-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
The increased transcription of the Cyp6g1 gene of Drosophila melanogaster, and consequent resistance to insecticides such as DDT, is a widely cited example of adaptation mediated by cis-regulatory change. A fragment of an Accord transposable element inserted upstream of the Cyp6g1 gene is causally associated with resistance and has spread to high frequencies in populations around the world since the 1940s. Here we report the existence of a natural allelic series at this locus of D. melanogaster, involving copy number variation of Cyp6g1, and two additional transposable element insertions (a P and an HMS-Beagle). We provide evidence that this genetic variation underpins phenotypic variation, as the more derived the allele, the greater the level of DDT resistance. Tracking the spatial and temporal patterns of allele frequency changes indicates that the multiple steps of the allelic series are adaptive. Further, a DDT association study shows that the most resistant allele, Cyp6g1-[BP], is greatly enriched in the top 5% of the phenotypic distribution and accounts for ∼16% of the underlying phenotypic variation in resistance to DDT. In contrast, copy number variation for another candidate resistance gene, Cyp12d1, is not associated with resistance. Thus the Cyp6g1 locus is a major contributor to DDT resistance in field populations, and evolution at this locus features multiple adaptive steps occurring in rapid succession.
Published in the journal: . PLoS Genet 6(6): e32767. doi:10.1371/journal.pgen.1000998
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000998Summary
The increased transcription of the Cyp6g1 gene of Drosophila melanogaster, and consequent resistance to insecticides such as DDT, is a widely cited example of adaptation mediated by cis-regulatory change. A fragment of an Accord transposable element inserted upstream of the Cyp6g1 gene is causally associated with resistance and has spread to high frequencies in populations around the world since the 1940s. Here we report the existence of a natural allelic series at this locus of D. melanogaster, involving copy number variation of Cyp6g1, and two additional transposable element insertions (a P and an HMS-Beagle). We provide evidence that this genetic variation underpins phenotypic variation, as the more derived the allele, the greater the level of DDT resistance. Tracking the spatial and temporal patterns of allele frequency changes indicates that the multiple steps of the allelic series are adaptive. Further, a DDT association study shows that the most resistant allele, Cyp6g1-[BP], is greatly enriched in the top 5% of the phenotypic distribution and accounts for ∼16% of the underlying phenotypic variation in resistance to DDT. In contrast, copy number variation for another candidate resistance gene, Cyp12d1, is not associated with resistance. Thus the Cyp6g1 locus is a major contributor to DDT resistance in field populations, and evolution at this locus features multiple adaptive steps occurring in rapid succession.
Introduction
The genetic basis of adaptation remains one of the central unresolved issues within evolutionary biology. From a genome-wide perspective, recent studies of Drosophila and bacteria suggest that a large proportion of nucleotide fixations in these genomes are adaptive, although their individual effects on fitness are typically small [1]–[5]. From the perspective of an individual adaptive trait, the number of genetic variants that contribute to the trait needs to be quantified, and for each variant, its phenotypic effect estimated, including any pleiotropic fitness costs [6], [7]. Ultimately a complete understanding of the genetics of adaptation requires the synthesis of both perspectives. Currently however there are few examples of adaptive traits in which the genetic basis is understood at a high resolution. In bacteria the best examples come from chemostat experiments [8]. In eukaryotes, the predominant examples come from Quantitative Trait Loci (QTL) studies, which suffer from biases, notably that genes of small effect will generally not be identified. Another bias is that a single Quantitative Trait Locus may result from multiple allelic substitutions and hence QTL studies by themselves could underestimate the number of changes in a bout of adaptive evolution. Here we use insecticide resistance as a model adaptive trait in a eukaryote organism. It offers an opportunity to view an adaptive response to a change in a single environmental component and it occurs on a timescale that allows multiple genetic changes to be observed.
Historically, insecticide resistance has provided a good model for adaptation because a novel selective agent is applied to large natural populations. A key insight provided by many insecticide resistance studies is the frequency of parallel mutation. Often, exactly the same mutations arise independently in a gene or in orthologous genes, as adaptive responses to insecticide selection. For example, at the Resistance to dieldrin (Rdl) locus, a single A302S substitution has independently occurred across a diverse array of insect species. In the case of Tribolium casteneum, this mutation has occurred multiple times in different geographically defined populations [9], [10]. Similarly, the L1014F mutation in the para voltage gate sodium channel, the molecular target of pyrethroids and Dichloro-Diphenyl-Trichloroethane (DDT), has arisen independently in numerous species [11]. Such parallel evolution is not restricted to the target molecules of insecticides, but is also seen in detoxifying enzymes, one example being the G137D change in the Resistance to organophosphate (Rop1) esterase locus in Lucilia cuprina and Musca domestica [12]. Parallel evolution is one manifestation of the limits to adaptation, because it suggests that there are a limited number of alternatives. Or more precisely, if there are alternatives, they are harder to reach via mutation, provide less of a benefit, or impose greater fitness costs.
The Cyp6g1 locus of Drosophila melanogaster and Drosophila simulans provides another example of parallel evolution. In D. melanogaster the insertion of transposable element sequence into the 5′ regulatory region correlates with increased transcription of the Cyp6g1 gene that encodes a cytochrome P450 enzyme capable of metabolising multiple insecticides, most notably DDT [13], [14]. The 491 bp insertion is derived from the long terminal repeat (LTR) of an Accord transposable element (TE), and lies 287 bp from the transcription start site [13]. Transgenic studies have demonstrated that the 491 bp Accord insertion can drive expression of a GFP reporter gene in detoxification tissues [15], implicating the insertion as the mutation that causes resistance and thereby providing a robust example of cis-regulatory adaptation [16]. Furthermore the Accord insertion is not found in flies collected before 1940 but is found at high frequency (32–100%) in 34 contemporary populations around the world, and its presence correlates with resistance [17]. In D.simulans, a 4,803 bp Doc element, a non-LTR TE, has inserted about 200 bp upstream of the putative transcription start site. This insertion correlates with a two-fold increase in transcription and appears to have recently swept to high frequency in a Californian population. Direct evidence of a phenotypic effect of the Doc insertion is equivocal, but the large window of reduced nucleotide variation around D.simulans Cyp6g1 is consistent with insecticide based selection [18].
DDT resistance in Drosophila is an engaging case study of adaptive evolution, partly because the results of genetic mapping studies have reached different conclusions about the basis of resistance. Most early research, including the classic studies of J.F. Crow, indicated that DDT resistance was polygenic with genes contributing to DDT resistance distributed among all three major chromosomes [19]–[22]. In contrast, work by Kikkawa in 1964 showed that the resistance in the Hikone-R strain was due to a single dominant locus, which mapped to 64.5 cM on chromosome II [23]. This was subsequently identified as Cyp6g1 [24]. Since the identification of Cyp6g1 as a resistance locus, researchers have identified the molecular mechanism of the upregulation [15], looked for selective sweeps centred at this locus [17], and have contended that other loci also play important roles in DDT resistance [25]. Other than Cyp6g1, two additional loci have been implicated in DDT resistance, and both are also cytochrome P450 genes. CYP6A2 has DDTase activity that has been shown to be reliant on three amino acid substitutions that distinguish a lab resistant strain from susceptible strains [26]. The other locus, Cyp12d1, is highly inducible by DDT [27]. Its overexpression in flies using transgenic techniques increases DDT resistance [28], and the locus exhibits high frequency copy number variation in natural populations. However Cyp6g1 is the only locus shown to have alleles that contribute to variation in DDT resistance in field populations of D. melanogaster.
We, and others [29], [30], have found copy number variation and extra alleles at the Cyp6g1 locus of D. melanogaster. This casts a new light on the nature of the resistance mutations, the phenotypic contribution to DDT resistance and the adaptive significance of Cyp6g1 variation. The aims of the research reported here are to: (i) characterise this molecular variation, focussing on additional transposable element insertions in the locus and gene copy number variation, (ii) determine if this molecular variation correlates with phenotypic variation in the form of DDT dosage-mortality relationships, (iii) assess whether the variation is adaptive by analysing the genotypic frequencies of this newly described variation in historical and contemporary populations of D. melanogaster and (iv) quantify the contribution that Cyp6g1 locus variation makes to overall DDT resistance in a natural population.
Results
Characterisation of Cyp6g1 copy number variation (CNV) and TE insertion complexity in the RK146 strain
We sequenced Cyp6g1 from seven D. melanogaster isochromosomal lines (chr.II), which revealed that six lines had double peaks on their sequence chromatograms. Since the isochromosomal lines should only have one allele for genes on the second chromosome we inferred that there must be CNV for Cyp6g1. We confirmed this by determining that both alternate states of sequence variants were passed to all individuals in the next generation (data not shown). To determine the details of this CNV we initially characterized the Cyp6g1 loci in a single isochromosomal line (RK146). In situ hybridisation of a Cyp6g1 probe to polytene chromosomes indicated that the copies of Cyp6g1 were within the same cytological band (Figure S1). Assuming that these were arrayed in a direct tandem manner, we performed a PCR and identified one breakpoint of the locus duplication, adjacent to the previously identified Accord LTR.
To ascertain the other breakpoint associated with this CNV, inverse PCR was performed. Unexpectedly this revealed a ∼7 kb insertion of an HMS-Beagle TE within another Accord insertion. Notably the size of the HMS-Beagle insertion means that it would not have been detected using the Accord spanning PCR assays of previous studies, thus the copy lacking HMS-Beagle is presumably the locus scored previously [13], [17], [31].
To determine the extent of Cyp6g1 CNV we generated a model of the locus using Southern blots that we tested using PCR. A primer located within the HMS-Beagle sequence was used in combination with primers within Cyp6g1 and the resulting amplicons were cloned and sequenced. The longest amplicon was 12.5 kb and spans the distance between the HMS-Beagle element in the first copy (hereafter called Cyp6g1-a) and the beginning of the second full copy of Cyp6g1 (hereafter referred to as Cyp6g1-b Figure 1A). A similar approach, using a primer within Accord was used to amplify all of Cyp6g1-b. In this case the HMS-Beagle insertion was used to advantage as it prevented amplification of Cyp6g1-a. Table S1 shows the differences between Cyp6g1-a and Cyp6g1-b, none of which alter the predicted amino acid sequence.
Fig. 1. The molecular structure of the Cyp6g1 locus. 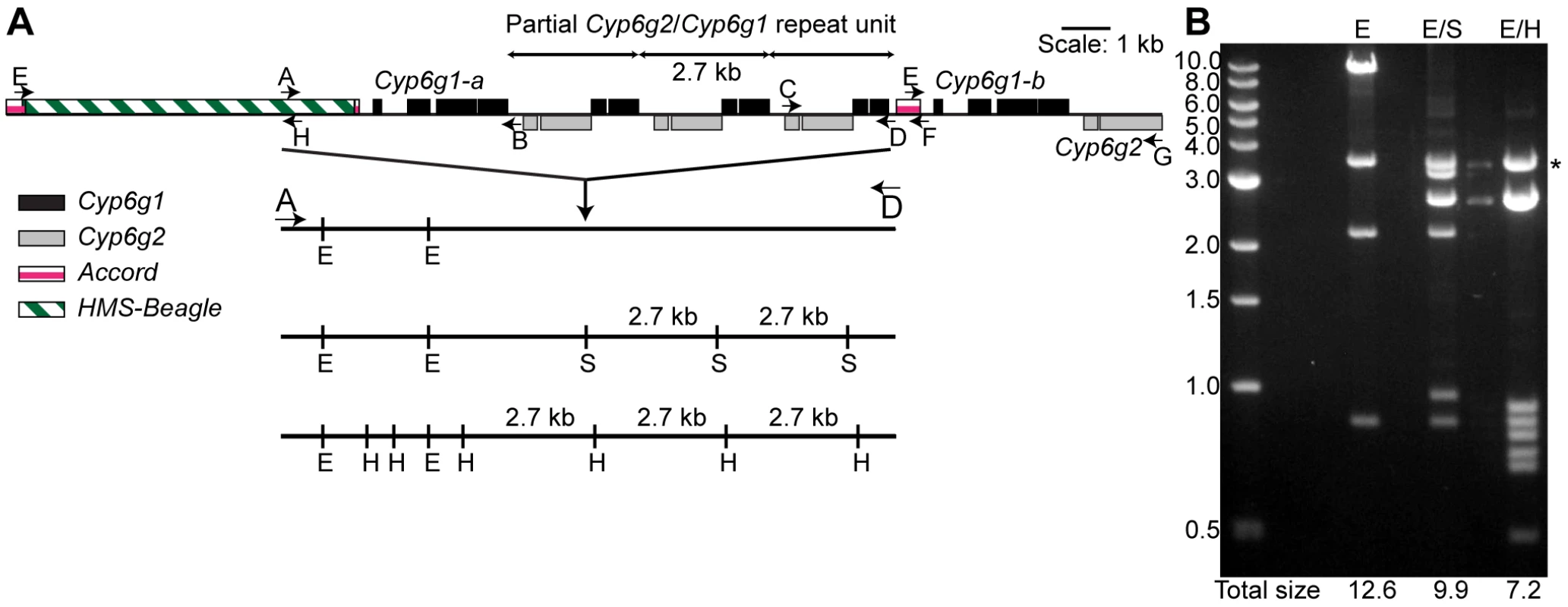
(A) The region encompassed by five overlapping PCR primer sets (E-H, A-B, A-D, C-D, E-G) that have been cloned are shown. Cyp6g1 exons are represented as black boxes above the line, Cyp6g2 exons as dark grey boxes below the line. The HMS-Beagle element and the Accord element sequences are represented as an empty box and as light grey boxes above the line respectively. (B) The complex structure of the locus is demonstrated by restriction digests (EcoRI; E, EcoRI-SacI; E/S and EcoRI-HindIII; E/H) of a plasmid containing the 12.5 kb A-D PCR product. The restriction sites for each digest are shown as lines drawn to scale with the locus structure. The 3.5 kb EcoRI-EcoR1 plasmid band is seen in all lanes. The sum of the molecular weights of the remaining fragments is shown at the bottom of each lane. The sums differ by intervals of 2.7 kb, which corresponds to the repeat unit represented in the top of the figure. * indicates 3.5 kb vector band. Sequences of E-H, A-B, C-F and E-G are lodged in Genbank with accessions HM214801, HM214799, HM214802 and HM214800 respectively. Aside from these two full-length copies of Cyp6g1 a repeat unit that contains a fusion of partial copies of both Cyp6g1 and Cyp6g2 was identified. Cyp6g1 and Cyp6g2 are transcribed from opposite strands and are arranged in a convergent orientation. The repeat unit starts at ∼codon 323 of Cyp6g1 and continues to about codon 73 of Cyp6g2. The repeat unit can be seen in Southern blots as a 2.7 kb fragment common to different restriction enzyme digests (Figure S2). It is also demonstrated by the multiple digests of the A-D amplicon clone shown in Figure 1B, where this structure is inferred by the consistent 2.7 kb size difference between different restriction enzyme digests.
While we are confident that the structure shown in Figure 1 occurs in the RK146 strain, it remains a formal possibility that there are Cyp6g1 associated elements outside this region.
Strain comparisons reveal an allelic progression
Previously a partial P element had been identified nested within the Accord element upstream of Cyp6g1 [17], [31]. We designed PCR assays to score flies for this partial P insertion, the Accord insertion, the HMS-Beagle insertion and the presence of the gene duplication. We could not develop a co-dominant assay for the duplication i.e. we could identify lines carrying the duplication with a PCR bridging a breakpoint but we could not develop an assay that uniquely amplified the single copy allele and not the double copy alleles. Therefore we performed PCR assays on isochromosomal lines, highly inbred lines and F1 crosses to a known genotype (Table S2 for details). These assays revealed even more variation; specifically alleles derived from the partial P-element insertion where most of that P-element had been removed leaving small scrambled portions of the terminal repeats (Figure S3). Thus we describe six alleles (M, A, AA, BA, BP, BPΔ) defined on the basis of this molecular variation (Figure 2). The facts that the two full length copies both have an Accord LTR insertion, some TEs are nested, and that all lines containing the P-element insertion also have the HMS-Beagle insertion, suggests an order to the molecular events at this locus, which is indicated in Figure 2.
Fig. 2. The six alleles of Cyp6g1. 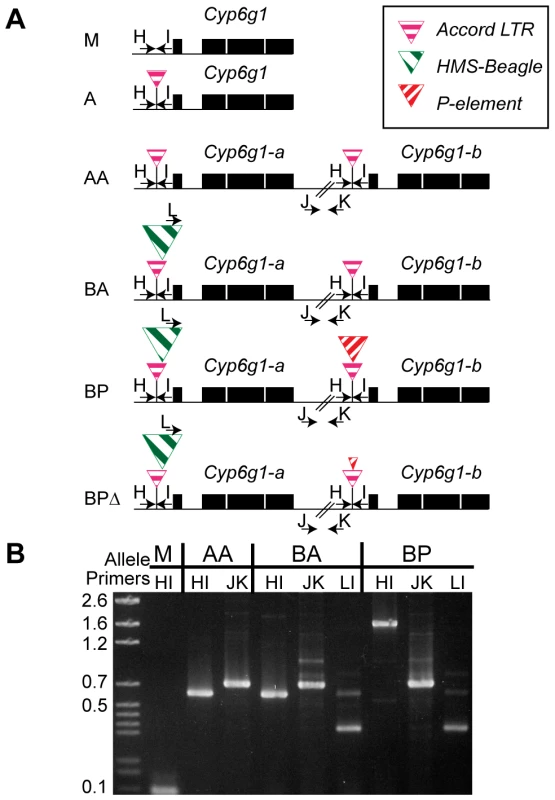
(A) The alleles are arranged from top to bottom in the order in which they arose. The M allele has a single copy of Cyp6g1 and lacks TE insertions. The A allele is single copy with an Accord insertion, the AA allele has two full length copies each with an Accord insertion. The BA allele has two full-length copies with the proximal copy containing HMS-Beagle. The BP allele has two full-length copies with the a copy containing HMS-Beagle and the b copy containing the P insertion. All of the lines assayed that have a P-element sequence in the Accord also have the HMS-Beagle insertion and so the BP class probably arose from the BA allele. The heterogeneous BPΔ class contains various low frequency variants that have scrambled P terminal repeats (Figure S3). PCR primers are shown as arrows and are named with a single letter. Note that primer L anneals to the HMS-Beagle sequence whereas primers H and I flank the transposable element insertion sites. (B) A gel demonstrating the diagnostic PCRs is shown on the right. Geographic and temporal patterns of allelic variation
We conducted three surveys of Cyp6g1 alleles in D. melanogaster populations: a survey from historical samples, a survey of lines from contemporary global populations, and a survey of wild males collected on the east coast of Australia (Figure 3). These studies included a re-analysis of lines scored for the Accord element in earlier studies to determine whether the lines had the duplicated locus or had other variants not originally described [13], [18]. Consistent with previous studies, the ancestral Cyp6g1-[M] allele is rare in most contemporary populations except for the population from Malawi, Africa [17], [18]. The historic samples show that the M allele was more frequent in the 1950–1980 samples and was the only allele observed among the lines established in the 1930s (Figure 3A). Surprisingly the Cyp6g1-[A] allele, which was the presumed state of previous studies, is not observed in three population surveys. Most flies in Europe, Asia, and the USA have Cyp6g1 duplicated and are of the Cyp6g1-[AA] or Cyp6g1-[BA] class. A re-analysis of 13 lines collected before 1966 that had been classified as having the Accord insertion [13], found that all contained the Cyp6g1 duplication and 11 also carried the “HMS-Beagle” insertion (Table S2). The latter includes Hikone-R, which is derived from flies collected in Japan in 1952 [32], and which was the DDT resistant strain that was initially used to map resistance to the map position where Cyp6g1 resides [21], [24].
Fig. 3. Temporal and geographic changes in Cyp6g1 allele frequencies. 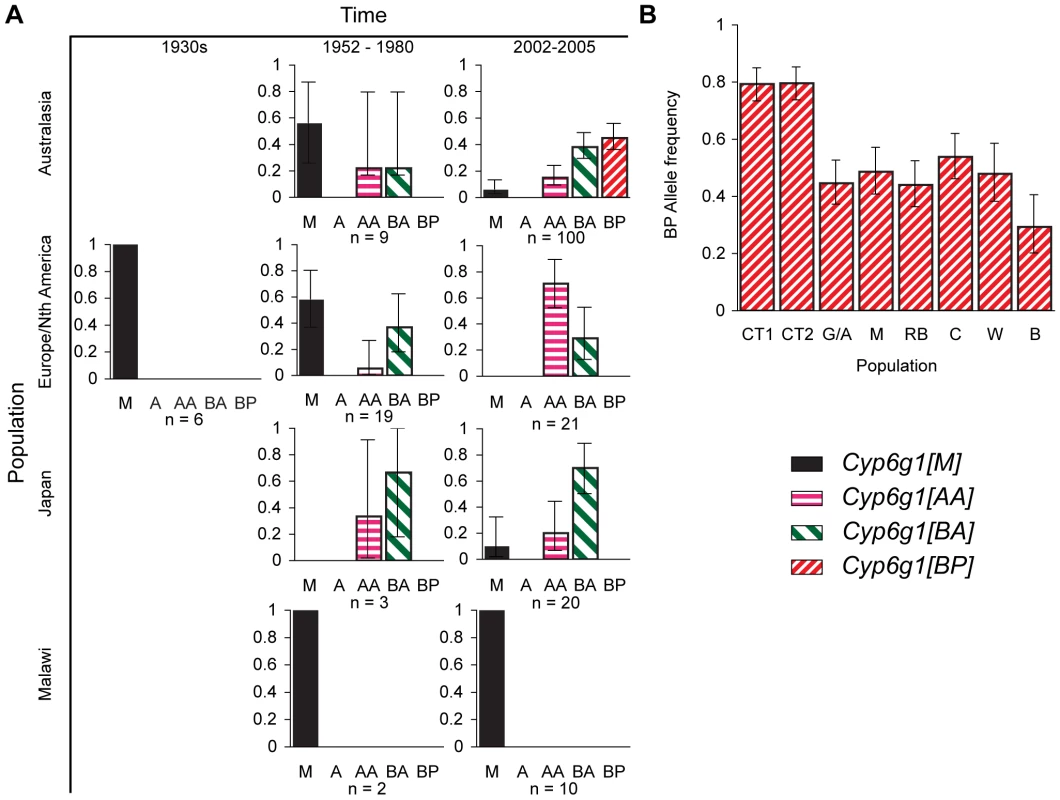
(A) Lines established between the 1930s and the present. Flies were typed using the PCR assays described in Figure 2. (B) The frequency of the BP allele was scored in >40 flies from each of eight locations along the east coast of Australia. The populations are from Cape Tribulation (CT1, CT2), Gladstone/Alstonville (G/A), Maryborough (M), Rainbow Beach (RB), Coffs Harbour (CH), Wollongong (W), Bega (B). Error bars in both (A,B) represent the 95% binomial confidence interval. Our survey of 190 alleles from historic and contemporary global populations found the Cyp6g1-[BP] allele only in current Australian populations (Figure 3A). The BP allele is not seen in any population before 1980 (n = 39), they are not seen in the non-Australian flies surveyed by us in 2002–2005 (n = 51), and Catania et al reports them at a frequency of 10/683 in a worldwide sample[17]. However the sample from Australia represented in Figure 3A shows that they are at a frequency of 40/100, which is highly significantly different from the Catania et al study (p<0.0001) and our own survey of non-Australian alleles (p = 0). Our third survey, which was of ∼40 males caught in each of eight populations, specifically addressed the frequency of the BP allele in Australia. The BP allele frequency ranged from 29–80% with the highest being in the most northerly populations (Figure 3B). This level of population differentiation between Australian and other populations is highly unusual in D. melanogaster and suggests positive selection has recently driven the BP allele to these high frequencies [33]–[35].
We identified two types of Cyp6g1-[BPΔ] alleles. Both were extremely rare being observed a combined total of three times; all from northern Australia. Because of their rarity we have not considered them further in this manuscript except to record their sequence in Figure S3.
Allelic variation at Cyp6g1 determines DDT resistance phenotype
For genetic variation to be adaptive it needs to contribute to phenotypic variance. So we asked whether the molecular variation described above contributes to DDT resistance – the phenotype originally attributed to this locus. To answer this, DDT resistance was calculated by contact assay [24] on four day old adults of 19 isochromosomal lines and 3 inbred lines, most of which were derived from Australian populations (Table S2 and Table S3). Males and females were assayed separately for at least five strains of each genotype: M, AA, BA and BP. Figure 4 shows that M strain males are on average 7 fold less resistant than AA strain males. For females there is 10 fold difference (see Table S3 for more details). Furthermore, with each new Cyp6g1 allele there is an increase in resistance level. There is about a 50% increase from AA to BA flies although this is only significant in the case of males. The males of the BP strains are on average 40 fold more resistant than the M strains and the females are 80 fold more resistant. The fact that the Cyp6g1 allelic classes strongly correlate with resistance, despite the diverse genetic background of the strains assayed, suggests Cyp6g1 is a major determining factor of the DDT resistance phenotype in these lines.
Fig. 4. DDT resistance correlates with Cyp6g1 allelic class. 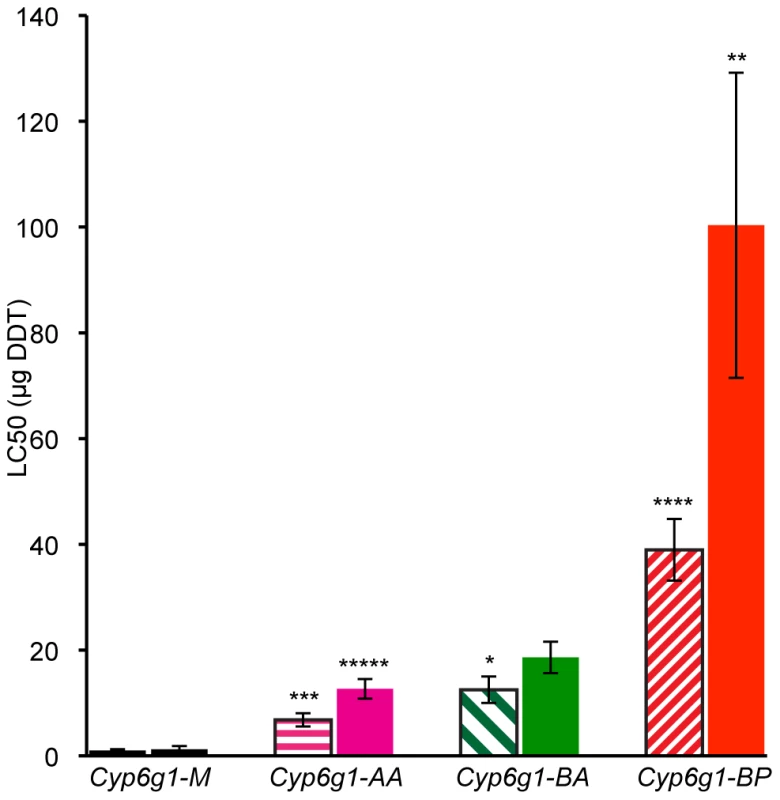
The resistance of lines isochromosomal for their second chromosome are shown grouped on the x-axis by Cyp6g1 alleles (M, AA, BA, BP). The error bars represent the standard errors of the mean LC50 of the lines. Significant differences with preceding classes are indicated e.g. BA vs. BP females (One-tailed t-test: ***** p = 0.0005, **** p = 0.002, *** p = 0.003, ** p = 0.019, * p = 0.044). Shaded bars represent data from males, solid bars from females. Transcription of Cyp6g1 genotypes
Transgenic studies have previously shown that the Accord LTR can act as a tissue specific enhancer of gene expression [15]. To test whether transcription levels correlate with the remaining steps in the allelic series, we analysed Cyp6g1 expression in 18 of the lines analysed for DDT resistance. The tissue and cell types where DDT detoxification occurs are currently unknown, but we chose to analyse the adult midgut and adult Malpighian tubules because a UAS-Cyp6g1 transgene confers resistance when over-expressed in these tissues using the Accord LTR Gal4 driver [28]. Furthermore, RNAi knockdown of Cyp6g1 in the tubules increases susceptibility of flies to DDT [36]. We found a positive and significant correlation between DDT resistance and transcription levels among the 18 lines assayed for both the midgut and the tubules (Spearmans rank; midgut = 0.62, p<0.02, tubule = 0.52, p<0.05). However when the 18 lines are grouped by allelic class, as shown in Figure 5, significant differences in transcription levels are observed for only some of the steps in the allelic progression. In the midgut, the derived alleles clearly exhibit higher gene expression than Cyp6g1-[M]; with AA having a 2.6 fold increase (P = 0.036, one tailed t-test) and BA and BP both showing a 5 fold increase. There is also a significant 2-fold increase of BA over AA (P = 0.0006, one tailed t-test). In tubule, only the step between M and AA results in a significant increase in transcription (P = 0.04, one tailed t-test; Figure 5B), and all three derived alleles exhibited ∼3 fold increases in gene expression.
Fig. 5. Cyp6g1 transcription in adult midgut and malphigian tubule. 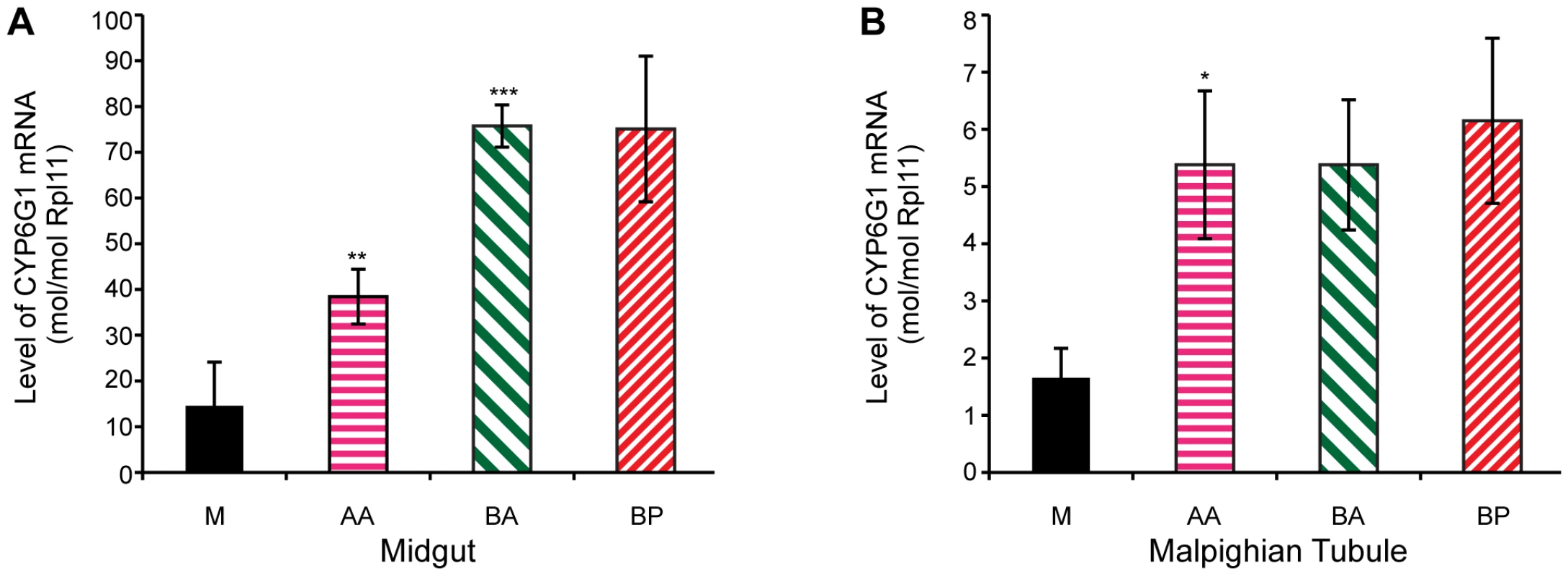
Derived alleles exhibit higher gene expression than Cyp6g1[M]. (A) The allelic progression results in increased gene expression in the midgut with AA having a 2.6 fold increase (p = 0.036, one tailed t-test) over M and BA and BP both showing a 5 fold increase. There is also a significant 2 fold increase of BA over AA (p = 0.0006, one tailed t-test). In contrast, only the step between M and AA results in a significant increase in transcription (p = 0.04, one tailed t-test) in adult tubule (B), and all three derived alleles exhibited ∼3 fold increases in gene expression. Cyp6g1 genotype determines resistance in field populations
There is a formal possibility that some other background genetic factor is by chance correlated with Cyp6g1 allele and that it contributes to the phenotypic trend shown among the lines we analysed for DDT resistance. This is unlikely, partly because most isochromosomal lines were generated in such a way that 100% of the X chromosome material and 75% of the third chromosome material would be derived from the Prl/CyO stock (Table S2). Nevertheless we decided to take a quantitative genetics association study approach to confirm and quantify the role of the most derived high frequency allele, Cyp6g1-[BP], in a field population.
The population for our association study consisted of 7500 non-virgin females that derived from 750 females caught in the field two generations earlier. A subset of our population was used to determine a dosage mortality curve that allowed an estimation of the population LC5 and LC95 values (i.e. the most susceptible 5% and most resistance 5% of the population to DDT exposure). The mortality of flies on a probit scale was close to linear to the log of the dose of DDT indicating that the underlying phenotype approximates a normal distribution (Figure 6A). Based on this we chose 2 µg/scintillation vial ( = 0.05 µg/cm2) of DDT to be the exposure that would only kill the individuals from the susceptible tail of the distribution and 120 µg/scintillation vial ( = 3.2 µg/cm2) to be the exposure to kill all but the most resistant individuals. Four replicates of 500 flies were then exposed to the lower dose of DDT and five replicates of 500 were exposed to the high dose of DDT. Nearly exactly 95% of the flies died (all but 124/2500) on the 120 µg treatment and close to 7% of flies died on the 2 µg treatment (133/2000). All the flies that survived the high dose and all those that died on the low dose, were genotyped for Cyp6g1 allele status. These were compared to 86 field caught males and to random samples of flies that lived on the low dose or died on the high dose. The BP allele is greatly enriched above field frequency among survivors of the high dose but was depleted among those that died at the low dose (Figure 6). Thus the association study confirmed the inbred and isochromosomal line analysis. Trend tests give a very significant association between BP and survival at LC95 (Cochrane Armitage test, chi square 42, p<<0.0001; [37]). In fact flies homozygous for the BP allele are twelve times more likely to survive than non BP homozygotes.
Fig. 6. Population analysis of DDT resistance. 
A DDT association study was conducted on 7500 females derived from a single population. (A) A dosage mortality analysis was undertaken to a identify dose that would kill the 5% of individual that were most susceptible to DDT and the dose that would kill all except the 5% of individuals that were most resistant to DDT. (B) The top three graphs show the allele frequencies of Cyp6g1-[BP] among the individuals that died on 2 ug of DDT (frequency of 0.15), among an unscreened field population sample (frequency of 0.31), and among the survivors of 120 ug of DDT (frequency of 0.58), respectively. Thus there is a strong association between DDT resistance and Cyp6g1-[BP]. The bottom three graphs show the frequency of a Cyp12d1 copy number variant among the same three groups (Cyp12d1 D/− = 0.81, 0.87 and 0.86) showing that it is not associated with field resistance. Note that flies carrying the Cyp12d1 duplication are denoted as D/−, whereas flies homozygous for a single copy are denoted as u/u. Error bars represent standard errors of the mean of biological replicates. For comparison we also genotyped the same flies for a CNV at another candidate DDT resistance locus, Cyp12d1, which is on chromosome 2 approximately 1 Mb away from Cyp6g1. The assay we used bridged the breakpoints of the distal and proximal copies of this locus, which is thus not a codominant assay and therefore individuals either heterozygous or homozygous for the duplication were not discriminated (Flybase release FB2010_01). Therefore a 2×2 test comparing the presence and absence of the Cyp12d1 duplication in resistant and susceptible flies was used which showed there is no correlation with the duplication of this locus and resistance or susceptibility (Fishers exact test P = 0.60, Figure 6).
These data can also be used to quantify the contribution of Cyp6g1 alleles to DDT resistance. Table 1 shows the survivorship of individuals partitioned by Cyp6g1-[BP] genotype. Heterozygotes are less than midway between the two homozygotes indicating this allele is slightly recessive at this dose (Figure S4). To calculate the contribution of this allele in the population, this recessiveness (k = −0.63), the survivorship difference between the two homozygotes (2a = 0.18), and the population frequency of Cyp6g1-[BP] (p = 0.28), need to be taken into account. Thus the average allelic effect of Cyp6g1-[BP] is to increase survivorship of a genotype by 6.5% ([38] equation 4.10b). The additive genetic variance attributable to this allele is 0.0017 ([38] equation 4.12a) and therefore the heritability on the observed scale, at an LC95 dose, attributable to this allele, is 3.7%. For a threshold trait such as this, we are observing binary phenotypes; alive or dead at a particular dose. However, as illustrated by the dose response relationship in Figure 6A, we can assume that there is a normal distribution underlying the DDT resistance trait across doses. Thus the heritability on the observed scale can be converted to an estimate of the heritability on the underlying scale and that indicates that approximately 16.5% of the ‘liability’ to survive is explained by the Cyp6g1-[BP] allele ([38] equation 25.8b). We have not calculated the variance of all genetic factors influencing DDT resistance, and thus do not know the heritability of the trait as a whole, but whatever that heritability is (it has to be between 0.16 and 1), the Cyp6g1-[BP] allele makes a major contribution to phenotypic variation in this population.
Tab. 1. Survivorship/mortality of <i>Cyp6g1-BP</i> genotypes on 120 µg of DDT. 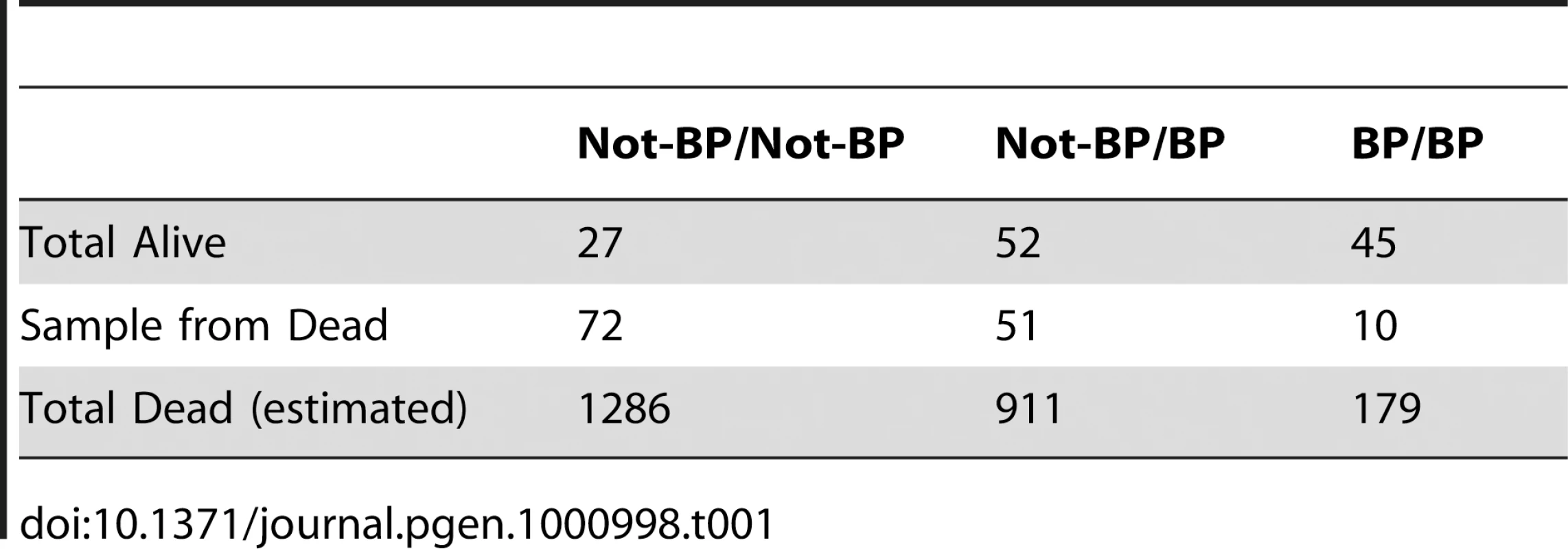
Discussion
An allelic succession at the Cyp6g1 locus increases DDT resistance
Recently, others have identified CNV at the Cyp6g1 locus using genome-wide tiling arrays [29], [30]. In the study of Emerson et al. the resolution of CNV boundaries is such that repeats containing a fusion of partial Cyp6g1 and Cyp6g2 genes were identified. Not only has the present work determined that the CNV represents at least two full-length copies of Cyp6g1, it has also established the way in which this locus has evolved. Furthermore, we have shown how this contributes to increasing DDT resistance using two separate approaches. Firstly we showed that the LC50 to DDT increases with the allelic progression in a set of 19 isochromosomal and 3 inbred lines (M<< AA< BA << BP). Secondly we performed an association study that clearly demonstrates that the BP allele makes a major contribution to the phenotypic variance in a single population where it is at high frequency.
The allelic succession is adaptive
D. melanogaster is not a pest and is generally not targeted by insecticide application. Could it be that variation we detect with our DDT assays is non-adaptive? As discussed in more detail below we present historical and geographic surveys of Cyp6g1 allelic variation that clearly demonstrate that at least two of the steps have been adaptive (the world-wide spread of the Accord bearing alleles and the spread of the BP alleles within Australia). Further support for adaptive change at this locus could come in the form of patterns in the patterns of polymorphism and fixation that support selective sweep models. In fact others have shown evidence for a selective sweep at the Cyp6g1 locus [17], [18], although they have not shown that there have been recurrent sweeps at this locus. The data presented here shows that an adaptive walk has occurred at the Cyp6g1 locus. Although not a classic adaptive walk, where evolution is conceived in coding sequence space [39], the allelic succession that we describe is explained as a sequential process where each new allele is derived directly from that preceding it. In the following we discuss each proposed step in this allelic succession.
The first step: insertion of partial Accord element
The first step of the walk would seem to have been the insertion of the Accord LTR into the Cyp6g1 promoter. As we did not detect the A allele (single copy Cyp6g1 with Accord LTR) in our sample, and thus cannot determine its phenotypic effect, the identification of the AA allele, in which both Cyp6g1 and the Accord LTR are duplicated, indicates that this insertion most likely occurred at or before the duplication event. A question that arises from the failure to detect the single copy A allele, is deciding which molecular variant, the partial Accord TE insertion or the CNV, was the first target of natural selection. A recent study using transgenic reporter genes showed that the Accord LTR acts as an enhancer increasing transcription in tissues consistent with the Cyp6g1 changes observed in ‘Accord flies’ [15]. This suggests the Accord insertion itself could cause the up-regulation and be the target of selection.
The second step: gene duplication
We propose that the second step was the duplication event producing two copies of Cyp6g1. Sequence analysis of cDNA from RK146 (not shown) indicates two full-length copies of Cyp6g1 are transcribed in adult flies, indicating that the duplication acts to increase transcriptional output. It is possible that the Accord insertion and the duplication happened from the one complex event. Thus a minimum of one selective sweep is required to explain the rapid change in frequency of the AA alleles. Alternatively it is possible that there have been two adaptive steps, one that is the Accord insertion and the other the generation of the CNV. In that scenario the A allele may never have reached high frequencies before being replaced by the AA allele. Whichever the case, the net result is a 7–10 fold increase in resistance phenotype in comparison to the ancestral M allele (Figure 4).
The third step: HMS-Beagle insertion
The third step in the walk involved the insertion of the HMS-Beagle insertion into the Accord LTR that lies proximal to Cyp6g1-a. From the DDT resistance data it is hard to determine whether the AA→BA step is adaptive, as the increase in DDT resistance phenotype is only significant for males in our current data set. Our population surveys confirm previous results, which suggest that the Accord alleles (which we now know as A, AA and BA alleles) were either absent or at low frequencies pre-DDT. They also show that both AA and BA alleles began to spread at the same time, and have now spread globally. Among the lines carrying the BA allele is Hikone-R, which was the DDT resistant strain that was initially used to map Cyp6g1-based resistance [21], [24]. Hikone-R was collected in Japan in 1952 and although the AA and BA alleles may have existed for some time before the 1950's, historic fly collections show they have only reached high frequencies recently (Figure 3). These results concur with previously reported skews in the polymorphism frequency spectrum around Cyp6g1, which suggests recent strong positive selection [17], [40].
The fourth step: partial P-element insertion
The fourth step in the walk is the insertion of a partial P-element into the Accord LTR that lies proximal to Cyp6g1-b. Since all flies that carry a P-element insertion also contain an HMS-Beagle element upstream of Cyp6g1-a we infer the insertion would have occurred in a Cyp6g1-[BA] background. This results in a significant increase in DDT resistance phenotype over AA and BA alleles with BP males 6 and 3 fold more resistant respectively. BP females are 8 and 5 fold more resistant than their AA and BA counterparts. Furthermore the association study shows a highly significant association between the BP allele and DDT resistance.
Curiously, the robust association between the BP allele and the DDT resistance phenotype is not reflected in our transcriptional analysis. This may be because the P-element insertion may simply be a marker in linkage disequilibrium with the causal variant – which could be an amino acid change in an uncharacterised copy of Cyp6g1. Another possibility is that the BP allele gives resistance by altering the transcript abundance in tissues that have not been assayed here. The possibility that there is tissue specific variation in transcript levels is illustrated by the observed differences in expression between Malpighian tubules and midgut. Thus it is possible, for instance, that Cyp6g1 transcription is higher in the head, fat body or reproductive tissues in BP lines.
Regardless of the exact details of the molecular mechanism of resistance we have no doubt that the fourth step is adaptive, as analysis of eight Australian populations suggests the Cyp6g1-[BP] variant has recently and rapidly increased to be the most frequent allele in Australia. Thus Australian flies are very different from other parts of the world where BP alleles were recorded in only 10 out of 683 lines [17]. The lack of BP alleles in fly lines established from Papua New Guinea and Australia in the 1980s (Figure 4A) supports this selection model as do reports that the P-element transposable element itself was only detected in Australia in the late 1970's [41].
Neither drift nor population bottlenecks can satisfactorily account for the high frequency of BP alleles in Australia. We conducted three independent surveys of contemporary Australian populations (our original survey, a survey of east Australian clinal samples and the association study collection) sampling from multiple locations spanning the east coast of Australia. Thus a local bottleneck (i.e. from a single collection site) could not explain the data. Similarly D. melanogaster populations from the east coast of Australia exhibit extensive gene flow and share the same diversity as non-African populations [35], [42], indicating populations are not isolated. There has also been enough time since their introduction to Australia for the establishment of strong latitudinal clines that parallel those found in other parts of the globe [43]–[45]. Furthermore the flies used here to survey allele frequency across the east coast of Australia have been previously characterized for many other loci and are consistent with other Australian surveys, ruling out the possibility that our samples are somehow biased, non-representative or corrupted[43]. Finally if the P element did not enter Australia until the 1970's [41] then the BP alleles of Cyp6g1 must have entered into established populations. There is no way that genetic drift could explain the frequencies of BP alleles, rather the BP alleles must have spread through these populations with positive selection. This raises the interesting proposition that the BP alleles may increase in frequency in other parts of the world in the future.
It is worth noting that DDT has been banned from use in most of the world including Australia since the 1980s [46] and yet we are postulating that the BP allele has risen to high frequency in Australian populations since then. Notwithstanding the possibility that DDT still persists in the environment, we also note that it is well established that Cyp6g1 upregulation provides resistance to a number of insecticides and other chemicals [13], [24]. Thus DDT resistance may be considered as a phenotypic marker of this allelic variation rather than the actual selective agent.
Further steps: more CNV and TE sequence deletion
Our population surveys also identified two different Cyp6g1-[BPΔ] alleles, formed by imprecise excision of the P-element insertion. These alleles are at low frequency, and we have not characterised their contribution to DDT resistance. Their formation questions the stability of the locus structure that we have defined. Over evolutionary time we would expect this structure to be simplified. For instance if the BPΔ alleles have the same phenotype as their BP parent, it may indicate that only discrete functional DNA sequences need be preserved, with the rest free to be deleted. Opposing this simplification is the instability introduced by the gene duplications, which may increase the rate of copy number variation by molecular slippage. We have shown that in the RK146 strain there are at least two full-length copies of Cyp6g1, but in light of the above it is possible that even more copies exist in other strains.
Precedence for allelic succession
Allelic succession, the process whereby different adaptive alleles are substituted sequentially, has also been characterised in several studies of insecticides resistance. In Culex pipiens mosquitoes, alleles at the Ester ‘super-locus’ (so called because some alleles contain CNV of more than one gene) confer organophosphate (OP) resistance. Ester1 and Ester4 both result in the overproduction of an insecticide metabolising esterase [47]. Ester1 was first detected in 1972, while Ester4 appeared over a decade later. Despite a moderately lower level of OP resistance Ester4 has replaced Ester1 due to a lower overall fitness cost [48]. In a second Culex example, the resistance allele of the target of OP insecticides, Ace1R, also confers a fitness cost, but this cost seems to have been reduced through gene duplication by the creation of a permanent heterozygous allele, consisting of a copy each of the susceptible and resistant alleles [49]. Allelic succession in both these cases appears to be driven by selection removing a fitness cost introduced with the preceding resistance allele.
Two previous studies have associated fitness costs with Cyp6g1 upregulation. A study in which males were selected for reduced competitive mating success indicated that Cyp6g1 was significantly upregulated [50]. In contrast, females seem to carry no cost for a range of fitness traits [51]. It would be worthwhile revisiting these earlier experiments in light of the complex variation we have shown to exist for Cyp6g1. However it is not necessary to invoke fitness costs, as the data shown in Figure 4 suggests that the allelic succession occurring at Cyp6g1 is driven by selection for ever-greater resistance.
Concluding remarks
Cyp6g1 has become a highly cited example of adaptive evolution [52]–[54]. Cyp6g1 resistance alleles are not only selected, in parallel, in sibling species, we show that they have also been repeatedly selected, in series, in D. melanogaster. These results are pertinent to a long-standing evolutionary debate concerning the number of steps that have to occur to move a species to a new adaptive optimum [6], [55]. Support for a model requiring only a moderate number of steps includes mapping experiments, where the number of loci that contribute to a given adaptive trait are calculated, and their relative phenotypic effects apportioned. However, as recently described by the careful dissection of a morphological trait that differs between species, mapping experiments may hide the number of steps that have occurred at a single locus over evolutionary time [56]. Here we have described at least four steps at a single locus that have occurred within 70 years. The intense selection of insecticides has provided the opportunity to see the adaptive process at a resolution invisible in many other examples of adaptation.
Materials and Methods
Fly stocks
The stocks used for the DDT toxicology experiment were made isochromosomal for the II chromosome by backcrossing to Prl/CyO flies (Bloomington Stock 3079). For stock list see Table S2.
Polytene chromosomes
Probes were made using the PCR DIG Probe Synthesis kit of Roche Boehringer Mannheim (version# 2003). The primers: 5′-CAGCCTAGAGAATCCCAACG-3′ and 5′GCCATGGCCACTATGTTCTT-3′ were used to amplify exon 3 and exon 4 from a Cyp6g1 subclone. The chromosomes were prepared following the method of Phillips et al [57].
Long PCR
Roche Expand High Fidelity PCR system was used to generate all PCR products greater than 2.5 kb following the manufacturers protocol except with the following cycling parameters: 94°C 2 mins, 10 cycles of 94°C 15s, 62°C* 30s, 68°C 10 mins, 30 cycles of 94°C 15s, 56°C 30s, 68°C 10# mins, and a final 60 min extension at 68°C. * A touchdown cycle, with the annealing temperature decreasing by 0.5°C per cycle. # Extension time increases 10 s per cycle. Primers used (refer Figure 2) listed 5′-3′: A CGTCTTAGAAAGAAACAGGAAACTG, B ACATTTGGGAGATGCCTTTG, C ATTAAACACAACCGGCTTTCTCG, D GTCTCACCACCCAGGAAAGA, E CTTTTTGTGTGCTATGGTTTAGTTAG, F GGGTGCAACAGAGTTTCAGGTA, G TTTCAGCCAGTTGGACATTG. PCR products were gel purified and cloned using the TOPO XL PCR Cloning Kit (Invitrogen) following manufacturers instructions.
Inverse PCR
125 ng of RK146 genomic DNA was digested with EcoR1 in a total volume of 100 uL, then 5 uL of the digest was diluted to 100 uL in a ligation reaction mix, left overnight at 14°C. This allowed linear EcoR1 fragments to circularise. PCR's using primers 5′-GATCCGCGGCTGAAGGACGA-3′ and 5′-TGCGGCGACCACCACAAAGA-3′ were conducted with the 30 cycles of: 94°C for 30 s, 62°C for 30 s and 68°C for 2minutes. A nested PCR was then performed using a new reverse primer (5′-TGCCAGTGCCCTCAGCATTATCTTATC-3′) and the original forward primer (5′-GATCCGCGGCTGAAGGACGA-3′). The product was cloned into pGEM-T easy and sequenced.
Allele scoring
DNA was prepared from single flies. Diagnostic assays to detect TE insertions and the Cyp6g1 gene duplication used standard PCR conditions with the following cycling parameters: 94°C 2 mins, 30 cycles of 94°C 15 s, *°C 30 s, 72°C # mins. Reactions used the following primers (refer Figure 2) listed 5′-3′: H GAAAGCCGGTTGTGTTTAATTAT, I CTTTTTGTGTGCTATGGTTTAGTTAG, J CGAGTACGAGAGCGTGGAG, K ATTAAACACAACCGGCTTTCTCG, L TGCGATCATCTGCACTTCTC. Annealing temperatures, *, and extension times, #, for each primer pair: HI 57°C 2 mins, JK 56°C 1 min, LI 58°C 45 secs.
DDT resistance assays
4 day old non-virgin male and female flies were treated separately. DDT was coated on the inside of glass scintillation vials by applying 200 µl of acetone containing varying concentrations of DDT and rolling the vial until the acetone had evaporated. 20 flies per vial were used with the vials plugged with cotton soaked in 5% sucrose. Mortality was scored after 24 h. LC50 estimation was performed using PriProbit[58], using five concentrations and three replicates per concentration.
Analysis of gene expression
Midguts and Tubules were dissected separately from 4 day old adult males, 5 strains per genotype, and pooled in groups of 6–10 tissues, for 3 biological replicates per strain. mRNA was extracted in 200 ul Trizol and 60 ul chloroform. After being pelleted all the extracted RNA from each sample was used in cDNA synthesis, and cDNA was reverse transcribed and quantified according to standard procedures. For quantitative PCR (qPCR), samples were split and amplified with Cyp6g1 primers, using Rpl11 as a reference gene.
DDT association study
750 isofemale lines were established from field caught females. At the F2, 10 4–8 day non-virgin females were collected from each line, in essence recapitulating the extant genetic variation of this population. Our experimental design involved comparing the two tails of the DDT resistance phenotype distribution. To this end a subset of the F2 flies were used to determine a dosage mortality curve for the population and allow estimation of the population LC5 and LC95 values (2 µg and 190 µg respectively). The large 95% confidence interval for the LC95 estimate suggested a dose of 190 µg would be inaccurate, so instead we used a dose of 120 µg, a dose slightly higher than the highest dose used in the DMC assay (112.5 µg) which had ∼92% mortality. These doses were scaled, by internal surface area, to allow exposure of 500 individuals in 2 L Schott bottles, which were stoppered by cotton wool wrapped around a 10 ml disposable pipette. After exposure for 24 hours a vacuum pump was attached to the pipette to remove the flies into separate dead and alive cohorts. For the LC5, all dead flies (133 in total) and an equivalent number of surviving flies were assayed. For the LC95 all surviving flies (124) and an equivalent number of dead flies were assayed. Flies were assayed using the HI primer pair to detect Accord/P-element status as described above. Determination of the Cyp12d1 duplication genotype status (u/u or D/−) of these flies utilised a four primer PCR reaction using the same general PCR conditions described above. Primers used were; Rout TCCTAAGAATTCCCACCATCAC, Rin GGTCCATCATCCCTACCATTT, Fout GGCCATTACGTTCCCCTTC and Fin GGTCTCGGAAAATGAGCAAC. The Rin/Fout and Rout/Fin primer pairs amplify products from both single copy and duplicated Cyp12d1 loci 767 bp 933 bp in length respectively. The pair Rout/Fout is specific to the presence of the gene duplication, and gives a product of 389 bp.
Supporting Information
Zdroje
1. AndolfattoP
2005
Adaptive evolution of non-coding DNA in Drosophila.
Nature
437
1149
1152
2. SmithNG
Eyre-WalkerA
2002
Adaptive protein evolution in Drosophila.
Nature
415
1022
1024
3. SellaG
PetrovDA
PrzeworskiM
AndolfattoP
2009
Pervasive natural selection in the Drosophila genome?
PLoS Genet
5
e1000495
doi:10.1371/journal.pgen.1000495
4. BarrickJE
YuDS
YoonSH
JeongH
OhTK
2009
Genome evolution and adaptation in a long-term experiment with Escherichia coli.
Nature
461
1243
1247
5. BegunDJ
HollowayAK
StevensK
HillierLW
PohYP
2007
Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans.
PLoS Biol
5
e310
doi:10.1371/journal.pbio.0050310
6. OrrHA
2005
The genetic theory of adaptation: a brief history.
Nature Reviews Genetics
6
119
127
7. GillespieJH
1991
The causes of Molecular Evolution: Oxford University Press Inc
8. PerfeitoL
FernandesL
MotaC
GordoI
2007
Adaptive mutations in bacteria: high rate and small effects.
Science
317
813
815
9. AndreevD
KreitmanM
PhillipsTW
BeemanRW
ffrench-ConstantRH
1999
Multiple origins of cyclodiene insecticide resistance in Tribolium castaneum (Coleoptera: Tenebrionidae).
JME
48
615
624
10. ffrench-ConstantRH
1994
The molecular and population genetics of cyclodiene insecticide resistance.
Insect Biochem Mol Biol
24
335
345
11. SoderlundDM
KnippleDC
2003
The molecular biology of knockdown resistance to pyrethroid insecticides.
Insect Biochem Mol Biol
33
563
577
12. ClaudianosC
RussellRJ
OakeshottJG
1999
The same amino acid substitution in orthologous esterases confers organophosphate resistance on the house fly and a blowfly.
Insect Biochemistry and Molecular Biology
29
675
686
13. DabornPJ
YenJL
BogwitzMR
Le GoffG
FeilE
2002
A single p450 allele associated with insecticide resistance in Drosophila.
Science
297
2253
2256
14. JoussenN
HeckelDG
HaasM
SchuphanI
SchmidtB
2007
Metabolism of imidacloprid and DDT by P450 CYP6G1 expressed in cell cultures of Nicotiana tabacum suggests detoxification of these insecticides in Cyp6g1-overexpressing strains of Drosophila melanogaster, leading to resistance.
Pest Manag Sci
15. ChungH
BogwitzMR
McCartC
AndrianopoulosA
Ffrench-ConstantRH
2007
Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1.
Genetics
175
1071
1077
16. HoekstraHE
CoyneJA
2007
The locus of evolution: Evo devo and the genetics of adaptation.
Evolution
61
995
1016
17. CataniaF
KauerMO
DabornPJ
YenJL
Ffrench-ConstantRH
2004
World-wide survey of an Accord insertion and its association with DDT resistance in Drosophila melanogaster.
Mol Ecol
13
2491
2504
18. SchlenkeTA
BegunDJ
2004
Strong selective sweep associated with a transposon insertion in Drosophila simulans.
Proc Natl Acad Sci U S A
101
1626
1631
19. CrowJF
1957
Genetics of insect resistance to chemicals.
Annual Reviews of Entomology
2
227
246
20. DapkusD
MerrellDJ
1977
Chromosomal analysis of DDT-resistance in a long-term selected population of Drosophila melanogaster.
Genetics
87
685
697
21. Kikkawa
1961
Genetical studies on the resistance to parathion in Drosophila melanogaster.
9
1
20
Annual Report of Scientific Works, Faculty of Science, Osaka University
22. HallstromI
1985
Genetic regulation of the cytochrome P-450 system in Drosophila melanogaster. II. Localization of some genes regulating cytochrome P-450 activity.
Chem Biol Interact
56
173
184
23. KikkawaH
1964
Genetical studies on the resistance to parathion in Drosophila melanogaster. II. Induction of a resistance gene from its susceptible allele.
Botyu-Kagaku
29
37
42
24. DabornP
BoundyS
YenJ
PittendrighB
ffrench-ConstantR
2001
DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid.
Mol Genet Genomics
266
556
563
25. PedraJH
McIntyreLM
ScharfME
PittendrighBR
2004
Genome-wide transcription profile of field - and laboratory-selected dichlorodiphenyltrichloroethane (DDT)-resistant Drosophila.
Proc Natl Acad Sci U S A
101
7034
7039
26. AmichotM
TaresS
Brun-BaraleA
ArthaudL
BrideJM
2004
Point mutations associated with insecticide resistance in the Drosophila cytochrome P450 Cyp6a2 enable DDT metabolism.
Eur J Biochem
271
1250
1257
27. BrandtA
ScharfM
PedraJH
HolmesG
DeanA
2002
Differential expression and induction of two Drosophila cytochrome P450 genes near the Rst(2)DDT locus.
Insect Mol Biol
11
337
341
28. DabornPJ
LumbC
BoeyA
WongW
Ffrench-ConstantRH
2007
Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression.
Insect Biochem Mol Biol
37
512
519
29. EmersonJJ
Cardoso-MoreiraM
BorevitzJO
LongM
2008
Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster.
Science
320
1629
1631
30. DopmanEB
HartlDL
2007
A portrait of copy-number polymorphism in Drosophila melanogaster.
Proc Natl Acad Sci U S A
31. PykeFM
BogwitzMR
PerryT
MonkA
BatterhamP
2004
The genetic basis of resistance to diazinon in natural populations of Drosophila melanogaster.
Genetica
121
13
24
32. OshimaC
1958
The resistance of strains of D. melanogaster to DDT and Dieldrin.
The American Naturalist
92
171
182
33. TaylorMF
HeckelDG
BrownTM
KreitmanME
BlackB
1993
Linkage of pyrethroid insecticide resistance to a sodium channel locus in the tobacco budworm.
Insect Biochem Mol Biol
23
763
775
34. DavidJR
CapyP
1988
Genetic variation of Drosophila melanogaster natural populations.
Trends Genet
4
106
111
35. TurnerTL
LevineMT
EckertML
BegunDJ
2008
Genomic analysis of adaptive differentiation in Drosophila melanogaster.
Genetics
179
455
473
36. YangJ
McCartC
WoodsDJ
TerhzazS
GreenwoodKG
2007
A Drosophila systems approach to xenobiotic metabolism.
Physiological Genomics
30
223
231
37. GibsonG
MuseS
2004
A primer of genome science.
Sunderland, , MA
Sinauer Associations
38. LynchM
WalshB
1998
Genetics and Analysis of Quantitative Traits.
Sunderland, , MA
Sinauer Associates.
980
39. SmithJM
1970
Natural selection and the concept of a protein space.
Nature
225
563
564
40. PanhuisTM
SwansonWJ
2006
Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila.
Genetics
173
2039
2047
41. BoussyIA
ItohM
RandD
WoodruffRC
1998
Origin and decay of the P element-associated latitudinal cline in Australian Drosophila melanogaster.
Genetica
104
45
57
42. KenningtonW
GockelJ
PartridgeL
2003
Testing for asymmetrical gene flow in a Drosophila melanogaster body-size cline.
Genetics
165
667
673
43. UminaP
WeeksA
KearneyM
McKechnieS
HoffmannA
2005
A rapid shift in a classic clinal pattern in Drosophila reflecting climate change.
Science
308
691
693
44. AndersonPR
OakeshottJG
1984
Parallel geographical patterns of allozyme variation in two sibling Drosophila species.
Nature
308
729
731
45. HoffmannAA
WeeksAR
2007
Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia.
Genetica
129
133
147
46. Australian GovernmentCPS
Department of the Environment, Water, Heritage, and the Arts
1997
Scheduled Wastes Fact Sheet.
In: Department of the Environment W, Heritage, and the Arts, editor
47. RookerS
GuillemaudT
BergeJ
PasteurN
RaymondM
1996
Coamplification of esterase A and B genes as a single unit in Culex pipiens mosquitoes.
Heredity
77 (Pt 5)
555
561
48. GuillemaudT
LenormandT
BourguetD
ChevillonC
PasteurN
1998
Evolution of resistance in Culex pipiens: Allele replacement and changing environment.
Evolution
52
443
453
49. LenormandT
GuillemaudT
BourguetD
RaymondM
1998
Appearance and sweep of a gene duplication: Adaptive response and potential for new functions in the mosquito Culex pipiens.
Evolution
52
1705
1712
50. DrnevichJM
ReedyMM
RuediEA
Rodriguez-ZasS
HughesKA
2004
Quantitative evolutionary genomics: differential gene expression and male reproductive success in Drosophila melanogaster.
Proc Biol Sci
271
2267
2273
51. McCartC
BucklingA
Ffrench-ConstantRH
2005
DDT resistance in flies carries no cost.
Curr Biol
15
R587
589
52. LiXC
SchulerMA
BerenbaumMR
2007
Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics.
Annual Review of Entomology
52
231
253
53. WrayGA
2007
The evolutionary significance of cis-regulatory mutations.
Nature Reviews Genetics
8
206
216
54. BrookfieldJF
2005
The ecology of the genome - mobile DNA elements and their hosts.
Nature Reviews Genetics
6
128
136
55. DeanAM
ThorntonJW
2007
Mechanistic approaches to the study of evolution: the functional synthesis.
Nature Reviews Genetics
8
675
688
56. McGregorAP
OrgogozoV
DelonI
ZanetJ
SrinivasanDG
2007
Morphological evolution through multiple cis-regulatory mutations at a single gene.
Nature
448
587
590
57. PhillipsAM
MartinJ
BedoDG
2000
In situ hybridization to polytene chromosomes of Drosophila melanogaster and other dipteran species.
Methods Mol Biol
123
83
102
58. SakumaM
1998
Probit analysis of preference data.
Applied Entomology and Zoology
33
339
347
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 6- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- Translational Selection Is Ubiquitous in Prokaryotes
- Whole-Genome Sequencing of a Single Proband Together with Linkage Analysis Identifies a Mendelian Disease Gene
- Web-Based, Participant-Driven Studies Yield Novel Genetic Associations for Common Traits
- The Met 470 Allele Is Associated with Lower Birth Rates in Fertile Men from a Population Isolate
- Contributions of Status and Allele Expression, But Not Copy Number Variation, to the Control of SIVmac251 Replication in Indian-Origin Rhesus Monkeys
- A Genome-Wide Association Study of Optic Disc Parameters
- Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid
- The Transcription Factor REST Is Lost in Aggressive Breast Cancer
- 14-3-3 Mediates Histone Cross-Talk during Transcription Elongation in
- Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
- Use of Genome-Wide Expression Data to Mine the “Gray Zone” of GWA Studies Leads to Novel Candidate Obesity Genes
- Genome-Wide RNAi Screen Identifies Multiple Regulators of HIF–Dependent Transcription in Hypoxia
- The CYCLIN-A CYCA1;2/TAM Is Required for the Meiosis I to Meiosis II Transition and Cooperates with OSD1 for the Prophase to First Meiotic Division Transition
- Inactivation of hnRNP K by Expanded Intronic AUUCU Repeat Induces Apoptosis Via Translocation of PKCδ to Mitochondria in Spinocerebellar Ataxia 10
- Mice with Alopecia, Osteoporosis, and Systemic Amyloidosis Due to Mutation in , a Gene Coding for Palmitoyl Acyltransferase
- siRNA–Mediated Methylation of Telomeres
- Chromosome 4 Replicates in Two Phases That Correlate with Chromatin State
- Dynamic Switch of Negative Feedback Regulation in Akt–TOR Signaling
- On the Use of Variance per Genotype as a Tool to Identify Quantitative Trait Interaction Effects: A Report from the Women's Genome Health Study
- and Deficiency Cooperate in the Progression of Mouse Prostate Tumourigenesis
- An Integration of Genome-Wide Association Study and Gene Expression Profiling to Prioritize the Discovery of Novel Susceptibility Loci for Osteoporosis-Related Traits
- Consent and Internet-Enabled Human Genomics
- Understanding Adaptation in Large Populations
- Identification of a Functional Genetic Variant at 16q12.1 for Breast Cancer Risk: Results from the Asia Breast Cancer Consortium
- Evidence that Adaptation in Is Not Limited by Mutation at Single Sites
- The IG-DMR and the -DMR at Human Chromosome 14q32.2: Hierarchical Interaction and Distinct Functional Properties as Imprinting Control Centers
- Cushing's Syndrome and Fetal Features Resurgence in Adrenal Cortex–Specific Knockout Mice
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Web-Based, Participant-Driven Studies Yield Novel Genetic Associations for Common Traits
- The IG-DMR and the -DMR at Human Chromosome 14q32.2: Hierarchical Interaction and Distinct Functional Properties as Imprinting Control Centers
- Amplification of a Cytochrome P450 Gene Is Associated with Resistance to Neonicotinoid Insecticides in the Aphid
- Copy Number Variation and Transposable Elements Feature in Recent, Ongoing Adaptation at the Locus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



