-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBias and Evolution of the Mutationally Accessible Phenotypic Space in a Developmental System
Genetic and developmental architecture may bias the mutationally available phenotypic spectrum. Although such asymmetries in the introduction of variation may influence possible evolutionary trajectories, we lack quantitative characterization of biases in mutationally inducible phenotypic variation, their genotype-dependence, and their underlying molecular and developmental causes. Here we quantify the mutationally accessible phenotypic spectrum of the vulval developmental system using mutation accumulation (MA) lines derived from four wild isolates of the nematodes Caenorhabditis elegans and C. briggsae. The results confirm that on average, spontaneous mutations degrade developmental precision, with MA lines showing a low, yet consistently increased, proportion of developmental defects and variants. This result indicates strong purifying selection acting to maintain an invariant vulval phenotype. Both developmental system and genotype significantly bias the spectrum of mutationally inducible phenotypic variants. First, irrespective of genotype, there is a developmental bias, such that certain phenotypic variants are commonly induced by MA, while others are very rarely or never induced. Second, we found that both the degree and spectrum of mutationally accessible phenotypic variation are genotype-dependent. Overall, C. briggsae MA lines exhibited a two-fold higher decline in precision than the C. elegans MA lines. Moreover, the propensity to generate specific developmental variants depended on the genetic background. We show that such genotype-specific developmental biases are likely due to cryptic quantitative variation in activities of underlying molecular cascades. This analysis allowed us to identify the mutationally most sensitive elements of the vulval developmental system, which may indicate axes of potential evolutionary variation. Consistent with this scenario, we found that evolutionary trends in the vulval system concern the phenotypic characters that are most easily affected by mutation. This study provides an empirical assessment of developmental bias and the evolution of mutationally accessible phenotypes and supports the notion that such bias may influence the directions of evolutionary change.
Published in the journal: . PLoS Genet 6(3): e32767. doi:10.1371/journal.pgen.1000877
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000877Summary
Genetic and developmental architecture may bias the mutationally available phenotypic spectrum. Although such asymmetries in the introduction of variation may influence possible evolutionary trajectories, we lack quantitative characterization of biases in mutationally inducible phenotypic variation, their genotype-dependence, and their underlying molecular and developmental causes. Here we quantify the mutationally accessible phenotypic spectrum of the vulval developmental system using mutation accumulation (MA) lines derived from four wild isolates of the nematodes Caenorhabditis elegans and C. briggsae. The results confirm that on average, spontaneous mutations degrade developmental precision, with MA lines showing a low, yet consistently increased, proportion of developmental defects and variants. This result indicates strong purifying selection acting to maintain an invariant vulval phenotype. Both developmental system and genotype significantly bias the spectrum of mutationally inducible phenotypic variants. First, irrespective of genotype, there is a developmental bias, such that certain phenotypic variants are commonly induced by MA, while others are very rarely or never induced. Second, we found that both the degree and spectrum of mutationally accessible phenotypic variation are genotype-dependent. Overall, C. briggsae MA lines exhibited a two-fold higher decline in precision than the C. elegans MA lines. Moreover, the propensity to generate specific developmental variants depended on the genetic background. We show that such genotype-specific developmental biases are likely due to cryptic quantitative variation in activities of underlying molecular cascades. This analysis allowed us to identify the mutationally most sensitive elements of the vulval developmental system, which may indicate axes of potential evolutionary variation. Consistent with this scenario, we found that evolutionary trends in the vulval system concern the phenotypic characters that are most easily affected by mutation. This study provides an empirical assessment of developmental bias and the evolution of mutationally accessible phenotypes and supports the notion that such bias may influence the directions of evolutionary change.
Introduction
A principal quest in biology is to disentangle the relative contribution and interplay of mutational versus selective forces in the evolutionary process [1]. While biological research is predominated by the search for adaptive explanation underlying phenotypic evolution, it is also of critical importance to study how the mutational process alone produces phenotypic variation. Such studies indicate which phenotypic space can actually be explored by mutation to generate variation for selection to act upon. The mutationally inducible phenotypic spectrum is thus the fundamentally limiting force constraining and biasing potential evolutionary trajectories of the phenotype.
Importantly, the mutational spectrum is multidimensional and quantitative in character, where certain regions of the phenotypic space may be easier to reach by mutation than others. In quantitative genetic terms, the mutational variance VM of the phenotype represents the amount of variation introduced into the population by mutation each generation and can be extended to a multidimensional phenotypic space, theoretically the M matrix of mutational variance-covariance between phenotypic traits [2]–[4]. The structure of the mutationally accessible space can be best determined through the use of mutation accumulation (MA) lines, where mutations are allowed to accumulate for many generations with minimal selection [5]. Although the importance of the multivariate mutational process is well-appreciated theoretically [6],[7], empirical data are limited and most studies have focused on complex, composite traits, particularly life-history traits [8]–[10]. To our knowledge, no study has attempted to characterize the multivariate mutational structure of a developmental system.
Developmental bias and evolution
It is evident that the genotype-phenotype map, encompassing organismal development, is highly non-linear, so that random mutation does not result in random phenotypic variation. For example, mutation may induce plentiful phenotypic variation for one trait but none for another. In the extreme case there is an absolute bias, so that certain phenotypes are impossible to generate though mutationally induced developmental changes, i.e. there is a developmental constraint [11],[12]. The phenomenon of developmental bias can be thought of as milder, relative constraint, where random mutational (or environmental) effects translate preferably into certain phenotypes [13]–[15]. Differences in such bias may be primarily quantitative and can be expressed as different probabilities of generating a given phenotypic spectrum upon random perturbation.
There is circumstantial evidence that developmental bias is common [13], [15]–[19]. In addition, experimental evidence suggests that genetic and developmental architecture bias the production of phenotypic variation. For example, repeated instances of parallel evolution indicate that evolution may follow a limited range of pathways [e.g. 20,21]. However, identifying the relative contribution of mutational versus selective forces in these comparative analyses remains challenging. Recent tests using experimental evolution approaches provide direct evidence on how genetic architecture may bias molecular variation made available to selection [22].
Overall, very few studies [e.g. 23] have quantified the inducible spectrum of phenotypic variation to evaluate whether “intrinsic” tendencies may influence the direction of phenotypic evolution. In general, as pointed out by Yampolsky & Stoltzfus (2001) there is little research focusing on experimental characterization of the spectrum of spontaneous variation and the underlying causes of molecular and developmental causes of any observed biases, which would allow testing the hypothesis that biases in the introduction of variation have influenced evolutionary patterns of the examined traits.
Genotype-dependence of developmental mutability
The mutational architecture may itself evolve, i.e. the regions of phenotypic space reached by mutation differ among genotypes. In other words, developmental bias is genotype-dependent. The inducible phenotypic spectrum for a given genotype has been referred to as “phenotypic neighbourhood” [24] or “local bias” [17]. Such evolutionary variation in mutational properties may be characterized by comparative quantitative analyses of mutation accumulation (MA) lines started from multiple distinct genotypes. Such studies show that mutational parameters may vary substantially between taxa and/or between genotypes of a single species [25]–[27]. We previously showed that mutational damage accumulates about twice as fast in C. briggsae as in C. elegans for lifetime reproductive output (≈“fitness”) [25],[28], body size [29], and at dinucleotide microsatellites [30]. These results reveal evolution of quantitative biases in the production of phenotypic variation (which could be due to evolution of mutation rates), but the underlying developmental and molecular causes of biases in the examined traits are so far unknown. To quantify and evaluate the significance of developmental bias and its genotype-dependence, analogous studies need to be carried out in simple, tractable developmental systems.
The study system: Caenorhabditis vulval cell fate patterning
C. elegans vulval cell fate patterning is a model system for the study of intercellular signalling events [31] and has also served to study developmental robustness, cryptic variation and evolution [32]–[35].
The C. elegans hermaphrodite vulva develops from a subset of ventral epidermal blast cells, the Pn.p cells. In wild-type animals, three neighbouring cells, P5.p, P6.p and P7.p adopt vulval cell fates in the sequence 2°−1°−2°. Furthermore, three additional Pn.p cells, P3.p, P4.p and P8.p, have the capacity to adopt a vulval cell fate, when one or more cells of P5.p to P7.p are missing [36]. The six cells, P3.p to P8.p, therefore constitute the vulval competence group. During the second and third larval stages, the vulval precursor cells adopt alternative cell fates governed by an intercellular signalling network of Ras, Notch and Wnt pathways (Figure 1). A correct fate pattern of three vulval precursor cells (2°−1°−2°) is required to form a functional vulva. Deviation from this pattern can cause a reduction in offspring number due to impaired egg laying capacity and may further prevent male mating [34].
Fig. 1. Caenorhabditis elegans vulval cell fate patterning. 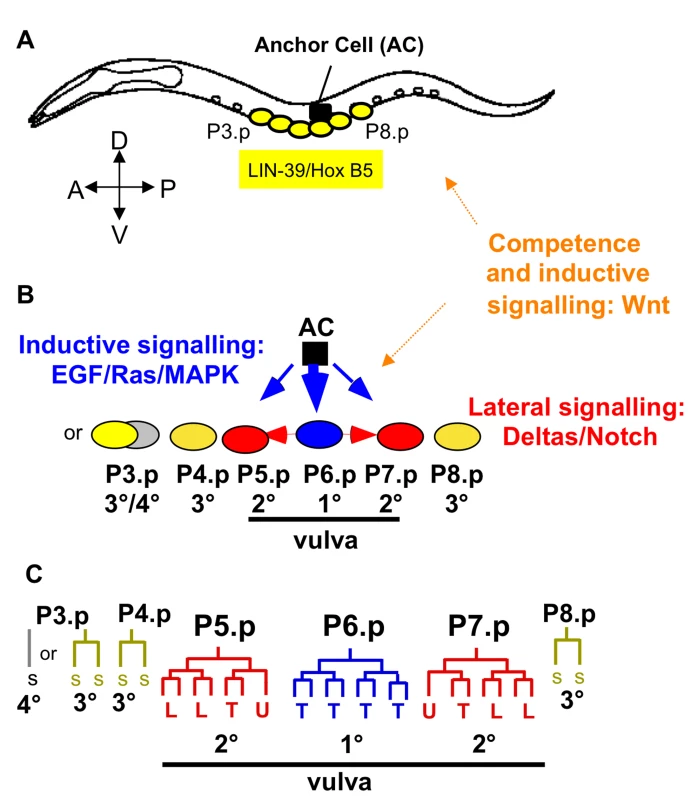
The different cell fates of P3.p to P8.p are characterized by their cell lineage, i.e. number and orientation of cell divisions. Only three precursor cells, P5.p, P6.p and P7.p, adopt a vulval fate: P6.p adopts the central 1° vulval fate, and P5.p and P7.p the outer 2° vulval fate. P4.p and P8.p adopt a non-vulval 3° fate. P3.p shows high variability in its cell fate: it may divide once (3° fate) or it directly fuses with the syncytium without division (4° fate). For the reference isolate N2, the ratio of individuals adopting the 3° versus the 4° fate is approximately 1:1 [37],[68]. The canonical C. elegans pattern for P3.p to P8.p is thus defined as follows: 3°/4°−3°−2°−1°−2°−3°. The vulval cell fate pattern is conserved in Caenorhabditis. The lineages and competence properties of P3.p, P4.p and P8.p, however, may vary within and between species. Specifically, P3.p divides less frequently and is less competent (to adopt a vulval fate) in C. briggsae; in addition, P4.p and P8.p do not always divide in C. briggsae, i.e. they do sometimes adopt a 4° instead of a 3° fate [34],[37],[38],[57]. (A) L1-L2 stages: Competence establishment and maintenance of the vulval competence group. (B) Early L3 stage: Specification of vulval precursor cell fates involving primarily EGF/Ras/MAPK and Delta/Notch pathways. (C) Late L3 stage: Cell lineages. Each vulval fate corresponds to an invariant cell division pattern executed during the late L3 stage, resulting in a total of 22 vulval cells. The cell lineages of P5.p to P7.p are identical in all Caenorhabditis species [38]. Vulval morphogenesis takes place during the L4 stage and the complete vulval organ develops by the final moult to the adult. AC: anchor cell, T: transverse (left-right) division, L: longitudinal (antero-posterior) division, U: undivided, SS: fusion to the epidermal syncytium (hyp7) after single division (3° fate); S: fusion to the syncytium in the L2 stage with no division (4° fate) (3° and 4° fates: non-vulval fates). Vulval cell fate patterning is conserved among Caenorhabditis species [37]–[39]: P5.p to P7.p adopt vulval fates with the pattern 2°−1°−2° while all other vulval precursor cells adopt non-vulval fates, either a 3° fate (the Pn.p cell divides once) or a 4° fate (the Pn.p cell fuses early to the epidermal syncytium hyp7 without division). Species, however, may differ in the frequency of 3° versus 4° fate adopted by P3.p, P4.p and P8.p [37] and in the replacement competence of these cells upon laser ablation [38].
We previously quantified the precision of vulval development of (isogenic) C. elegans and C. briggsae isolates in multiple experimental environments [34],[37]. The results suggest that vulval development is robust to environmental and stochastic perturbations: apparent vulval defects occur in approximately 1 out of 1000 animals [34]. In contrast, developmental defects and variants increased significantly in mutation accumulation lines derived from a single C. elegans isolate, N2 [40], thus degrading the precision of vulval cell fate patterning [34]. This result indicates that mutation accumulation represents a feasible approach to quantify largely unbiased, mutationally induced phenotypic variation of this developmental system.
In this study, we examined the variation in mutational responses of the vulval developmental system within and between related species. We used mutation accumulation (MA) lines derived from two C. briggsae (HK104 and PB800) and two C. elegans (N2 and PB306) wild isolates that had accumulated mutations over approximately 250 generations [25]. We focused on quantifying and characterizing the spectrum of vulval developmental variants induced by spontaneous random mutation to address the following questions:
1) Does developmental precision decay upon mutation for all four isolates, and if so, can the action of natural selection be inferred by comparison of the degree of precision among wild isolates?
2) Does the vulval developmental system show a bias in its mutational response, i.e. are certain developmental variants more likely to occur than others? Which phenotypic characters of the developmental systems show maximal mutability?
3) Do the degree and spectrum of mutationally induced developmental variation vary between genotypes, i.e. to what extent is developmental bias genotype-dependent? How does the degree of mutability of a given developmental phenotype relate to its actual evolutionary variation within and between species?
Results
The canonical vulval cell fate pattern in C. elegans and C. briggsae ancestral controls is 3°−2°−1°−2°−3° (P4.p to P8.p), whereas the most anterior P3.p cell adopts either a 3° or a 4° fate (Figure 1). The MA lines showed a consistently increased proportion of diverse variants (Figure 2), although the canonical P(4–8).p pattern remained the most frequent. Based on the observed variation in MA lines, we distinguished 13 distinct non-canonical cell fate variants deviating from the canonical vulval pattern (Material and Methods; Figure 2 legend). For some tests, these 13 variants were placed into three classes of decreasing order of vulva pattern disruption (A, B, and C). All variants were expressed in proportion of animals adopting the corresponding pattern.
Fig. 2. Variant cell fate patterns of vulval precursor cells (P3.p to P8.p). 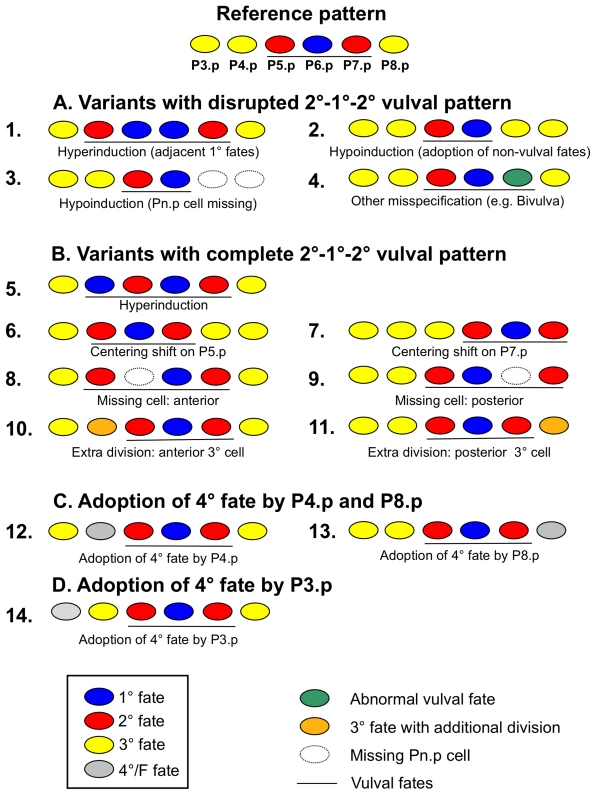
We distinguish three classes of variant vulval patterns in decreasing order of vulva pattern disruption: Variants with disrupted 2°−1°−2° pattern (Class A: “defects”); variants with complete 2°−1°−2° pattern and altered vulval vs. non-vulval fates for the remaining cells (Class B), variants with complete 2°−1°−2° pattern and variable adoption of 3° versus 4° fate by P4.p and P8.p (Class C). 13 non-canonical subcategories of variants are further defined by their deviant cell fate pattern in P(4–8).p (see Material and Methods). Finally we present results on a highly variant trait, P3.p fate: 4° versus 3° (Class D), yet do not include this trait in the analysis of vulva precision. The reference (canonical) pattern for this figure (top) is arbitrarily shown with P3.p adopting a 3° fate. Note that not all variant patterns are mutually exclusive, so that a given individual may adopt multiple variants. (A) Variants with disrupted 2°−1°−2° vulval pattern (Class A). This class groups variant patterns that cause defects in the final vulval structure, likely leading to a reduction in offspring production [34]. (1) Hyperinduction: more than three vulval precursor cells adopt a vulval cell fate (1° or 2° fate), preventing the formation of a complete vulva. For example, P8.p is induced and displaces P7.p progeny from the main invagination. Such cases of hyperinduction are often observed in the presence of an additional anchor cell. (2) Hypoinduction due to adoption of 3° or 4° non-vulval fates: fewer than three cells adopt a vulval cell fate (1° or 2° fate) because of a fate change from vulval to non-vulval. Example: P7.p adopts a 3° non-vulval fate instead of a 2° vulval fate. (3) Hypoinduction due to missing cells: Fewer than three cells adopt a vulval cell fate because one or several Pn.p cells are missing. Example: P7.p and P8.p are missing and only two cells, P5.p and P6.p, adopt vulval cell fates. (4) Misspecification of vulval fates (other than hyper- and hypoinduction): three cells adopt vulval fates but their cell lineages deviate from the canonical pattern. Example: P7.p is misspecified (in green) and adopts the lineage LLTU instead of UTLL. Such a defect in lineage orientation causes a ventral protrusion and is referred to as Bivulva phenotype [69]. Although this specific case of fate misspecification need not always disrupt functionality of the vulva, it eliminates the capacity to mate with males [70]. (B) Variants with complete 2°−1°−2° pattern and altered vulval versus non-vulval fates for the remaining cells (Class B). Such variant patterns do not obviously disrupt the formation of a functional vulval organ; however, whether certain variants negatively impact egg laying or other functions is unclear [34]. (5) Hyperinduction: more than three cells adopt vulval cell fates. Example: P4.p adopts a 2° vulval cell fate instead of a 3° non-vulval cell fate. (6–7) Vulval centering shifts: the three cells adopting vulval fates are shifted to the anterior (centering on P5.p) or posterior (centering on P7.p). Example: P5.p adopts a 1° vulval fate while P4.p and P6.p adopt a 2° fate, with the anchor cell being attached to P5.p progeny. (8–9) Missing cells: One or more vulval precursor cells are missing. Example: P7.p is missing and P8.p adopts a 2° vulval fate instead. Note that in our experiments we could not distinguish whether this variant was due to a missing P7.p or P8.p cell. Therefore, we distinguish only whether one or more anterior cell (P3.p to P5.p) or a posterior cell (P7.p and P8.p) was missing. (10–11) Supernumerary cell divisions: Anterior (P3.p or P4.p) or posterior (P8.p) cells divide more than once, generating three or four cell progeny that fuse with the hypodermis. Example: P4.p (shown in pink) divides twice instead (lineage “ssss” instead of “SS”). (C) Adoption of 3° versus 4° fate of P4.p and P8.p (Class C). This class includes: (12) P4.p adopts the 4° fate or F fate, fusing with the hypodermis without prior division. (13) P8.p adopts the 4° fate. (D) Adoption of 3° versus 4° fate by P3.p (Class D). (14) P3.p adopts the 4° fate (frequent in the wild type). Mutational decay of developmental precision
Proportions of variant vulval cell fate patterns (p)
(Table 1) - The frequency of non-canonical vulval variants was very low in the ancestral controls, approximately 0.4% in C. briggsae and 0.05% in C. elegans, averaged over variants #1–13 and isolates. The tenfold difference in the frequency of variants between the two species was almost entirely due to variants in the adoption of 3° versus 4° fate by P4.p and P8.p (class C, Table 1). Overall, for each of the four tested isolates, defective and other vulval variant patterns were more frequent in MA lines than in ancestral controls.
Tab. 1. Proportion p of individuals exhibiting variant vulval phenotypes. 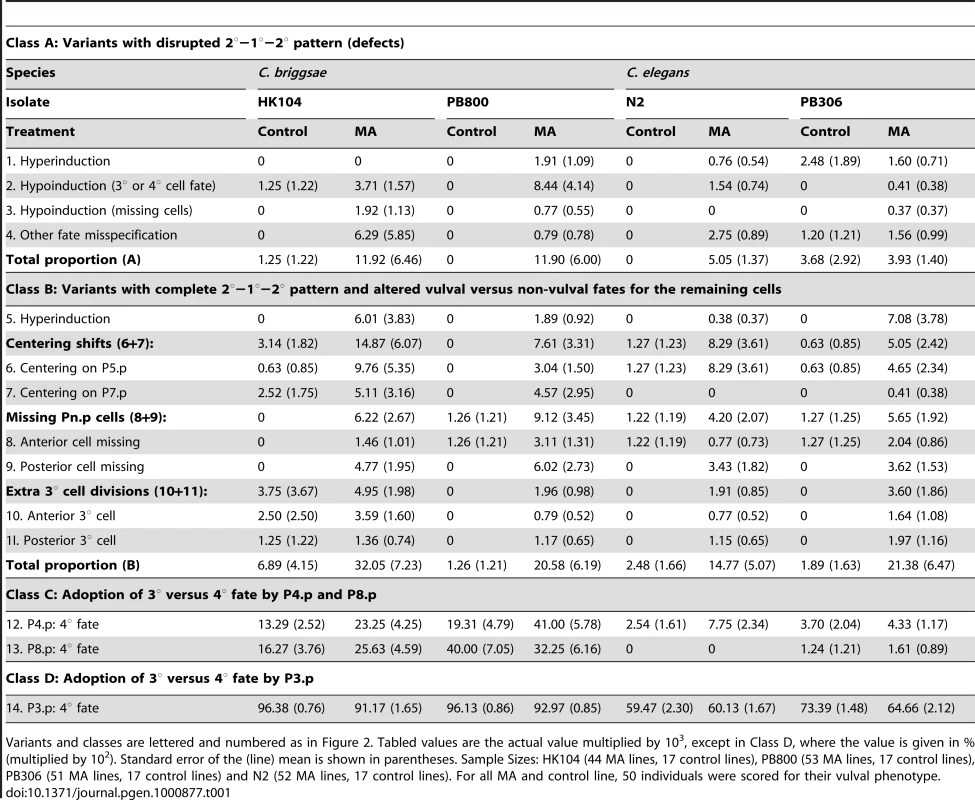
Variants and classes are lettered and numbered as in Figure 2. Tabled values are the actual value multiplied by 103, except in Class D, where the value is given in % (multiplied by 102). Standard error of the (line) mean is shown in parentheses. Sample Sizes: HK104 (44 MA lines, 17 control lines), PB800 (53 MA lines, 17 control lines), PB306 (51 MA lines, 17 control lines) and N2 (52 MA lines, 17 control lines). For all MA and control line, 50 individuals were scored for their vulval phenotype. Change in mean frequency of variant vulval cell fate patterns (Rm)
(Table S2) - Summing over the variants #1–13, point estimates of Rm, the rate of change in trait mean frequency, were positive in all four isolates. Mean change values in the two isolates of C. briggsae (HK104 Rm = 22.3×10−5/gen, PB800 Rm = 19.0×10−5) were about twice those of the two C. elegans isolates (N2 Rm = 9.3×10−5, PB306 Rm = 7.9×10−5). The individual Rm values for each deviant pattern were overall positive, indicating that most deviant patterns increased in frequency upon mutation accumulation in all four isolates.
Change in the among-line variance (ΔV)
(Table S3) - The among-line variance in mean values of p increased with mutation. The differences among species, isolates, and variants closely mirrored changes in the mean. Summing over all variants #1–13 (Table S3, classes A–C), point estimates of ΔV in the two isolates of C. briggsae (HK104 = 11.0×10−6, PB800 = 13.5×10−6) were about 5X greater than those of the two C. elegans isolates (N2 = 1.9×10−6, PB306 = 2.9×10−6).
Mutational correlations
Correlations of line means between two categories of non-canonical variant patterns (Class A and B) and two categories of fitness-related traits (W, CVE,W) are reported in Table S4. Given the number of variant categories and examined isolates, these tests are not powerful, but several trends emerged from the pattern of correlations. First, the correlation between class A variants (disrupted 2°−1°−2° pattern, likely resulting in defects) and other variants with complete 2°−1°−2° (class B+C) was positive in all isolates. The strength of the correlation between defects and variants was dependent on the starting genotype but was not species-specific: the correlation was strong and significant in C. briggsae PB800 and C. elegans PB306, but much weaker in the other isolates of each species. Second, the correlation between fitness traits and variants with complete 2°−1°−2° pattern (class B+C), but not variants with disrupted 2°−1°−2° pattern (class A), was stronger in C. elegans than in C. briggsae. In particular, the correlation between variant classes B+C and the within-line variance in fitness was uniformly strong and positive in C. elegans (∼0.5) and much weaker in C. briggsae (not significantly different from zero). The correlation in the VEL N2 lines was less than in the CFB N2 lines (∼0.2; not significantly different from zero). Third, all correlations were uniformly weak in the HK104 isolate of C. briggsae, a result we have consistently observed in this isolate [29].
Comparison of mutational variance (VM) and standing genetic variance (VG)
To compare the mutational variance (VM) for variant vulval phenotypes with the standing genetic variance (VG), we analyzed data on developmental precision obtained from 10 C. briggsae and 25 C. elegans isogenic wild isolates (Nindividuals = 8′460). Wild isolate data are presented in Table S6, showing the proportion of variants for classes A, B, and C. Point estimates of the variance in line means (V L̂) were very low (∼10−5) for class A variants (strongly disrupted vulval patterns, defects) and for the pool of class B + C variant categories, and jackknife 95% confidence limits included zero in both categories in all isolates. Further, when isolates for which multiple estimates of p were available were considered, the maximum likelihood estimates for the among-isolate (genetic) component of variance were zero for both categories in both species. Thus, vulval development was highly invariant in both C. elegans and C. briggsae wild isolates, and most of the variant patterns observed were limited to variants of class C (3° to 4° transformation of P4.p/P8.p), in C. briggsae.
Developmental bias: random mutation induces vulval variants at different frequencies
Across all four sets of MA lines, the different vulval variant patterns were observed at unequal frequencies (Table 1). Vulval precursor cells adopting a non-vulval 3° fate (P3.p, P4.p and P8.p) showed overall more variability than the cells adopting a vulval cell fate (P5.p to P7.p). Specifically, we found that the developmental phenotype with the highest mutational variance is that already showing high variability in the ancestral controls, i.e. P3.p division frequency (3° versus 4° fate; variant #14; class D) (Table 1 and Figure 3; note change of scale for this variant). The second most common variants concern P4.p and P8.p division frequency (variant #12 and 13; class C). Behind comes a subset of the variant patterns that affect the vulval fates such as centering shifts (class B), hyperinduction (class A or B) or missing precursor cells (class B). Therefore, variants causing likely defects in vulval function (class A) were overall less frequent than variants in classes B or C. That different sub-traits of the vulval developmental system degrade at different rates is further confirmed by the mixed-model analysis of the rate of change in the trait mean frequency Rm (see below).
Fig. 3. Per-generation change in frequency Rm for variant vulval phenotypes. 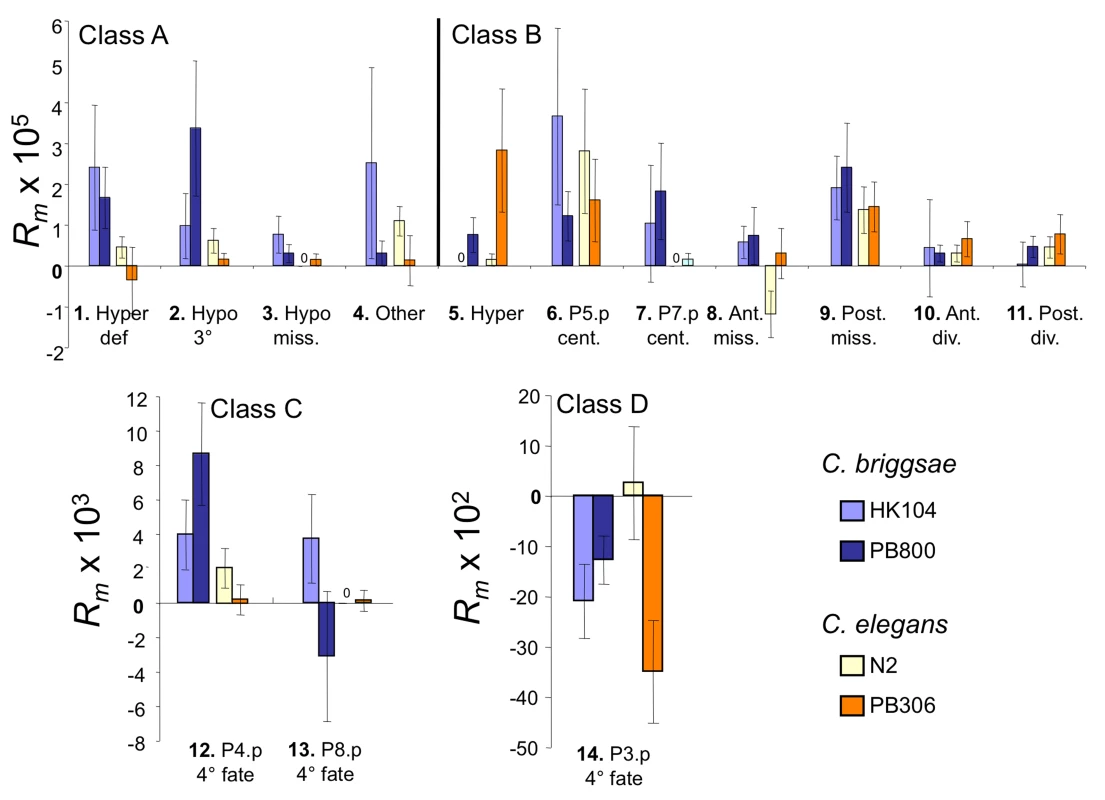
Mean per-generation change in variant frequency Rm in mutation accumulation lines started from four C. elegans and C. briggsae isolates (colour-coded). Variants are numbered and placed in four classes (A–D) as in Figure 2. Note the different vertical scales of the graphs. Sample Sizes: HK104 (44 MA lines, 17 control lines), PB800 (53 MA lines, 17 control lines), PB306 (51 MA lines, 17 control lines) and N2 (52 MA lines, 17 control lines). For each MA and control line, 50 individuals were scored for their vulval phenotype. Error bars indicate standard errors of the (line) mean. Genotype-dependence of developmental mutability
To detect evolutionary variation in the mutability of the vulval developmental system, we tested for an overall interaction between variant vulval phenotype and ancestral genotype in an analysis of variance framework. The mixed-model analysis of the rate of change in the trait mean frequency Rm confirmed a substantial main effect of trait (nominal P<0.0001) and the expected large main effect of species (nominal P<0.002) (Table S5). Thus, the rate of change in mean frequency during mutation accumulation depended on the variant trait and the species. The main effects of isolate (nominal P>0.8) and trait x isolate (nominal P>0.10) were not significant. However, note that several of the most extreme differences in mutational induction of specific vulval variants occurred between the isolates of the same species rather than between species (see below).
Below we report specific examples of genotypic biases in mutationally induced phenotypic variants. Note that because of low frequency of developmental variants and multiple comparisons, the significance level of given comparisons may be poor (the critical experiment-wide 5% significance level for thirteen individual comparisons is P<0.0038).
Class A: variants with disrupted 2°−1°−2° vulval pattern
(Table 1 and Figure 3A).
The propensity to generate a specific defective pattern (hypoinduction, variant #2) varied among isolates. This variant was found at the highest frequency in C. briggsae PB800-derived MA lines (8/53), and was very rare in C. elegans PB306-derived MA lines (1/51 lines: a single individual in the affected line; Table S1) (Fisher's Exact Test, P = 0.036), whereas this variant was never found in the control lines of either of these genotypes. Thus, hypoinduction variants were easier to induce by mutation in C. briggsae PB800 than in C. elegans PB306.
Class B: variants with complete 2°−1°−2° vulval pattern (yet altered vulval vs. non-vulval fates)
(Table 1 and Figure 3B)–Mutational induction of several of these variants showed biases depending on the genotype. Vulval centering shifts on P7.p (variant #7) were most frequent in MA lines of the two C. briggsae isolates but never found in MA lines of C. elegans N2. Conversely, the induction of excessive vulval cells (hyperinduction, variant #5) occurred more frequently in C. elegans N2 than in C. elegans PB306 and the two C. briggsae lines. Within C. elegans, this hyperinduced variant occurred frequently in MA lines derived from PB306 (8/51 MA lines) but not in MA lines derived from N2 (0/52 MA lines) (Fisher's Exact Test, P = 0.005); and this variant was not found in the control lines of either of these two isolates. We further consider this induction variant below.
Class C: adoption of 4° versus 3° fate (P4.p, P8.p)
(Table 1 and Figure 3C) - These cell division phenotypes were the most variable traits in C. briggsae MA lines. The induction of this variant differed in frequency between the two species: relative to C. elegans, C. briggsae MA lines as well as ancestral controls showed increased frequency and variability in P4.p and P8.p adopting 4° versus 3° fates. For P8.p, the adoption of the 4° fate never occurred in MA lines derived from C. elegans N2, while this variant occurred in MA lines of the other three isolates, with a particularly high frequency for the two C. briggsae isolates.
Class D: adoption of 4° versus 3° fate (P3.p)
(Table 1 and Figure 3D) - Unlike P(4–8).p, P3.p has a highly variable fate in ancestral controls. In the ancestral controls, P3.p adopted a 4° fate more frequently in C. briggsae than in C. elegans. After MA, the proportion of P3.p with a 3° fate was increased for C. briggsae and C. elegans PB306 but not for N2, which showed the highest frequency of P3.p adopting a 3° fate in ancestral controls.
Causes underlying genotype-dependence of developmental mutability
The clearest examples of intraspecific variation in the mutational pattern are the hyper - and hypo-induction variants in C. elegans: MA lines displayed more hyperinduction variants and less hypoinduction variants in the PB306 isolate compared to the reference isolate N2. One hypothetical scenario to explain the elevated propensity to generate hyperinduced variants upon mutation accumulation in PB306 might be an increased activity of inductive vulval signalling, already present in the ancestral (wild type) genotype. In this scenario, such a difference would rarely be phenotypically expressed in the ancestral genetic background, but become more prevalent in MA lines due to mutational perturbations. To test this hypothesis, we asked whether the activity of the main signalling cascade inducing vulval cell fates, the EGF/RAS/MAPK cascade, was higher in PB306 than in N2. We introgressed an integrated construct containing a transcriptional Ras reporter, egl-17::cfp [41], into the two isolates to examine Ras activity levels during the vulval patterning process from mid-L2 to early-L3 stage (see Materials and Methods). Consistent with the hypothesis, PB306 showed a significantly higher Ras pathway activity in the relevant vulval precursor cell, P6.p, during mid-L2 and early L3 stages compared to N2 (Figure 4). Thus, the difference in the mutational accessibility of hyperinduced variants between PB306 and N2 may result through variation in the activity of the Ras pathway, which is phenotypically silent (cryptic) under normal conditions.
Fig. 4. Comparison of Ras pathway activity in isolates of C. elegans (N2 versus PB306). 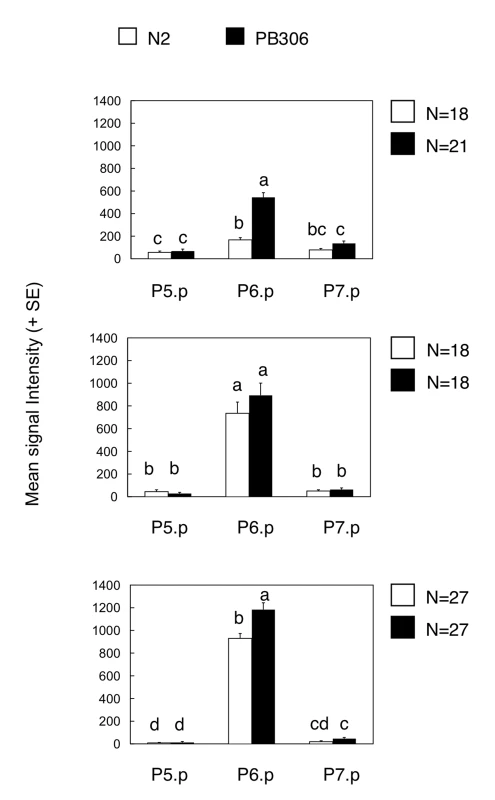
Quantification of wild genetic background effects on Ras reporter activity in animals carrying an integrated Ras pathway reporter transgene (egl-17::cfp-lacZ), at three distinct developmental stages during vulval induction. Bars labelled with the same letter did not show significant differences in expression levels (Tukey's HSD, P<0.05). Error bars indicate standard error of the mean. For ANOVA results, see Table S7. Discussion
Mutational decay of developmental precision
The developmental system underlying Caenorhabditis vulva precursor cell fate patterning was consistently degraded in mutation accumulation (MA) lines derived from all four isolates. In contrast to previously examined traits, such as body size, a quantitative trait varying along a single axis [29],[42], the variation is here practically absent among and within ancestral controls and mutational challenges induce novel variants. Vulval patterning variants almost always had a very low penetrance in a given mutation accumulation line. Many MA lines showed multiple, distinct variants and we never found a line in which a specific variant pattern was fixed.
The observed mutational pattern of small-effect variants may either be explained by non-null mutations in structural genes or mutations in regulatory regions with effects too small to be retained in conventional genetic screens. The core genetic elements of the vulval signalling network amount to approximately 30 genes [31], covering an estimated 150 kb. A conservative estimate of the mutation rate is one mutation per genome per generation in C. elegans [43], so that tested MA lines exhibit an average of 250 mutations per genome (100 Mb). Assuming that about a third of the nucleotide sites are susceptible to mutations having some phenotypic effect, the probability of mutating such a site in this category of “identified vulva genes” is 0.125 for a given MA line. This is consistent with the frequency of defects that we observe; however, this estimate is highly speculative, in particular, because we have no information on the distribution of mutational effects at a given locus. Moreover, it appears likely that several of the mutationally induced vulval variants may have been triggered by mutations of genes not directly involved in the vulval signalling network. Diverse developmental mutations primarily affecting body size and shape have the potential to disrupt the spatial and temporal integrity of the vulval induction process [44], and we have observed that many of these mutations (e.g., dpy, lon, sma, unc) show diverse low-penetrance vulval variants and defects similar to the ones observed in MA lines.
One consequence of the induction of deviation from an invariant pattern is an increase in the within-line component of variance. We previously demonstrated that the environmental (within-line) component of variance (VE) consistently increases with mutation accumulation for W, total lifetime fecundity, and body volume in these same lines [45]. We interpreted this result as evidence that spontaneous mutations de-canalize the phenotype, but could not completely rule out the possibility that that result was an artefact of the way in which these data were scaled. In contrast, the increase in vulval variants and defects with MA is most straightforwardly interpreted as an increase in the environmental component of variance, i.e., de-canalization, and it cannot be attributed to scaling. Thus, mutation accumulation increases the sensitivity of the vulval developmental system to stochastic (micro-environmental) perturbations [46].
Comparison of mutational variance (VM) and standing genetic variance (VG): strong purifying selection
We calculated an estimate of the standing genetic variance (VG) for variant vulval phenotypes using data from 25 C. elegans and 10 C. briggsae wild isolates. At mutation-selection balance in a large population, the ratio of the mutational variance to the standing genetic variance provides an estimate of the strength of purifying selection of mutations affecting the trait, i.e., S≈VM/VG, where S is the average selection coefficient against a new mutation. Using the point estimate of of the wild isolates as a surrogate for VG and the point estimate of ΔV as a surrogate for VM, the average selection coefficient against mutations affecting Class A variants inferred from the ratio VM/VG ( = S) is on the order of 10% or larger (for C. briggsae the point estimate of S = 0.30; for C. elegans S = 0.16). Conversely, the ratio VG/VM can be interpreted as the “persistence time” of a new mutation, i.e., the expected number of generations the mutation segregates before it is lost [47]. Thus, as expected, new mutations that cause Class A variants segregate for only a very few generations before they are removed by selection (Class A variants in the system are clearly deleterious in laboratory conditions, because they prevent egg-laying and reduce progeny number [34]). By way of comparison to life history traits in the same species, selection coefficients inferred in this way for W, body volume, and lifespan are on the order of 1–5% [48],[49]. This result confirms that vulval development is under strong purifying selection to maintain an invariant phenotypic output. The observed selection thus very likely corresponds to the type of stabilizing selection, as defined by Schmalhausen [50], and canalizing selection [51].
Concerning other variant classes, comparison of the genetic variance among wild isolates and after spontaneous mutation accumulation with minimal selection provides indirect evidence of their elimination by selection in natural populations. Especially in class B, the frequency of developmental variants was very low in the four controls as well as in a large set of wild isolates of C. elegans and C. briggsae covering a much larger range of genetic variation than the MA lines [43],[52] (Table S6). Averaged over variants and species, the ratio VM/VG ( = S) of Class B variants is again on the order of 10%, very similar to Class A variants (for C. briggsae the point estimate of S = 0.12, for C. elegans S≈1). Among the class B variants, variants with vulva centering shifts or missing Pn.p cells (variants #6–9) form a complete vulva due to cell fate regulation among the vulva competence group (cells that can adopt a vulval fate through expression of the lin-39/Hox gene [31]). Importantly, this result strongly argues for strong selection against class B variants in natural populations although these variants do not disrupt functionality of the vulval organ and show no fitness effects in the laboratory [34]. By contrast, selection against class C variants appears much weaker (S on the order of 0.1%). Class C variants describe variation in non-vulval fates of P4.p and P8.p, which normally do not affect P(5–7).p vulval fates. When adopting the variant pattern (i.e. adoption of the 4° fate), P4.p and P8.p fuse to epidermal syncytium without division in the L2 stage [53], so that the cells lose their competence to respond to late inductive vulval signalling. Nevertheless, these cells may still be able to respond to Wnt or EGF signalling earlier before hypodermal fusion, and thus to replace one of the P(5–7).p cells in the case of co-occurrence of a class B variant.
Developmental bias: differential mutational accessibility of phenotypic variants
In contrast to classic mutagenesis screens selecting for developmental mutants with high penetrance phenotypes, the screening of the phenotypic spectrum of MA lines is largely unbiased and representative of the phenotypic spectrum induced by spontaneous random mutation. We found that MA induced certain phenotypic variants much more readily than others, demonstrating biases in the mutational accessibility of phenotypic variants. The vulval trait with the highest mutational variance is that already showing high variability in the ancestral controls (P3.p division frequency, variant #14), followed by P4.p and P8.p division frequency (variant #12 and #13; class C). Variants causing likely defects in vulval function (class A) were overall less frequent than variants in classes B or C. In addition, several of these variant patterns have not been found by mutagenesis in the laboratory, presumably because they were too subtle for efficient phenotypic scoring. On the other hand, we did not uncover all possible variant vulval patterns, which suggests that certain of these variants are either fully lethal and could not be propagated in MA lines, or their appearance through mutational effects is too improbable. Such variants include lateral inhibition defects with vulval cells showing adjacent 1° fates as seen in lin-12/Notch mutants [54]. Although a fully penetrant loss of lateral inhibition may be lethal, it is interesting that we did not find this variant at low penetrance like other fate pattern variants. This suggests that the mutational target size for this variant (relying on Notch pathway regulation) is small. Taken together, these observations provide clear examples of developmental bias [13]–[15],[18],[19], with certain phenotypic variants being more easily induced by mutation than others.
Genotype-dependence of developmental mutability
Several results show that biases in the production of vulval variants are genotype-dependent. First, overall rates of mutational decay differ among ancestral controls, most likely due to higher molecular mutation rates in the C. briggsae isolates compared to the C. elegans isolates [25],[30]. The approximately two-fold greater change in the trait mean in C. briggsae was roughly consistent with previous results concerning other traits [28],[29]. Second, we observed differences in the relative mutability of the same canonical pattern to different types of variant pattern. These differences in the mutationally inducible phenotypic spectra may be explained by one of two possible mechanisms. First, the mutation rate at specific loci may vary among wild isolates. For example, a microsatellite repeat present at these loci in some isolates and absent in others may dramatically change mutation rates at the locus [55]. Second, a distinct bias in the developmental system may occur if the internal system variables are slightly offset in some isolates towards the production of a given variant pattern. For example, C. elegans PB306 may mutate more frequently to genotypes producing hyperinduction defects if the Ras pathway involved in vulval induction is in average slightly more active in individuals of this isolate (compared to other wild isolates). More mutations of small effect on the system may then tip the balance towards hyperinduction when acting on the C. elegans PB306 isolate, and remain silent in other isolates. In this case, the different relative mutability to the hyperinduced phenotype of different starting genotypes may thus depend on cryptic genetic variation causing variation in system parameters, also termed intermediate phenotypes [32].
Apparent cryptic variation in such a quantitative developmental parameter may be confirmed by introgression of mutations or by measurements of signalling pathway activity. A higher Ras pathway activity in the C. elegans PB306 isolate is indeed supported by the higher induction index of let-60(n1046gf/ras, lin-3(n378rf)/egf mutants and of the ark-1(sy247lf); gap-1(n1691lf) double mutant [35]. Our present results using a reporter gene further confirm that the Ras pathway is significantly more active in C. elegans PB306 compared to C. elegans N2 (Figure 4). This result demonstrates the presence of intraspecific variation in the activity of vulval signalling pathways and agrees with the proposed second mechanism of evolution of the mutational variance through a bias in mutational effects. In the future, the determination of the molecular lesions and their introgression in different genetic backgrounds may definitively answer whether this difference accounts for the increased frequency of hyperinduced variants in PB306.
Mutational and environmental perturbations can both cause de-canalization of the phenotype [56]. Yet, there is limited experimental evidence whether these two sources of variation also affect the same elements of developmental systems. When comparing the phenotypic effects of mutational vs. environmental perturbation, analyses are often restricted to a single or few environmental conditions using a single or few genetic variants. MA lines provide a more extensive and unbiased sampling of genotypic space. Yet, unlike mutation, environments cannot be systematically sampled. We therefore limit our comparison to six environments examined in an earlier study [34], showing that certain vulval variants are specifically generated in certain environments and genotypes. Several of these previously observed variant patterns were also frequently found after MA. Specifically, vulval centering shift variants on P7.p were never found in C. elegans N2 MA lines, but occurred often in MA lines derived from the other three ancestral genotypes. Similarly, N2 never generated P7.p centering shifts under starvation stress, while C. briggsae showed increased and increased frequency of this variant pattern. Mutational perturbations therefore may mirror environmental perturbations, so that both sources of variation reveal the genotype-dependence of developmental bias.
Bias in developmental mutability and evolutionary trends
Examination of different Caenorhabditis MA lines allows us to detect axes of high mutational variability in the vulval developmental system. Whether or not such high mutational variance translates into actual evolution then depends on selection. Some of these phenotypic axes of least resistance upon mutation may correspond to traits under purifying selection. In this case, the available mutational variance does not result in phenotypic evolution. For other variant types, however, the high mutational variance may correspond to phenotypic evolution observed in the species or among closely related species. In the Caenorhabditis genus, intra - and interspecific variation in vulval patterning traits is limited to the frequency of P3.p adopting a 3° versus 4° fate, and to a lesser extent that of P4.p [37],[39],[57]. For these two vulval phenotypes we also found the greatest mutational variance. The mutational bias and the evolutionary trend in the vulva system thus mainly affect the same trait. At a larger evolutionary scale, a similar match between mutational pattern and evolution is found in the Oscheius genus, but for vulva variants that concern the second round of 3° cell divisions (variants #10–11). In this case, the mutational variance in the occurrence of the second round of 3° cell divisions appears high in Oscheius tipulae CEW1 (from EMS-induced mutant lines) [58] and the same trait varies greatly within the Oscheius genus [24],[37],[39]. By contrast, we found very little mutational variation in the occurrence of a second division round for the 3° cells (variants #10–11), and these traits are invariant within the Caenorhabditis genus, presumably because of developmental constraints. Such studies of relative trait mutability are thus crucial to understand variation in evolutionary trends between taxa and thereby bridge the gap between micro - and macro-evolutionary variation.
In conclusion, our results provide an empirical view on the developmental variation induced by spontaneous random mutation. In the case of the highly canalized vulval developmental system, this variation is generally very subtle and difficult to quantify. In addition, the induced phenotypic variation is very complex despite the seeming molecular and developmental simplicity of this process. Nonetheless, we could uncover a number of developmental and genetic biases in the introduction of phenotypic variation, supporting the notion that such asymmetries bias the range of phenotypes available for selection to act upon [11]–[15],[18],[19]. Many more studies characterizing biases in the production spontaneous phenotypic variation (and its correspondence to evolutionary variation of the studied phenotypes) are required to evaluate whether such asymmetries play important roles as direction-giving forces in the evolutionary process.
Materials and Methods
Mutation accumulation lines
The main set of mutation accumulation (MA) lines in this study is that of Baer et al. [25] (called CFB lines). The lines were originated from a single highly inbred individual from each of two isogenic wild isolates of C. elegans (N2 and PB306 isolates) and C. briggsae (HK104 and PB800 isolates). Criteria for choice of these isolates are given in [25]. The mutation accumulation experiments began with 100 replicate MA lines per isolate. Details of the mutation accumulation protocols are given in the original paper. Briefly, highly inbred stocks of each isolate were replicated 100 times and perpetuated by single-hermaphrodite transfer for 250 generations. This protocol results in a genetic effective population size of Ne≈1 (the approximation is the result of occasionally having to use backup stocks of worms when the original worm did not survive), thereby minimizing the efficiency of natural selection and ensuring that all but the most deleterious mutations behave according to neutral dynamics. Worm stocks, including G0 ancestral controls and ultimate generation MA lines, were cryopreserved using standard methods [59].
Wild isolates
Wild isolates of C. elegans (N = 25) and C. briggsae (N = 10) used in this study are listed in Table S6. Both species display a high selfing rate in natural populations [52],[60]. The (isogenic) wild isolates were originally established by selfing populations derived from a single individual isolated from the wild.
Scoring of vulval cell fates
Worms were kept on Petri dishes (55 mm diameter) filled with NGM (Nematode Growth Medium) agar, seeded with approximately 200 µl bacterial suspension of the E. coli strain OP50. All experiments were carried out at 20°C. For each of three experimental blocks, a random set of MA lines and the four ancestral controls were thawed (for samples size, see below). To eliminate potential genetic variation in the stock culture, a single individual from each line was selected to initiate the experimental populations. After population expansion, 20–30 adult hermaphrodites per line were hypochlorite treated to clear individuals form potential microbial contaminations. (At this time, for each of the four ancestral controls, multiple replicates were established except for the first block). The resulting eggs were allowed to develop into adults at which stage 20 hermaphrodites (from the same NGM plate) were transferred to a new NGM plate. When the majority of the offspring had reached the L4 stage (after approximately 2–5 days depending on the line), 50 offspring/line were randomly selected to score their vulval phenotype. The vulval cell phenotype was determined during the early to mid L4 stage using Nomarski microscopy on individuals anaesthetized with sodium azide [59]. We counted induced cells and determined the fates of the cells P3.p to P8.p as described previously [44]. MA and control lines underwent approximately 4–6 generations on NGM plates (at low densities) between thawing and scoring.
We defined different types of vulval developmental variants (shown in Figure 2) by taking into account developmental features of the system. Note that due to replacement regulation between vulval precursor cells [31], the fate of each individual cell is not independent from that of the other cells. For example, when the anchor cell is positioned on P5.p, the entire pattern is displaced anteriorly and four Pn.p cell fates are affected simultaneously; if P5.p is missing, P4.p adopts a 2° fate; if the anchor cell is missing, the fates of P(5–7).p switch to a 3° fate, etc. Defining 14 distinct variant types allowed us to greatly lower the number of variant types compared to the combination of each fate for each cell (1°/2°/3°/4°/missing x 6 = 30 classes). Some of these variants correspond to changes due to independent developmental events as defined by mutational analysis [24],[53],[61]. For example, hypoinduction phenotypes through cell fate change from a vulval fate to a non-vulval fate (trait #2) likely occur through low activities of Ras or possibly Wnt pathways (Induction Vulvaless in [61]). In contrast, hypoinduction phenotypes arising by lack of Pn.p cells (trait #3) occur because of cell death or earlier switch in cell fate (Generation Vulvaless in [61]).
Sample sizes
The following number of MA and control lines were analyzed for each isolate: HK104 (44 MA lines, 17 control lines), PB800 (53 MA lines, 17 control lines), PB306 (51 MA lines, 17 control lines) and N2 (52 MA lines, 17 control lines). For each MA and control line, 50 individuals were scored for their vulval phenotype.
Data analysis: MA lines
There are two fundamental observable quantities of interest in a MA experiment—the change in the trait mean and the change in the variance. In this study, vulval character state is a binary random variable X with state 0 = wild-type and state 1 = non-canonical” (for traits 1–13). The data are binomially-distributed with parameter p = Pr(X = 1). Within a genotype/treatment group (“treatment” = MA or G0 ancestral control), each line provides a single independent estimate of p.
(i) Change in the mean (Rm)
The per-generation change in the trait mean can be considered either on the raw scale (Rm, the slope of the regression of the trait value against time, measured in generations of MA) or scaled as a fraction of the generation 0 mean (ΔM = Rm/M0, where M0 is the ancestral mean). ΔM is typically the more meaningful of the two because the average mutational effect is meaningful only relative to the starting phenotype, but the interpretation of ΔM breaks down when M0 is close to zero. In the extreme case of a mutation that increases the frequency of a variant phenotype from 0 to 1/n, ΔM is infinite for all n. In this study we use Rm as the measure of the change in the frequency of variant vulva phenotypes because of the very low frequency of variant phenotypes in the ancestral C. elegans controls.
We first tested for an effect of assay block using a general linear mixed model as implemented in SAS v. 9.2 PROC GLIMMIX, testing each isolate individually and employing a logit link function (http://support.sas.com/rnd/app/papers/glimmix.pdf). Block, treatment (MA vs. control) and their interaction are considered fixed effects; significance of approximate F-tests for fixed effects is determined by the residual pseudo-likelihood method [62]; error degrees of freedom are calculated by the Kenward-Rogers method. The model is pT = Block + Treatment + Block x Treatment + error, where pT is the binomial parameter. In no case was there a significant main effect of or interaction with block (P>0.1 in all cases), so data were pooled over blocks for subsequent analyses.
To assess the statistical significance of differences between groups in Rm, we used a bootstrap resampling protocol, as follows. A pseudo-dataset was constructed by resampling the data with replacement at the level of line, maintaining the same number of control and MA lines as in the original data set. The mean binomial parameter p was calculated for control and MA lines separately and Rm estimated as (pMA-p0)/t, where t is the number of generations of MA. This procedure was repeated 1000 times; the upper and lower 2.5% of pseudo-estimates establish approximate 95% confidence limits on Rm [63]. Differences between groups are considered significant if the 95% confidence intervals do not overlap.
To investigate the possibility that the variation among traits in Rm may vary between species and/or isolates - that is, that there is a trait x taxon (here species or isolate) interaction in the variable Rm - we employed a general linear mixed model as implemented in SAS. v. 9.2 PROC MIXED. We first calculated a line-specific value of Rm for each trait j for each MA line i (Rm,ij) by subtracting the control mean value of p from each line-specific value of p, i.e., Rm,ji = p ij −p̂0,j (we omit the number of MA generations, t, from this calculation for convenience). We then analyzed the linear model Rm = Species + Trait + Trait x Species + Isolate(Species) + Trait x Isolate(Species) + Error. Six of the 3000 data points were identified as high outliers by visual inspection of a Q-Q plot and removed from the analysis. Residual (error) variance was estimated separately for each trait/species combination via the GROUP option in PROC MIXED; the model failed to converge when residuals were estimated for each trait/isolate combination.
The above analysis is potentially compromised in two ways. First, the analysis is strictly valid only when the data are normally distributed; the data in this case deviate substantially from normality and cannot be transformed to meet the assumption of normality. To assess the sensitivity of the analysis to violation of distributional assumptions we performed randomization tests using the aforementioned linear model, with data randomly permuted over traits within each isolate. If the test is robust, the frequency of a particular outcome in randomly permuted data should be approximately equivalent to its theoretical probability of occurrence given the assumptions (i.e., its P-value). In every case we examined, the distribution of P-values was almost identical to the theoretical expectation.
Second, the analysis treats the control mean for each trait/isolate combination as a parameter of the model rather than a random variable. Therefore, the P-values associated with the pseudo-F-tests [62] are inflated to some degree. We report “nominal” P-values, which are useful for comparison of the relative magnitudes of the effects within the model but cannot be taken at face value. However, estimates of control means are based on many more measurements (usually 17 times, i.e. 850 individuals) than estimates for any given MA line (50 individuals), so the sampling variance of the control mean should be much less than the within-line variance of any MA line.
(ii) Change in the among-line variance (ΔV)
The mutational variance, VM, is typically estimated from the per-generation change in the among-line component of variance [64]. However, it is not possible to estimate meaningful within - and among-line components of variance from these MA data because we have only a single independent estimate from each line and the within-line variance [ = p(1-p)] is a function of the mean (p). Instead, we consider the change in the variance in line means, i.e., the variance in the binomial parameter pj, where pj is the frequency of variant phenotypes in line j. The change in the variance is calculated as ΔV = (VMA−V0)/t, where VMA is the variance in p among MA line means, V0 is the variance in p among ancestral control line means, and t is the number of generations of MA. If the ancestral control is homozygous at all loci as assumed, V0 provides an estimate of the within-line variance and ΔV/2 provides an estimate of the mutational variance. We use the term ΔV rather than VM to emphasize that the per-generation increase in variance is not calculated from variance components. Note that although the within-line variance is a defined function of the mean [ = p(1-p)], the variance in line means is not. Differences among groups in ΔV were assessed using the same bootstrap protocol as described above for Rm. For each pseudo-dataset we calculated the variance in p for control and MA lines and then calculated ΔV. Confidence intervals and significance criterion for ΔV were determined as for Rm.
(iii) Mutational correlations
We estimated mutational correlations of vulval development (p) with two fitness-related traits that we previously assayed in these MA lines [28],[45]. The first trait is lifetime reproductive output (called “Total Fitness”, W, in [28]), which is closely correlated with demographic fitness (Pearson's r≈0.9; Baer, unpublished data). The second trait is the environmental (here meant as within-line) coefficient of variation in W (CVE,W), which provides an estimate of environmental canalization of W [45]. Because we cannot estimate (co)variance components for vulval development, we report correlations calculated from (co)variances of line means, which will generally underestimate the absolute value of the among-line correlation [29]. W and CVE,W are not independent so we do not report the correlation between those variables.
To accommodate among-block variation in W and CVE,W, we first defined a new variable wijk as the proportional deviation of an individual worm's W from the block mean W of the ancestral control, i.e., , where Wijk is the value of individual i in MA line j in experimental block k and is the mean of the ancestral controls in block k. We next calculated line means and within-line CVs, CVE,j. We then estimated the (co)variances of pj and wj and CVE, j. Finally, we calculated a corrected correlation , where x represents the relevant variable (wj or CVE,j), tx is the number of MA generations at the time x was measured and tv is the number of MA generations at the time vulval development was measured. The term enters the denominator because, if two traits are measured at different generations, mutations that occur after the first trait was measured cannot contribute to the correlation between the two traits; Var(p) is multiplied by this fraction - rather than Var(w) - because vulval development was measured at a later generation than was fitness. Fitness variables were measured at 200 and 220 generations; we used the average value of 210 generations for tx. (Co)variances were estimated by REML using SAS v. 9.2 PROC MIXED with unstructured covariance (TYPE = UN option).
Data analysis: wild isolates
If wild isolates are homozygous at all loci (a plausible approximation for a highly-selfing species; see above), the standing genetic variance (VG) can be estimated from the among-line component of variance [65]. However, for 22/25 wild isolates of C. elegans, we only have a single estimate of the binomial parameter p and therefore cannot meaningfully partition the variance in p into within and among-isolate components. Instead, we use the variance in isolate means V L̂ as an upper bound on VG. Using ΔV and V L̂ to approximate the mutational variance VM and VG, respectively, the relationship VG≈VM/S provides an estimate of the strength of selection against new mutations (S), provided the system is at mutation-(purifying) selection balance (MSB) [47]. For the isolates for which we have multiple independent estimates of p, we partitioned the variance into within - and among-isolate components using REML as implemented in the MIXED procedure of SAS v. 9.2. We can then compare the variance components of these isolates to V L̂ to gain a rough idea of the relative fraction of the variance that is among isolates.
To establish confidence intervals on ΔV and V L̂ we used a delete-one jackknife method [66] to estimate the standard error of the statistic, which was then used in the standard Student's-t calculation of the 95% confidence limits [67],
Ras pathway activity measurements using transcriptional reporter egl-17::cfp
To estimate Ras pathway activity level in the C. elegans N2 and PB306 isolates, we used a previously generated transgenic strain containing an integrated transcriptional reporter construct for the LET-60/Ras pathway, egl-17::cfp-lacZ (strain GS3582) [41]. This construct contains a nuclear localization sequences upstream of the CFP coding sequence and was generated using the isolate N2 [41]. We then generated the egl-17::cfp-lacZ strain JU480 from the strain GS3582 by genetically removing the transformation marker unc-4(e120). Each integrated transgenic array generated in the N2 background was outcrossed ten times to PB306, by crossing at each generation the male progeny to wild hermaphrodites. After ten backcrosses, the introgressed line was made isogenic by selfing for several generations, yielding strain JU488.
The CFP fluorescence quantification experiment was performed as described in [34] in standard conditions at 20°C, for JU480 and JU488 in parallel. For each individual/image, we quantified signal (pixel) intensity of P5.p, P6.p and P7.p. For each examined developmental stage, we carried out an ANOVA (JMP 7.0 for Mac) testing for the fixed effects of isolate, individual (nested in isolate), cell, and the interaction between isolate and cell type using mean signal intensity as a response variable. The inclusion of the effect individual(isolate) allowed us to control for the non-independence between measures of P5.p, P6.p, and P7.p taken from a single individual. Post-hoc tests (Tukey's HSD) were then performed to determine differences in signal expression between isolates and cells (P5.p. P6.p, P7.p).
Supporting Information
Zdroje
1. LynchM
2007 The evolution of genetic networks by non-adaptive processes. Nat Rev Genetics 8 803 813
2. LandeR
1980 Maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet Res 26 221 235
3. SteppanSJ
PhillipsPC
HouleD
2002 Comparative quantitative genetics: evolution of the G matrix. TREE 17 320 327
4. EstesS
PhillipsPC
2006 Variation in pleiotropy and the mutational underpinnings of the G-matrix. Evolution 60 2655 2660
5. KeightleyPD
Eyre-WalkerA
1999 Terumi Mukai and the riddle of deleterious mutation rates. Genetics 153 515 523
6. JonesAG
ArnoldSJ
BürgerR
2007 The mutation matrix and the evolution of evolvability. Evolution 61 727 745
7. JonesAG
ArnoldSJ
BuergerR
2003 Stability of the G-matrix in a population experiencing pleiotropic mutation, stabilizing selection, and genetic drift. Evolution 57 1747 1760
8. BeginM
SchoenDJ
2007 Transposable elements, mutational correlations, and population divergence in Caenorhabditis elegans. Evolution 61 1062 1070
9. HouleD
HughesKA
HoffmasterDK
IharaJ
AssimacopolousS
1994 The effects of spontaneous mutation on quantitative traits. 1. Variances and covariances of life-history traits. Genetics 138 773 785
10. EstesS
AjieBC
LynchM
PhillipsPC
2005 Spontaneous mutational correlations for life-history, morphological and behavioral characters in Caenorhabditis elegans. Genetics 170 645 653
11. GouldSJ
1977 Ontogeny and Phylogeny. Cambridge, MA Belknap Press
12. Maynard SmithJ
BurianR
KauffmanS
AlberchP
CampbellJ
1985 Developmental constraints and evolution. Q Rev Biol 60 265 287
13. ArthurW
2004 Biased embryos and evolution. Cambridge Cambridge University Press
14. ThomsonKS
1985 Essay review: The relationship between development and evolution. Oxford Surveys in Evolutionary Biology 2 220 233
15. YampolskyLY
StoltzfusA
2001 Bias in the introduction of variation as an orienting factor in evolution. Evol Dev 3 73 83
16. ArthurW
2004 The effect of development on the direction of evolution: toward a twenty-first century consensus. Evol Dev 6 282 288
17. PsujekS
BeerRD
2008 Developmental bias in evolution: evolutionary accessibility of phenotypes in a model evo-devo system. Evol Dev 10 375 390
18. StoltzfusA
2006 Mutationism and the dual causation of evolutionary change. Evol Dev 8 304 317
19. StoltzfusA
YampolskyLY
2009 Climbing mount probable: mutation as a cause of nonrandomness in evolution. J Hered 100 637 647
20. SchluterD
1996 Adaptive radiation along genetic lines of least resistance. Evolution 50 1766 1774
21. SternDL
OrgogozoV
2009 Is genetic evolution predictable? Science 323 746 751
22. McDonaldMJ
GehrigSM
MeintjesPL
ZhangX-X
RaineyPB
2009 Adaptive divergence in experimental populations of Pseudomonas fluorescens. IV. Genetic constraints guide evolutionary trajectories in a parallel adaptive radiation. Genetics 183 1041 1053
23. AlberchP
GaleE
1985 A developmental analysis of an evolutionary trend: Digital reduction in amphibians. Evolution 39 8 23
24. Dichtel-DanjoyM-L
FélixM-A
2004 Phenotypic neighborhood and micro-evolvability. TIG 20 268 276
25. BaerCF
ShawF
StedingC
BaumgartnerM
HawkinsA
2005 Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc Natl Acad Sci USA 102 5785 5790
26. WoodruffRC
ThompsonJN
SeegerMA
SpiveyWE
1984 Variation in spontaneous mutation and repair in natural population lines of Drosophila melanogaster. Heredity 53 223 234
27. MukaiT
BabaM
AkiyamaM
UowakiN
KusakabeS
1985 Rapid change in mutation rate in a local population of Drosophila melanogaster. Proc Natl Acad Sci USA 82 7671 7675
28. BaerCF
PhillipsN
OstrowD
AvalosA
BlantonD
2006 Cumulative effects of spontaneous mutations for fitness in Caenorhabditis: role of genotype, environment and stress. Genetics 174 1387 1395
29. OstrowD
PhillipsN
AvalosA
BlantonD
BoggsA
2007 Mutational bias for body size in rhabditid nematodes. Genetics 176 1653 1661
30. PhillipsN
SalomonM
CusterA
OstrowD
BaerC
2009 Spontaneous Mutational and Standing Genetic (Co)variation at Dinucleotide Microsatellites in Caenorhabditis briggsae and Caenorhabditis elegans. Molecular Biology and Evolution 26
31. SternbergPW
2005 Vulval development. Wormbook. ed. The C. elegans Research Community. doi/10.1895/wormbook.1.6.1, http://www.wormbook.org
32. FélixM-A
WagnerA
2008 Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity 100 132 140
33. BraendleC
MillozJ
FélixM-A
2008 Mechanisms and evolution of environmental responses in Caenorhabditis elegans. Curr Topics Dev Biol 80 171 207
34. BraendleC
FélixM-A
2008 Plasticity and errors of a robust developmental system in different environments. Dev Cell 15 714 724
35. MillozJ
DuveauF
NuezI
FélixM-A
2008 Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev 22 3064 3075
36. SulstonJE
WhiteJG
1980 Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev Biol 78 577 597
37. DelattreM
FélixM-A
2001 Polymorphism and evolution of vulval precursor cell lineages within two nematode genera, Caenorhabditis and Oscheius. Curr Biol 11 631 643
38. FélixM-A
2007 Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr Biol 17 103 114
39. KiontkeK
BarrièreA
KolotuevI
PodbilewiczB
SommerRJ
2007 Trends, stasis and drift in the evolution of nematode vulva development. Curr Biol 17 1925 1937
40. VassilievaLL
LynchM
1999 The rate of spontaneous mutation for life-history traits in Caenorhabditis elegans. Genetics 151 119 129
41. YooAS
BaisC
GreenwaldI
2004 Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science 303 663 666
42. AzevedoRBR
KeightleyPD
Laurén-MäättäC
VassilievaLL
LynchM
2002 Spontaneous mutational variation for body size in Caenorhabditis elegans. Genetics 162 755 765
43. DenverDR
MorrisK
LynchM
ThomasWK
2004 High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430 679 682
44. SternbergPW
HorvitzHR
1986 Pattern formation during vulval development in Caenorhabditis elegans. Cell 44 761 772
45. BaerCF
2008 Quantifying the decanalizing effects of spontaneous mutations in rhabditid nematodes. Am Nat 172 272 281
46. DworkinI
2005 Canalization, cryptic variation, and developmental buffering: a critical examination and analytical perspective.
HallgrimssonB
HallBK
Variation - A central concept in biology: Elsevier Inc 131 158
47. HouleD
MorikawaB
LynchM
1996 Comparing mutational variabilities. Genetics 143 1467 1483
48. SalomonMP
OstrowD
PhillipsN
BlantonD
BourW
2009 Comparing mutational and standing genetic variability for fitness and size in Caenorhabditis briggsae and C. elegans. Genetics 183 685 692
49. Joyner-MatosJ
UpadhyayA
SalomonMP
GrigaltchikV
BaerCF
2009 Genetic (Co)variation for life span in rhabditid nematodes: role of mutation, selection, and history. J Gerontol A Biol Sci Med Sci 1134 1145
50. SchmalhausenII
1949 Factors of Evolution: The Theory of Stabilizing Selection. Chicago Univ of Chicago Press
51. WagnerGP
BoothG
Bagheri-ChaichianH
1997 A population genetic theory of canalization. Evolution 51 329 347
52. BarrièreA
FélixM-A
2005 Natural variation and population genetics of C. elegans. Wormbook. ed. The C. elegans Research Community, doi/10.1895/wormbook.1.43.1, http://www.wormbook.org
53. EisenmannDM
MaloofJN
SimskeJS
KenyonC
KimSK
1998 The β-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125 3667 3680
54. GreenwaldIS
SternbergPW
HorvitzHR
1983 The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34 435 444
55. FondonJWI
GarnerHR
2004 Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci USA 101 18058 18063
56. FlattT
2005 The evolutionary genetics of canalization. Q Rev Biol 80 287 316
57. DolginES
FélixM-A
CutterAD
2008 Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity 100 304 315
58. DichtelM-L
Louvet-ValléeS
VineyME
FélixM-A
SternbergPW
2001 Control of vulval cell division number in the nematode Oscheius/Dolichorhabditis sp. CEW1. Genetics 157 183 197
59. WoodWB
1988 The nematode Caenorhabditis elegans. Cold Spring Harbor, New York Cold Spring Harbor Laboratory
60. CutterAD
FélixM-A
BarrièreA
CharlesworthD
2006 Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics 173 2021 2031
61. FergusonEL
SternbergPW
HorvitzHR
1987 A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326 259 267
62. WolfingerR
O'ConnellM
1993 Generalized linear mixed models: a pseudo-likelihood approach. Journal of Statistical Computation and Simulation 4 233 243
63. EfronB
TibshiraniRJ
1993 An introduction to the bootstrap. New York Chapman and Hall
64. LynchM
WalshB
1998 Genetics and analysis of quantitative traits: Sinauer Associates, Inc.
65. FalconerDS
1989 Introduction to Quantitative Genetics. New York John Wiley Sons
66. KnappSJ
BridgesWCJr
YangMH
1989 Nonparametric confidence interval estimators for heritability and expected selection response. Genetics 121 891 898
67. SokalRR
RohlfFJ
1981 Biometry, 3nd edition. New York W.H. Freeman and Cie
68. SulstonJ
HorvitzHR
1977 Postembryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol 56 110 156
69. InoueT
OzHS
WilandD
GharibS
DeshpandeR
2004 C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell 118 795 806
70. FergusonE
HorvitzHR
1985 Identification and characterization of 22 genes that affect the vulval cell lineages of Caenorhabditis elegans. Genetics 110 17 72
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 3- Akutní intermitentní porfyrie
- Farmakogenetické testování pomáhá předcházet nežádoucím efektům léčiv
- Růst a vývoj dětí narozených pomocí IVF
- IVF a rakovina prsu – zvyšují hormony riziko vzniku rakoviny?
- Pilotní studie: stres a úzkost v průběhu IVF cyklu
-
Všechny články tohoto čísla
- Parental Genome Dosage Imbalance Deregulates Imprinting in
- Identification and Functional Analysis of the Vision-Specific BBS3 (ARL6) Long Isoform
- HAP2(GCS1)-Dependent Gamete Fusion Requires a Positively Charged Carboxy-Terminal Domain
- Initial Genomics of the Human Nucleolus
- Role of RecA and the SOS Response in Thymineless Death in
- PPS, a Large Multidomain Protein, Functions with Sex-Lethal to Regulate Alternative Splicing in
- Mislocalization of XPF-ERCC1 Nuclease Contributes to Reduced DNA Repair in XP-F Patients
- Transgenic Rat Model of Neurodegeneration Caused by Mutation in the Gene
- Human Population Differentiation Is Strongly Correlated with Local Recombination Rate
- Local-Scale Patterns of Genetic Variability, Outcrossing, and Spatial Structure in Natural Stands of
- Arginylation-Dependent Neural Crest Cell Migration Is Essential for Mouse Development
- HP1 Recruitment in the Absence of Argonaute Proteins in
- MiR-218 Inhibits Invasion and Metastasis of Gastric Cancer by Targeting the Robo1 Receptor
- Bias and Evolution of the Mutationally Accessible Phenotypic Space in a Developmental System
- Papillorenal Syndrome-Causing Missense Mutations in / Result in Hypomorphic Alleles in Mouse and Human
- Rapid Assessment of Genetic Ancestry in Populations of Unknown Origin by Genome-Wide Genotyping of Pooled Samples
- Regulation of Lifespan, Metabolism, and Stress Responses by the SH2B Protein, Lnk
- KRAB–Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading
- Identification of the Regulatory Logic Controlling Pathoadaptation by the SsrA-SsrB Two-Component System
- Drosophila Xpd Regulates Cdk7 Localization, Mitotic Kinase Activity, Spindle Dynamics, and Chromosome Segregation
- Multiple Signals Converge on a Differentiation MAPK Pathway
- In the Tradition of Science: An Interview with Victor Ambros
- Association of the Polymorphism His615Arg with Melanin Content in East Asian Populations: Further Evidence of Convergent Evolution of Skin Pigmentation
- Fatal Cardiac Arrhythmia and Long-QT Syndrome in a New Form of Congenital Generalized Lipodystrophy with Muscle Rippling (CGL4) Due to Mutations
- Deciphering Normal Blood Gene Expression Variation—The NOWAC Postgenome Study
- Derepression of the Plant Chromovirus Induces Germline Transposition in Regenerated Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Deciphering Normal Blood Gene Expression Variation—The NOWAC Postgenome Study
- Transgenic Rat Model of Neurodegeneration Caused by Mutation in the Gene
- Papillorenal Syndrome-Causing Missense Mutations in / Result in Hypomorphic Alleles in Mouse and Human
- Fatal Cardiac Arrhythmia and Long-QT Syndrome in a New Form of Congenital Generalized Lipodystrophy with Muscle Rippling (CGL4) Due to Mutations
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



