-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Antimicrobial Functions of Lactoferrin Promote Genetic Conflicts in Ancient Primates and Modern Humans
Immunity genes can evolve rapidly in response to antagonism by microbial pathogens, but how the emergence of new protein functions impacts such evolutionary conflicts remains unclear. Here we have traced the evolutionary history of the lactoferrin gene in primates, which in addition to an ancient iron-binding function, acquired antimicrobial peptide activity in mammals. We show that, in contrast to the related gene transferrin, lactoferrin has rapidly evolved at protein domains that mediate iron-independent antimicrobial functions. We also pinpoint signatures of natural selection acting on lactoferrin in human populations, suggesting that lactoferrin genetic diversity has impacted the evolutionary success of both ancient primates and humans. Our work demonstrates how the emergence of new host immune protein functions can drastically alter evolutionary and molecular interactions with microbes.
Published in the journal: . PLoS Genet 12(5): e32767. doi:10.1371/journal.pgen.1006063
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1006063Summary
Immunity genes can evolve rapidly in response to antagonism by microbial pathogens, but how the emergence of new protein functions impacts such evolutionary conflicts remains unclear. Here we have traced the evolutionary history of the lactoferrin gene in primates, which in addition to an ancient iron-binding function, acquired antimicrobial peptide activity in mammals. We show that, in contrast to the related gene transferrin, lactoferrin has rapidly evolved at protein domains that mediate iron-independent antimicrobial functions. We also pinpoint signatures of natural selection acting on lactoferrin in human populations, suggesting that lactoferrin genetic diversity has impacted the evolutionary success of both ancient primates and humans. Our work demonstrates how the emergence of new host immune protein functions can drastically alter evolutionary and molecular interactions with microbes.
Introduction
Genetic conflicts between microbes and their hosts are an important source of evolutionary innovation [1]. Selective forces imposed by these antagonistic interactions can give rise to dramatic bouts of adaptive gene evolution through positive selection. J.B.S. Haldane originally speculated on the importance of infectious disease as an “evolutionary agent” over 60 years ago [2], and the Red Queen hypothesis later posited that predators and their prey (or pathogens and their hosts) must constantly adapt in order to sustain comparative fitness [3,4]. More recent studies have demonstrated how evolutionary conflicts progress at the single gene or even single nucleotide level, as molecular interfaces between host and microbial proteins can strongly impact virulence and immunity [5–7]. Host-pathogen interactions thus provide fertile ground for studying rapid gene evolution and acquisition of novel molecular traits [8].
Lactoferrin presents a compelling model for investigating adaptation from an ancestral “housekeeping” function to a specialized immunity factor. Lactoferrin arose from a duplication of the transferrin gene in the ancestor of eutherian mammals roughly 160 million years ago [9]. A fundamental and shared feature of these proteins is the presence of two evolutionary and structurally homologous iron binding domains, the N and C lobes, each of which chelates a single iron ion with high affinity. Iron binding by these proteins can effectively starve microbes of this crucial metal, a protective effect termed nutritional immunity [10,11]. Microbes in turn actively scavenge iron from these and other host proteins in order to meet their nutrient requirements [12,13]. The importance of iron in human infectious disease is highlighted by genetic disorders of iron overload, such as hereditary hemochromatosis, which render affected individuals highly susceptible to bacterial and fungal infections [14,15]. In addition to its role in nutritional immunity, lactoferrin has acquired new immune functions independent of iron binding following its emergence in mammals. Lactoferrin is expressed in a variety of tissues and fluids including breast milk, colostrum, saliva, tears, mucous, as well as the secondary granules of neutrophils and possesses broad antimicrobial activity [16]. Portions of the lactoferrin N lobe are highly cationic, facilitating interaction with and disruption of microbial membranes. Two regions of the lactoferrin N lobe in particular, lactoferricin and lactoferrampin, can be liberated from the lactoferrin polypeptide by proteolytic cleavage and exhibit potent antimicrobial activity against bacteria, fungi, and viruses [17,18]. Lactoferrin, as well as lactoferricin alone, can directly bind the lipid A component of lipopolysaccharide (LPS) as well as lipoteichoic acid, contributing to interactions with surfaces of Gram-negative and Gram-positive bacteria [19,20]. Lactoferrin thus poses a unique challenge for microbes—while its ability to bind iron makes it an attractive target for “iron piracy,” lactoferrin surface receptors could render cells more susceptible to associated antimicrobial activity. Despite a growing appreciation for lactoferrin’s immune properties, the evolutionary implications of these unique functions remain unclear. In the present study we decipher recent signatures of natural selection acting on lactoferrin in primates as well as modern humans to understand the evolutionary consequences of a newly acquired antimicrobial activity from a distinct ancestral function.
Results
Positive selection has shaped the lactoferrin N lobe in primates
To assess the evolutionary history of lactoferrin in primates, we assembled gene orthologs from publicly available databases and cloned lactoferrin complementary DNA (cDNA) prepared from primary cell lines. In total, we compared 15 lactoferrin orthologs from hominoids, Old World, and New World monkeys, representing roughly 40 million years of primate divergence (Fig 1A and S1 Fig). We then used maximum likelihood-based phylogenetic approaches (performed with the PAML and HyPhy software packages) to calculate nonsynonymous to synonymous substation rate ratios (dN/dS) across this gene phylogeny [21–23]. For our study we included the N-terminal 19 amino acid positions of the full-length lactoferrin protein, which are removed during processing of the mature polypeptide in humans. Our analysis indicated that lactoferrin has evolved under episodic positive selection in the primate lineage, consistent with a history of evolutionary conflict with microbes (Fig 1A and S1–S7 Tables). These findings are also in line with previous genome-wide scans for positive selection in primates which identified the lactoferrin gene (LTF) among other candidate loci [24]. We next determined signatures of selection across individual codons in lactoferrin. In total, 17 sites displayed strong evidence of positive selection (posterior probability >0.95 from Naïve Empirical Bayes and Bayes Empirical Bayes analyses in PAML), with 13 of the 17 sites found in the N lobe (Fig 1B and 1C and S1 Fig and S2, S4, S5 and S6 Tables). This observation was notably dissimilar from a parallel analysis of primate serum transferrin, where sites under positive selection were restricted to the C lobe (Fig 1B and 1C and S3 Table). These results are further consistent with our previous work indicating that rapid evolution in primate transferrin is likely due to antagonism by the bacterial iron acquisition receptor TbpA, which exclusively binds the transferrin C lobe [25–28]. Thus, while lactoferrin and transferrin both exhibit signatures of positive selection in primates, patterns of selection across the two proteins are highly discordant.
Fig. 1. Dynamic evolution of the lactoferrin N lobe in primates. 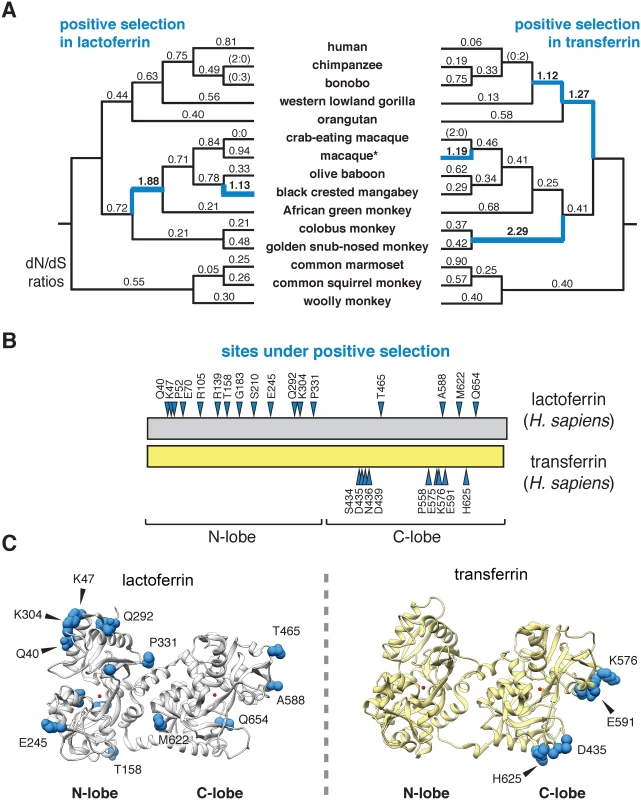
A. Paired primate phylograms showing signatures of positive selection in lactoferrin and transferrin. dN/dS ratios along each lineage are shown, with ratios greater than 1 (indicative of positive selection) shown in blue. Branches with no silent or nonsynonymous mutations display ratios in parentheses. *For lactoferrin analyses the sequence of the Taiwanese macaque was used, whereas for transferrin rhesus macaque was included. This difference does not change the topology of the primate phylogram. B. Sites subject to positive selection in lactoferrin and transferrin are shown (blue arrows) along a schematic of the two proteins (phylogenetic analysis by maximum likelihood, posterior probability >0.95 by Naïve and Bayes Empirical Bayes analyses). The relative positions of the N and C lobes are shown. C. Ribbon diagrams for crystal structures of diferric lactoferrin (PDB: 1LFG) and transferrin (PDB: 3V83), with side chains of sites under positive selection calculated in B shown in blue. Iron in the N and C lobes is shown in red. Evolution and diversity of lactoferrin in modern humans
Evidence of episodic positive selection in primate lactoferrin led us to more closely investigate variation of this gene across human populations. Data from the 1000 Genomes Project revealed six nonsynonymous polymorphisms at greater than 1% allele frequency in humans (S8 Table). Of the 17 sites we identified as rapidly evolving across primate species, amino acid position 47 overlapped with a high frequency arginine (R) to lysine (K) substitution in the N lobe of lactoferrin in humans (Fig 2A and S8 and S9 Tables). This position is markedly polymorphic between populations; while individuals of African ancestry carry the K47 allele at about 1% frequency, this variant is found in non-African populations at roughly 30–65% allele frequency, with the highest frequencies observed among Europeans (Fig 2B and S9 Table). The presence of R47 in related great apes combined with its high frequency in African populations suggests that R47 is in fact the ancestral allele in humans. Data from the Neanderthal genome browser (http://neandertal.ensemblgenomes.org) further revealed lysine to be the consensus residue at position 47 in recently sequenced Neanderthals. The presence of the lactoferrin K47 allele in Neanderthal and non-African human populations and its near absence in Africans suggests one of several intriguing genetic models for the history of this variant, including long-term allelic diversity in hominins, convergent evolution, or introgression from Neanderthals into modern humans.
Fig. 2. Diversity and evolution of human lactoferrin. 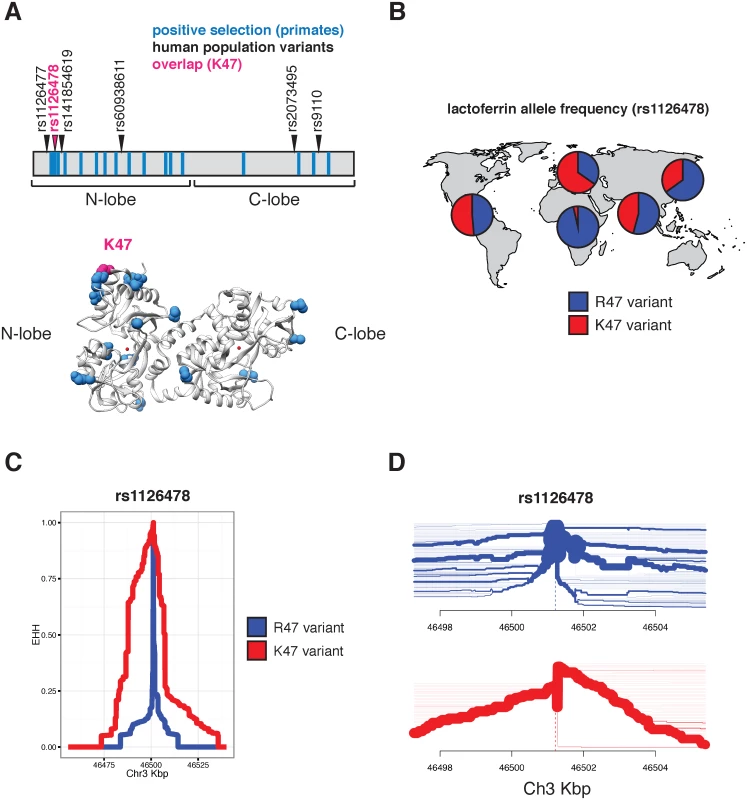
A. Schematic representation of the lactoferrin protein showing positions of abundant (>1% allele frequency) nonsynonymous polymorphisms found in humans (arrows). Sites previously identified under positive selection across primates are shown as blue bars. The position of one variant, rs1126478 at amino acid position 47, which is also rapidly evolving in primates, is shown in magenta. The position of lysine 47 (K47) is also shown in the lactoferrin crystal structure (bottom panel). B. Relative allele frequencies of the R47 (blue) and K47 (red) lactoferrin variants shown as pie charts across human populations. Data were obtained from the 1000 Genomes Project Phase III. C. Extended haplotype homozygosity (EHH) plot around the lactoferrin for the R47 (blue) and K47 (red) around the variable position site, showing the extended haplotype around the K47 variant. D. Haplotype bifurcation plot showing breakdown of linkage disequilibrium in individuals carrying the lactoferrin R47 (blue) and K47 (red) alleles around the variant position. Thickness of the line corresponds to the number of individuals with shared haplotypes. Given the shared variation at position 47 between primate species and among human populations, we sought to determine whether lactoferrin exhibits signatures of positive selection in modern humans. Calculation of pairwise FST between a subset of human populations identified an elevated signal of differentiation between European (CEU) and African (YRI) populations [29], consistent with observed differences in allele frequencies between these groups (S2 Fig). The FST at rs1126478 was 0.70 (empirical p-value < 0.001), 0.30, and 0.03 for CEU-YRI, CEU-CHB, and CEU-FIN, respectively. Single nucleotide variants neighboring rs1126478 also showed signs of elevated FST suggesting that a shared CEU haplotype was driving the signal of differentiation (S2 Fig).
We next applied measures of haplotype homozygosity to assess the possibility that the K47 haplotype has been subject to natural selection in humans. Linkage around R47 alleles breaks down rapidly within a few kilobases, while the K47 variant possesses an extended haplotype (homozygosity of 0.5 at 21,913 bases), consistent with the possibility of an adaptive sweep in this genomic region (Fig 2C). A selective sweep is also consistent with bifurcation plots around position 47, where the K47 haplotypes possess increased homogeneity relative to R47 haplotypes (Fig 2D). We observed a slight an elevation of the genome-wide corrected integrated haplotype score (iHS) for the K47 allele (-1.40136) and a depletion of observed heterozygosity (S2, S3 and S4 Figs). We also examined the patterns of cross population extended haplotype homozygosity (XP-EHH). Consistent with the FST and EHH results, the XP-EHH score was elevated at the K47 position when CEU individuals were compared against YRI (1.1; p-value: 0.129) or CHB (3.1; p-value: 0.003)(S5 Fig). While XP-EHH between CEU and YRI was moderate, surrounding SNPs less than 3 kilobases away had values as high as 2.89 (rs189460549; p-value: 0.01). Genome-wide, the K47 XP-EHH signal is moderate compared to other loci. Next we compared the joint distribution of the p-values from dN/dS analyses [24] with the empirical p-values from the CEU-CHB XP-EHH analyses (S6 Fig). The previous genome-wide rank for lactoferrin, from dN/dS analyses, was 226 before considering the joint distribution and 156 after. The top 20 genes with the greatest change in rank (dN/dS p-value < 0.01) include BLK, DSG1, FAS, SLC15A1, GLMN, SULT1C3, WIPF1, and LTF. This meta-analysis highlights candidate genes that have undergone species-level as well as population-level selection in primates and humans, respectively. By integrating molecular phylogenetic analyses and population genetics approaches, we pinpointed signatures of positive selection associated with an abundant human lactoferrin polymorphism.
Rapid evolution of lactoferrin-derived antimicrobial peptides
Signatures of positive selection in the lactoferrin N lobe among diverse primates, including position 47 in humans, led us to more closely investigate evolutionary pressures that have influenced variation in this region. After gene duplication from ancestral transferrin, lactoferrin gained potent antimicrobial activities independent of iron binding through cationic domains capable of disrupting microbial membranes. Two portions of the lactoferrin N lobe in particular, termed lactoferricin (amino acids 20–67 in full-length protein; 1–48 in mature protein) and lactoferrampin (amino acids 288–304 in full-length protein; 269–285 in mature protein), have been implicated in these antimicrobial functions [18,30].
Phylogenetic analysis revealed that several sites corresponding to lactoferricin and lactoferrampin display signatures of positive selection (Fig 3A and 3B). Notably, positive selection in lactoferricin localized to sites harboring cationic (lysine, arginine) or polar uncharged residues (asparagine), which could mediate membrane disruption and regulate antimicrobial activity. Position 47, which exhibits signatures of selection in humans as well as other primates, also lies within the lactoferricin peptide region. In contrast, hydrophobic tryptophan residues proposed to mediate insertion into microbial membranes are completely conserved among primates, as are cysteine residues that participate in intramolecular disulfide bond formation (Fig 3A). We also observed rapid evolution of the position immediately C-terminal to the pepsin cleavage site in lactoferrampin (Fig 3A), suggesting that the precise cleavage site in this peptide may be variable among species. Notably, the proteases responsible for lactoferrin processing in mucosal secretions and neutrophils remain elusive; identification of such factors will assist in revealing the consequences of genetic variation proximal to cleavage sites. Expanding our phylogenetic analysis to other mammalian taxa, we found that lactoferrin also exhibits signatures of positive selection in rodents and carnivores (S7 Fig and S10 Table). While the specific positions that contribute most strongly to these signatures could not be resolved with high confidence, N-terminal regions corresponding to lactoferricin in primates are absent in several rodent and carnivore transcripts, suggesting that this activity may have been lost or modified in divergent mammals. These observations are further consistent with previous work which identified signatures of positive selection in lactoferrin antimicrobial peptide domains across diverse mammals [31]. Together these results demonstrate that lactoferrin-derived cationic peptides of the N lobe are rapidly evolving at sites critical for antimicrobial action.
Fig. 3. Rapid evolution of lactoferrin-derived antimicrobial peptides and pathogen binding interfaces. 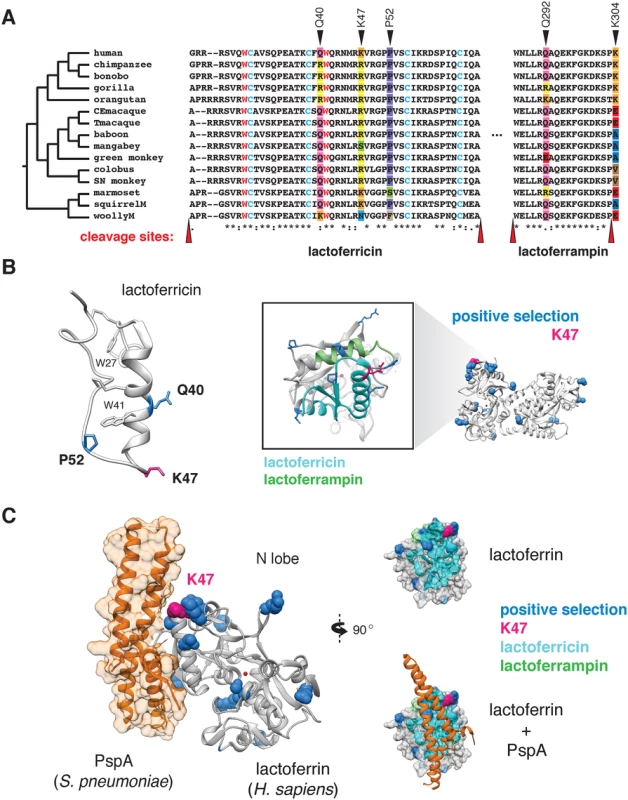
A. Amino acid alignment of the lactoferricin and lactoferrampin peptide sequences across primates. Sites under positive selection are denoted with black arrows, with amino acids at these positions color-coded. Conserved tryptophan (red) and cysteine (blue) residues are highlighted, which contribute to target membrane interactions and disulfide bond formation respectively. The reported cleavage sites of the two peptides are denoted with red arrows. B. Left: solution structure of the free human lactoferricin peptide (PDB: 1Z6V), with sites under positive selection (blue), including position 47 (magenta) indicated. Conserved tryptophan and cysteine residues highlighted in A are also shown. Right: enlarged view of the human lactoferrin N lobe highlighting sequences corresponding to lactoferricin (cyan) and lactoferrampin (green) antimicrobial peptides. Sites previously identified under positive selection in primates are shown in blue, with the position 47 variant shown in magenta. C. Crystal structure (PDB: 2PMS) of human lactoferrin N lobe (gray) bound to PspA from Streptococcus pneumoniae (orange). Side chains of sites under positive selection (blue), including position 47 (magenta) are shown. Distinct microbial interfaces are subject to positive selection in lactoferrin
While rapid evolution of the lactoferrin N lobe may reflect selection for improved targeting of microbial surfaces, it could also represent adaptations that prevent binding by inhibitors encoded by bacteria. For example, pneumococcal surface protein A (PspA) is a crucial virulence determinant of Streptococcus pneumoniae, and several studies have demonstrated that PspA specifically binds and inhibits antimicrobial portions of the lactoferrin N lobe [32]. Consistent with an important evolutionary impact for this interaction, numerous sites under positive selection in the lactoferrin N lobe lie proximal to the PspA binding interface [33], including those corresponding to the lactoferricin peptide (Fig 3C). These data suggest that adaptive substitutions in lactoferrin could negate PspA binding, leading to enhanced immunity against S. pneumoniae or related pathogens.
Many strains of pathogenic Neisseria, which cause the sexually transmitted disease gonorrhea as well as acute meningitis, encode lactoferrin binding proteins (LbpA and LbpB) which mediate iron acquisition from lactoferrin [34,35]. Of four sites identified under positive selection in the lactoferrin C lobe, at least two appear proximal to the proposed Neisseria LbpA binding interface based on recent molecular modeling studies (S8 Fig) [36]. One of these, position 589, also aligns to a region under strong positive selection in transferrin (position 576 in humans) which directly contacts the related bacterial receptor TbpA (Fig 1B) [28]. These findings suggest that, similarly to transferrin, antagonism by bacterial Lbp proteins may have promoted natural selection in the lactoferrin C lobe. Signatures of selection at distinct lactoferrin-pathogen interfaces thus highlight the diverse conflicts that have arisen during the evolution of this unique immunity factor.
Discussion
Together our results suggest that the emergence of novel antimicrobial activity in the N lobe of lactoferrin strongly influenced host-microbe interactions in primates, including modern humans (Fig 4). High disparity in sites under positive selection between the N and C lobes of lactoferrin and transferrin indicate that distinct selective pressures influenced these proteins during primate evolution. We previously demonstrated that primate transferrin has been engaged in recurrent evolutionary conflicts with the bacterial receptor, TbpA [25]. This receptor is an important virulence factor in several Gram-negative opportunistic pathogens including Neisseria gonorrhoeae, Neisseria meningitidis, Haemophilus influenzae, as well as related animal pathogens [26,37–39]. Notably, TbpA binds and extracts iron exclusively from the C lobe of transferrin, and signatures of positive selection in transferrin are almost entirely restricted to the TbpA binding interface (Fig 1) [25]. The fact that transferrin family proteins are recurrently targeted by microbes for iron acquisition may have provided the selective advantage for antimicrobial functions that arose in the lactoferrin N lobe.
Fig. 4. Model of lactoferrin evolution and genetic conflicts with pathogens. 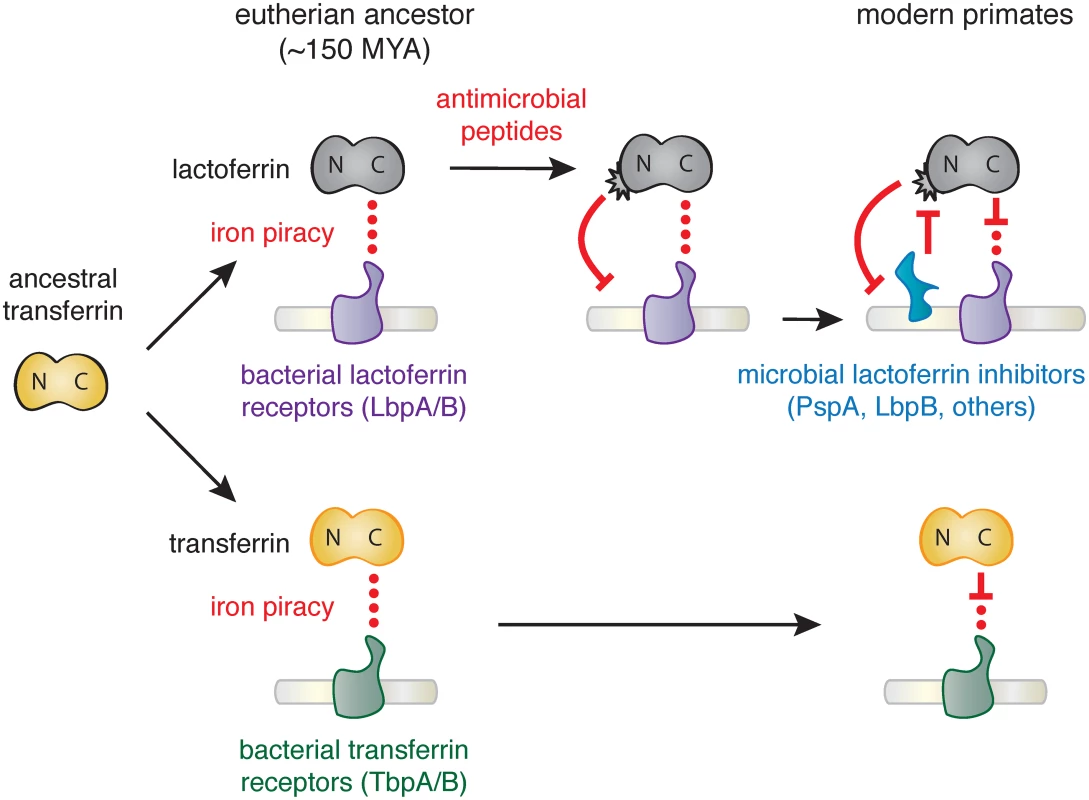
Following a duplication of the transferrin gene in the ancestor of eutherian mammals, interactions between the transferrin (yellow) C lobe and the bacterial transferrin receptors such as TbpA (green) led to the emergence of a molecular arms race. In contrast, while lactoferrin has likely also been engaged in evolutionary conflicts with pathogen iron acquisition receptors like LbpA (purple), the emergence of antimicrobial peptide activity in the N lobe would have provided novel defense activity against pathogens targeting lactoferrin as an iron source. This function would have led to the emergence of pathogen inhibitors of lactoferrin antimicrobial peptide activity (such as PspA or LbpB), which have dominated subsequent evolutionary conflicts localized to the lactoferrin N lobe. Our results suggest at least two non-mutually exclusive scenarios for evolutionary conflicts involving the lactoferrin N lobe. Positive selection in this region could reflect adaption of lactoferrin for enhanced targeting of variable pathogen surfaces. Lactoferricin is capable of binding the bacterial LPS, which itself is heavily modified in many human-associated bacteria to mediate immune evasion and could provoke counter-adaptations at this interface. Conversely, variation in the lactoferrin N lobe could negate interactions with bacterial inhibitory proteins such as PspA encoded by S. pneumoniae. Lactoferrin binding activity has also been identified in several other important bacterial pathogens including Treponema pallidum [40], Staphlococcus aureus [41], and Shigella flexneri [42], raising the possibility of multiple independent evolutionary conflicts playing out at the lactoferrin N lobe. Iron-loaded lactoferrin could further be viewed as a “Trojan horse,” where microbes that target it as a nutrient iron source may be more susceptible to antimicrobial peptides. Consistent with this hypothesis, recent work has suggested that Neisseria encoded LbpB recognizes the lactoferrin N lobe, in contrast to its homolog TbpB which selectively interacts with the iron-loaded C lobe of transferrin [35,43,44]. LbpB binding to the lactoferrin N lobe could thus provide a counter-adaptation with dual benefits by neutralizing lactoferrin antimicrobial activity through negatively charged protein surfaces while simultaneously promoting iron acquisition by its co-receptor, LbpA [43]. These observations point to adaptations involving de novo protein functions on both sides of this molecular interface.
It is important to note that many “pathogenic” bacteria that routinely encounter lactoferrin in the respiratory mucosa are generally commensals that rarely cause disease. For example, H. influenzae colonizes a huge proportion of the human population but typically only causes disease in young children who lack a robust immune response. In addition, the dual functions of lactoferrin likely have pleiotropic effects on complex microbial communities in the host mucosa, with inhibition of some members creating new niches for others. Thus, the evolutionary forces acting on lactoferrin and the consequences for positive selection are likely more nuanced than a two-dimensional host-pathogen arms race. Future studies aimed at understanding the functional impact of lactoferrin variation will assist in understanding such complex biological effects.
Our results raise the possibility that the lactoferrin K47 variant introgressed into humans from Neanderthals at some point after the out-of-Africa expansion [45]. An alternative explanation could be convergent evolution of lactoferrin in distinct lineages of early hominins for enhanced immune function. Recent reports indicate that the human lactoferrin K47 variant, within the N lobe lactoferricin peptide, may have a protective effect against dental cavities associated with pathogenic bacteria [46]. Moreover, saliva isolated with patients homozygous for the K47 variant possesses enhanced antibacterial activity against oral Streptococci relative to homozygous R47 individuals [47]. Future analysis of lactoferrin sequence in archaic humans could provide additional insight on the history and functional properties of this variant. Together these studies provide a direct link between variation in the lactoferrin N lobe and protection against disease-causing bacteria, consistent with adaptive evolution of lactoferrin in humans and other primates.
Notably, the lactoferrin gene, LTF, is located only ~60 kilobases away from CCR5, a chemokine receptor which is also an entry receptor for HIV [48–52]. A 32-base pair deletion in CCR5 (CCR5-Δ32) confers resistance to HIV infection, and is present at a high frequency in northern Europeans while absent from African populations [53]. Although early evidence suggested that CCR5-Δ32 might itself be subject to positive selection in humans, more recent studies have concluded that these signatures are more consistent with neutral evolution [54]. It is intriguing that, like CCR5-Δ32, the lactoferrin K47 variant exhibits increased allele frequency in European populations relative to Africans. However, the presence of the K47 variant at high frequencies in Asian and American populations points to a much earlier origin for this variant than CCR5-Δ32. Moreover, EHH and bifurcation analyses indicate that the haplotypes associated with the lactoferrin K47 variant do not encompass CCR5, suggesting that variation at the CCR5 locus is unlikely to contribute to signatures of selection in LTF (Fig 2B and 2C and S9 Table). The proximity of the LTF and CCR5 genes combined with their high degree of polymorphism and shared roles in immunity suggest the potential for genetic interactions relating to host defense. Future studies could reveal functional or epidemiological links between these two factors in human immunity.
In summary, we have discovered that lactoferrin constitutes a crucial node of host-microbe evolutionary conflict based on signatures of natural selection across primates, including humans. Our findings suggest an intriguing mechanism for molecular arms race dynamics where adaptations and counter-adaptations rapidly emerge at the level of new protein functions in addition to recurrent amino acid substitutions at a single protein interface (Fig 4). Our evolutionary analyses highlight how the process of gene duplication and subfunctionalization can drastically alter the progression of host-microbe genetic conflicts.
Materials and Methods
Primate genetic sources
RNA was obtained from the following species via the Coriell Cell Repositories where sample codes are indicated: Homo sapiens (human; primary human foreskin fibroblasts; gift from A. Geballe), Gorilla gorilla (western lowland gorilla; AG05251), Papio anubis (olive baboon; PR00036), Lophocebus albigena (grey-cheeked mangabey; PR01215), Cercopithecus aethiops (African green monkey; PR01193), Colobus guereza (colobus monkey; PR00240), Callithrix geoffroyi (white-fronted marmoset; PR00789), Lagothrix lagotricha (common woolly monkey; AG05356), Saimiri sciureus (common squirrel monkey; AG05311). Gene sequences from additional primate, rodent, and carnivore species were obtained from Genbank.
cDNA cloning and sequencing
RNA (50 ng) from each primate cell line was prepared (RNeasy kit; Qiagen) and used as template for RT–PCR (SuperScript III; Invitrogen). Primers used to amplify lactoferrin cDNA were as follows: GTGGCAGAGCCTTCGTTTGCC (LF-forward; oMFB256) and GACAGCAGGGAATTGTGAGCAGATG (LF-rev; oMFB313). PCR products were TA-cloned into pCR2.1 (Invitrogen) and directly sequenced from at least three individual clones. Gene sequences have been deposited in Genbank (KT006751 –KT006756).
Phylogenetic analyses and structural observations
DNA multiple sequence alignments were performed using MUSCLE and indels were manually trimmed based on amino-acid comparisons. A generally accepted primate species phylogeny [55] (Fig 1A) was used for evolutionary analysis. A gene tree generated from the alignment of lactoferrin corresponded to this species phylogeny (PhyML; http://atgc.lirmm.fr/phyml/). Maximum-likelihood analysis of the lactoferrin and transferrin data sets was performed with codeml of the PAML software package [21]. A free-ratio model allowing dN/dS (omega) variation along branches of the phylogeny was employed to calculate dN/dS values between lineages. Two-ratio tests were performed using likelihood models to compare all branches fixed at dN/dS = 1 or an average dN/dS value from the whole tree applied to each branch to varying dN/dS values according to branch.
Positive selection in lactoferrin was assessed by fitting the multiple alignment to either F3X4 or F61 codon frequency models. Likelihood ratio tests (LRTs) were performed by comparing pairs of site-specific models (NS sites): M1 (neutral) with M2 (selection), M7 (neutral, beta distribution of dN/dS<1) with M8 (selection, beta distribution, dN/dS>1 allowed). Additional LRTs from the HyPhy software package that also account for synonymous rate variation and recombination (FUBAR, REL, FEL, MEME, BUSTED) were performed [22,23]. Molecular structures of lactoferrin, transferrin and associated proteins were visualized using Chimera (http://www.cgl.ucsf.edu/chimera/).
Human population genetics analysis
For variant-based analyses we used genotype calls from the 1000 Genomes project (release: 20130502, shapeit2 phased). Weir and Cockerham’s Fst estimator [29] was used for the population comparisons, implemented in GPAT++. EHH and the bifurcation diagrams were calculated using the [R] package REHH [56]. Genome-wide iHS scans were performed using GPAT++ and XPEHH plots were generated previously published datasets [57,58].
Supporting Information
Zdroje
1. Daugherty MD, Malik HS. Rules of Engagement: Molecular Insights from Host-Virus Arms Races. Annu Rev Genet. 2012;46 : 677–700. doi: 10.1146/annurev-genet-110711-155522 23145935
2. Haldane J. Disease and evolution. La Ricerca Scientifica Supplemento. 1949;: 1–11.
3. Van Valen L. A new evolutionary law. Evol Theory. 1973;1 : 1–30.
4. Hamilton WD, Axelrod R, Tanese R. Sexual reproduction as an adaptation to resist parasites (a review). Proc Natl Acad Sci USA. 1st ed. National Academy of Sciences; 1990;87 : 3566–3573.
5. Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. National Acad Sciences; 2005;102 : 2832–2837.
6. Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457 : 485–489. doi: 10.1038/nature07529 19043403
7. Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS, Vance RE. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Molecular Cell. 2014;54 : 17–29. doi: 10.1016/j.molcel.2014.02.018 24657167
8. Barber MF, Elde NC. Buried Treasure: Evolutionary Perspectives on Microbial Iron Piracy. Trends in Genetics. Elsevier Ltd; 2015;31 : 627–636.
9. Lambert LA. Molecular evolution of the transferrin family and associated receptors. Biochimica et Biophysica Acta. 2012;1820 : 244–255. doi: 10.1016/j.bbagen.2011.06.002 21693173
10. Weinberg ED. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA. 1975;231 : 39–41. 1243565
11. Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Micro. 2012;10 : 525–537.
12. Cassat JE, Skaar EP. Iron in infection and immunity. 2013;13 : 509–519.
13. Morgenthau A, Pogoutse A, Adamiak P, Moraes TF, Schryvers AB. Bacterial receptors for host transferrin and lactoferrin: molecular mechanisms and role in host-microbe interactions. Future Microbiol. 2013;8 : 1575–1585. doi: 10.2217/fmb.13.125 24266357
14. Andrews NC. Disorders of iron metabolism. N Engl J Med. 1999;341 : 1986–1995. 10607817
15. Weinberg ED. Iron withholding: a defense against infection and neoplasia. Physiol Rev. 1984;64 : 65–102. 6420813
16. García-Montoya IA, Cendón TS, Arévalo-Gallegos S, Rascón-Cruz Q. Lactoferrin a multiple bioactive protein: an overview. Biochimica et Biophysica Acta. 2012;1820 : 226–236. doi: 10.1016/j.bbagen.2011.06.018 21726601
17. Berlutti F, Pantanella F, Natalizi T, Frioni A, Paesano R, Polimeni A, et al. Antiviral properties of lactoferrin—a natural immunity molecule. Molecules. 2011;16 : 6992–7018. doi: 10.3390/molecules16086992 21847071
18. Haney EF, Nazmi K, Lau F, Bolscher JGM, Vogel HJ. Novel lactoferrampin antimicrobial peptides derived from human lactoferrin. Biochimie. 2009;91 : 141–154. doi: 10.1016/j.biochi.2008.04.013 18534196
19. Elass-Rochard E, Roseanu A, Legrand D, Trif M, Salmon V, Motas C, et al. Lactoferrin-lipopolysaccharide interaction: involvement of the 28–34 loop region of human lactoferrin in the high-affinity binding to Escherichia coli 055B5 lipopolysaccharide. Biochem J. 1995;312 (Pt 3): 839–845. 8554529
20. Vorland LH, Ulvatne H, Rekdal O, Svendsen JS. Initial binding sites of antimicrobial peptides in Staphylococcus aureus and Escherichia coli. Scand J Infect Dis. 1999;31 : 467–473. 10576125
21. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. Oxford University Press; 2007;24 : 1586–1591.
22. Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21 : 676–679. 15509596
23. Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26 : 2455–2457. doi: 10.1093/bioinformatics/btq429 20671151
24. George RD, McVicker G, Diederich R, Ng SB, MacKenzie AP, Swanson WJ, et al. Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Research. Cold Spring Harbor Lab; 2011;21 : 1686–1694.
25. Barber MF, Elde NC. Escape from bacterial iron piracy through rapid evolution of transferrin. Science. 2014;346 : 1362–1366. doi: 10.1126/science.1259329 25504720
26. Cornelissen CN, Biswas GD, Tsai J, Paruchuri DK, Thompson SA, Sparling PF. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. journal of bacteriology. 1992;174 : 5788–5797. 1325963
27. Moraes TF, Yu R-H, Strynadka NCJ, Schryvers AB. Insights into the Bacterial Transferrin Receptor: The Structure of Transferrin-Binding Protein B from Actinobacillus pleuropneumoniae. Molecular Cell. 2009;35 : 523–533. doi: 10.1016/j.molcel.2009.06.029 19716795
28. Noinaj N, Easley NC, Oke M, Mizuno N, Gumbart J, Boura E, et al. Structural basis for iron piracy by pathogenic Neisseria. Nature. 2012;483 : 53–58. doi: 10.1038/nature10823 22327295
29. Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38 : 1358.
30. Yamauchi K, Tomita M, Giehl TJ, Ellison RT. Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infection and Immunity. 1993;61 : 719–728. 8423097
31. Liang GM, Jiang XP. Positive selection drives lactoferrin evolution in mammals. Genetica. Springer Netherlands; 2010;138 : 757–762.
32. Shaper M, Hollingshead SK, Benjamin WH, Briles DE. PspA Protects Streptococcus pneumoniae from Killing by Apolactoferrin, and Antibody to PspA Enhances Killing of Pneumococci by Apolactoferrin. Infection and Immunity. 2004;72 : 5031–5040. 15321996
33. Senkovich O, Cook WJ, Mirza S, Hollingshead SK, Protasevich II, Briles DE, et al. Structure of a complex of human lactoferrin N-lobe with pneumococcal surface protein a provides insight into microbial defense mechanism. Journal of Molecular Biology. 2007;370 : 701–713. 17543335
34. Schryvers AB, Morris LJ. Identification and characterization of the human lactoferrin-binding protein from Neisseria meningitidis. Infection and Immunity. 1988;56 : 1144–1149. 3128478
35. Brooks CL, Arutyunova E, Lemieux MJ. The structure of lactoferrin-binding protein B from Neisseria meningitidis suggests roles in iron acquisition and neutralization of host defences. Acta Crystallogr F Struct Biol Commun. 2014;70 : 1312–1317. doi: 10.1107/S2053230X14019372 25286931
36. Noinaj N, Cornelissen CN, Buchanan SK. Structural insight into the lactoferrin receptors from pathogenic Neisseria. Journal of Structural Biology. 2013;184 : 83–92. doi: 10.1016/j.jsb.2013.02.009 23462098
37. Cornelissen CN, Kelley M, Hobbs MM, Anderson JE, Cannon JG, Cohen MS, et al. The transferrin receptor expressed by gonococcal strain FA1090 is required for the experimental infection of human male volunteers. Molecular Microbiology. 1998;27 : 611–616. 9489672
38. Gray-Owen SD, Loosmore S, Schryvers AB. Identification and characterization of genes encoding the human transferrin-binding proteins from Haemophilus influenzae. Infection and Immunity. American Society for Microbiology (ASM); 1995;63 : 1201–1210.
39. Silva LP, Yu R, Calmettes C, Yang X, Moraes TF, Schryvers AB, et al. Conserved interaction between transferrin and transferrin-binding proteins from porcine pathogens. Journal of Biological Chemistry. 2011;286 : 21353–21360. doi: 10.1074/jbc.M111.226449 21487007
40. Deka RK, Brautigam CA, Tomson FL, Lumpkins SB, Tomchick DR, Machius M, et al. Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin. J Biol Chem. 2007;282 : 5944–5958. 17192261
41. Naidu AS, Miedzobrodzki J, Musser JM, Rosdahl VT, Hedström SA, Forsgren A. Human lactoferrin binding in clinical isolates of Staphylococcus aureus. J Med Microbiol. 1991;34 : 323–328. 2056516
42. Tigyi Z, Kishore AR, Maeland JA, Forsgren A, Naidu AS. Lactoferrin-binding proteins in Shigella flexneri. Infection and Immunity. Am Soc Microbiol; 1992;60 : 2619–2626.
43. Morgenthau A, Beddek A, Schryvers AB. The negatively charged regions of lactoferrin binding protein B, an adaptation against anti-microbial peptides. PLOS one. 2014.
44. Yu RH, Schryvers AB. Regions located in both the N-lobe and C-lobe of human lactoferrin participate in the binding interaction with bacterial lactoferrin receptors. Microb Pathog. 1993;14 : 343–353. 8396192
45. Ségurel L, Quintana-Murci L. Preserving immune diversity through ancient inheritance and admixture. Current Opinion in Immunology. 2014;30 : 79–84. doi: 10.1016/j.coi.2014.08.002 25190608
46. Azevedo LF, Pecharki GD, Brancher JA, Cordeiro CA, Medeiros KGDS, Antunes AA, et al. Analysis of the association between lactotransferrin (LTF) gene polymorphism and dental caries. J Appl Oral Sci. 2010;18 : 166–170. 20485928
47. Fine DH, Toruner GA, Velliyagounder K, Sampathkumar V, Godboley D, Furgang D. A lactotransferrin single nucleotide polymorphism demonstrates biological activity that can reduce susceptibility to caries. Infection and Immunity. 2013;81 : 1596–1605. doi: 10.1128/IAI.01063-12 23460521
48. Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381 : 661–666. 8649511
49. Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381 : 667–673. 8649512
50. Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, et al. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. 1996;85 : 1149–1158.
51. Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272 : 1955–1958. 8658171
52. Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. 1996;85 : 1135–1148.
53. Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382 : 722–725. 8751444
54. Sabeti PC, Walsh E, Schaffner SF, Varilly P, Fry B, Hutcheson HB, et al. The case for selection at CCR5-Δ32. Plos Biology. 2005;3.
55. Perelman P, Johnson WE, Roos C, Seuánez HN, Horvath JE, Moreira MAM, et al. A molecular phylogeny of living primates. Plos Genetics. 2011;7: e1001342. doi: 10.1371/journal.pgen.1001342 21436896
56. Gautier M, Vitalis R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics. 2012;28 : 1176–1177. doi: 10.1093/bioinformatics/bts115 22402612
57. Prendergast J, Maclean CA, Chue Hong N. hapbin: An efficient program for performing haplotype based scans for positive selection in large genomic datasets. Dataset. 2015.
58. Pybus M, Dall'Olio GM, Luisi P, Uzkudun M, Carreño-Torres A, Pavlidis P, et al. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Research. 2014;42: D903–9. doi: 10.1093/nar/gkt1188 24275494
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2016 Číslo 5- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Animal Models Are Valid to Uncover Disease Mechanisms
- Hypothalamic Leptin Resistance: From BBB to BBSome
- Spermatogenesis Studies Reveal a Distinct Nonsense-Mediated mRNA Decay (NMD) Mechanism for mRNAs with Long 3′UTRs
- Parental Origin of Interstitial Duplications at 15q11.2-q13.3 in Schizophrenia and Neurodevelopmental Disorders
- Antimicrobial Functions of Lactoferrin Promote Genetic Conflicts in Ancient Primates and Modern Humans
- Genomic Imprinting: A New Epigenetic Perspective of Sleep Regulation
- Embryonic Lethality of Mitochondrial Pyruvate Carrier 1 Deficient Mouse Can Be Rescued by a Ketogenic Diet
- Bayesian Inference of Reticulate Phylogenies under the Multispecies Network Coalescent
- A Conserved DNA Repeat Promotes Selection of a Diverse Repertoire of Surface Antigens from the Genomic Archive
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Animal Models Are Valid to Uncover Disease Mechanisms
- Embryonic Lethality of Mitochondrial Pyruvate Carrier 1 Deficient Mouse Can Be Rescued by a Ketogenic Diet
- Hypothalamic Leptin Resistance: From BBB to BBSome
- Parental Origin of Interstitial Duplications at 15q11.2-q13.3 in Schizophrenia and Neurodevelopmental Disorders
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



