-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Egyptian H5N1 Influenza Viruses—Cause for Concern?
article has not abstract
Published in the journal: . PLoS Pathog 8(11): e32767. doi:10.1371/journal.ppat.1002932
Category: Opinion
doi: https://doi.org/10.1371/journal.ppat.1002932Summary
article has not abstract
Highly pathogenic avian H5N1 influenza viruses are now enzootic in parts of Southeast Asia and the Middle East. Occasionally, these viruses transmit to humans and cause severe respiratory disease and fatalities. Currently, these viruses are not efficiently transmitted from person to person, although limited human-to-human transmission may have occurred [1]–[4]. A major determinant of influenza virus host range is the viral hemagglutinin (HA) protein: avian virus HA binds preferentially to sialic acid linked to the penultimate galactose residue by an α2,3-linkage (Siaα2,3Gal) [5]–[7], as found for sialic acid–containing receptors of the epithelial cells in duck intestine [8], the site of avian influenza virus replication. By contrast, human virus HA has higher affinity for Siaα2,6Gal [5]–[7], the main sialyloligosaccharide on the epithelial cells of the human upper respiratory tract [9], [10].
Recently, Herfst et al. [11] and Imai et al. [12] identified H5 HA-possessing viruses that transmit via respiratory droplets among ferrets, an established animal model for influenza virus transmission studies. The H5N1 transmissible virus identified by Herfst et al. [11] possesses three mutations that were intentionally introduced (PB2-627K, which confers efficient replication in mammals [13], and HA-Q222L/G224S (H5 numbering), which confer human-type receptor-binding specificity [14], [15]). The “Herfst virus” also possessed two mutations that emerged during virus passages in ferrets. One of these, HA-T156A, results in the loss of a glycosylation site on the head of the HA; the other, HA-H103Y, localizes to the HA trimer interface. The transmissible virus identified by Imai et al. [12] possesses a mutant HA gene of an avian H5N1 virus and the remaining seven viral genes of a prototypic pandemic 2009 (H1N1) virus. Random mutagenesis of the HA globular head identified two mutations in HA (HA-N220K and HA-Q222L; note that the latter is identical to one of the mutations identified in the “Herfst virus”) that conferred human-type receptor-binding specificity. Virus passages in ferrets resulted in the selection of two additional mutations in HA. One of these, HA-N154D, resulted in the loss of the same glycosylation site as HA-T156A in the “Herfst virus”; the other, T314I, affected HA stability [12]).
Although the Herfst and Imai studies used different experimental strategies and tested viruses of HA/H5 clade 2 or 1, respectively, the results were remarkably similar: the transmissible mutant H5 viruses bound to human-type receptors, lost the glycosylation site at HA-154–156, and acquired an additional mutation in HA that likely increased the protein's stability. Moreover, both studies, and findings by others [16]–[18], suggest that a shift towards human-type receptor-binding specificity may be necessary, but not sufficient, for H5N1 virus transmissibility in mammals.
The loss of a glycosylation site at HA-154–156, using two different mutations, is particularly notable. Amino acids 154–156 of many H5 HAs encode an N-glycosylation site (N-X(except P)-S/T), which is located near the receptor-binding pocket. Loss of this glycosylation site enhances H5N1 virus binding to Siaα2,6Gal (conferred by the Q226L/G228 mutations [19]) and is critical for H5N1 virus transmissibility in guinea pigs [20]. In the Herfst and Imai studies, loss of this glycosylation site occurred during the first virus passages in ferrets, suggesting that this trait is essential for H5 virus transmissibility in ferrets.
Since lack of the HA154–156 glycosylation site appears to be critical for H5 virus transmission in mammals, we inspected avian H5N1 viruses for this feature. A phylogenetic tree of publicly available H5 HA sequences showed that a substantial number of these viruses, distributed across time and geography, lack this glycosylation site. Closer inspection of 2009–2011 H5N1 viruses from Vietnam, Indonesia, and Egypt (i.e., countries with appreciable numbers of human H5N1 infections) revealed that ∼25% of Vietnamese, 0% of Indonesian, but >70% of Egyptian isolates lack the HA154–156 glycosylation site. The H5N1 viruses currently circulating in Egypt are descendants of the so-called Qinghai Lake viruses that killed wild birds (which typically do not succumb to influenza virus infections) at Qinghai Lake, China, in 2005 [21]–[23], and have now spread through Europe to the Middle East and Africa.
Next, we looked for a correlation between the HA154–156 glycosylation site and recent human H5N1 virus infections. Human H5N1 infections in Vietnam and Indonesia from 2009 to 2011 were mostly (Vietnam) or exclusively (Indonesia) caused by viruses with the HA154–156 glycosylation site. However, all 46 H5N1 viruses isolated in 2009–2011 from infected individuals in Egypt lacked the HA154–156 glycosylation site, while 28% of H5N1 viruses circulating in avian species in Egypt in 2009–2011 possessed this site (Table 1). Phylogenetic analysis further suggested that mutations resulting in loss of the glycosylation site occurred in birds and that these variants subsequently transmitted to humans. Although speculative at this point, this finding might suggest that avian H5N1 viruses lacking the HA154–156 glycosylation site transmit to humans more readily than those that possess the glycosylation site, at least in the genetic background of Egyptian H5N1 viruses.
Tab. 1. HA154–156 glycosylation site in Egyptian H5N1 influenza viruses. 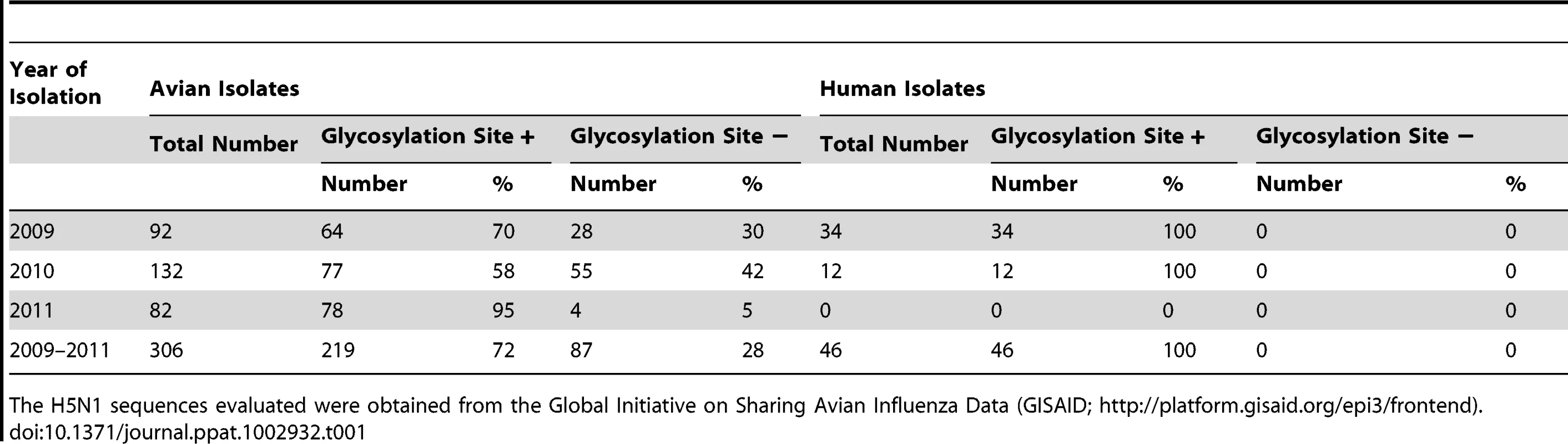
The H5N1 sequences evaluated were obtained from the Global Initiative on Sharing Avian Influenza Data (GISAID; http://platform.gisaid.org/epi3/frontend). In addition to mutations in HA, other amino acid changes may be critical to confer transmissibility in humans. One such mutation may be the glutamic acid-to-lysine mutation at position 627 of the PB2 polymerase protein (PB2-627K), which confers efficient replication in mammals [13] and is a recognized host determinant of influenza viruses [24]. Herfst introduced this mutation into their virus; the “Imai virus” possesses the 2009 pandemic PB2 gene, in which a basic amino acid at position 591 compensates for the lack of PB2-627K [25], [26]. The mammalian-adapting PB2-627K mutation also emerged in the Qinghai Lake viruses [21]–[23] and has been maintained in viruses of this lineage to this day, with the exception of a few revertants. Most Egyptian H5N1 viruses, which descend from the Qinghai Lake viruses, thus possess two mutations that may facilitate transmissibility in mammals: a mutation in HA (resulting in the lack of a glycosylation site) that is critical for H5N1 virus transmissibility in ferrets and guinea pigs, and a mutation in PB2 that confers efficient replication in mammals. In addition, some Egyptian H5N1 viruses have acquired increased affinity for human-type receptors [27].
The data presented here are invaluable for monitoring circulating viruses for variants with increased potential to acquire transmissibility to mammals. Our database searches identified two H5N1 viruses that encode HA-220K and have lost the HA154–156 glycosylation site (A/muscovy duck/Vietnam/NCVD-11/2007; A/duck/Egypt/10185SS/2010), indicating that only two additional mutations are needed to create variants with the “transmissibility features” identified in the Kawaoka study. Because the outbreak of H5N1 viruses in Egypt is extensive, Egyptian H5N1 viruses may, therefore, present a far greater pandemic risk than H5N1 viruses circulating in other countries.
Zdroje
1. KandunIN, WibisonoH, SedyaningsihER, Yusharmen, HadisoedarsunoW, et al. (2006) Three Indonesian Clusters of H5N1 Virus Infection in 2005. N Engl J Med 355 : 2186–2194.
2. OlsenSJ, UngchusakK, SovannL, UyekiTM, DowellSF, et al. (2005) Family clustering of avian influenza A (H5N1). Emerg Infect Dis 11 : 1799–1801.
3. TranTH, NguyenTL, NguyenTD, LuongTS, PhamPM, et al. (2004) Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med 350 : 1179–1188.
4. UngchusakK, AuewarakulP, DowellSF, KitphatiR, AuwanitW, et al. (2005) Probable person-to-person transmission of avian influenza A (H5N1). N Engl J Med 352 : 333–340.
5. RogersGN, PaulsonJC (1983) Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127 : 361–373.
6. ConnorRJ, KawaokaY, WebsterRG, PaulsonJC (1994) Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205 : 17–23.
7. StevensJ, BlixtO, GlaserL, TaubenbergerJK, PaleseP, et al. (2006) Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol 355 : 1143–1155.
8. ItoT, KawaokaY (2000) Host-range barrier of influenza A viruses. Vet Microbiol 74 : 71–75.
9. CouceiroJN, PaulsonJC, BaumLG (1993) Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res 29 : 155–165.
10. MatrosovichMN, MatrosovichTY, GrayT, RobertsNA, KlenkHD (2004) Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A 101 : 4620–4624.
11. HerfstS, SchrauwenEJ, LinsterM, ChutinimitkulS, de WitE, et al. (2012) Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336 : 1534–1541.
12. ImaiM, WatanabeT, HattaM, DasSC, OzawaM, et al. (2012) Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486 : 420–428.
13. HattaM, GaoP, HalfmannP, KawaokaY (2001) Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293 : 1840–1842.
14. VinesA, WellsK, MatrosovichM, CastrucciMR, ItoT, et al. (1998) The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol 72 : 7626–7631.
15. ChutinimitkulS, van RielD, MunsterVJ, van den BrandJM, RimmelzwaanGF, et al. (2010) In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol 84 : 6825–6833.
16. MainesTR, ChenLM, MatsuokaY, ChenH, RoweT, et al. (2006) Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 103 : 12121–12126.
17. MainesTR, ChenLM, Van HoevenN, TumpeyTM, BlixtO, et al. (2011) Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology 413 : 139–147.
18. ChenLM, BlixtO, StevensJ, LipatovAS, DavisCT, et al. (2012) In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology 422 : 105–113.
19. WangW, LuB, ZhouH, SuguitanALJr, ChengX, et al. (2010) Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol 84 : 6570–6577.
20. GaoY, ZhangY, ShinyaK, DengG, JiangY, et al. (2009) Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog 5: e1000709 doi:10.1371/journal.ppat.1000709
21. ChenH, SmithGJ, ZhangSY, QinK, WangJ, et al. (2005) Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436 : 191–192.
22. ChenH, LiY, LiZ, ShiJ, ShinyaK, et al. (2006) Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J Virol 80 : 5976–5983.
23. LiuJ, XiaoH, LeiF, ZhuQ, QinK, et al. (2005) Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309 : 1206.
24. MatrosovichM, StechJ, KlenkHD (2009) Influenza receptors, polymerase and host range. Revue scientifique et technique 28 : 203–217.
25. MehleA, DoudnaJA (2009) Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A 106 : 21312–21316.
26. YamadaS, HattaM, StakerBL, WatanabeS, ImaiM, et al. (2010) Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog 6: e1001034 doi:10.1371/journal.ppat.1001034
27. WatanabeY, IbrahimMS, EllakanyHF, KawashitaN, MizuikeR, et al. (2011) Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog 7: e1002068 doi:10.1371/journal.ppat.1002068
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Egyptian H5N1 Influenza Viruses—Cause for Concern?
- Epigenetics of Host–Pathogen Interactions: The Road Ahead and the Road Behind
- Environmental Regulation of Prions in Yeast
- : Dissecting the Molecular Interface between Pathogen and Plant
- A Wolf in Sheep's Clothing: SV40 Co-opts Host Genome Maintenance Proteins to Replicate Viral DNA
- Exploring New Biological Functions of Amyloids: Bacteria Cell Agglutination Mediated by Host Protein Aggregation
- A Trade-off between the Fitness Cost of Functional Integrases and Long-term Stability of Integrons
- Intracellular Vesicle Acidification Promotes Maturation of Infectious Poliovirus Particles
- Distinct Effects on Diversifying Selection by Two Mechanisms of Immunity against
- Whole Genome Sequencing Reveals Local Transmission Patterns of in Sympatric Cattle and Badger Populations
- Early Mechanisms of Pathobiology Are Revealed by Transcriptional Temporal Dynamics in Hippocampal CA1 Neurons of Prion Infected Mice
- Revised Phylogeny and Novel Horizontally Acquired Virulence Determinants of the Model Soft Rot Phytopathogen SCC3193
- : Where Does It Live?
- The Role of Auxin-Cytokinin Antagonism in Plant-Pathogen Interactions
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Wolf in Sheep's Clothing: SV40 Co-opts Host Genome Maintenance Proteins to Replicate Viral DNA
- Intracellular Vesicle Acidification Promotes Maturation of Infectious Poliovirus Particles
- Revised Phylogeny and Novel Horizontally Acquired Virulence Determinants of the Model Soft Rot Phytopathogen SCC3193
- The Role of Auxin-Cytokinin Antagonism in Plant-Pathogen Interactions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



