-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Conservative Sex and the Benefits of Transformation in
Natural transformation has significant effects on bacterial genome evolution, but the evolutionary factors maintaining this mode of bacterial sex remain uncertain. Transformation is hypothesized to have both positive and negative evolutionary effects on bacteria. It can facilitate adaptation by combining beneficial mutations into a single individual, or reduce the mutational load by exposing deleterious alleles to natural selection. Alternatively, it may expose transformed cells to damaged or otherwise mutated environmental DNA and is energetically expensive. Here, we examine the long-term effects of transformation in the naturally competent species Streptococcus pneumoniae by evolving populations of wild-type and competence-deficient strains in chemostats for 1000 generations. Half of these populations were exposed to periodic mild stress to examine context-dependent benefits of transformation. We find that competence reduces fitness gain under benign conditions; however, these costs are reduced in the presence of periodic stress. Using whole genome re-sequencing, we show that competent populations fix fewer new mutations and that competence prevents the emergence of mutators. Our results show that during evolution in benign conditions competence helps maintain genome stability but is evolutionary costly; however, during periods of stress this same conservativism enables cells to retain fitness in the face of new mutations, showing for the first time that the benefits of transformation are context dependent.
Published in the journal: . PLoS Pathog 9(11): e32767. doi:10.1371/journal.ppat.1003758
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003758Summary
Natural transformation has significant effects on bacterial genome evolution, but the evolutionary factors maintaining this mode of bacterial sex remain uncertain. Transformation is hypothesized to have both positive and negative evolutionary effects on bacteria. It can facilitate adaptation by combining beneficial mutations into a single individual, or reduce the mutational load by exposing deleterious alleles to natural selection. Alternatively, it may expose transformed cells to damaged or otherwise mutated environmental DNA and is energetically expensive. Here, we examine the long-term effects of transformation in the naturally competent species Streptococcus pneumoniae by evolving populations of wild-type and competence-deficient strains in chemostats for 1000 generations. Half of these populations were exposed to periodic mild stress to examine context-dependent benefits of transformation. We find that competence reduces fitness gain under benign conditions; however, these costs are reduced in the presence of periodic stress. Using whole genome re-sequencing, we show that competent populations fix fewer new mutations and that competence prevents the emergence of mutators. Our results show that during evolution in benign conditions competence helps maintain genome stability but is evolutionary costly; however, during periods of stress this same conservativism enables cells to retain fitness in the face of new mutations, showing for the first time that the benefits of transformation are context dependent.
Introduction
Natural transformation is an important cause of genome evolution in bacteria, but the evolutionary factors maintaining natural transformation, or competence, in bacteria remain uncertain [1], [2], [3], [4]. Transformation is widely believed to have evolved to facilitate adaptation, especially in a clinical context where transformation occurs at a high rate and may allow pathogens to evade antibiotics or immune surveillance [2], [5], [6], [7]. However, transformation can both be beneficial and costly to bacterial cells. It can speed up adaptation by combining non-antagonistically epistatic beneficial mutations into a single individual [8], [9], [10], [11], similar to the Fisher-Muller effect in Eukaryotes. It can also reduce the mutation load by combining deleterious alleles into a common background, which more efficiently exposes these mutations to natural selection [12], [13], [14], [15]. Similarly, transformation can eliminate deleterious alleles if these are replaced by transformed DNA with the wild-type sequence [12]. Such a function has been inferred in naturally transforming Neisseria, where high rates of transformation with ‘self-DNA’ leads to conservation of core regions of the genome [16]. Alternatively, transformation can impose significant fitness costs because it is energetically expensive and bacterial cells may incorporate damaged or mutated environmental DNA that reduces bacterial fitness [12]. Thus, although the mechanisms that regulate bacterial transformation are well understood, the evolutionary factors that maintain this process are not.
Experiments designed to quantify the evolutionary effects of bacterial transformation have been equivocal. While Baltrus et al. [17] showed that transformable strains of Helicobactor pylori evolved more rapidly than non-transformable strains, Bacher et al. [18] found the reverse in Acinetobacter baylyi. Indeed, wild-type strains in this experiment evolved lower rates of transformation during laboratory culture. Although these experimental differences may have been caused by details particular to the species investigated, they may have also been caused by experimental approaches that inadvertently exposed different benefits or costs of competence. For example, studying yeast, Saccharomyces cerivisiae, Gray and Goddard [19] showed that sex only increased adaptation in a stressful environment, and that this was especially pronounced in populations of cells with a high mutation rate. Similar studies designed to partition the effects of these experimental factors have thus far not been attempted using bacteria.
Here, we use an experimental evolution approach to examine the long-term effects of transformation on the naturally competent opportunistic pathogen Streptococcus pneumoniae. Replicate cultures of a competent strain and an isogenic mutant unable to become competent were evolved for 1000 generations in two different chemostat environments. Half of the populations were evolved in a constant benign environment, while the other half was exposed to twice-weekly pulses of a sub-minimum inhibitory concentration (MIC) of kanamycin. This treatment was used to examine context-dependent effects of transformation. Although the concentration of kanamycin we used did not influence the growth rate of cells in our experimental populations (Figure S1), it is sufficient to induce cellular stress and also induce natural competence [20]. In brief, we found that competent cells evolved in a benign environment increased in fitness less than non-competent cells; however, this cost of competence was alleviated when cells evolved in an environment where they were challenged with periodic stress. Using whole genome sequencing of evolved isolates, we show that non-competent populations fixed more mutations than competent ones and are furthermore were more likely to evolve to become mutators. Our results provide direct experimental evidence that the effects of transformation are context dependent. In benign conditions the effects of transformation are conservative and evolutionarily costly; however, this same conservativism benefits cells living in stressful environments.
Results
Conditional effects of competence
To quantify the effects of competence on the evolution of S. pneumoniae we evolved replicate populations of a competent wild-type and an isogenic non-competent mutant in chemostats for 1000 generations. We introduced periodic stress in half of the evolving populations by applying sub-MIC concentrations of kanamycin to chemostat vessels twice each week. Kanamycin was added as a single injection to the chemostat sampling port in order to achieve a final concentration of 5 ug/mL, which is approximately 20-fold below the MIC for this strain. At the chemostat flow rates used, the kanamycin was eliminated within 7 hours. For the majority of time, these populations therefore experienced the same environment as the unstressed populations. At the start of this experiment, all ancestral populations exhibited pronounced oscillations of several orders of magnitude in cell density (between 109/ml and 105/ml), as previously described with this species [21]. However, within 500 generations these fluctuations were uniformly lost [22], and all populations retained a stable density of 109 cells/ml. As a consequence of this change, which made it impossible to directly compete evolved and ancestral cells, fitness differences between treatments after experimental evolution were estimated using pair-wise competition assays between differentially marked terminal populations [23]. This allowed us to directly estimate fitness differences between evolved strains, and to quantify the relative fitness of competent and non-competent populations under benign and periodically stressed conditions.
In contrast to the expectation that competence accelerates adaptation, we found instead that evolved competent populations of S. pneumoniae were significantly less fit than non-competent populations (Fig. 1, restricted maximum likelihood (REML) mixed model compared to 0: t = −2.22; p<0.001). This evolutionary cost of competence corresponds to a fitness difference between evolved populations of ∼0.098/hr, which implies that under benign conditions the effects of competence on adaptation are significantly negative. By contrast, the relative fitness of competent and non-competent populations evolved in the presence of periodic stress was indistinguishable (Fig. 1, REML mixed model compared to 0: t = −0.18; p = 0.741) indicating that these conditions significantly offset the evolutionary costs of competence (REML mixed model: F = 2.077; p = 0.017).
Fig. 1. Fitness differences between experimental treatments estimated from direct competition assays between evolved populations. 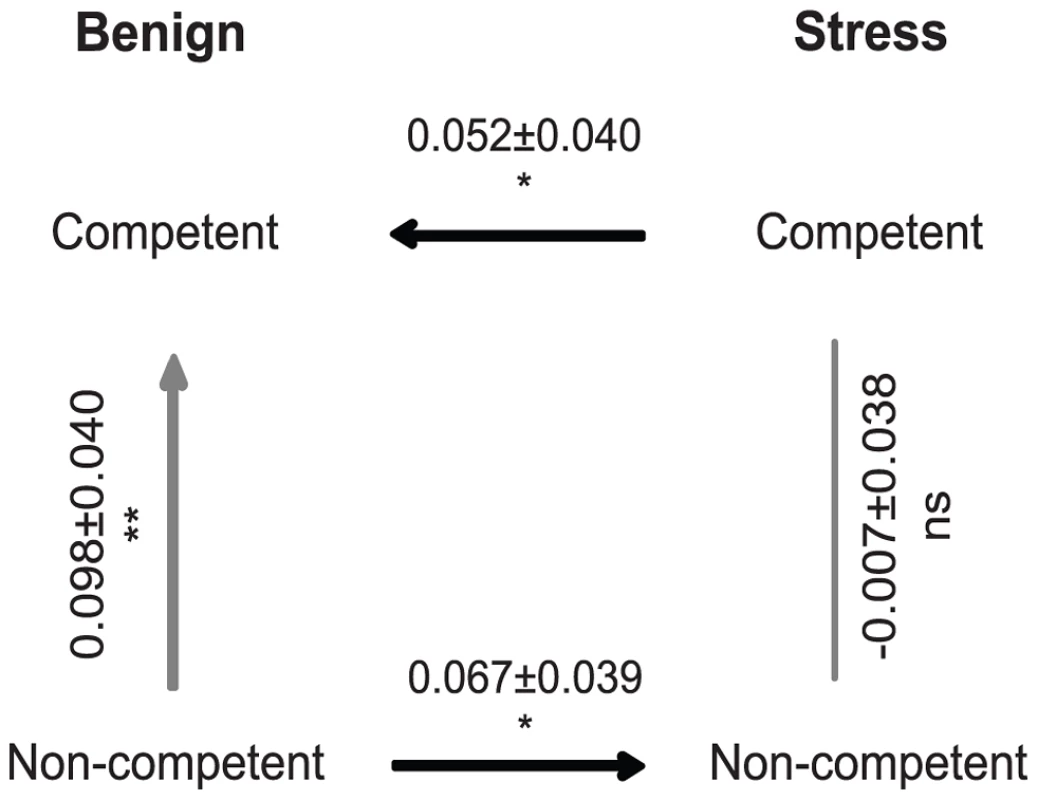
Values represent the mean selection rate constant/hour (± standard error). Ns: non-significant; *: P<0.05; **: P<0.01 compared to the null expectation of 0 (i.e. equal fitness). Exposure to periodic stress could offset the costs of competence by increasing adaptation of competent cells and/or by reducing the rate of adaptation of non-competent cells. The results from competition assays support both explanations. When we estimated the relative fitness of stressed versus unstressed competent cells we found that stressed populations were significantly more fit (∼0.052/hr) than populations evolved in an unstressed environment (Fig. 1, REML mixed model compared to 0: t = 1.27; one-tailed p = 0.045). By contrast, we found the reverse relationship for non-competent cells, and of roughly equal magnitude; stressed non-competent populations were significantly less fit (∼0.067/hr) than populations evolved in an unstressed environment (Fig. 1, REML mixed model compared to 0: t = −1.67; one-tailed p = 0.012). The sum of these two effects on the rate of adaptation, i.e. the benefit for stressed competent populations and the cost for stressed non-competent populations, is similar to the ∼0.098/hr cost for competent cells noted above (Fig. 1). The benefits to competence of stress therefore arise in two ways: by increasing adaptation of competent cells and by enabling competent cells to avoid fitness reductions attributable to exposure to stress.
Genomic effects of competence
To analyze the influence of competence and stress on genome evolution during experimental evolution we obtained the complete genome sequences of evolved clones from each of the 16 populations together with their respective ancestors. Competence could potentially alter mutation fixation in two ways, both caused by the fact that competence unites mutations from separate cells into a common genetic background. First, if recombined mutations are beneficial either alone or in combination, competence could increase the fixation rate because recombinant cells would be predicted to increase in frequency. Second, if recombined mutations are deleterious, alone or in combination, competence could reduce the fixation rate because recombinant cells would be exposed to natural selection and eliminated from the experimental population. A similar reduction in fixation rate would be anticipated if competence replaces new mutations in the host genome with donor DNA containing the wild-type allele, thereby “correcting” mutations. We furthermore predict that sub-MIC antibiotic stress will have a general increase on mutation fixation, owing to potentially mutagenic effects of kanamycin [24]. Sequencing of evolved genomes identified a total of 421 synonymous mutations and 1282 non-synonymous mutations across all evolved lines. Substitutions were not evenly distributed across treatments and populations, however. Consistent with the second possibility outlined above, we found that the total number of mutations in competent populations was significantly lower than in non-competent populations (Fig. 2A; GLM: z = −9.344, df = 1 p<0.0001). Moreover, the total number of mutations was significantly higher in populations experiencing periodic stress for both competent and non-competent populations (Fig. 2A; GLM: z = 7.379, df = 1 p<0.0001).
Fig. 2. Number of mutations and mutation rates of evolved populations. 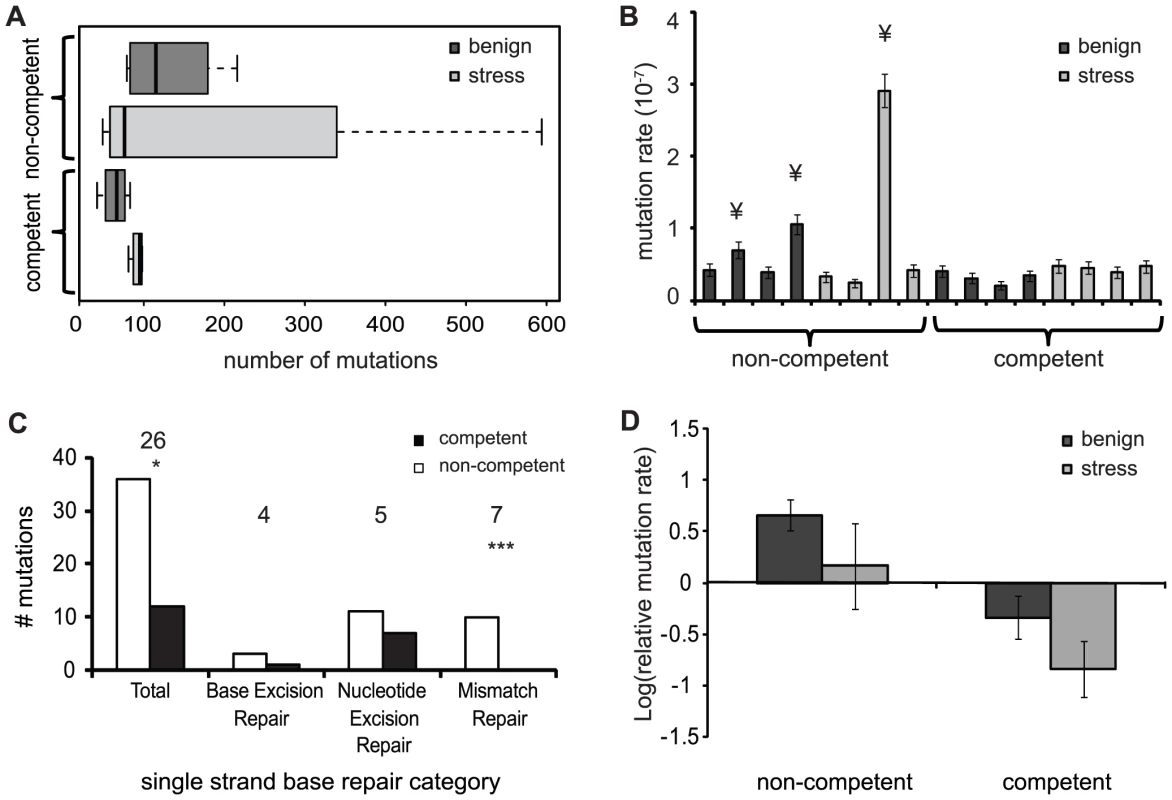
Dark grey are populations evolved in the benign environment and light grey are the populations evolved in the periodic stress environment. A) The average number of mutations found per evolved population in each treatment. B) Mutation rate for each evolved population calculated from the total number of mutations. ¥ indicates populations with a mutation rate of 2–10-fold above average. Error bars are 95% confidence intervals C) Mutations found in genes for DNA repair for non-competent (white) and competent (black) populations. Numbers above each category indicate the number of mutated genes in that category. * Indicates significant difference between competent and non-competent populations. D) Mean of log-transformed relative mutation rates of the evolved lines compared to the ancestor. Error bars are SE of the mean. We next determined the mutation rate of each population based on the total number of mutations. This method assumes that the mutation rate was constant over the period of evolution and that there are no back mutations. We found that three of eight non-competent populations substituted between two and eight-fold more mutations than the other populations (Fig. 2B), suggesting that these lineages had evolved to become genetic mutators. However, because this estimate assumes a constant mutation rate during the 1000 generations of experimental evolution, rather than a changing rate due to the fixation of mutator alleles, mutators emerging at the end of experimental evolution may not be identified. To address this limitation, we first searched for mutations in genes diagnostic for bacterial mutators (e.g. DNA-repair genes polC, dnaQ, rpoABC and mutL [25], see Table S1 for a comprehensive list). We focused here on non-synonymous mutations because these are more likely to cause functional defects in the relevant genes. Next we determined the mutation rate of all evolved lineages relative to their respective ancestor using a phenotypic assay designed to detect the frequency of spontaneous mutants to resistance to either rifampicin or streptomycin [26]. Using the first approach, we detected significantly more non-synonymous mutations in DNA repair genes in non-competent than in competent populations (Fig. 2C: GLM one-tailed: χ2 = 162.00, df = 1, p = 0.0478), an effect that is most pronounced in genes for mismatch repair (Fig. 2B; GLM: Mismatch repair: χ2 = 9.875, df = 1, p = 0.001). Next, using a phenotypic assay, we found that the mutation frequency of non-competent lineages was significantly higher than competent populations (Fig. 2D; Two-way ANOVA: F1,11 = 10.189; p = 0.009), although the mutation frequency of ancestral strains were indistinguishable (t-test with unequal variances: t = 1.1336, df = 7.112, p = 0.2937). In contrast to re-sequencing results, these assays found no overall effect of stress on the mutation frequency (Fig. 2D; Two-way ANOVA: F1,11 = 2.699; p = 0.129), nor an interaction between stress and competence (Fig. 2C; Two-way ANOVA: F1,11 = 0.0003; p = 0.9876). Thus both at the genetic and phenotypic levels, our data support a model where competence reduces mutation fixation and limits the emergence of mutator phenotypes, but this conservatism comes possibly at the expense of reduced adaptation under benign growth conditions.
Discussion
Transformation can dramatically benefit S. pneumoniae by facilitating the evolution of drug resistance and the emergence of novel modes of virulence [27], [28], [29]. However, these benefits in pathogenic bacterial lineages under strong antibiotic selection tell only part of the story, and may not reflect the effects of transformation more broadly. Using an experimental evolution approach, we found that competence benefited cells by reducing the mutation load and limiting the emergence of mutators (Fig. 2). Additionally, competent populations reached higher fitness when evolving in the presence of periodic stress; equally, exposure to periodic stress decreased the rate of evolution of non-competent populations (Fig. 1). Although we applied an extremely mild stress in our experiment (Figure S1), it is notable that the kanamycin concentration we used is sufficient to induce competence in wild-type strains [20]. It is therefore possible that benefits to competence in populations that experienced drug-stress was the result of increased recombination, which could have off-set the cost of transformation in a benign environment by slightly increasing their rate of adaptation. By contrast, non-competent cells exposed to kanamycin may face greater costs because kanamycin causes an inability to repair ribosomal decoding errors, which can subsequently lead to DNA damage and increase the mutation rate [24]. These stress-dependent benefits of competence may be particularly important in the human nasopharynx, where S. pneumoniae is exposed to unpredictable and severe stress from drug exposure, immune surveillance and from coexisting bacterial competitors.
Transformation is predicted to benefit bacterial species with high mutation rates by reducing their mutation load [12]. Using complete genome sequences, we estimate that the average mutation rate in S. pneumoniae is ∼3.8×10−8 per bp per generation. This corresponds to U = 0.08 mutations per genome per generation, or about 200-fold higher than Escherichia coli [30], yet similar to other naturally transformable opportunistic pathogens, such as H. pylori and H. influenzae [30], [31], [32], [33]. Despite these high rates of mutation we were surprised to find that some of the non-competent strains evolved even higher rates of mutation than their ancestor during this long-term experiment (Figs. 2A and 2B). These genotypic results were confirmed phenotypically (Fig. 2D), and suggest that our genomic data underestimates the difference in fixation rates in competent and non-competent populations. Although we are uncertain what caused the difference in mutation rates between competent and non-competent lineages to arise, one strong possibility is that transformation separates mutator alleles from the mutations they cause. Thus while mutations in DNA repair genes leading to mutators may arise equally in both competent and non-competent cells, they are lost before they become common in competent lineages [34]. Accordingly, competent lineages fix fewer mutations overall. Under benign conditions this may limit adaptation while causing minimal harm to non-competent populations. However, non-competent cells suffer to a greater degree when faced with stress, because they cannot revert to a less loaded state, and because stress may exacerbate the negative fitness effects of new mutations [35], [36]. In a similar recent study with the yeast Saccharomyces cerevisiae sex neutralised the deleterious effects of hyper mutation on the rate of adaptation [19]. The neutralisation of potentially deleterious mutations, e.g. those that lead to hyper mutation, is an example of how transformation can function to conserve genome integrity. Similar effects are inferred in the naturally transformable bacterial genus Neisseria where the number of species-specific DNA uptake sequences (i.e. small sequence tags that identify that a DNA fragment is derived from a particular species) are more frequent in the core genome. This indicates that these core genes are often the target of ‘selfing’ events, by which. transformation stabilizes the integrity of key genes [16]. Although S. pneumoniae is much more promiscuous than Neisseria when it comes to environmental DNA choice, the mechanism that induces competence is assumed to lead to the uptake of DNA released from lysed cells of the same and/or very closely related species [37], which suggests that the conservative benefits we observe from competence in our study may extend more broadly to pneumococci in the natural environment.
Bacteria in nature face unpredictable patterns of stress and mutation. Our results suggest that these conditions, together with an intrinsically high mutation rate, favour the maintenance of transformation while infrequent stress may facilitate its loss. Notably, surveys of naturally competent species such as H. influenzae and B. subtilis have revealed that transformation rates among clones within species can vary by over 6 orders of magnitude [38], [39], [40]. Similar variation exists in S. pneumoniae [41], [42], [43], indicating that competence is readily gained and lost in this species. In summary, we conclude that competence in S. pneumoniae is a conservative process acting to preserve alleles, rather than an innovative one that persists because of benefits it provides by recombining beneficial mutations.
Materials and Methods
Strains, culture conditions and chemostats
Strains used in this study were derived from Rx1 and its isogenic non-competent derivative FP5, which is unable to secrete the competence stimulating peptide, CSP [42]. Spontaneous rifampicin or streptomycin resistant mutants were isolated from each strain, and then four independent colonies of each type were further sub-cloned and stored at −80°C. These four independent ancestors were selected based on similar growth rates (Figure S2) and mutation rates (mean rates ±95% CI Non-competent: 1.84*10−7±1.56*10−7, competent: 8.41*10−8±7.51*10−8; t-test with unequal variances: t = 1.1336, df = 7.112, p = 0.2937). These 16 total clones (2 strains×2 drug resistance types×4 replicates) represented the ancestral populations for experimental evolution. Cultures were grown in ¼ CTM pH 7.8 (Complete Transformation Medium), which per litre consists of: 7.5 g Tryptic Soy Broth; 0.25 g yeast extract; 6 g NaCl at pH 7.8. This environment supported high levels of transformation (Figure S3). Blood agar plates (Tryptic Soy Agar (TSA)+3% horse blood), supplemented, where necessary, with either 4 µg/mL rifampicin or 100 µg/mL streptomycin, were used to enumerate cell density within chemostats and for colony counting during competition assays.
Experimental evolution and competition assays were performed in custom-made chemostats with a 25 mL working volume and a flow rate of 4 mL/hr, whilst maintained at 37°C [29]. Chemostat cultures were inoculated and maintained as described previously [21], and sampled every 50 generations of growth. Samples were stored at −80°C as freezer stocks in ¼ CTM pH 7.8+25% v/v glycerol at an OD600 of 0.20, corresponding to a density of 2×108 cells mL−1.
Long-term evolution experiment
Sixteen chemostat populations were inoculated with independently picked clones from the original antibiotic resistant strain to generate 4 replicates each of a 2*2 treatment design pairing competence and stress. The replicates in each treatment were equally split between the two differently marked versions of Rx1 (competent strain) and Fp5 (non-competent strain). Half of the populations were exposed twice a week to low doses of kanamycin introduced directly into the chemostat to simulate short periods of stress. Kanamycin concentrations were 5 µg/mL upon introduction, but declined with the normal outflow rate of the chemostat. This concentration of kanamycin had no effect on the growth rate of cells (Figure S1), but is sufficient to cause ribosomal decoding errors during protein production, which promotes the induction of competence [20], [44]. For simplicity, this treatment is referred to as “periodic stress” and the basal treatment as “benign”. Each strain was evolved independently, thereby avoiding potential effects of cross-induction of competence or competence-induced cell-lysis [45], [46]. Every week, after approximately 50 generations, a 1 mL sample was taken from each population and tested for the presence of the correct marker and absence of the opposite marker. Contaminated populations were restarted with 50 µL of the previous sampled uncontaminated time point. Populations were maintained for 20 weeks, which corresponds to about 1,000 generations.
Fitness assays
Fitness was determined by comparing the change in relative densities of two reciprocally marked evolved populations in a chemostat in mixed culture over a 32-hour span. This time period was chosen because it is within the period that the periodically stressed populations spend in the benign environment between doses of kanamycin. Competition assays were initiated by inoculating chemostats with equal densities of each competitor. Chemostats were sampled immediately and then again after 32 hours to determine the relative densities of each competitor. The Malthusian parameters per hour were then calculated for each strain based on the density of each strain at the start and end of the competition, as described previously [47]. The selection rate constant was then calculated as the difference between Malthusian parameters as described previously [23]. First, we tested for a significant fitness difference between competitors for each treatment by comparing a restricted maximum likelihood mixed model against an intercept of zero, corresponding to equal fitness. In the mixed model, replicate fitness assays of competitor combinations were nested as a random effect within the fixed effect of treatments (absence/presence periodic stress and absence/presence competence). Second, we used the restricted maximum likelihood (REML) mixed model, again with replicate fitness assays as a random factor within treatments, to test for fitness differences between treatments (periodic stress or competence as a fixed factor). All analyses were done in R with package LME4. P-values were estimated by MCMC simulation with 10,000-fold replication using the p.vals command from the languageR package.
DNA isolation and sequencing
Clonal isolates from each of the 16 evolved populations as well as all four ancestral strains were sequenced using the SOLiD4 platform at the University of Manchester genomics facility. Genomic DNA was obtained using phenol-chloroform isolation and ethanol precipitation [48]. SOLiD data were normalised to an equal number of reads (8,315,863 per strain) for each sample using a custom perl script, that randomly sampled the reads from the original dataset (getRandomTags_Index_fastq.pl) developed by I. Donaldson. The normalisation equalized the size of the datasets to the strain with the lowest number of reads thereby normalising the quality of the consensus sequences. The normalised reads were then mapped against the fully sequenced reference strain S. pneumoniae R6 (genbank accession: NC_003098 = AE007317, an easily accessible version of the genome database can be found at http://www.streppneumoniae.com) using BFAST (0.6.4e) using default colour space methodology giving an average coverage depth of 151-fold. Mapped reads were then locally realigned around INDELs using SRMA (0.1.15). SNPs and small INDELs were then determined from the resulting BAM-files using the Geneious package (Geneious 5.4, Auckland, New Zealand; [49]. A SNP or INDEL was called when the change to the reference was supported in 60% of the reads with at least a coverage depth of 20 reads using the variant calling tool in Geneious 5.4 to minimise false positives and negative SNP calls. The 60% support is slightly less stringent than previous studies, but this is compensated by the high average coverage depth of 151 (±SE 2.53) reads/base [50], [51]. Subsequently, variant tables extracted from Geneious were used in the Galaxy online tool set [52], [53] to identify mutations for each evolved clone compared to its ancestor. Parallel changes were then double checked by hand in the UCSC microbial genome browser [54] to eliminate false positives. The resulting mutation tables were used for further analysis.
Mutations and mutation rate
To determine the effect of periodic stress and competence the total numbers of mutations were compared in a GLM model with a Poisson distribution using R. Subsequently, two different methods were used to determine the mutation rate of evolved populations. First, the per base per generation mutation rate was calculated from genome data from the total number of mutations (See Table S2 for mutation rates based on coding mutations and synonymous mutations only); 95% confidence intervals for these mutation rates were determined in R using a Poisson distribution. This analysis assumes that the number of mutations is relatively small, that there are no back mutations, and that the mutation rate was constant over the period of evolution.
Second, the mutation frequency of terminal lineages (i.e. the number of spontaneous mutants with either rifampicin or streptomycin resistance/total population density) was determined following the methods in [26] to estimate the mutation rate of terminal populations isolated after their final generation of experimental evolution. Each strain and its corresponding ancestor was grown overnight at 37°C+5% CO2. Cells were then washed and concentrated by centrifugation and re-suspended in 100 µL 0.8% NaCl. 10 µL spots at several different dilutions (five spots per dilution) were plated onto blood agar plates supplemented with either 4 µg/mL rifampicin or 100 µg/mL streptomycin, whilst total cell densities were determined on unsupplemented plates. Mutation frequency was estimated as the ratio of the number of mutants to the total population size. Assays were performed in triplicate for each genotype, and the relative mutation frequency was determined as the ratio of the mutation frequency of each evolved lineage to its corresponding ancestor. Mean relative mutation frequencies were log-transformed before analysis using a two-way ANOVA.
Finally, parallel changes in DNA repair genes were examined at the level of each gene and functional group, as determined from the KEGG classifications for S. pneumoniae R6 (http://www.genome.jp/dbget-bin/www_bget?gn:T00060). The table of non-synonymous SNPs was used to create a SNP-by-gene table by scoring presence/absence of at least one SNP in a given gene for each strain (see Table S1 for a detailed summary of the genes involved and the amino acid changes found per evolved line). Functional group associations were created from the total SNP-by-gene table by summarising presence and absence of SNPs for genes associated with a functional group according to the KEGG-database. Generalised linear models were used to test for differences between treatments for parallel non-synonymous mutations in functional groups.
Supporting Information
Zdroje
1. OchmanH, LawrenceJG, GroismanEA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405 : 299–304.
2. VosM (2009) Why do bacteria engage in homologous recombination? Trends in Microbiology 17 : 226–232.
3. RedfieldRJ (2001) Do bacteria have sex? Nature Reviews: Genetics 2 : 634–639.
4. MichodRE, BernsteinH, NedelcuAM (2008) Adaptive value of sex in microbial pathogens. Infection, Genetics and Evolution 8 : 267–285.
5. EshelI, FeldmanMW (1970) On the evolutionary effect of recombination. Theoretical Population Biology 1 : 88–100.
6. LevinBR (1981) Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics 99 : 1–23.
7. GogartenJP, DoolittleWF, LawrenceJG (2002) Prokaryotic evolution in light of gene transfer. Molecular Biology and Evolution 19 : 2226–2238.
8. MullerHJ (1932) Some genetic aspects of sex. American Naturalist 66 : 118–138.
9. Fisher RA (1930) The genetical theory of natural selection. Oxford: Oxford University Press.
10. FlynnKM, CooperTF, MooreFBG, CooperVS (2013) The environment affects epistatic interactions to alter the topology of an empirical fitness landscape. PLoS Genetics 9: e1003426.
11. KhanAI, DinhDM, SchneiderD, LenskiRE, CooperTF (2011) Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332 : 1193–1196.
12. RedfieldRJ (1988) Evolution of bacterial transformation: is sex with dead cells ever better than no sex at all? Genetics 119 : 213–221.
13. HaighJ (1978) The accumulation of deleterious genes in a population–Muller's Ratchet. Theoretical Population Biology 14 : 251–267.
14. MullerHJ (1964) The relation of recombination to mutational advance. Mutation Research 106 : 2–9.
15. KondrashovAS (1988) Deleterious mutations and the evolution of sexual reproduction. Nature 336 : 435–440.
16. TreangenTJ, AmburOH, TonjumT, RochaEP (2008) The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome biology 9: R60.
17. BaltrusDA, GuilleminK, PhillipsPC (2008) Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62 : 39–49.
18. BacherJM, MetzgarD, de Crecy-LagardV (2006) Rapid evolution of diminished transformability in Acinetobacter baylyi. Journal of Bacteriology 188 : 8534–8542.
19. GrayJC, GoddardMR (2012) Sex enhances adaptation by unlinking beneficial from detrimental mutations in experimental yeast populations. BMC Evolutionary Biology 12 : 43.
20. PrudhommeM, AttaiechL, SanchezG, MartinB, ClaverysJP (2006) Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313 : 89–92.
21. CornejoOE, RozenDE, MayRM, LevinBR (2008) Oscillations in continuous culture populations of Streptococcus pneumoniae: population dynamics and the evolution of clonal suicide. Proceedings of the Royal Society of London, Series B: Biological Sciences 276 : 999–1008.
22. EngelmoerDJP (2012) The evolution of natural competence in Streptococcus pneumoniae. Faculty of Life Sciences PhD 152.
23. TravisanoM, LenskiRE (1996) Long-term experimental evolution in Escherichia coli .4. Targets of selection and the specificity of adaptation. Genetics 143 : 15–26.
24. CharpentierX, PolardP, ClaverysJP (2012) Induction of competence for genetic transformation by antibiotics: convergent evolution of stress responses in distant bacterial species lacking SOS? Current Opinion in Microbiology 15 : 570–576.
25. HorstJP, WuTH, MarinusMG (1999) Escherichia coli mutator genes. Trends in Microbiology 7 : 29–36.
26. BjedovI, TenaillonO, GerardB, SouzaV, DenamurE, et al. (2003) Stress-induced mutagenesis in bacteria. Science 300 : 1404–1409.
27. HillerNL, AhmedA, PowellE, MartinDP, EutseyR, et al. (2010) Generation of Genic Diversity among Streptococcus pneumoniae Strains via Horizontal Gene Transfer during a Chronic Polyclonal Pediatric Infection. Plos Pathogens 6: e1001108.
28. DowsonCG, HutchisonA, BranniganJA, GeorgeRC, HansmanD, et al. (1989) Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proceedings of the National Academy of Sciences of the United States of America 86 : 8842–8846.
29. CoffeyTJ, EnrightMC, DanielsM, MoronaJK, MoronaR, et al. (1998) Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Molecular Microbiology 27 : 73–83.
30. WielgossS, BarrickJE, TenaillonO, CruveillerS, Chane-Woon-MingB, et al. (2011) Mutation rate inferred form synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3: Genes, Genomes, Genetics 1(3): 183–186.
31. BjorkholmB, SjolundM, FalkPG, BergOG, EngstrandL, et al. (2001) Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proceedings of the National Academy of Sciences of the United States of America 98 : 14607–14612.
32. OliverA, BaqueroF, CantonR, CampoP, BlazquezJ (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288 : 1251–1253.
33. WatsonME, SmithAL, BurnsJL (2004) Hypermutable Haemophilus influenzae with mutations in mutS are found in cystic fibrosis sputum. Microbiology-Sgm 150 : 2947–2958.
34. TenaillonO, Le NagardH, GodelleB, TaddeiF (2000) Mutators and sex in bacteria: Conflict between adaptive strategies. Proceedings of the National Academy of Sciences, USA 97 : 10465–10470.
35. CooperTF, LenskiRE, ElenaSF (2005) Parasites and mutational load: an experimental test of a pluralistic theory for the evolution of sex. Proceedings of the Royal Society of London Series B: Biological Sciences 272 : 311–317.
36. SzafraniecK, BortsRH, KoronaR (2001) Environmental stress and mutational load in diploid strains of the yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America 98 : 1107–1112.
37. MajewskiJ, ZawadzkiP, PickerillP, CohanFM, DowsonCG (2000) Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. Journal of Bacteriology 182 : 1016–1023.
38. MaughanH, RedfieldRJ (2009) Extensive variation in natural competence in Haemophilus influenzae. Evolution 63 : 1852–1866.
39. DuitmanEH, WyczawskiD, BovenLG, VenemaG, KuipersOP, et al. (2007) Novel methods for genetic transformation of natural Bacillus subtilis isolates used to study the regulation of the mycosubtilin and surfactin synthetases. Applied and Environmental Microbiology 73 : 3490–3496.
40. LiYH, LauPC, LeeJH, EllenRP, CvitkovitchDG (2001) Natural genetic transformation of Streptococcus mutans growing in biofilms. Journal of Bacteriology 183 : 897–908.
41. PozziG, MasalaL, IannelliF, ManganelliR, HavarsteinLS, et al. (1996) Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. Journal of Bacteriology 178 : 6087–6090.
42. IannelliF, OggioniMR, PozziG (2005) Sensor domain of histidine kinase ComD confers competence pherotype specificity in Streptoccoccus pneumoniae. FEMS Microbiology Letters 252 : 321–326.
43. EvansBA, RozenDE (2013) Significant variation in transformation frequency in Streptococcus pneumoniae. ISME Journal 7(4): 791–9.
44. StevensKE, ChangD, ZwackEE, SebertME (2011) Competence in Streptococcus pneumoniae is regulated by the rate of ribosomal decoding errors. MBio 2: e00071-11.
45. SteinmoenH, KnutsenE, HavarsteinLS (2002) Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proceedings of the National Academy of Sciences, USA 99 : 7681–7686.
46. SteinmoenH, TeigenA, HavarsteinLS (2003) Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. Journal of Bacteriology 185 : 7176–7183.
47. LenskiRE, RoseMR, SimpsonSC, TadlerSC (1991) long-term experimental evolution in Escherichia coli .1. Adaptation and divergence during 2,000 generations. American Naturalist 138 : 1315–1341.
48. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual. New York: Cold Spring Harbor Laboratory.
49. Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, et al.. (2011) Geneious v5.4. http://wwwgeneiouscom.
50. HarrisSR, FeilEJ, HoldenMTG, QuailMA, NickersonEK, et al. (2010) Evolution of MRSA during hospital transmission and intercontinental spread. Science 327 : 469–474.
51. CroucherNJ, HarrisSR, FraserC, QuailMA, BurtonJ, et al. (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331 : 430–434.
52. BlankenbergD, Von KusterG, CoraorN, AnandaG, LazarusR, et al. (2010) Galaxy: a web-based genome analysis tool for experimentalists. Current Protocols in Molecular Biology Chapter 19: Unit 19 10 11–21.
53. GoecksJ, NekrutenkoA, TaylorJ, TeamG (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology 11: R86.
54. SchneiderKL, PollardKS, BaertschR, PohlA, LoweTM (2006) The UCSC Archaeal Genome Browser. Nucleic Acids Research 34: D407–410.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Baculoviruses: Sophisticated Pathogens of Insects
- How Do Viruses Avoid Inhibition by Endogenous Cellular MicroRNAs?
- The Regulation of Trypanosome Gene Expression by RNA-Binding Proteins
- DNA Damage Repair and Bacterial Pathogens
- Disease to Dirt: The Biology of Microbial Amyloids
- Fungal Immune Evasion in a Model Host–Pathogen Interaction: Versus Macrophages
- Infectious Prions Accumulate to High Levels in Non Proliferative C2C12 Myotubes
- The Biology and Taxonomy of Head and Body Lice—Implications for Louse-Borne Disease Prevention
- Antibodies Trap Tissue Migrating Helminth Larvae and Prevent Tissue Damage by Driving IL-4Rα-Independent Alternative Differentiation of Macrophages
- The Effects of Somatic Hypermutation on Neutralization and Binding in the PGT121 Family of Broadly Neutralizing HIV Antibodies
- Natural Selection Promotes Antigenic Evolvability
- Type I Interferon Protects against Pneumococcal Invasive Disease by Inhibiting Bacterial Transmigration across the Lung
- Mode of Parainfluenza Virus Transmission Determines the Dynamics of Primary Infection and Protection from Reinfection
- Type I and Type III Interferons Drive Redundant Amplification Loops to Induce a Transcriptional Signature in Influenza-Infected Airway Epithelia
- Unraveling a Three-Step Spatiotemporal Mechanism of Triggering of Receptor-Induced Nipah Virus Fusion and Cell Entry
- A Novel Membrane Sensor Controls the Localization and ArfGEF Activity of Bacterial RalF
- Macrophage and T Cell Produced IL-10 Promotes Viral Chronicity
- Global Rescue of Defects in HIV-1 Envelope Glycoprotein Incorporation: Implications for Matrix Structure
- Turning Defense into Offense: Defensin Mimetics as Novel Antibiotics Targeting Lipid II
- The Neonatal Fc Receptor (FcRn) Enhances Human Immunodeficiency Virus Type 1 (HIV-1) Transcytosis across Epithelial Cells
- Brd4 Is Displaced from HPV Replication Factories as They Expand and Amplify Viral DNA
- A Viral Genome Landscape of RNA Polyadenylation from KSHV Latent to Lytic Infection
- The Cytotoxic Necrotizing Factor of (CNF) Enhances Inflammation and Yop Delivery during Infection by Activation of Rho GTPases
- The Inflammatory Kinase MAP4K4 Promotes Reactivation of Kaposi's Sarcoma Herpesvirus and Enhances the Invasiveness of Infected Endothelial Cells
- Conservative Sex and the Benefits of Transformation in
- Microbial Endocrinology in the Microbiome-Gut-Brain Axis: How Bacterial Production and Utilization of Neurochemicals Influence Behavior
- Colonization Resistance: Battle of the Bugs or Ménage à Trois with the Host?
- Intracellular Interferons in Fish: A Unique Means to Combat Viral Infection
- SPOC1-Mediated Antiviral Host Cell Response Is Antagonized Early in Human Adenovirus Type 5 Infection
- Involvement of the Cellular Phosphatase DUSP1 in Vaccinia Virus Infection
- Killer Bee Molecules: Antimicrobial Peptides as Effector Molecules to Target Sporogonic Stages of
- A Unique SUMO-2-Interacting Motif within LANA Is Essential for KSHV Latency
- A Role for Host Activation-Induced Cytidine Deaminase in Innate Immune Defense against KSHV
- Haploid Genetic Screens Identify an Essential Role for PLP2 in the Downregulation of Novel Plasma Membrane Targets by Viral E3 Ubiquitin Ligases
- A Small Molecule Glycosaminoglycan Mimetic Blocks Invasion of the Mosquito Midgut
- Identification of the Adenovirus E4orf4 Protein Binding Site on the B55α and Cdc55 Regulatory Subunits of PP2A: Implications for PP2A Function, Tumor Cell Killing and Viral Replication
- Can Non-lytic CD8+ T Cells Drive HIV-1 Escape?
- Deletion of the α-(1,3)-Glucan Synthase Genes Induces a Restructuring of the Conidial Cell Wall Responsible for the Avirulence of
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Baculoviruses: Sophisticated Pathogens of Insects
- Identification of the Adenovirus E4orf4 Protein Binding Site on the B55α and Cdc55 Regulatory Subunits of PP2A: Implications for PP2A Function, Tumor Cell Killing and Viral Replication
- A Unique SUMO-2-Interacting Motif within LANA Is Essential for KSHV Latency
- Natural Selection Promotes Antigenic Evolvability
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



