-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Culture-induced recurrent epigenetic aberrations in human pluripotent stem cells
hPSCs were shown to acquire genetic aberrations during their growth in culture. The aberrations are non-random and positively selected, by altering multiple cellular phenotypes. Similarly to genetic mutations, epigenetic aberrations may also change gene expression levels, leading to altered cellular behaviors, as seen in tumors. In this study we focus on methylation changes, and we showed that there is a set of genes which are recurrently hypermethylated and silenced in high passage cells. One of these genes, TSPYL5, was shown to be downregulated by hypermethylation in multiple cancer types. We showed that upon its silencing in hPSC, differentiation genes and tumor-suppressor genes are downregulated, and pluripotency - and growth promoting genes are upregulated, which drive the positive selection. These results highlight another challenge faced by hPSC in regard to maintenance of intact gene expression program, and emphasize the role of epimutations described in cancer cells also to hPSCs.
Published in the journal: . PLoS Genet 13(8): e32767. doi:10.1371/journal.pgen.1006979
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1006979Summary
hPSCs were shown to acquire genetic aberrations during their growth in culture. The aberrations are non-random and positively selected, by altering multiple cellular phenotypes. Similarly to genetic mutations, epigenetic aberrations may also change gene expression levels, leading to altered cellular behaviors, as seen in tumors. In this study we focus on methylation changes, and we showed that there is a set of genes which are recurrently hypermethylated and silenced in high passage cells. One of these genes, TSPYL5, was shown to be downregulated by hypermethylation in multiple cancer types. We showed that upon its silencing in hPSC, differentiation genes and tumor-suppressor genes are downregulated, and pluripotency - and growth promoting genes are upregulated, which drive the positive selection. These results highlight another challenge faced by hPSC in regard to maintenance of intact gene expression program, and emphasize the role of epimutations described in cancer cells also to hPSCs.
Introduction
Human pluripotent stem cells (hPSCs) play a major role in current genetic research and in future regenerative medicine, since they are capable of both self-renewal and differentiation into multiple cell types. The persistence for genetically intact cells is crucial in order to ensure safe hPSC-based treatments as well as reliable analysis of their characteristics. However, prolonged culturing of these cells, alongside their unique cell-cycle checkpoints, rapid cell cycle and high levels of replication stress, place them under constant selection pressures[1,2].
During prolonged culturing of PSCs, the cells might acquire genomic changes of variable magnitudes, ranging from full trisomies to copy number changes and point mutations in a process termed culture adaptation[3]. Among the recurrent genetic changes are trisomies of chromosomes 1,12,17 and X, and duplication of a specific region in chromosome 20[2–5]. These aberrations influence central properties such as cell growth, differentiation and susceptibility to apoptosis, and they are positively selected for and propagated until taking over the entire culture[2,6–8].
While gene expression levels are greatly affected by copy number variations or genetic mutations, they may also be modulated by epigenetic changes. Epigenetic mechanisms such as DNA methylation have crucial roles in maintaining gene expression patterns, hence, alterations in DNA methylation patterns may lead to erroneous gene expression. This phenomenon is well-documented in cancers, where epigenetic aberrations may contribute to tumor formation and growth. Global DNA hypomethylation is a common feature of both benign and malignant tumors[9]. It occurs mainly at repetitive sequences, coding sequences and promoters, leading to chromosomal instability, loss of imprinting and oncogenes activation. DNA hypermethylation occurs mainly in gene promoters leading to silencing of tumor suppressor genes[9,10].
Here we show that as in cancer, epigenetic aberrations also occur during culturing of pluripotent cells. We define a set of genes that are recurrently silenced due to DNA hypermethylation at CpG islands in their promotor. Specifically, we focus on testis-specific Y-encoded like protein 5 (TSPYL5), a gene which was previously shown to be hypermethylated and suppressed in multiple types of cancers[11–14]. We study the DNA methylation dynamics of TSPYL5 during culturing of hPSCs, and demonstrate its function in suppressing differentiation and upregulating growth-related genes.
Results
Identification of recurrent epigenetic changes in hPSCs
To evaluate whether recurrent epigenetic aberrations occur during culture adaptation of hPSCs, we have analyzed a database that assessed the DNA methylation state alongside gene expression levels in samples of different passage by methylation and expression arrays[15] (S1 File). Samples harboring large chromosomal aberrations, as detected by e-Karyotyping analysis[5,16] were discarded (S1 Fig). The remaining diploid samples were divided into two groups of low-passage cell lines (p25 or below) and high-passage cell lines (p50 or higher). Using this criterion, both groups have high number of samples with distinct separation regarding the passage numbers. Unbiased hierarchical clustering of the expression profiles showed that most high-passage samples clustered together, indicative of gene expression changes during culturing (Fig 1A and S2 Fig). Methylation β values are measured as the ratio of the methylated bead intensity from the total locus intensity. Unbiased hierarchical clustering of the DNA methylation profiles also resulted in segregation of the samples by the passage number, hinting for unstable methylation during cell culture (Fig 1A and S2 Fig). This patterns of clustering were independent of the gender of the samples or the clustering methodology (S3 and S4 Figs). To gain a better understanding of this phenomenon we plotted the methylation β-value difference between the low - and high-passage samples for each probe, versus the expression change of the gene related to it. This analysis assess the differences in the average expression and methylation levels between the low - and high passage groups. We used a cutoff of methylation difference of 0.2 and expression fold change of 1.5 as this levels of change are statistically significant (less the 5% of the probes). Using this analysis, we detected 34 probes that were hypermethylated in the high-passage group, corresponding to 19 downregulated genes (Fig 1B and 1C). Importantly, there was a negative correlation between the expression and DNA methylation levels of these genes. (Fig 1B and 1C). These results were maintained even when removing the human induced pluripotent samples (hiPSCs) and using only samples of human embryonic stem cells (hESCs) (S5 Fig).
Fig. 1. Changes in expression and methylation during prolonged culturing of hPSCs. 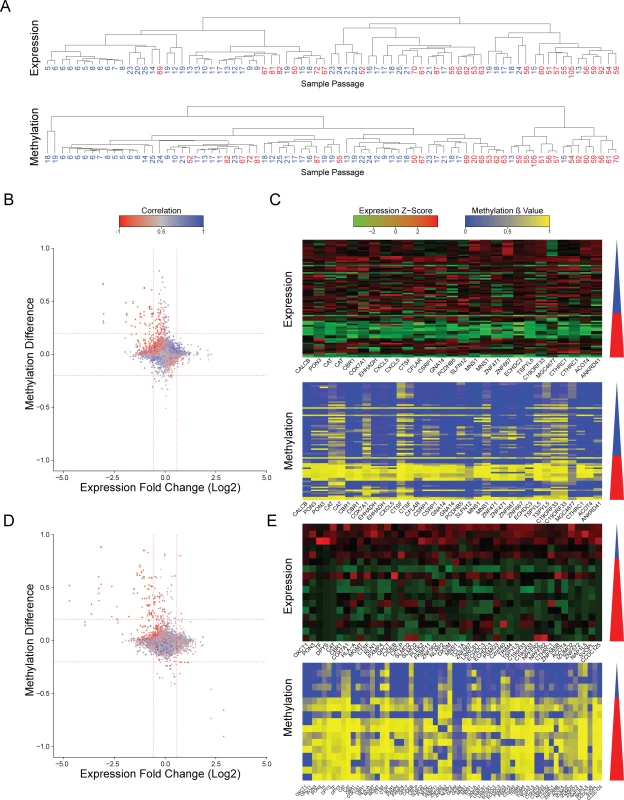
(A) Hierarchical clustering of expression patterns (upper panel) and methylation patterns (lower panel) of the same samples. Number shown are the passage number of each sample. Clustering was performed using Pearson correlation and complete linkage. (B) Scatter plots of expression and methylation differences between high passage and low passage samples of data from Nazor et al[15]. Values represent the change between the averages of the high passage group and the low passage group. Colors indicate the correlation between the expression and methylation. Vertical red lines represent expression change of 1.5 fold. Horizontal red lines indicated methylation change of 0.2. (C) Expression and methylation heatmaps of the probes that showed hypermethylation and reduced expression at the high passage samples in data from Nazor et al[15]. The triangle to the right of each heatmap represents the increasing passage number, blue denotes the low passage samples and red denotes the high passage sample. (D and E) Same as (B and C) but for data from Mallon et al[17]. To validate our results, we repeated the analysis using an independent database of matched expression and methylation profiles of pluripotent cell lines at different passages[17] (S1 File). Since this database is smaller, and it has the bias that all low passage samples are hiPSCs and all high passage samples are hESCs, we used this analysis as validation for the main analysis. Again, we detected a group of 68 methylation probes that were hypermethylated in the high-passage cell lines, correlating with reduced expression of the 40 corresponding expression probes (Fig 1D and 1E). Collectively, we have detected 10 candidate genes that were identified as hypermethylated and silenced in a passage-dependent manner by both assays (Fig 2A, random probability of 1.07×10−21)
Fig. 2. Recurrent hypermethylation during culturing occurs at regulatory CpG Islands. 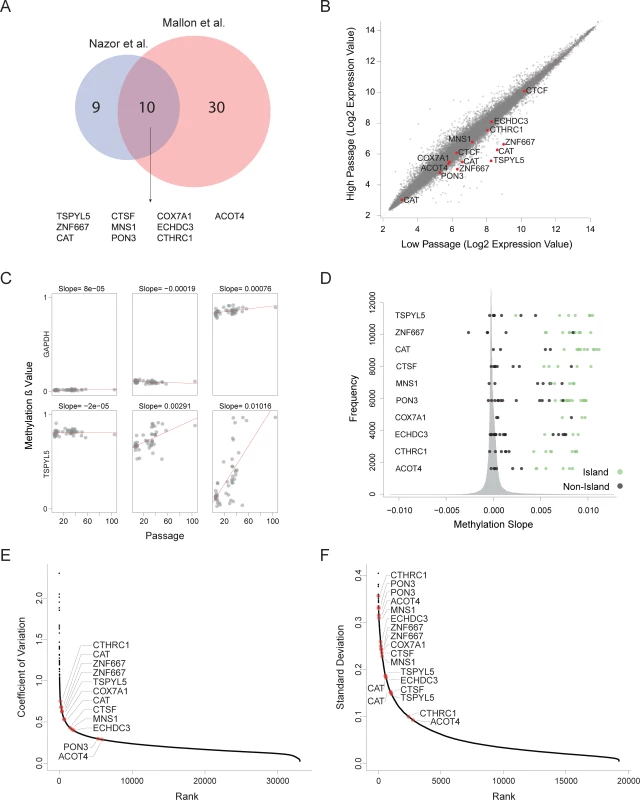
(A) Venn diagram defining 10 genes that undergo hypermethylation and silencing in both analyzed datasets. (B) Expression changes between high passage and low passage hESCs in large expression database of samples with given passage. Shown are the average values of each probe in each group. (C) Examples of methylation levels of six probes in different samples from different passages. In red are the linear regression lines. The slopes of the trend lines are presented above each graph. (D) Histogram describing the distribution of the methylation slopes of all the methylation probes. The methylation sloped of the probes of the 10 candidate genes are shown. (E-F) Variation in expression levels (E) and methylation values (F) in data sets of multiple hESCs without known passage number. Methylation probes shown are only those located in CpG islands. For further validation of our findings, we assembled a large database of 68 expression profiles of hESCs with reported passage numbers from the Gene Expression Omnibus (GEO) database[18] (S1 File). We divided the samples to low-passage group (p25 or below) and a high-passage group (p25 and higher), and compared average expression levels between those groups. In this analysis we considered any sample above p25 as high passage sample since there were not enough samples with reported passage above p50. Out of the 10 genes detected by our previous analyses, 8 genes had probes showing statistically significant reduction in expression in the high-passage group compared with the low-passage group (FDR corrected Wilcoxon tests P<0.05, except ECHDC3 and CTCF) (Fig 2B).
To extend the validation of the DNA methylation data, we have utilized 48 samples analyzed using Illumina Infinium 450K Methylation BeadChips[15], enabling a much denser coverage (S1 File). For each probe, we modeled the relationship between the methylation state and passage number by linear regression, and extracted the slope of the fitted line. (Fig 2C). Methylation slope near zero implies a stable methylation state during culturing, while positive or negative methylation slope values imply gain or loss of methylation during culturing. Compared with all the assayed probes (365,146 probes linked to a gene), most of the probes that correspond to CpG islands of the 10 candidate genes ranked very highly, among the 1% most hypermethylated CpGs. (Fig 2D and S1 Table). Probes located outside of the CpG Islands where relatively stable. These results support the notion that DNA hypermethylation of specific genes during culturing may turn off gene expression.
Since our data suggest that the expression and methylation states of certain candidate genes are not stable during prolonged culturing, we hypothesized that these genes will demonstrate high degree of variability in their methylation and expression levels relative to other genes. To examine our hypothesis, we collected a dataset of 126 expression microarray profiles and 46 Illumina 27k methylation arrays of hESCs (S1 File). Analysis of the variation levels revealed that the candidate genes are among the most variable genes in both expression and methylation (Fig 2E and 2F). Collectively, our analysis pointed to 10 genes whose silencing by aberrant hypermethylation is selected during prolonged culturing of hPSCs.
Hypermethylation and downregulation of TSPYL5 in hPSCs during prolonged culturing
Out of the 10 hypermethylated genes, we focused on TSPYL5 (Fig 3A) since it was shown to be hypermethylated in various types of cancers including glioma[13], glioblastoma[13], astrocytomas[13], gastric cancer[11], esophageal squamous cell carcinomas[14], hepatocellular carcinoma[19] and endometrial cancer[20]. In some of these tumors, hypermethylation was correlated with reduced TSPYL5 expression[11,13]. Furthermore, re-expression of TSPYL5 in glioma and gastric cell lines, in which the endogenous TSPYL5 promoter was shown to be methylated, suppressed cell growth in culture[11,13]. This gene was also shown to be involved in resistance to γ-radiation in lung adenocarcinoma cells[12]. The observed hypermethylation in multiple types of cancers suggests that TSPYL5 may play a role in cell growth regulation and survival.
Fig. 3. TSPYL5 undergoes hyper methylation and silencing in culture. 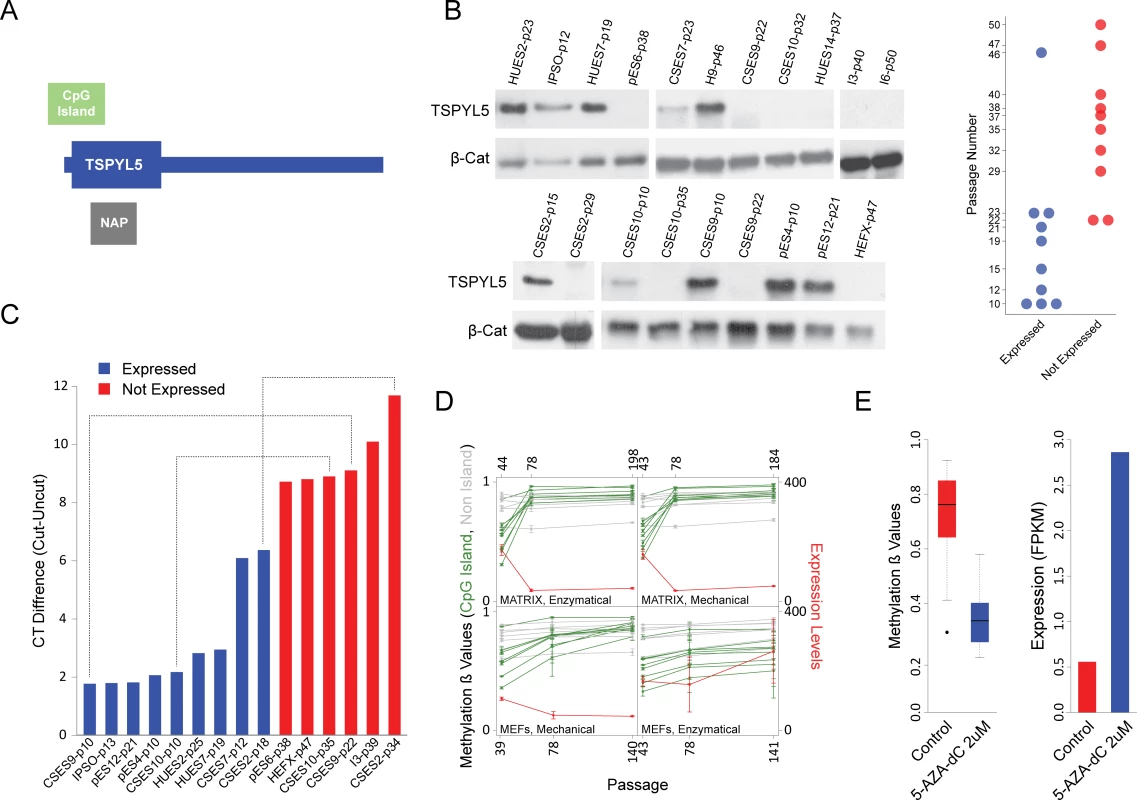
(A) Schematic representation of TSPYL5 gene. (B) Western blot analysis for TSPYL5 of samples from multiple hPSCs cell lines at different passages. The plot at the right panel represents the TSPYL5 expression state of each sample as detected in the western blot analysis. (C) Analysis of methylation levels using the methylation sensitive enzyme McrBC. Shown are differences between quantitative-PCR (qPCR) results of digested and undigested DNA. Blue samples express TSPYL5 while red samples do not. Samples of the same cell line are connected with dashed line. (D) TSPYL5 expression and methylation changes during continuous passaging of the WA09 cell line. Samples were grown either on extracellular matrix or on MEFs, and passaged mechanically or enzymatically. (E) Changes in TSPYL5 Expression and methylation levels in pES6 cells after 5 days treatment with 5-AZA-dC. To understand the role of TSPYL5 in hPSCs, we have analyzed the pattern of TSPYL5 gene expression during early human development, from zygote to early blastocyst and low-passage hESCs, using microarray data[21]. Comparison of TSPYL5 expression to known maternal genes (FIGLA and ZP2) shows that TSPYL5 is a zygotic gene that is actively expressed in early stage hESCs (S6A Fig). Additionally, TSPYL5 is highly expressed in naïve hPSCs[22], which represent the ground state of pluripotency, and during the transition from primed to naïve state it is induced by 10 folds (S6B Fig). These results suggest that TSPYL5 is normally expressed in hESCs, and that its disappearance during prolonged culturing is aberrant.
To confirm these observations, we examined TSPYL5 protein levels in 12 different hPSC lines from various passages. Indeed, the majority of low-passage cell lines expressed TSPYL5, whereas high-passage samples did not (Fig 3B). Importantly, in the cell lines CSES2, CSES9 and CSES10, expression of TSPYL5 was detected at low passage, but not at latter passages (Fig 3B). To determine if silencing of TSPYL5 is indeed due to hypermethylation at the promoter CpG island, we used the McrBC restriction enzyme, whose activity depends on methylated CpG sites. Cell lines that expressed TSPYL5 protein showed hypomethylation in its DNA sequences (Fig 3C). To further support the relation between DNA methylation and expression of TSPYL5, we re-analyzed expression and methylation data of the WA09 hESC line grown continuously for more than 100 passages[23]. This analysis showed that, TSPYL5 was indeed expressed at the beginning of the experiment and subsequently silenced after intensive passaging, accompanied by hypermethylation of the CpG island (Fig 3D). Overall, our results demonstrate that the methylation and silencing of TSPYL5 are not limited to specific cell lines, but is a general culture-induced process.
To show that the methylation state of the promoter of TSPYL5 determines its expression, we treated the TSPYL5-non-expressing cell line pES6 with the demethylating agent 5-aza-2'-deoxycytidine (5-aza-dC). As expected, the treatment caused massive demethylation at the TSPYL5 locus, which was sufficient to reactivate its expression, pointing to the importance of methylation in the regulation of this gene (Fig 3E).
TSPYL5 silencing modifies expression of differentiation, pluripotency and growth related genes
TSPYL5, which is located on chromosome 8, was suggested to contain a nucleosome assembly protein (NAP) domain[11], and was shown to bind gene promoters[24] and may thus affect gene expression[24,25]. To examine the effects of TSPYL5 silencing in culture we used shRNAs to knockdown TSPYL5 expression in a TSPYL5-expressing cell line (Fig 4A). Analysis of gene expression by RNA sequencing showed the expected downregulation of TSPYL5 in the knockdown cell lines (Fig 4B). Then, we selected genes whose mean expression changed after TSPYL5 knockdown, identifying 126 differentially expressed genes (fold change between averages>2, and both repeats have fold change>1.5) (Fig 4C). Gene set enrichment analysis (GSEA) revealed a significant enrichment for genes involved in differentiation among the downregulated genes (Fig 4D and 4E and S2 Table), while the upregulated genes were enriched for chromatin-related genes (Fig 4D and S2 Table). The upregulated gene set also included known genes related to pluripotency and growth, such as KLF5, FGF4 and GDF3 (Fig 4F), and 8 downregulated genes were known tumor suppressors (Fig 4G). Interestingly, five of the upregulated genes were histone coding genes (Fig 4H).
Fig. 4. Gene expression analysis upon TSPYL5 knockdown shows its importance in differentiation. 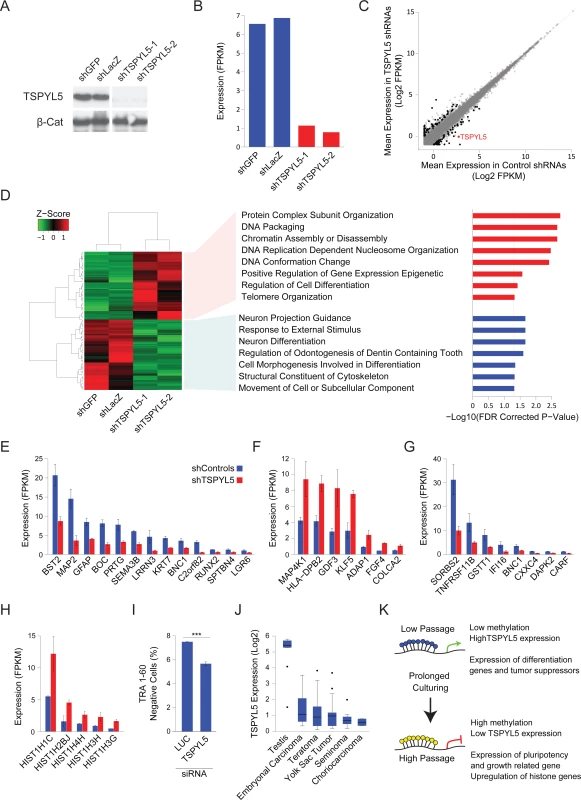
(A-B) Western blot analysis (A) and FPKM expression levels (B) for TSPYL5 for CSES7 cells after knockdown with shRNAs or control constructs. (C) Scatter plot of gene expression levels of shTSPYL5 cells versus controls. Values are the mean of both repeats. Genes marked in black are those with two folds change, and both repeats have at least 1.5 folds change. (D) Heatmap of the genes that showed significant change. For the up- or down-regulated genes, GO Enrichment categories, analyzed in GSEA are shown. (E-H) Bar plots of expression levels of differentiation genes (E), pluripotency and growth related genes (F), tumor suppressor genes (G), and histone genes (H). (I) Percentage of TRA-1-60 negative cells after treatment with siRNA for Luciferase (used as control) or TSPYL5. Statistical significance was analyzed with Student’s t-test; ***p<0.001 (J) TSPYL5 expression levels in multiple types of GCTs and testis as a control. (K) A model describing the observed changes in methylation during prolonged culturing of hPSCs. To support the notion that the downregulation of many differentiation-related genes upon TSPYL5 silencing may affect the extent of spontaneous differentiation, we knockdown TSPYL5 with siRNAs in low-passage TSPYL5-expressing cell line. We then assessed the percentage of differentiated cells by immunostaining for the pluripotency marker TRA-1-60, and analyzing the cells with fluorescence-activated cell sorting (FACS). In concordance with the transcriptomic analysis, knockdown of TSPYL5 results in less spontaneous differentiation (Fig 4I).
Lastly, we asked whether TSPYL5 silencing also occurs in germ cell tumors (GCT), which are tumors of germ cells origin that initiate from primordial germ cells at different developmental stages[26]. For this analysis, we assembled microarray expression data from two independent studies of different GCTs and normal testis (S7 Fig). Similar to the detected silencing in hPSCs grown in culture, TSPYL5 expression was reduced in all GCTs samples compared to normal testis control (Fig 4J), suggesting that TSPYL5 silencing can have advantage also in-vivo.
Discussion
The identification of specific chromosomal aberrations that are recurrently acquired and propagated in hPSC cultures, have highlighted the importance of frequent examination of the genetic integrity of these cells. Our work demonstrates that the same approach may also apply for epigenetic aberrations, were a defined set of loci gain DNA methylation that reduces gene expression. These stochastic changes are propagated by positive selection forces conferred by alterations in pathways that regulate cellular properties such as differentiation and pluripotency, as we show here for TSPYL5 (Fig 4K). Further research is needed in order to assess the importance of this phenomenon to downstream applications of hPSCs.
The growth of cells in culture has many similarities to cancerous processes. Both processes are characterized by accumulation of mutations that provide a growth advantage to the aberrant cells. Genetic mutations of different magnitudes, ranging from the chromosome scale to single nucleotides, have been widely analyzes in hPSCs[2,6,7,27]. Here we showed that epimutations, in the form of aberrant DNA methylation, could also occur during culturing, altering known pathways that are often modulated during tumorigenesis.
While the mechanisms enabling the acquisition of aberrant DNA methylation requires further study, it is important to note that hPSCs had been reported to have defects in the maintenance of intact methylation patterns regarding various processes. Loss of the monoallelic patterns of expression at imprinted loci was repeatedly documented in hPSC lines[15,28,29]. This altered pattern of expression was attributed to aberrant methylation[15,28,29]. In addition, X-chromosome inactivation, which is initiated by the expression of XIST RNA and maintained by DNA methylation and heterochromatinization, was shown to vary among female hPSC cell lines[30,31]. This phenotype was shown to be related to changes in DNA methylation in the X chromosome[15]. The methylation instability in hPSCs can be attributed to the dynamic nature of the methylation maintenance mechanisms in pluripotent cells[32], coupled with the expression of the de-novo methyltransferases DNMT3A and DNMT3B[33].
In our large-scale analysis, we have suggested 10 candidate genes that are frequently undergo epigenetic silencing during passaging. Although our analysis was comprehensive, there may be additional genes that become silenced during culturing, but are not represented in the platforms used for analyzing DNA methylation and gene expression levels. Using analysis of bisulfite sequencing together with RNA-seq may be beneficial in this regard.
We chose to focus on the role of TSPYL5 in culture adaptation due to the multiple reports showing hypermethylation and silencing of TSPYL5 in various tumors. Through their interaction with chromatin, NAP domain proteins such as TSPYL5 were shown to alter gene expression, apoptosis and the cell cycle[25]. Indeed, we demonstrated that TSPYL5 silencing may cause reduction in expression of differentiation-promoting genes and tumor suppressors, and upregulation in pluripotency - and growth-related genes. These changes can translate into less spontaneous differentiation in culture. Thus, our data suggest positive selection forces when TSPYL5 is silenced, and support its relevance to tumor progression.
Lastly, we showed that TSPYL5 is also downregulated in GCT samples. It was shown that similar chromosomal aberrations are gained in tumors and in cultured stem cells of the same tissue[34]. The most prominent example is the enrichment of trisomy 12 in both hPSCs and GCT. Here, we show that TSPYL5 expression is markedly reduced also in both high-passage hPSCs and GCTs, supposedly due to common selection forces. Overall, we demonstrate an additional epigenetic maintenance challenge faced by hPSCs grown in culture, and emphasize its potential importance in regard to tumorigenesis.
Materials and methods
Microarray expression and methylation profiles analysis
Published expression microarray profiles were downloaded from the Gene Expression Omnibus database[18] and RMA normalized using Affymetrix Expression Console. Expression values below a certain threshold were raised to that fixed threshold level to reduce noise of non-expressing genes. The original dataset contains 88 samples with methylation and expression arrays, of cell lines passages < = 25 and > = 50. 11 samples were excluded from the analysis due to large chromosomal aberration or noisy transcriptome as determined by e-Karyotyping[16]. None of the samples contained deletion or amplifications of chromosome 8. Methylation profile were downloaded as β values from the GEO database. Methylation β values of each probe from Nazor et al. were obtained by range-scaling to control samples of unmethylated, completely methylated and half-methylated-DNA[15].
For detection of methylation and expression changes during culturing only samples that had both methylation and expression data where included in this analysis. Sample were divided into two groups by their passage, a group of samples below or equal to passage 25 and a group of samples of passage equal or above 50. Probes on the sex chromosomes where discarded as well as genes unexpressed in more than 80% of the samples in both groups. The expression fold between the two groups was calculated by dividing the median values. Methylation difference where calculated by subtracting the ß-values. The correlation between the expression and methylation was also calculated for each probe. Genes were considered hypermethylated and silenced if the changes in expression were above 1.5 folds, and methylation differences were above 0.2.
For the analysis of multiple expression profiles collected from different studies, samples were divided into low passage samples (passage 25 or lower) and high passage samples (passage 25 and more). In this analysis we were less restrictive in the high passage cutoff due to lack of samples with very high passage number.
For the analysis of Infinium 450K Methylation beadChips (Illumina), for each probe we preformed linear regression between the methylation values and the passage numbers and extracted the slope values. This slope values were considered as an estimate for the methylation change over time in culture.
Cell culture
hESCs were cultured on mouse embryonic fibroblast (MEF) treated with mitomycin-C. Growth medium contained KnockOut Dulbecco's modified Eagle's medium (Gibco-Invitrogen, CA) supplemented with 15% KnockOut-SR (Gibco-Invitrogen, CA), 1mM glutamine, 0.1mM ß-mercaptoethanol (Sigma-Aldrich, MO), 1% nonessential amino acids stock (Gibco-Invitrogen, CA), penicillin (50units/ml), streptomycin (50μg/ml), and 8ng/ml FGF2 (Gibco-Invitrogen, CA). 10μM ROCK inhibitor (Y27632) was supplemented to the medium in the first 24h after thawing cells. Cells were passaged using short treatment with trypsin or trypsin-EDTA (Biological Industries, Beit Haemek, Israel) maintaining the cells as clumps of multiple cells.
Methylation analysis by McrBC digestion
The analysis is based on previously described method[35]. Genomic-DNA was extracted using NucleoSpin Tissue kit (Macherey-Nagel). 500ng of genomic-DNA was digested in 30μl reaction using McrBC enzyme (NEB) for 3h at 37°C and hit-inactivated at 65°C for 20min. As a control, parallel reaction without McrBC was performed. The restriction products diluted to 90μl and 5μl served as a temple for Real-time PCR analysis with primers surrounding the CpG island promoter region. Product could be amplified only if there was no restriction, meaning all the amplicon was unmethylated. qPCR was performed using PerfeCTa SYBR Green SuperMix (Quantabio) with the following conditions: 2min 50°C, 10min 95°C, 45 cycles of 15sec 95°C, 1min 65°C. Primers used are CCCCGAGACTCTGGTACTGT and GAGTCCGCGCGAGATGG.
5-Aza-2'-deoxycytidine treatments
pES6 cells were plated in a density of 100,000 cells per well in a six well plate and cultured for 24h before the beginning of the treatment. 5-Aza-dC (Sigma-Aldrich) was administered for 5 days in a final concentration of 2μM. Cell media was exchanged every day, supplemented with fresh 5-aza-dC. At the end of the experiment, genomic-DNA was extracted and analyzed by Infinium 450K Methylation beadChips (Illumina) and RNA was extracted and analyzed by RNA-Sequencing.
DNA methylation analysis by methylation arrays
DNA methylation analysis was performed on genomic DNA using Infinium 450K Methylation beadChips (Illumina) following the Infinium HD Methylation Protocol as was previously described[28]. Data was processed and normalized by using subset-quantile within array normalization (SWAN) and adjusted for batch effects using the R package ChAMP (version 1.4.0).
Western-blot analysis
Polyacrylamide gel (10%) was used for protein separation. Gels were transferred to a nitrocellulose membrane, and antibody hybridization and chemiluminescence were performed according to standard procedures. The primary antibodies used in this analysis were rabbit anti-TSPYL5 (N15) sc98186 (Santa Cruz Biotechnology) and mouse anti-ß-Catenin (AC-15) sc69879 (Santa Cruz Biotechnology). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies were obtained from Jackson ImmunoResearch Laboratories.
TSPYL5 knockdown experiments
For stable knockdown of TSPYL5 we used shRNAs plasmids obtained from the MISSION shRNAs collection (Sigma-Aldrich). TSPYL5 constructs used are TRCN0000322652 and TRCN0000322581. LacZ and GFP shRNA constructs used were TRCN0000072224 and TRCN0000072194, respectively. Plasmids were co-transfected together with packaging plasmids using polyethylenimine (PEI) into 293T cells. The medium was filtered through 45μM filter and stored at -80°C. CSES7 cells were infected with the lentiviruses during splitting. Puromycin selection (0.34μM) was initiated 2 days after infection. Surviving cells were pulled together.
For Transient knockdown, we used MISSION esiRNA (Sigma-Aldrich) for TSPYL5 (EHU046361) and Renilla Luciferase (EHURLUC). siRNAs were reverse-transfected into low-passage CSES10 cells. For single well of 12 well plate, 30-50nM siRNA was mixed with 1.4μl of DharmaFECT-1 transfection reagent (Dharmacon) in 250μl Opti-MEM I reduced serum media (Thermo Fisher Scientific) for 30 minutes. The transfection mix was then added to 40000 cells plated on Matrigel-coated plates (Corning) cultured in mTeSR medium (STEMCELL Technologies). The cells were grown for 3 days with fresh mTeSR medium supplemented every 24 hours.
RNA extraction, sequencing and analysis
Total RNA was extracted using NucleoSpin RNA Plus kit (Marcherey-Nagel). RNA integrity (RIN>9) was validated using Bioanalyzer (Agilent Technologies). mRNA was enriched by Poly-A selection and sequencing libraries were prepared using TruSeq RNA Library Prep Kit v2 (Illumina). Single-end 85bp sequencing was performed using Illumina Next-Seq500.
Raw sequencing reads were aligned to HG19 reference genome using TopHat2[36] and Cufflinks was used to obtain normalized FPKM values for each sample[37]. Expression threshold was defined as 0.5, elevating genes with a lower expression to this level. To select genes with changed expression levels between the TSPYL5 shRNA and the controls, we set a threshold of two folds change. We also required that both repeats would have fold change of 1.5 over the two controls. GO enrichment analysis for upregulated or downregulated genes was performed in the Broad Institute’s Gene Set Enrichment Analysis (GSEA) tool (http://software.broadinstitute.org/gsea/)[38]
TRA-1-60 immunocytochemistry
Cells were trypsinized using TrypLE Select (Thermo Fisher Scientific) in cold PBS containing 10% KSR, centrifuged and resuspended in 200 μl PBS containing 10% KSR. Staining was performed with PE-conjugated mouse anti-human TRA-1-60 antibody (1 : 40, BD Biosciences) for 30 minutes at 4°C with rotation. The cells were then washed with PBS containing 10% KSR, centrifuged at 300g at 4°C and resuspended in PBS with 10% KSR. Stained cells were filtered through a 70-μm cell strainer and analyzed in BD FACSAria III for the proportion of TRA-1-60-negative and positive cells.
Statistics
All analyses were performed in R statistical software (http://www.r-project.org/). Unsupervised Hierarchical clustering analyses were performed using Pearson correlation or Euclidean distances and complete linkage. To test statistical significance of the clustering, the two main branched were analyzed for the proportion of low - and high passage samples by two-tailed Fisher’s exact test.
Supporting Information
Zdroje
1. Lamm N, Ben-David U, Golan-Lev T, Storchová Z, Benvenisty N, Kerem B. Genomic Instability in Human Pluripotent Stem Cells Arises from Replicative Stress and Chromosome Condensation Defects. Cell Stem Cell. 2016;18 : 253–261. doi: 10.1016/j.stem.2015.11.003 26669899
2. Weissbein U, Benvenisty N, Ben-David U. Genome maintenance in pluripotent stem cells. J Cell Biol. 2014;204 : 153–163. doi: 10.1083/jcb.201310135 24446481
3. Baker DEC, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25 : 207–215. doi: 10.1038/nbt1285 17287758
4. Werbowetski-Ogilvie TE, Bossé M, Stewart M, Schnerch A, Ramos-Mejia V, Rouleau A, et al. Characterization of human embryonic stem cells with features of neoplastic progression. Nat Biotechnol. 2009;27 : 91–97. doi: 10.1038/nbt.1516 19122652
5. Mayshar Y, Ben-David U, Lavon N, Biancotti J-C, Yakir B, Clark AT, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. Elsevier Inc.; 2010;7 : 521–531. doi: 10.1016/j.stem.2010.07.017 20887957
6. Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. Nature Publishing Group; 2011;11 : 268–277. doi: 10.1038/nrc3034 21390058
7. Lund RJ, Närvä E, Lahesmaa R. Genetic and epigenetic stability of human pluripotent stem cells. Nat Rev Genet. Nature Publishing Group; 2012;13 : 732–744. doi: 10.1038/nrg3271 22965355
8. Ben-David U, Arad G, Weissbein U, Mandefro B, Maimon A, Golan-Lev T, et al. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat Commun. Nature Publishing Group; 2014;5 : 4825. doi: 10.1038/ncomms5825 25198699
9. Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7 : 21–33. doi: 10.1038/nrg1748 16369569
10. Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16: R50–R59. doi: 10.1093/hmg/ddm018 17613547
11. Jung Y, Park J, Bang Y-J, Kim T-Y. Gene silencing of TSPYL5 mediated by aberrant promoter methylation in gastric cancers. Lab Invest. 2008;88 : 153–160. doi: 10.1038/labinvest.3700706 18059362
12. Kim EJ, Lee SY, Kim TR, Choi SI, Cho EW, Kim KC, et al. TSPYL5 is involved in cell growth and the resistance to radiation in A549 cells via the regulation of p21(WAF1/Cip1) and PTEN/AKT pathway. Biochem Biophys Res Commun. Elsevier Inc.; 2010;392 : 448–453. doi: 10.1016/j.bbrc.2010.01.045 20079711
13. Kim T-Y, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66 : 7490–7501. doi: 10.1158/0008-5472.CAN-05-4552 16885346
14. Oka D, Yamashita S, Tomioka T, Nakanishi Y, Kato H, Kaminishi M, et al. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: a target for risk diagnosis and prevention of esophageal cancers. Cancer. 2009;115 : 3412–3426. doi: 10.1002/cncr.24394 19472401
15. Nazor KL, Altun G, Lynch C, Tran H, Harness J V, Slavin I, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10 : 620–634. doi: 10.1016/j.stem.2012.02.013 22560082
16. Ben-David U, Mayshar Y, Benvenisty N. Virtual karyotyping of pluripotent stem cells on the basis of their global gene expression profiles. Nat Protoc. Nature Publishing Group; 2013;8 : 989–997. doi: 10.1038/nprot.2013.051 23619890
17. Mallon BS, Chenoweth JG, Johnson KR, Hamilton RS, Tesar PJ, Yavatkar AS, et al. StemCellDB: the human pluripotent stem cell database at the National Institutes of Health. Stem Cell Res. Elsevier B.V.; 2013;10 : 57–66. doi: 10.1016/j.scr.2012.09.002 23117585
18. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41: D991–D995. doi: 10.1093/nar/gks1193 23193258
19. Qiu X, Hu B, Huang Y, Deng Y, Wang X, Zheng F. Hypermethylation of ACP1, BMP4, and TSPYL5 in Hepatocellular Carcinoma and Their Potential Clinical Significance. Dig Dis Sci. 2016;61 : 149–157. doi: 10.1007/s10620-015-3878-3 26386860
20. Witek Ł, Janikowski T, Bodzek P, Olejek A, Mazurek U. Expression of tumor suppressor genes related to the cell cycle in endometrial cancer patients. Adv Med Sci. Medical University of Bialystok; 2016;61 : 317–324. doi: 10.1016/j.advms.2016.04.001 27218895
21. Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138 : 3699–3709. doi: 10.1242/dev.064741 21775417
22. Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. Elsevier Inc.; 2014;15 : 471–487. doi: 10.1016/j.stem.2014.07.002 25090446
23. Garitaonandia I, Amir H, Boscolo FS, Wambua GK, Schultheisz HL, Sabatini K, et al. Increased Risk of Genetic and Epigenetic Instability in Human Embryonic Stem Cells Associated with Specific Culture Conditions. Roh T-Y, editor. PLoS One. 2015;10: e0118307. doi: 10.1371/journal.pone.0118307 25714340
24. Liu M, Ingle JN, Fridley BL, Buzdar AU, Robson ME, Kubo M, et al. TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Mol Endocrinol. 2013;27 : 657–670. doi: 10.1210/me.2012-1397 23518928
25. Park Y-J, Luger K. Structure and function of nucleosome assembly proteins. Biochem Cell Biol. 2006;84 : 549–558. doi: 10.1139/o06-088 16936827
26. Oosterhuis JW, Looijenga LHJ. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer. 2005;5 : 210–222. doi: 10.1038/nrc1568 15738984
27. Peterson SE, Loring JF. Genomic Instability in Pluripotent Stem Cells: Implications for Clinical Applications. J Biol Chem. 2014;289 : 4578–4584. doi: 10.1074/jbc.R113.516419 24362040
28. Johannesson B, Sagi I, Gore A, Paull D, Yamada M, Golan-Lev T, et al. Comparable Frequencies of Coding Mutations and Loss of Imprinting in Human Pluripotent Cells Derived by Nuclear Transfer and Defined Factors. Cell Stem Cell. 2014;15 : 634–642. doi: 10.1016/j.stem.2014.10.002 25517467
29. Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone - and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27 : 2686–2690. doi: 10.1002/stem.205 19711451
30. Bruck T, Benvenisty N. Meta-analysis of the heterogeneity of X chromosome inactivation in human pluripotent stem cells. Stem Cell Res. Elsevier B.V.; 2011;6 : 187–193. doi: 10.1016/j.scr.2010.12.001 21276761
31. Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. Elsevier Inc.; 2012;10 : 595–609. doi: 10.1016/j.stem.2012.02.014 22560080
32. Shipony Z, Mukamel Z, Cohen NM, Landan G, Chomsky E, Zeliger SR, et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. Nature Publishing Group; 2014;513 : 115–119. doi: 10.1038/nature13458 25043040
33. Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. Nature Publishing Group; 2015;47 : 469–478. doi: 10.1038/ng.3258 25822089
34. Ben-David U, Mayshar Y, Benvenisty N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell. Elsevier Inc.; 2011;9 : 97–102. doi: 10.1016/j.stem.2011.06.013 21816361
35. Tesarova L, Simara P, Stejskal S, Koutna I. The Aberrant DNA Methylation Profile of Human Induced Pluripotent Stem Cells Is Connected to the Reprogramming Process and Is Normalized During In Vitro Culture. Lako M, editor. PLoS One. 2016;11: e0157974. doi: 10.1371/journal.pone.0157974 27336948
36. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. BioMed Central Ltd; 2013;14: R36. doi: 10.1186/gb-2013-14-4-r36 23618408
37. Trapnell C, Williams B a, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. Nature Publishing Group; 2010;28 : 511–515. doi: 10.1038/nbt.1621 20436464
38. Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34 : 267–273. doi: 10.1038/ng1180 12808457
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2017 Číslo 8- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
-
Všechny články tohoto čísla
- Deciphering the genetic control of gene expression following antigen stimulation
- Culture-induced recurrent epigenetic aberrations in human pluripotent stem cells
- Positional cloning of quantitative trait nucleotides for blood pressure and cardiac QT-interval by targeted CRISPR/Cas9 editing of a novel long non-coding RNA
- PRRG4 function reveals that Robo trafficking is evolutionarily conserved
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Deciphering the genetic control of gene expression following antigen stimulation
- Culture-induced recurrent epigenetic aberrations in human pluripotent stem cells
- Positional cloning of quantitative trait nucleotides for blood pressure and cardiac QT-interval by targeted CRISPR/Cas9 editing of a novel long non-coding RNA
- PRRG4 function reveals that Robo trafficking is evolutionarily conserved
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



