffered saline (PBS), and collected by centrifugation at 1,000×g for 5 min. The cells were resuspended in 250 µl of Cytofix/Cytoperm solution (BD Biosciences) and incubated at 4°C for 20 min in the dark. All subsequent wash and staining steps were performed in Perm/Wash buffer (BD Biosciences). The cells were washed twice and incubated in 200 µl of mouse α-JEV antiserum (ATCC, 1∶500) for 1 h at 4°C. Subsequently, the cells were washed twice and incubated in 200 µl of Alexa Fluor 488 goat α-mouse IgG (Molecular Probes, 1∶1,000) for 1 h at 4°C. The cells were then washed twice and resuspended in 200 µl of Perm/Wash buffer. The samples were analyzed on a FACSAriaIII cell sorter with Diva 6.1.3 software (BD Biosciences). For each sample, 50,000 events were collected within the linear range of detection.
Sequence analysis
The full genome sequences of SA14-14-2 and its neurovirulent variants were determined as described [77]. Sequencing of the prM-E coding region of the E-244 mutants was done as follows: (i) amplification of a 2,069-bp cDNA by RT-PCR using a set of three primers (prMErt, prMEfw, and prMErv; see Table S4); (ii) cloning of a 2,057-bp XhoI-SacII fragment into the pRS2 vector; and (iii) sequencing of ∼30 randomly picked independent clones containing the insert. Multiple sequence alignments were performed using ClustalX [116].
Mouse infection
Female 3-week-old ICR mice (Charles River) were used. Groups of 10 or 20 mice were inoculated IC (20 µl), IM (50 µl), or IP (50 µl) with 10-fold serial dilutions of virus stock in α-MEM. Mice were monitored for any JEV-induced clinical signs or death every 12 h for 24 days. The LD50 values were determined as described [75], [77]. In all mice, viral replication in brain tissue was confirmed by plaque titration and/or RT-PCR [75].
Ethics statement
All animal studies were conducted in strict accordance with the regulations in the Guide for the Care and Use of Laboratory Animals issued by the Ministry of Health and Welfare of the Republic of Korea. The protocol was approved by the Institutional Animal Care and Use Committee of the Chungbuk National University Medical School (Permit Number: LML08-73). All mice were housed in our animal facility located at the Chungbuk National University Medical School, and every effort was made to minimize suffering.
Immunohistochemistry
Groups of 3-week-old female ICR mice (n = 15 per group) were infected IC with 103 PFU of virus in 20 µl of α-MEM; 10 control mice were inoculated IC with an equivalent volume of supernatant from uninfected control BHK-21 cell cultures at comparable dilution. At 3, 5, and 7 dpi, five randomly selected mice were transcardially perfused with ice-cold PBS, followed by 4% paraformaldehyde (PFA). Brains were fixed in 4% PFA, embedded in paraffin, and cut into 6-µm sections. Brain sections were treated in microwave for antigen retrieval and incubated with 1% H2O2 in ice-cold methanol for 30 min to block endogenous peroxidase. They were then blocked with 1% normal goat serum and incubated with rabbit α-NS1 antiserum (1∶200) for 12 h at 4°C, followed by incubation with biotinylated α-rabbit IgG plus the avidin-biotin-peroxidase complex (Vector). Signals were visualized by staining with 3,3′-diaminobenzidine solution containing 0.003% H2O2 and counterstaining with hematoxylin.
Homology modeling
The sequence and structure of the E ectodomain of WNV NY99 (PDB accession code 2HG0) was used as template for the homology modeling. The sequence alignment was done using the online version of ClustalW2 [117]. Protein structure homology modeling was performed using the SWISS-MODEL Workspace, accessible via the ExPASy web server [118]. The generated model was visualized using UCSF Chimera 1.5.3. The model is in agreement with a recent crystal structure of the E ectodomain of JEV SA14-14-2 (PDB accession code 3P54) [80].
Supporting Information
Zdroje
1. EndyTP, NisalakA (2002) Japanese encephalitis virus: ecology and epidemiology. Curr Top Microbiol Immunol 267 : 11–48.
2. MackenzieJS, BarrettAD, DeubelV (2002) The Japanese encephalitis serological group of flaviviruses: a brief introduction to the group. Curr Top Microbiol Immunol 267 : 1–10.
3. MackenzieJS, GublerDJ, PetersenLR (2004) Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med 10: S98–109.
4. MackenzieJS, JohansenCA, RitchieSA, van den HurkAF, HallRA (2002) Japanese encephalitis as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia. Curr Top Microbiol Immunol 267 : 49–73.
5. GublerDJ (2007) The continuing spread of West Nile virus in the western hemisphere. Clin Infect Dis 45 : 1039–1046.
6. NashD, MostashariF, FineA, MillerJ, O'LearyD, et al. (2001) The outbreak of West Nile virus infection in the New York City area in 1999. N Engl J Med 344 : 1807–1814.
7. WeaverSC, BarrettAD (2004) Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol 2 : 789–801.
8. NettRJ, CampbellGL, ReisenWK (2009) Potential for the emergence of Japanese encephalitis virus in California. Vector Borne Zoonotic Dis 9 : 511–517.
9. WeaverSC, ReisenWK (2010) Present and future arboviral threats. Antiviral Res 85 : 328–345.
10. Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, et al.. (2011) Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ 89: : 766–774, 774A–774E.
11. MonathTP (2002) Japanese encephalitis vaccines: current vaccines and future prospects. Curr Top Microbiol Immunol 267 : 105–138.
12. SolomonT, VaughnDW (2002) Pathogenesis and clinical features of Japanese encephalitis and West Nile virus infections. Curr Top Microbiol Immunol 267 : 171–194.
13. SolomonT (2006) Control of Japanese encephalitis-within our grasp? N Engl J Med 355 : 869–871.
14. MisraUK, KalitaJ (2010) Overview: Japanese encephalitis. Prog Neurobiol 91 : 108–120.
15. DoklandT, WalshM, MackenzieJM, KhromykhAA, EeKH, et al. (2004) West Nile virus core protein: tetramer structure and ribbon formation. Structure 12 : 1157–1163.
16. MaL, JonesCT, GroeschTD, KuhnRJ, PostCB (2004) Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci U S A 101 : 3414–3419.
17. KuhnRJ, ZhangW, RossmannMG, PletnevSV, CorverJ, et al. (2002) Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108 : 717–725.
18. MukhopadhyayS, KimBS, ChipmanPR, RossmannMG, KuhnRJ (2003) Structure of West Nile virus. Science 302 : 248.
19. ZhangW, ChipmanPR, CorverJ, JohnsonPR, ZhangY, et al. (2003) Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol 10 : 907–912.
20. ChenY, MaguireT, HilemanRE, FrommJR, EskoJD, et al. (1997) Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med 3 : 866–871.
21. DavisCW, NguyenHY, HannaSL, SanchezMD, DomsRW, et al. (2006) West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J Virol 80 : 1290–1301.
22. Navarro-SanchezE, AltmeyerR, AmaraA, SchwartzO, FieschiF, et al. (2003) Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep 4 : 723–728.
23. PokidyshevaE, ZhangY, BattistiAJ, Bator-KellyCM, ChipmanPR, et al. (2006) Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124 : 485–493.
24. TassaneetrithepB, BurgessTH, Granelli-PipernoA, TrumpfhellerC, FinkeJ, et al. (2003) DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med 197 : 823–829.
25. ChuJJ, NgML (2004) Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J Virol 78 : 10543–10555.
26. van der SchaarHM, RustMJ, WaartsBL, van der Ende-MetselaarH, KuhnRJ, et al. (2007) Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J Virol 81 : 12019–12028.
27. van der SchaarHM, RustMJ, ChenC, van der Ende-MetselaarH, WilschutJ, et al. (2008) Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog 4: e1000244.
28. BressanelliS, StiasnyK, AllisonSL, SturaEA, DuquerroyS, et al. (2004) Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J 23 : 728–738.
29. LiaoM, Sanchez-San MartinC, ZhengA, KielianM (2010) In vitro reconstitution reveals key intermediate states of trimer formation by the dengue virus membrane fusion protein. J Virol 84 : 5730–5740.
30. ModisY, OgataS, ClementsD, HarrisonSC (2004) Structure of the dengue virus envelope protein after membrane fusion. Nature 427 : 313–319.
31. NayakV, DessauM, KuceraK, AnthonyK, LedizetM, et al. (2009) Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J Virol 83 : 4338–4344.
32. StiasnyK, KosslC, LepaultJ, ReyFA, HeinzFX (2007) Characterization of a structural intermediate of flavivirus membrane fusion. PLoS Pathog 3: e20.
33. HarrisonSC (2008) Viral membrane fusion. Nat Struct Mol Biol 15 : 690–698.
34. Lindenbach BD, Thiel HJ, Rice CM (2007) Flaviviridae: the viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA et al.., editors. Fields virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins Publishers. pp. 1101–1152.
35. BrintonMA (2002) The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu Rev Microbiol 56 : 371–402.
36. ParanjapeSM, HarrisE (2010) Control of dengue virus translation and replication. Curr Top Microbiol Immunol 338 : 15–34.
37. VillordoSM, GamarnikAV (2009) Genome cyclization as strategy for flavivirus RNA replication. Virus Res 139 : 230–239.
38. WestawayEG, MackenzieJM, KhromykhAA (2002) Replication and gene function in Kunjin virus. Curr Top Microbiol Immunol 267 : 323–351.
39. GillespieLK, HoenenA, MorganG, MackenzieJM (2010) The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84 : 10438–10447.
40. HsuNY, IlnytskaO, BelovG, SantianaM, ChenYH, et al. (2010) Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141 : 799–811.
41. WelschS, MillerS, Romero-BreyI, MerzA, BleckCK, et al. (2009) Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5 : 365–375.
42. LiL, LokSM, YuIM, ZhangY, KuhnRJ, et al. (2008) The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science 319 : 1830–1834.
43. ZhangY, CorverJ, ChipmanPR, ZhangW, PletnevSV, et al. (2003) Structures of immature flavivirus particles. EMBO J 22 : 2604–2613.
44. LorenzIC, AllisonSL, HeinzFX, HeleniusA (2002) Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J Virol 76 : 5480–5491.
45. StadlerK, AllisonSL, SchalichJ, HeinzFX (1997) Proteolytic activation of tick-borne encephalitis virus by furin. J Virol 71 : 8475–8481.
46. YuIM, ZhangW, HoldawayHA, LiL, KostyuchenkoVA, et al. (2008) Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319 : 1834–1837.
47. ZhangY, ZhangW, OgataS, ClementsD, StraussJH, et al. (2004) Conformational changes of the flavivirus E glycoprotein. Structure 12 : 1607–1618.
48. Gubler DJ, Kuno G, Markoff L (2007) Flaviviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA et al.., editors. Fields virology. 5th ed. Philadelphia, PA: Lippincott Williams & Wilkins Publishers. pp. 1153–1252.
49. BeasleyDW, LewthwaiteP, SolomonT (2008) Current use and development of vaccines for Japanese encephalitis. Expert Opin Biol Ther 8 : 95–106.
50. Wilder-SmithA, HalsteadSB (2010) Japanese encephalitis: update on vaccines and vaccine recommendations. Curr Opin Infect Dis 23 : 426–431.
51. FischerM, LindseyN, StaplesJE, HillsS (2010) Centers for Disease Control and Prevention (2010) Japanese encephalitis vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 59 : 1–27.
52. PlesnerAM (2003) Allergic reactions to Japanese encephalitis vaccine. Immunol Allergy Clin North Am 23 : 665–697.
53. YuY (2010) Phenotypic and genotypic characteristics of Japanese encephalitis attenuated live vaccine virus SA14-14-2 and their stabilities. Vaccine 28 : 3635–3641.
54. HalsteadSB, ThomasSJ (2010) Japanese encephalitis: new options for active immunization. Clin Infect Dis 50 : 1155–1164.
55. JelinekT (2009) Ixiaro: a new vaccine against Japanese encephalitis. Expert Rev Vaccines 8 : 1501–1511.
56. KollaritschH, Paulke-KorinekM, Dubischar-KastnerK (2009) IC51 Japanese encephalitis vaccine. Expert Opin Biol Ther 9 : 921–931.
57. Centers for Disease Control and Prevention (2011) Recommendations for use of a booster dose of inactivated vero cell culture-derived Japanese encephalitis vaccine: advisory committee on immunization practices, 2011. MMWR Morb Mortal Wkly Rep 60 : 661–663.
58. Centers for Disease Control and Prevention (2011) Update on Japanese encephalitis vaccine for children: United States, May 2011. MMWR Morb Mortal Wkly Rep 60 : 664–665.
59. ChambersTJ, NestorowiczA, MasonPW, RiceCM (1999) Yellow fever/Japanese encephalitis chimeric viruses: construction and biological properties. J Virol 73 : 3095–3101.
60. MonathTP, LevenbookI, SoikeK, ZhangZX, RatterreeM, et al. (2000) Chimeric yellow fever virus 17D-Japanese encephalitis virus vaccine: dose-response effectiveness and extended safety testing in rhesus monkeys. J Virol 74 : 1742–1751.
61. HalsteadSB, ThomasSJ (2011) New Japanese encephalitis vaccines: alternatives to production in mouse brain. Expert Rev Vaccines 10 : 355–364.
62. AppaiahgariMB, VratiS (2012) Clinical development of IMOJEV (R)-a recombinant Japanese encephalitis chimeric vaccine (JE-CV). Expert Opin Biol Ther 12 : 1251–1263.
63. AiharaS, RaoCM, YuYX, LeeT, WatanabeK, et al. (1991) Identification of mutations that occurred on the genome of Japanese encephalitis virus during the attenuation process. Virus Genes 5 : 95–109.
64. NiH, ChangGJ, XieH, TrentDW, BarrettAD (1995) Molecular basis of attenuation of neurovirulence of wild-type Japanese encephalitis virus strain SA14. J Gen Virol 76 : 409–413.
65. NitayaphanS, GrantJA, ChangGJ, TrentDW (1990) Nucleotide sequence of the virulent SA-14 strain of Japanese encephalitis virus and its attenuated vaccine derivative, SA-14-14-2. Virology 177 : 541–552.
66. CeciliaD, GouldEA (1991) Nucleotide changes responsible for loss of neuroinvasiveness in Japanese encephalitis virus neutralization-resistant mutants. Virology 181 : 70–77.
67. ChambersTJ, DrollDA, JiangX, WoldWS, NickellsJA (2007) JE Nakayama/JE SA14-14-2 virus structural region intertypic viruses: biological properties in the mouse model of neuroinvasive disease. Virology 366 : 51–61.
68. HasegawaH, YoshidaM, ShiosakaT, FujitaS, KobayashiY (1992) Mutations in the envelope protein of Japanese encephalitis virus affect entry into cultured cells and virulence in mice. Virology 191 : 158–165.
69. LeeE, HallRA, LobigsM (2004) Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J Virol 78 : 8271–8280.
70. NiH, BarrettAD (1996) Molecular differences between wild-type Japanese encephalitis virus strains of high and low mouse neuroinvasiveness. J Gen Virol 77 : 1449–1455.
71. NiH, BarrettAD (1998) Attenuation of Japanese encephalitis virus by selection of its mouse brain membrane receptor preparation escape variants. Virology 241 : 30–36.
72. ShahPS, TanakaM, KhanAH, MathengeEG, FukeI, et al. (2006) Molecular characterization of attenuated Japanese encephalitis live vaccine strain ML-17. Vaccine 24 : 402–411.
73. SumiyoshiH, TignorGH, ShopeRE (1995) Characterization of a highly attenuated Japanese encephalitis virus generated from molecularly cloned cDNA. J Infect Dis 171 : 1144–1151.
74. WuSC, LinCW, LeeSC, LianWC (2003) Phenotypic and genotypic characterization of the neurovirulence and neuroinvasiveness of a large-plaque attenuated Japanese encephalitis virus isolate. Microbes Infect 5 : 475–480.
75. KimJM, YunSI, SongBH, HahnYS, LeeCH, et al. (2008) A single N-linked glycosylation site in the Japanese encephalitis virus prM protein is critical for cell type-specific prM protein biogenesis, virus particle release, and pathogenicity in mice. J Virol 82 : 7846–7862.
76. YunSI, ChoiYJ, SongBH, LeeYM (2009) 3' cis-acting elements that contribute to the competence and efficiency of Japanese encephalitis virus genome replication: functional importance of sequence duplications, deletions, and substitutions. J Virol 83 : 7909–7930.
77. SongBH, YunGN, KimJK, YunSI, LeeYM (2012) Biological and genetic properties of SA14-14-2, a live-attenuated Japanese encephalitis vaccine that is currently available for humans. J Microbiol 50 : 698–706.
78. YunSI, KimSY, RiceCM, LeeYM (2003) Development and application of a reverse genetics system for Japanese encephalitis virus. J Virol 77 : 6450–6465.
79. NybakkenGE, NelsonCA, ChenBR, DiamondMS, FremontDH (2006) Crystal structure of the West Nile virus envelope glycoprotein. J Virol 80 : 11467–11474.
80. LucaVC, AbiMansourJ, NelsonCA, FremontDH (2012) Crystal structure of the Japanese encephalitis virus envelope protein. J Virol 86 : 2337–2346.
81. NiH, BurnsNJ, ChangGJ, ZhangMJ, WillsMR, et al. (1994) Comparison of nucleotide and deduced amino acid sequence of the 5' non-coding region and structural protein genes of the wild-type Japanese encephalitis virus strain SA14 and its attenuated vaccine derivatives. J Gen Virol 75 : 1505–1510.
82. World Health Organization (2005) Global advisory committee on vaccine safety, 9–10 June 2005. Wkly Epidemiol Rec 80 : 242–247.
83. MukhopadhyayS, KuhnRJ, RossmannMG (2005) A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3 : 13–22.
84. StiasnyK, HeinzFX (2006) Flavivirus membrane fusion. J Gen Virol 87 : 2755–2766.
85. ReyFA, HeinzFX, MandlC, KunzC, HarrisonSC (1995) The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375 : 291–298.
86. AllisonSL, SchalichJ, StiasnyK, MandlCW, HeinzFX (2001) Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol 75 : 4268–4275.
87. LeeE, LobigsM (2002) Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. J Virol 76 : 4901–4911.
88. LeeJW, ChuJJ, NgML (2006) Quantifying the specific binding between West Nile virus envelope domain III protein and the cellular receptor αVβ3 integrin. J Biol Chem 281 : 1352–1360.
89. BeasleyDW, BarrettAD (2002) Identification of neutralizing epitopes within structural domain III of the West Nile virus envelope protein. J Virol 76 : 13097–13100.
90. CrillWD, RoehrigJT (2001) Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol 75 : 7769–7773.
91. KaufmannB, NybakkenGE, ChipmanPR, ZhangW, DiamondMS, et al. (2006) West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc Natl Acad Sci U S A 103 : 12400–12404.
92. WuKP, WuCW, TsaoYP, KuoTW, LouYC, et al. (2003) Structural basis of a flavivirus recognized by its neutralizing antibody: solution structure of the domain III of the Japanese encephalitis virus envelope protein. J Biol Chem 278 : 46007–46013.
93. StiasnyK, BressanelliS, LepaultJ, ReyFA, HeinzFX (2004) Characterization of a membrane-associated trimeric low-pH-induced form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystallization. J Virol 78 : 3178–3183.
94. ReyFA (2003) Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry. Proc Natl Acad Sci U S A 100 : 6899–6901.
95. KaufmannB, ChipmanPR, HoldawayHA, JohnsonS, FremontDH, et al. (2009) Capturing a flavivirus pre-fusion intermediate. PLoS Pathog 5: e1000672.
96. LinSR, ZouG, HsiehSC, QingM, TsaiWY, et al. (2011) The helical domains of the stem region of dengue virus envelope protein are involved in both virus assembly and entry. J Virol 85 : 5159–5171.
97. PangerlK, HeinzFX, StiasnyK (2011) Mutational analysis of the zippering reaction during flavivirus membrane fusion. J Virol 85 : 8495–8501.
98. SchmidtAG, YangPL, HarrisonSC (2010) Peptide inhibitors of dengue virus entry target a late-stage fusion intermediate. PLoS Pathog 6: e1000851.
99. ChuJJ, NgML (2004) Interaction of West Nile virus with αvβ3 integrin mediates virus entry into cells. J Biol Chem 279 : 54533–54541.
100. HurrelbrinkRJ, McMinnPC (2001) Attenuation of Murray Valley encephalitis virus by site-directed mutagenesis of the hinge and putative receptor-binding regions of the envelope protein. J Virol 75 : 7692–7702.
101. LeeE, LobigsM (2000) Substitutions at the putative receptor-binding site of an encephalitic flavivirus alter virulence and host cell tropism and reveal a role for glycosaminoglycans in entry. J Virol 74 : 8867–8875.
102. GotoA, YoshiiK, ObaraM, UekiT, MizutaniT, et al. (2005) Role of the N-linked glycans of the prM and E envelope proteins in tick-borne encephalitis virus particle secretion. Vaccine 23 : 3043–3052.
103. HannaSL, PiersonTC, SanchezMD, AhmedAA, MurtadhaMM, et al. (2005) N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J Virol 79 : 13262–13274.
104. LorenzIC, KartenbeckJ, MezzacasaA, AllisonSL, HeinzFX, et al. (2003) Intracellular assembly and secretion of recombinant subviral particles from tick-borne encephalitis virus. J Virol 77 : 4370–4382.
105. TajimaS, NeromeR, NukuiY, KatoF, TakasakiT, et al. (2010) A single mutation in the Japanese encephalitis virus E protein (S123R) increases its growth rate in mouse neuroblastoma cells and its pathogenicity in mice. Virology 396 : 298–304.
106. MonathTP, ArroyoJ, LevenbookI, ZhangZX, CatalanJ, et al. (2002) Single mutation in the flavivirus envelope protein hinge region increases neurovirulence for mice and monkeys but decreases viscerotropism for monkeys: relevance to development and safety testing of live, attenuated vaccines. J Virol 76 : 1932–1943.
107. ArroyoJ, GuirakhooF, FennerS, ZhangZX, MonathTP, et al. (2001) Molecular basis for attenuation of neurovirulence of a yellow fever virus/Japanese encephalitis virus chimera vaccine (ChimeriVax-JE). J Virol 75 : 934–942.
108. ChenLK, LinYL, LiaoCL, LinCG, HuangYL, et al. (1996) Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology 223 : 79–88.
109. ZhaoZ, DateT, LiY, KatoT, MiyamotoM, et al. (2005) Characterization of the E-138 (Glu/Lys) mutation in Japanese encephalitis virus by using a stable, full-length, infectious cDNA clone. J Gen Virol 86 : 2209–2220.
110. MelianEB, HinzmanE, NagasakiT, FirthAE, WillsNM, et al. (2010) NS1' of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J Virol 84 : 1641–1647.
111. FirthAE, AtkinsJF (2009) A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1' may derive from ribosomal frameshifting. Virol J 6 : 14.
112. YeQ, LiXF, ZhaoH, LiSH, DengYQ, et al. (2012) A single nucleotide mutation in NS2A of Japanese encephalitis-live vaccine virus (SA14-14-2) ablates NS1' formation and contributes to attenuation. J Gen Virol 93 : 1959–1964.
113. EckelsKH, YuYX, DuboisDR, MarchetteNJ, TrentDW, et al. (1988) Japanese encephalitis virus live-attenuated vaccine, Chinese strain SA14-14-2; adaptation to primary canine kidney cell cultures and preparation of a vaccine for human use. Vaccine 6 : 513–518.
114. SigelMM (1952) Influence of age on susceptibility to virus infections with particular reference to laboratory animals. Annu Rev Microbiol 6 : 247–280.
115. Monath TP (1986) Pathobiology of the flaviviruses. In: Schlesinger S, Schlesinger MJ, editors. The Togaviridae and Flaviviridae. New York, NY: Plenum. pp. 375–440.
116. ThompsonJD, GibsonTJ, PlewniakF, JeanmouginF, HigginsDG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 : 4876–4882.
117. GoujonM, McWilliamH, LiW, ValentinF, SquizzatoS, et al. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38: W695–699.
118. KieferF, ArnoldK, KunzliM, BordoliL, SchwedeT (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37: D387–392.
119. ReedLJ, MuenchH (1938) A simple method of estimating fifty percent endpoints. Am J Hyg 27 : 493–497.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDSČlánek The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV InfectionČlánek Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 7- Stillova choroba: vzácné a závažné systémové onemocnění
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Bacteriophages as Vehicles for Antibiotic Resistance Genes in the Environment
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
- Defensins and Viral Infection: Dispelling Common Misconceptions
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- The Wide World of Ribosomally Encoded Bacterial Peptides
- Microbial Egress: A Hitchhiker's Guide to Freedom
- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- HIV-1 Capture and Transmission by Dendritic Cells: The Role of Viral Glycolipids and the Cellular Receptor Siglec-1
- Tetherin Can Restrict Cell-Free and Cell-Cell Transmission of HIV from Primary Macrophages to T Cells
- The Frustrated Host Response to Is Bypassed by MyD88-Dependent Translation of Pro-inflammatory Cytokines
- Larger Mammalian Body Size Leads to Lower Retroviral Activity
- The Semen Microbiome and Its Relationship with Local Immunology and Viral Load in HIV Infection
- Lytic Gene Expression Is Frequent in HSV-1 Latent Infection and Correlates with the Engagement of a Cell-Intrinsic Transcriptional Response
- Phase Variation of Poly-N-Acetylglucosamine Expression in
- A Screen of Mutants Reveals Important Roles for Dot/Icm Effectors and Host Autophagy in Vacuole Biogenesis
- Structure of the Trehalose-6-phosphate Phosphatase from Reveals Key Design Principles for Anthelmintic Drugs
- The Impact of Juvenile Coxsackievirus Infection on Cardiac Progenitor Cells and Postnatal Heart Development
- Vertical Transmission Selects for Reduced Virulence in a Plant Virus and for Increased Resistance in the Host
- Characterization of the Largest Effector Gene Cluster of
- Novel Drosophila Viruses Encode Host-Specific Suppressors of RNAi
- Pto Kinase Binds Two Domains of AvrPtoB and Its Proximity to the Effector E3 Ligase Determines if It Evades Degradation and Activates Plant Immunity
- Genetic Analysis of Tropism Using a Naturally Attenuated Cutaneous Strain
- Plasmacytoid Dendritic Cells Suppress HIV-1 Replication but Contribute to HIV-1 Induced Immunopathogenesis in Humanized Mice
- A Novel Mouse Model of Gastroenteritis Reveals Key Pro-inflammatory and Tissue Protective Roles for Toll-like Receptor Signaling during Infection
- Pathogenicity of Is Expressed by Regulating Metabolic Thresholds of the Host Macrophage
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Independent Bottlenecks Characterize Colonization of Systemic Compartments and Gut Lymphoid Tissue by
- Peptidoglycan Recognition Proteins Kill Bacteria by Inducing Oxidative, Thiol, and Metal Stress
- G3BP1, G3BP2 and CAPRIN1 Are Required for Translation of Interferon Stimulated mRNAs and Are Targeted by a Dengue Virus Non-coding RNA
- Cytolethal Distending Toxins Require Components of the ER-Associated Degradation Pathway for Host Cell Entry
- The Machinery at Endoplasmic Reticulum-Plasma Membrane Contact Sites Contributes to Spatial Regulation of Multiple Effector Proteins
- Arabidopsis LIP5, a Positive Regulator of Multivesicular Body Biogenesis, Is a Critical Target of Pathogen-Responsive MAPK Cascade in Plant Basal Defense
- Plant Surface Cues Prime for Biotrophic Development
- Real-Time Imaging Reveals the Dynamics of Leukocyte Behaviour during Experimental Cerebral Malaria Pathogenesis
- The CD27L and CTP1L Endolysins Targeting Contain a Built-in Trigger and Release Factor
- cGMP and NHR Signaling Co-regulate Expression of Insulin-Like Peptides and Developmental Activation of Infective Larvae in
- Systemic Hematogenous Maintenance of Memory Inflation by MCMV Infection
- Strain-Specific Variation of the Decorin-Binding Adhesin DbpA Influences the Tissue Tropism of the Lyme Disease Spirochete
- Distinct Lipid A Moieties Contribute to Pathogen-Induced Site-Specific Vascular Inflammation
- Serovar Typhi Conceals the Invasion-Associated Type Three Secretion System from the Innate Immune System by Gene Regulation
- LANA Binds to Multiple Active Viral and Cellular Promoters and Associates with the H3K4Methyltransferase hSET1 Complex
- A Molecularly Cloned, Live-Attenuated Japanese Encephalitis Vaccine SA-14-2 Virus: A Conserved Single Amino Acid in the Hairpin of the Viral E Glycoprotein Determines Neurovirulence in Mice
- Illuminating Fungal Infections with Bioluminescence
- Comparative Genomics of Plant Fungal Pathogens: The - Paradigm
- Motility and Chemotaxis Mediate the Preferential Colonization of Gastric Injury Sites by
- Widespread Sequence Variations in VAMP1 across Vertebrates Suggest a Potential Selective Pressure from Botulinum Neurotoxins
- An Immunity-Triggering Effector from the Barley Smut Fungus Resides in an Ustilaginaceae-Specific Cluster Bearing Signs of Transposable Element-Assisted Evolution
- Establishment of Murine Gammaherpesvirus Latency in B Cells Is Not a Stochastic Event
- Oncogenic Herpesvirus KSHV Hijacks BMP-Smad1-Id Signaling to Promote Tumorigenesis
- Human APOBEC3 Induced Mutation of Human Immunodeficiency Virus Type-1 Contributes to Adaptation and Evolution in Natural Infection
- Innate Immune Responses and Rapid Control of Inflammation in African Green Monkeys Treated or Not with Interferon-Alpha during Primary SIVagm Infection
- Chitin-Degrading Protein CBP49 Is a Key Virulence Factor in American Foulbrood of Honey Bees
- Influenza A Virus Host Shutoff Disables Antiviral Stress-Induced Translation Arrest
- Nsp9 and Nsp10 Contribute to the Fatal Virulence of Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus Emerging in China
- Pulmonary Infection with Hypervirulent Mycobacteria Reveals a Crucial Role for the P2X7 Receptor in Aggressive Forms of Tuberculosis
- Syk Signaling in Dendritic Cells Orchestrates Innate Resistance to Systemic Fungal Infection
- A Repetitive DNA Element Regulates Expression of the Sialic Acid Binding Adhesin by a Rheostat-like Mechanism
- T-bet and Eomes Are Differentially Linked to the Exhausted Phenotype of CD8+ T Cells in HIV Infection
- Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health
- Influence of ND10 Components on Epigenetic Determinants of Early KSHV Latency Establishment
- Antibody to gp41 MPER Alters Functional Properties of HIV-1 Env without Complete Neutralization
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Molecular and Cellular Mechanisms of KSHV Oncogenesis of Kaposi's Sarcoma Associated with HIV/AIDS
- Holobiont–Holobiont Interactions: Redefining Host–Parasite Interactions
- BCKDH: The Missing Link in Apicomplexan Mitochondrial Metabolism Is Required for Full Virulence of and
- Helminth Infections, Type-2 Immune Response, and Metabolic Syndrome
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



