-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Very Long O-antigen Chains Enhance Fitness during -induced Colitis by Increasing Bile Resistance
Intestinal inflammation changes the luminal habitat for microbes through mechanisms that have not been fully resolved. We noticed that the FepE regulator of very long O-antigen chain assembly in the enteric pathogen Salmonella enterica serotype Typhimurium (S. Typhimurium) conferred a luminal fitness advantage in the mouse colitis model. However, a fepE mutant was not defective for survival in tissue, resistance to complement or resistance to polymyxin B. We performed metabolite profiling to identify changes in the luminal habitat that accompany S. Typhimurium-induced colitis. This analysis suggested that S. Typhimurium-induced colitis increased the luminal concentrations of total bile acids. A mutation in fepE significantly reduced the minimal inhibitory concentration (MIC) of S. Typhimurium for bile acids in vitro. Oral administration of the bile acid sequestrant cholestyramine resin lowered the concentrations of total bile acids in colon contents during S. Typhimurium infection and significantly reduced the luminal fitness advantage conferred by the fepE gene in the mouse colitis model. Collectively, these data suggested that very long O-antigen chains function in bile acid resistance of S. Typhimurium, a property conferring a fitness advantage during luminal growth in the inflamed intestine.
Published in the journal: . PLoS Pathog 8(9): e32767. doi:10.1371/journal.ppat.1002918
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002918Summary
Intestinal inflammation changes the luminal habitat for microbes through mechanisms that have not been fully resolved. We noticed that the FepE regulator of very long O-antigen chain assembly in the enteric pathogen Salmonella enterica serotype Typhimurium (S. Typhimurium) conferred a luminal fitness advantage in the mouse colitis model. However, a fepE mutant was not defective for survival in tissue, resistance to complement or resistance to polymyxin B. We performed metabolite profiling to identify changes in the luminal habitat that accompany S. Typhimurium-induced colitis. This analysis suggested that S. Typhimurium-induced colitis increased the luminal concentrations of total bile acids. A mutation in fepE significantly reduced the minimal inhibitory concentration (MIC) of S. Typhimurium for bile acids in vitro. Oral administration of the bile acid sequestrant cholestyramine resin lowered the concentrations of total bile acids in colon contents during S. Typhimurium infection and significantly reduced the luminal fitness advantage conferred by the fepE gene in the mouse colitis model. Collectively, these data suggested that very long O-antigen chains function in bile acid resistance of S. Typhimurium, a property conferring a fitness advantage during luminal growth in the inflamed intestine.
Introduction
Salmonella enterica serotype Typhimurium (S. Typhimurium) is an important cause of human gastroenteritis [1]. Upon ingestion, a fraction of the S. Typhimurium population enters intestinal epithelial cells using the invasion-associated type III secretion system (T3SS-1) [2], which is followed by macrophage survival mediated by a second type III secretion system (T3SS-2) [3]. The deployment of T3SS-1 and T3SS-2 triggers acute intestinal inflammation [4] and the resulting changes in the environment enhance growth of the luminal fraction of the S. Typhimurium population [5], [6], [7], [8] despite the fact that concentrations of antimicrobial substances are elevated in this habitat [9], [10], [11].
The surface of S. Typhimurium, which is exposed to antimicrobial substances in the inflamed gut, is decorated with lipopolysaccharide (LPS). The oligosaccharide core connects the lipid A moiety, which anchors the LPS molecule in the outer membrane, to O-antigen repeat units that extend from the bacterial surface. Each O-antigen repeat unit is composed of a trisaccharide backbone, consisting of α-D-mannose-(1,4)-α-L-rhamnose-(1,3)-α-D-galactose, and a branching sugar (abequose) that is α-(1,3)-linked to D-mannose in the backbone. S. Typhimurium can produce short LPS species containing between 1 and 15 O-antigen repeat units. Additionally, S. Typhimurium synthesizes LPS species containing a greater number of O antigen repeat units, which requires the length regulators WzzB [12], [13] and FepE [14]. The WzzB protein regulates the assembly of long LPS species carrying between 16 and 35 O-antigen repeat units [15] while the FepE protein controls the biosynthesis of very long LPS species with more than 100 O-antigen repeat units [14]. These regulatory mechanisms give rise to a tri-modal distribution in LPS length.
There is general agreement that WzzB-dependent assembly of long O-antigen chains (16–35 repeat units) provides a fitness advantage by conferring resistance to complement [14], [16]. In contrast, the function of very long O-antigen chains (>100 repeat units) during host microbe interaction remains unclear. The absence of very long O-antigen species in a fepE mutant reduces in vitro translocation of SipA [16], a T3SS-1 effector protein that significantly contributes to the induction of inflammation in the mouse colitis model [17]. However, in vivo the O-antigen is not required for T3SS-1-dependent induction of intestinal inflammatory responses [18]. These data indicate that SipA translocation proceeds through an O-antigen-independent pathway in vivo. One study suggests that very long O-antigen inhibits macrophage phagocytosis [19], while others observed no inhibition [16]. A fepE mutant and its S. Typhimurium wild-type parent are equally resistant to complement in vitro, although a wzzB fepE mutant is more serum sensitive than a wzzB mutant [14], [16], [20]. Importantly, an S. Typhimurium fepE mutant retains full virulence in the mouse typhoid model [14], suggesting that complement resistance and resistance to phagocyte-mediated killing mechanisms are independent of very long O-antigen chains in vivo. Since no phenotype has been described for a S. Typhimurium fepE mutant in mouse models, it remains unclear which selective forces prevent loss of very long O-antigen chains.
In the absence of selective forces to maintain the costly production of very long O-antigen chains, the fepE gene is predicted to accumulate point mutations and eventually become inactive. Interestingly, fepE is a pseudogene in S. enterica serotype Typhi, the causative agent of typhoid fever [21]. We thus reasoned that fepE might be involved in aspects of host pathogen interaction that are important during gastroenteritis (caused by S. Typhimurium) but dispensable during typhoid fever (caused by S. Typhi). While S. Typhimurium transmission requires maximum growth in the lumen of the inflamed intestine [22], S. Typhi persists in the human population through chronic gall bladder carriage [23], [24]. Based on these considerations, we proposed that very long O-antigen chains might enhance the fitness of S. Typhimurium in the environment of the inflamed intestine during gastroenteritis. We tested this hypothesis using the mouse colitis model and elucidated the underlying mechanism.
Results
Very long O-antigen chains confer a luminal growth advantage in the inflamed gut
To study the role of very long O-antigen chains, we generated a fepE mutant of the S. Typhimurium wild type strain IR715. The fepE mutant (RC31) was deficient for producing very long O-antigen chains, and this defect could be restored by introducing the cloned fepE gene (pCR37) on a plasmid (Fig. S1A). Consistent with previous reports [14], [16], the S. Typhimurium wild type (IR715) and the fepE mutant (RC31) exhibited similar levels of serum resistance (Fig. S1B). In contrast, a wzzB mutant that lacked long O-antigen chains but retained the ability to produce very long O-antigen chains (Fig. S1A) exhibited significantly (P<0.01) reduced serum resistance (Fig. S1B).
Genetically resistant mice infected with S. Typhimurium exhibit little intestinal inflammation during the first three days after inoculation (mouse typhoid model) [25]. In contrast, mice treated with streptomycin develop severe acute cecal inflammation within 2 days after infection (mouse colitis model). To determine whether very long O-antigen chains confer a fitness advantage during conditions of acute intestinal inflammation, we performed a pilot experiment in which streptomycin pre-treated, genetically susceptible mice (C57BL/6J) were infected intragastrically with an equal mixture of the S. Typhimurium wild type (IR715) and a fepE mutant (RC31) (mouse colitis model). By day four after infection, mice became moribund and the wild type was recovered in higher numbers than the fepE mutant from intestinal contents, but not from liver and spleen (Fig. S2).
To determine whether the luminal fitness advantage conferred by the fepE gene would increase over time, groups of streptomycin pre-treated, genetically resistant mice (129/SvJ) were infected intragastrically with an equal mixture of the S. Typhimurium wild type (IR715) and a fepE mutant (RC31) (mouse colitis model). For comparison, mice (129/SvJ) that had not been pre-treated with streptomycin were infected with the same mixture of S. Typhimurium strains (mouse typhoid model). In the mouse typhoid model, the wild type did not exhibit a luminal growth advantage over the fepE mutant as indicated by recovery of bacteria from feces (Fig. 1A) or colon contents (Fig. 1B). In contrast, the wild type was recovered in significantly higher numbers (P<0.05) from feces starting at day 3 after infection in the mouse colitis model (Fig. 1A). By day 7 after infection, the wild type was recovered in greater than 100-fold excess over the fepE mutant (Fig. 1B). In both the mouse typhoid model and in the mouse colitis model, the S. Typhimurium wild type and fepE mutant were recovered in similar numbers from the liver and spleen (Fig. 1C), suggesting that very long O-antigen chains are dispensable for growth in tissue. As expected, histopathogical analysis revealed a greater severity of cecal inflammation in the mouse colitis model compared to the mouse typhoid model seven days after infection (Fig. 1 and E). Bacteria recovered from intestinal contents of streptomycin pretreated mice exhibited strong expression of very long O-antigen chains, as indicated by Western blot analysis (Fig. S3).
Fig. 1. Very long O-antigen chains confer a fitness advantage in the lumen of the inflamed gut. 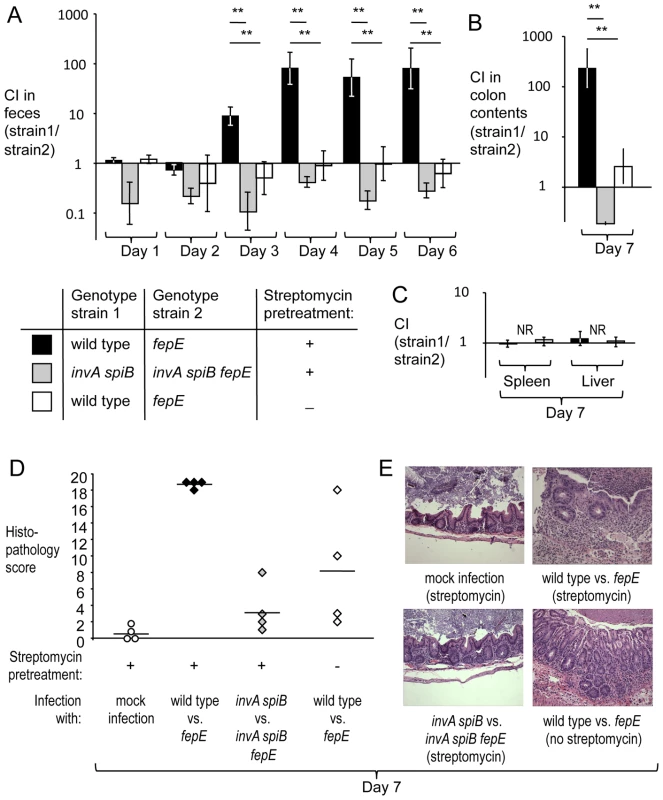
Mice (129/SvJ) were treated with streptomycin or were left untreated and were subsequently infected with equal mixtures of the indicated S. Typhimurium strains. (A) Competitive indices (CI) recovered from feces over time. (B) Competitive indices recovered from colon contents. (C) Competitive indices recovered from the liver and spleen. (A–C) Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01, NR, no bacteria recovered. (D) Combined histopathology score of pathological changes observed in sections from the cecum. Lines represent averages. Each circle or diamond represents the combined histopathology score from an individual animal. (E) Representative images of histopathological changes. Production of very long O-antigen chains had a similar effect on bacterial numbers in the mouse colitis model when we used CBA/J mice, a lineage that is also genetically resistant to S. Typhimurium infection (Fig. S4A and S4B). To assess complementation in vivo, streptomycin pre-treated CBA/J mice were infected with an equal mixture of a S. Typhimurium fepE mutant (RC61) and a derivative carrying the fepE gene inserted upstream of the phoN gene (phoN::fepE) (RC62). The complemented strain (RC62) was recovered in higher numbers from feces and colon contents (Fig. S4C), suggesting that an intact chromosomal copy of the fepE gene could restore the fitness defect of a fepE mutant. Next, we investigated whether the fitness advantage conferred by very long O-antigen chains was apparent in mice infected with individual S. Typhimurium strains (single infection design). Streptomycin pre-treated CBA/J mice were infected with either the S. Typhimurium wild type (IR715) or a fepE mutant (RC31) (mouse colitis model). Mice infected with the S. Typhimurium wild type shed significantly (P<0.01) higher bacterial numbers with their feces than mice infected with the fepE mutant (Fig. S4E), while similar bacterial loads were recovered from liver and spleen (Fig. S4D).
T3SS-1 and T3SS-2 cooperate to induce intestinal inflammation in the mouse colitis model [26], [27]. Inactivation of both T3SS-1 (through a mutation in invA) and T3SS-2 (through a mutation in spiB) renders the resulting S. Typhimurium double mutant strain unable to trigger intestinal inflammation [9]. To further investigate why very long O-antigen chains conferred a fitness advantage in the mouse colitis model, streptomycin pre-treated mice (129/SvJ) were infected with an equal mixture of an invA spiB mutant (SPN452) and an invA spiB fepE mutant (RC55). Mice infected with this mixture did not developed intestinal inflammation (Fig. 1A and 1B) and the invA spiB mutant did not exhibit a luminal growth advantage over the invA spiB fepE mutant (Fig. 1A and 1B). Due to the attenuation caused by mutations in invA and spiB, neither strain was recovered from the liver and spleen of mice seven days after infection (Fig. 1C). Collectively, these data pointed to the lumen of the acutely inflamed intestine as the habitat conferring a fitness advantage to S. Typhimurium expressing very long O-antigen chains. However, the mechanism by which very long O-antigen chains would provide a benefit in the environment of the inflamed gut remained unclear.
Metabolite profiling of luminal changes during S. Typhimurium induced colitis
To identify potential small molecule candidates that might explain the fitness advantage conferred by very long O-antigen chains in the inflamed gut, groups of streptomycin pre-treated mice (n = 4) were inoculated with sterile LB broth (mock infection) or S. Typhimurium. Four days after infection, the ceca were collected and metabolites extracted from the mucosa with water. Metabolites in these cecal washes were analyzed after removing bacteria by filtration to avoid contamination of samples with bacterial intracellular metabolites. Samples underwent hydrophilic interaction chromatography - liquid chromatography/mass spectrometry (HILIC-LC/MS) metabolic profiling. Peak metabolite values were measured for cecal washes of each animal and data were transformed logarithmically (log2) for statistical analysis and determination of false discovery rates. More than 800 components were detected by HILIC-LC/MS. Principle component analysis revealed group clustering of samples from mock-infected mice and S. Typhimurium-infected mice (Fig. S5), suggesting that S. Typhimurium infection was accompanied by characteristic changes in the luminal environment.
We were able to assign metabolite identities to 67 components (Table S1). A total of 20 components met the criteria for at least a 2-fold change between samples from mock-infected and S. Typhimurium-infected mice and a P value of less than 0.1, which is illustrated in a volcano plot in Figure 2A. Of these, 15 components were considered significantly changed (P<0.05; q<0.1) between both groups. We were able to assign metabolite identities to four of the six components that were significantly (P<0.05; q<0.1) increased in samples from S. Typhimurium-infected mice compared to mock-infected animals (Fig. 2B). Interestingly, one of these metabolites, phosphatidylinositol 3-phosphate, is produced in host cells by the T3SS-1 effector protein SopB [28]. Two metabolites were breakdown products of phosphatidylethanolamine (lysophosphatidylethanolamine [14∶1]) and phosphatidylcholine (lysophosphatidylcholine [16∶1]), respectively. The fourth metabolite with significantly increased abundance (P<0.05; q<0.1) in samples from S. Typhimurium infected mice was the bile acid tauromurocholate. These data indicated that increased levels of bile acids might accompany S. Typhimurium infection, thereby identifying a potential candidate for an anti-microbial metabolite that is elevated during colitis.
Fig. 2. Bile acids are elevated during S. Typhimurium-induced colitis. 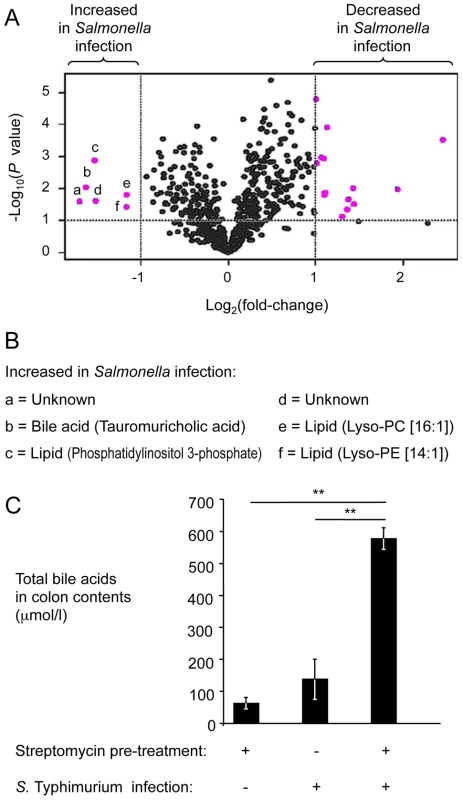
(A) Groups of streptomycin pre-treated mice were inoculated with sterile LB broth (mock infection, n = 4) or with S. Typhimurium (n = 4). Four days after infection, water extracts of metabolites from the cecal mucosa were analyzed by HILIC-LC/MS metabolite profiling. For each component detected by HILIC-LC/MS, peak areas from four mock-infected animals were averaged and compared to the average peak areas from four S. Typhimurium-infected animals. Each dot in the volcano plot represents the fold-change between mock-infected and S. Typhimurium-infected mice for one component. Components with at least a 2-fold change and a P value of less than 0.1 are indicated in pink. (B) List of metabolites showing at least a 2-fold increase during S. Typhimurium infection. Letters correspond to letters in panel A. Lys-PE, lysophosphatidylethanolamine; Lys-PC, lysophosphatidylcholine. (C) Determination of concentrations of total bile acids in colon contents. Bars represent geometric means ± standard error. **, P<0.01. To validate results from metabolite profiling, the concentration of total bile acids was determined in colon contents recovered from the mouse typhoid model and from the mouse colitis model using an enzymatic assay. The concentration of total bile acids was significantly (P<0.01) increased in the mouse colitis model, compared to the mouse typhoid model or to mock-infected mice (Fig. 2C). These data suggested that the concentration of total bile acids was increased during S. Typhimurium-induced colitis. Furthermore, these data established an inverse correlation between luminal concentrations of total bile acids (Fig. 2C) and the luminal fitness of the fepE mutant (Fig. 1A and 1B) but fell short of establishing cause and effect.
Very long O-antigen chains increasing resistance to bile
To investigate whether an increased concentration of total bile acids during S. Typhimurium-induced colitis was the mechanism responsible for the fitness advantage conferred by very long O-antigen chains (Fig. 1), we first determined the minimal inhibitory concentration (MIC) of S. Typhimurium strains to ox bile extract (sodium choleate) in vitro (Table 1). The MIC of sodium choleate was identical for the S. Typhimurium wild type (IR715) and a wzzB mutant (RC46). However, compared to the wild type, the MIC of sodium choleate for the fepE mutant (RC31) was reduced. Introduction of the cloned fepE gene (pCR37) into the fepE mutant restored resistance to sodium choleate. Lack of very long O-antigen chains in the fepE mutant did not reduce the MIC for the anionic detergent sodium dodecyl sulfate (SDS) or for the cationic antimicrobial peptide polymyxin B. In contrast, a mutation in pmrE, a gene necessary for 4-aminoarabinose lipid A modification [29], increased sensitivity to the antimicrobial peptide polymyxin B, but not to the anionic detergent SDS. Collectively, these data suggested that very long O-antigen chains, but not long O-antigen chains, are specifically required for full resistance to bile acids.
Tab. 1. Very long O-antigen chains confer resistance to bile acids. 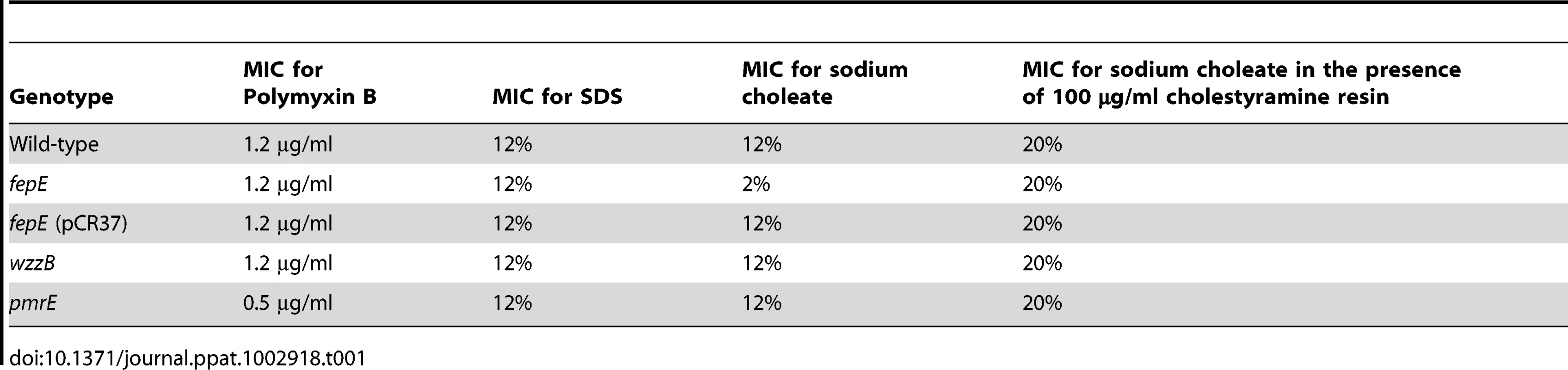
Since the MIC of the fepE mutant for sodium choleate (2%) was nearly 100-fold higher than the concentration of bile acids measured in intestinal contents (Fig. 2C), we compared growth of the S. Typhimurium wild type and a fepE mutant in medium containing different concentrations of bile acids. As expected, medium containing 3% sodium choleate supported growth of the S. Typhimurium wild type (MIC 12%) but fully suppressed growth of the fepE mutant (MIC 2%) (Fig. S6A). Growth of the fepE mutant was retarded compared to that of the S. Typhimurium wild type in medium containing 0.3% sodium choleate (Fig. S6B) or 0.03% sodium choleate (Fig. S6C). Finally, both strains grew equally in medium containing 0.003% sodium choleate (Fig. S6D). These data suggested that very long O-antigen chains conferred a fitness advantage at concentrations of bile acids that were approximately two orders of magnitude below the MIC of the fepE mutant (Table 1).
The concentration of total bile acids in intestinal contents can be lowered by bile sequestrants, such as cholestyramine resin, a compound used clinically to restrict cholesterol and fat intake. To investigate whether bile sequestrants could rescue growth of the fepE mutant in medium containing sodium choleate, MIC values were determined in vitro in the presence of cholestyramine resin (Table 1). The S. Typhimurium wild type and the fepE mutant exhibited identical MIC values for sodium choleate in the presence of cholestyramine resin.
To test whether increased concentrations of total bile acids are responsible for the fitness advantage conferred by very long O-antigen chains in the mouse colitis model, animals fed rodent chow containing cholestyramine resin or control chow were pretreated with streptomycin and inoculated with sterile LB broth (mock infection) or with an equal mixture of the S. Typhimurium wild type (IR715) and a fepE mutant (RC31). While the S. Typhimurium wild type was recovered in significantly higher numbers from feces (Fig. 3A) and colon contents (Fig. 3B) of mice fed control chow, this competitive advantage was abrogated in mice fed chow containing cholestyramine resin. Concomitantly, diet containing cholestyramine resin significantly (P<0.05) reduced the concentration of total bile acids in colon contents present during S. Typhimurium infection (Fig. 3C) but did not reduce the severity of intestinal inflammation (Fig. 3D and 3E). These data provided direct support for the idea that increased concentrations of total bile acids during S. Typhimurium-induced colitis reduced the fitness of bacteria lacking very long O-antigen chains.
Fig. 3. The bile sequestrant cholestyramine resin reduces the fitness advantage conferred by very long O-antigen chains in the mouse colitis model. 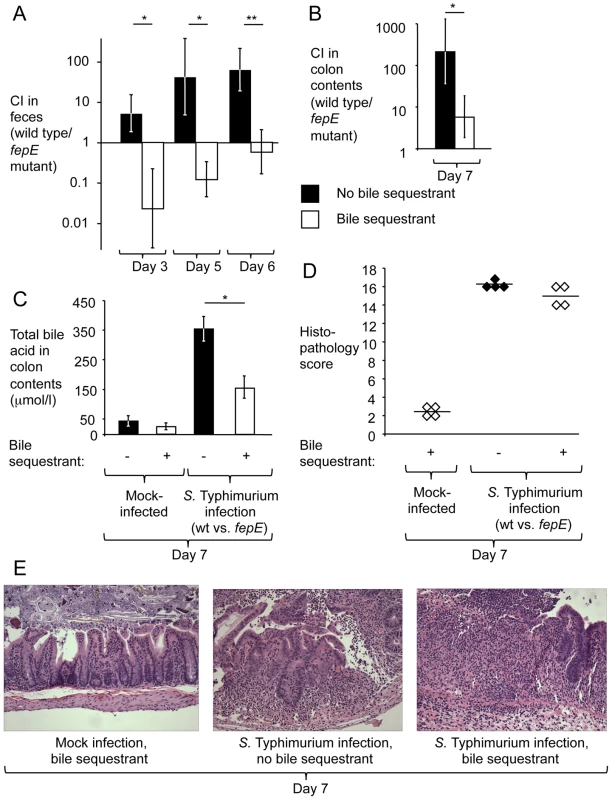
Mice (129/SvJ) receiving chow containing cholestyramine resin or control chow were pre-treated with streptomycin and infected with an equal mixture of the S. Typhimurium wild type and a fepE mutant. (A) Competitive indices recovered from feces over time. (B) Competitive indices recovered from colon contents. (C) Determination of concentrations of total bile acids in colon contents. (A–C) Bars represent geometric means ± standard error. *, P<0.05; **, P<0.01. (D) Combined histopathology score of pathological changes observed in sections from the cecum. Lines represent averages. Each circle or diamond represents the combined histopathology score from an individual animal. (E) Representative images of histopathological changes. Discussion
S. Typhimurium-induced colitis is accompanied by elevated luminal concentrations of several antimicrobial proteins, including lipocalin-2 [9], calprotectin [11], regenerating islet-derived 3 gamma (RegIIIγ) [30] and RegIIIβ [10]. In some cases, specific resistance mechanisms against these antimicrobial proteins have been shown to confer a fitness advantage in the lumen of the inflamed gut. For example, the ability to synthesize the siderophore salmochelin confers resistance to lipocalin-2 [31], a property that boosts luminal growth of S. Typhimurium in the mouse colitis model [9]. Similarly, the high affinity zinc transporter ZnuABC confers resistance to the zinc-binding protein calprotectin, thereby enhancing luminal growth of S. Typhimurium during colitis [11].
Here we used metabolite profiling to determine whether small antimicrobial metabolites are elevated during S. Typhimurium-induced colitis. Our results identify bile acids as a class of small antimicrobial molecules whose concentration is elevated in the mouse colitis model. A previous metabolite profiling study detected elevated bile acid biosynthesis in the mouse typhoid model by analyzing acetonitrile extracts from feces [32]. Total concentrations of bile acids in feces and fecal water are elevated two to five times in patients with inflammatory bowel disease [33]. However, fecal concentrations of total bile acids are significantly decreased during dextran sulfate sodium (DSS)-induced colitis in rats [34]. Thus, an increased concentration of bile acids is not likely to be a general feature of intestinal inflammation, but might involve mechanisms that are only triggered in a subset of inflammatory disorders, including S. Typhimurium-induced colitis. It should be noted that due to absorption in the large intestine, bile concentrations might be higher in the ileum, the primary site of S. Typhimurium infection in humans. The mouse is not an ideal model to study luminal growth in the ileum, because the bulk of luminal S. Typhimurium are present in the cecum.
The absence of acute colitis during typhoid fever might explain why fepE is a pseudogene in S. Typhi [21]. However, S. Typhi requires bile resistance for persistence in the gall bladder. It is possible that other long surface carbohydrates, such as the Vi-capsular polysaccharide or the O-antigen capsule [35], [36], can compensate for the lack of very long O-antigen chains during growth in the environment of the gall bladder. Nonetheless, our data show that very long O-antigen chains increased the fitness of S. Typhimurium in the lumen of the acutely inflamed intestine by mediating bile resistance, which represents the first function ascribed to this surface structure. Inactivation of yrbK and rlpB, two genes implicated in LPS transport, increases bile resistance of S. Typhimurium and correlates with differences in the oligosaccharide units that form long-chain LPS [37]. However, our data suggest that long O-antigen chains do not confer resistance to bile. Conversely, long O-antigen chains confer serum resistance, while very long O-antigen chains are dispensable for this trait [14], [16]. Thus, long O-antigen chains and very long O-antigen chains do not have redundant functions.
The picture emerging from these studies is that very long O-antigen is important for growth of S. Typhimurium in the lumen of the inflamed intestine. Maximum growth in the intestinal lumen is of relevance, because it enhances transmission by the fecal-oral route [22], which is predicted to confer a selective advantage. We conclude that the role of very long O-antigen chains in bile resistance likely represents one of the selective forces that help to maintain the fepE gene in the genomes of non-typhoidal Salmonella serotypes.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are presented in Table 2. Cultures of S. Typhimurium and Escherichia coli were routinely incubated with aeration at 37°C in Luria-Bertani (LB) broth (10 g tryptone, 5 g yeast extract, and 10 g NaCl per liter) or on LB agar plates unless indicated otherwise. Antibiotics were added as necessary at appropriate concentrations: chloramphenicol (Cm), 0.03 mg/ml; carbenicillin (Cb), 0.1 mg/ml; kanamycin (Kan), 0.05 mg/ml; and nalidixic acid (Nal), 0.05 mg/ml.
Tab. 2. Bacterial strains and plasmids used in this study. 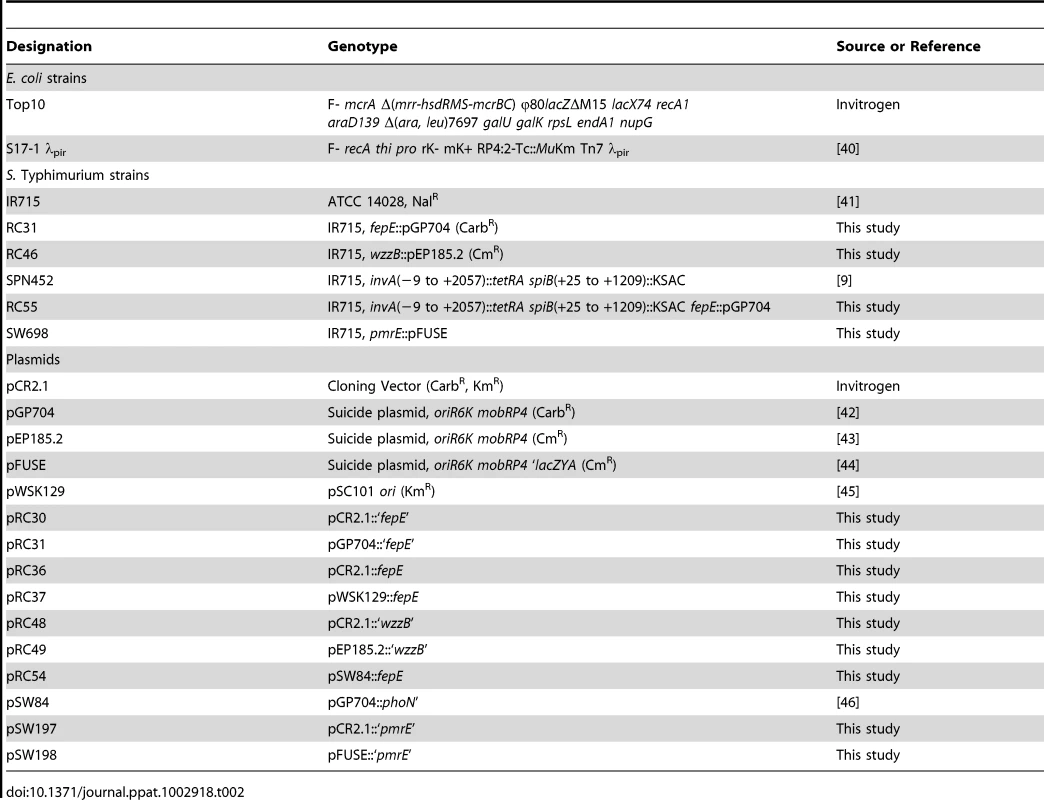
Construction of mutants and plasmids
Nucleotide sequences of oligonucleotide primers used for cloning are listed in Table 3. Internal fragment of S. Typhimurium the fepE and wzzB genes were polymerase chain reaction (PCR) amplified using primer pairs 62/63 and 150/151, respectively, and cloned into pCR2.1 (Invitrogen) to give rise to plasmids pRC30 and pRC48. The inserts were excised using restriction endonuclease digestion with SmaI and SalI (pRC30) or SacII and XhoI (pRC48) and cloned into suicide vector pGP704 digested with SmaI and SalI or into suicide vector pEP185.2 digested with SacII and XhoI to yield plasmids pRC31 and pRC49, respectively. Plasmids pRC31 and pRC49 were introduced into S. Typhimurium IR715 by conjugation with E. coli S17-1 λpir and exconjugants were designated RC31 and RC46, respectively. The wild-type fepE gene was PCR amplified using the primer pair 52/53 and cloned into pCR2.1 to yield plasmid pRC36. The insert of pCR36 was excised with EcoRI and cloned into low copy vector pWSK129 to yield pRC37. To construct a pmrE mutant, an internal fragment of the S. Typhimurium pmrE gene was PCR amplified using primers 263/264, cloned into pCR2.1 to yield plasmid pSW197. The insert of pSW197 was cloned into suicide plasmid pFUSE at XbaI and SmaI restriction sites to yield plasmid pSW198. Plasmid pSW198 was introduced into S. Typhimurium IR715 by conjugation to yield strain SW698. The fepE::pGP704 mutation was transduced from RC31 into the S. Typhimurium invA spiB mutant SPN452 using transduction with phage P22 HT int-105 to yield strain RC55.
Tab. 3. Oligonucleotide primers used in this study. 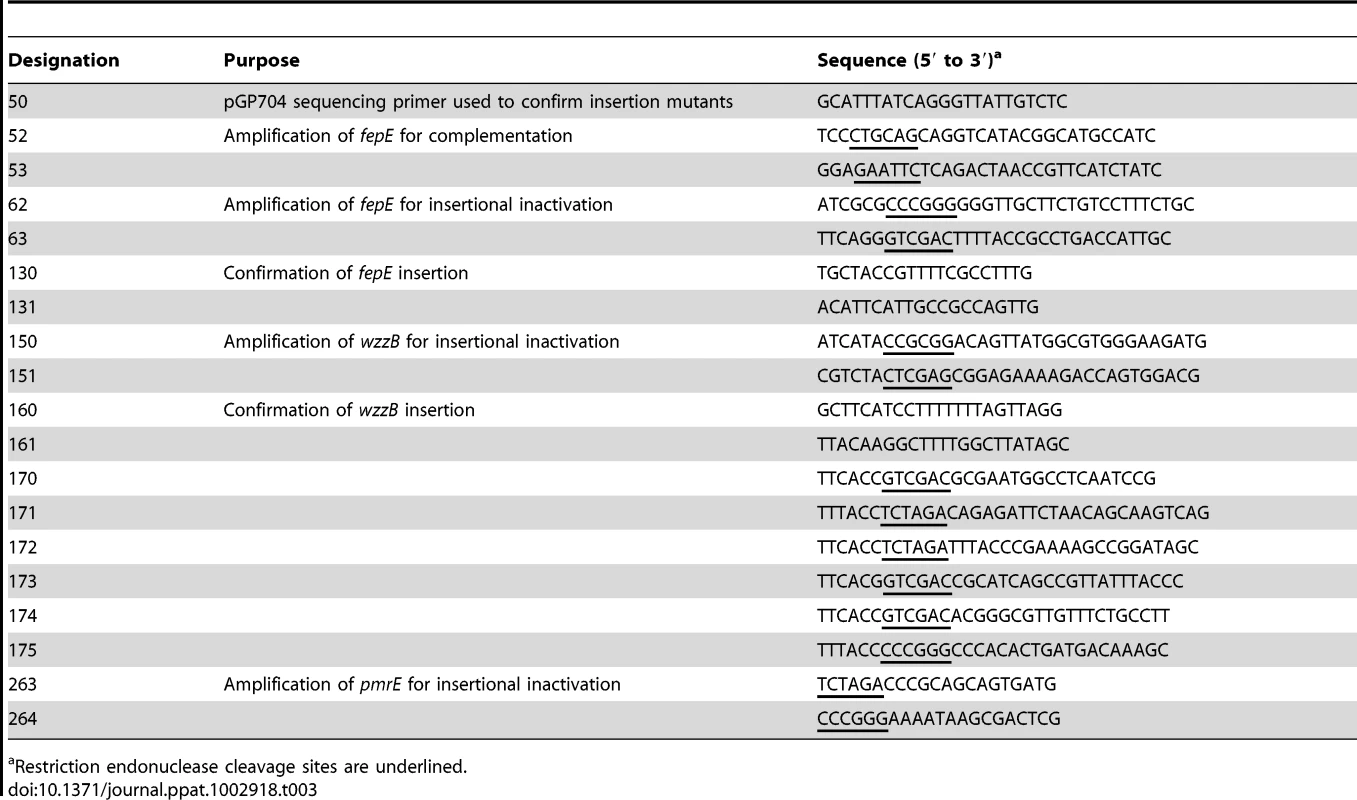
Restriction endonuclease cleavage sites are underlined. To construct a fepE deletion mutant, primers 170 and 171 were used to PCR amplify the region upstream of S. Typhimurium fepE (flanking region 1) from base 3537 of entF to base −48 of fepE. Primers 172 and 173 were used to PCR amplify the downstream region of S. Typhimurium fepE (flanking region 2) from base 1136 of fepE to base 322 of fepC. PCR products for the fepE upstream and downstream regions were gel purified (Qiagen), digested with XbaI, and ligated together with T4 DNA ligase (NEB). The ligation reaction served as a template for PCR amplification of the fepE flanking region construct using primers 170 and 173, and this product was gel purified and cloned into pCR2.1 (Invitrogen) to generate pRC50. The cassette containing fepE flanking regions was excised from pRC50 using SalI, ligated into SalI-digested pRDH10 using the Quick T4 DNA Ligase kit (NEB) to generate pRC51. The KSAC cassette from pBS34 was excised by XbaI digestion and ligated into the XbaI site of pRC51 to yield pRC52. Plasmid pRC52 was conjugated into S. Typhimurium IR715. An exconjugant selected on plates containing LB+Nal+Km was designated RC61 and deletion of fepE was confirmed by PCR. For complementation, the S. Typhimurium wild type fepE allele was PCR amplified using primers 174 and 175 from base −350 of fepE to base 1136 of fepE, and cloned into pCR2.1 to give rise to plasmid pRC53. The insert was excised using restriction endonuclease digestion with SalI and SmaI and ligated into SalI - and SmaI-digested pSW84, a suicide plasmid carrying a DNA fragment upstream of the phoN open reading frame, to yield pRC54. Plasmid pRC54 was introduced into RC61 by conjugation with E. coli S17-1 λpir and an exconjugant carrying pRC54 integrated into the phoN locus was designated RC62.
Analysis of LPS
Bacterial cultures grown 16 hours at 37°C in LB broth were suspended in 1 ml Dulbecco's phosphate buffered saline (DPBS, Gibco) at a concentration of approximately 1×109 CFU/ml. Samples were washed three times in DPBS (12,000× g for 4 minutes). The pellet was resuspended in 0.1 ml lysis buffer (0.5 M Tris-HCl at pH 6.8, 4% sodium dodecyl sulfate [SDS], 40% glycerol, 0.2% bromophenol blue, and 10% β-mercaptoethanol) and boiled for 10 minutes. We then added 0.1 ml of proteinase K solution (Invitrogen) (20 mg proteinase K/ml in 10 mM Tris-HCl, 50% glycerol, and 20 mM CaCl2) and incubated for 3 hours at 65°C. LPS preparations were boiled again for 10 minutes, and 0.02 ml of each sample loaded on a 15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by silver staining as described previously [38].
LPS purified from colon and cecal contents was probed with anti-Salmonella O:4 serum (Becton, Dickinson and Company) for production of the somatic O antigen by Western blot analysis. CFU of S. Typhimurium were determined for each sample and samples loaded in each lane were normalized to represent equivalent amounts of bacteria. Preparations were separated by 15% SDS-PAGE, transferred to an Immobilon PVDF membrane (Millipore) using a Trans-Blot semidry transfer apparatus (Bio-Rad), and detected with anti-Salmonella O:4 serum and an alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Bio-Rad Laboratories). Bands were visualized following exposure and development on a BioSpectrum AC Imaging System (Ultra-Violet Products).
Determination of MIC values
To determine MIC values of S. Typhimurium strains against polymyxin B (Sigma Aldrich), SDS (USB Corporation), sodium choleate (a crude ox bile extract from Sigma Aldrich, which contains the sodium salts of taurocholic, glycocholic, deoxycholic, and cholic acids), or with a combination of sodium choleate and cholestyramine resin (DOW Chemical Company), overnight cultures were washed twice in DPBS, diluted to 1×103 cells per ml with antimicrobial compounds in sterile LB broth (polymyxin B: 0–1.5 µg/ml, increments of 0.1 µg/ml; SDS and sodium choleate: 0–20% w/v, increments of 2%) and incubated for 15 hours at 37°C with aeration to determine the lowest concentration of each antimicrobial compound that prevented bacterial growth. Each experiment was repeated three times independently.
Serum sensitivity
For analyses of serum sensitivity, 1×107 bacteria from cultures grown to stationary phase were washed three times in DPBS and incubated in 10% human serum for one hour at 37°C with slight agitation. The number of viable bacteria was quantified by spreading serial 10-fold dilutions on LB agar plates containing the appropriate antibiotics.
Animal experiments
For competitive infection experiments, groups (n = 4) of female mice (C57BL/6J or 129/svJ Jackson Laboratory) aged 6 to 8 weeks were inoculated intragastrically with 20 mg streptomycin in a volume of 0.2 ml 48 hours prior to intragastric inoculation with either 0.1 ml sterile LB broth or 1×107 CFU of the indicated mixtures of S. Typhimurium strains. Alternatively, animals were inoculated intragastrically with either 0.1 ml sterile LB broth or 1×107 CFU of the indicated mixtures of S. Typhimurium strains without being pretreated with streptomycin. Fecal pellets were collected and organs were harvested at day 4 (C57BL/6J) or day 7 (129/svJ) after infection. Fecal and tissue samples were homogenized in PBS, serially diluted and plated on selective media to enumerate CFU. The competitive index (CI) was calculated by dividing the ratio of wild-type to mutant bacteria in homogenates (output) by the ration present in the inoculum (input). In some experiments, 129/SvJ mice were fed a normal rodent diet (Harlan-Teklad) supplemented with the bile acid sequestrant cholestyramine resin (2%, The Dow Chemical Company) starting the day before streptomycin treatment and continuing throughout the experiment.
For metabolite profiling, groups of 4 mice (C57BL/6J) received 20 mg streptomycin in a 0.1 ml volume intragastrically and were inoculated intragastrically 24 hours later with either 0.1 ml sterile LB broth or 1×109 CFU of S. Typhimurium in LB broth. The cecum was collected four days after infection, contents were removed and metabolites from the mucosal surface were extracted by five washes with 0.2 ml sterile UltraPure distilled water (Gibco). Debris was removed by centrifugation at 20,000× g for 5 minutes, the supernatant was filter sterilized to remove bacteria and submitted to metabolite profiling analysis.
Ethics statement
All animal experiments were performed according to Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines. All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of California, Davis.
Histopathology
Cecal tissues were formalin fixed, sectioned, stained with hematoxylin and eosin (H&E), and submitted to a veterinary pathologist for blinded scoring using a scale described previously [8]. Representative images of tissue sections were taken using an Olympus BX41 microscope.
Metabolite profiling
Reagents and standards: Liquid chromatography/mass spectrometry (LC/MS) grade acetonitrile and acetone (Burdick and Jackson, VWR International, West Chester, PA, USA) and extra pure ammonium formate (EMD, Gibbstown, NJ, USA) were purchased. Ultrapure water was supplied by an in house Millipore system (Billerica, MA, USA). Each lot of organic solvents was investigated by LC/MS infusion. Utilizing ultrapure water, aqueous buffer for liquid chromatography/electrospray ionization mass spectrometry (LC/ESI-MS) and a stock solution (1 mg/ml) of tuning reference compound was prepared freshly on a daily basis in a solvent system identical to the initial mobile phase composition of LC.
Hydrophilic interaction chromatography (HILIC)-LC/ESI-MS profiling: HILIC-LC/ESI-MS analysis [39] was performed with the use of a modified silica-based column (Luna HILIC Diol, 150_3 mm, 3 mm particle size; Phenomenex, Torrance, CA, USA). The mobile phases were 100 mM ammonium formate (pH 4.0) (A) and acetonitrile (B) (flow rate 0.4 mL/min at 40°C). After a 2 minute isocratic run at 3% A, a sequential ramping scheme was followed up to 40% A for total injection time of 20 min. The injection volume was set to 10 ml. The entire effluent from the high-pressure liquid chromatography (HPLC) column was directed into the ESI source of a linear trap quadrupole (LTQ) linear ion trap (LIT) mass spectrometer (Thermo Fisher, San Jose, CA, USA) operated under Xcalibur software (v1.4, Thermo Fisher). The electrospray voltage was set to 5 kV. Nitrogen sheath and auxiliary gas flow were set at 60 and 20 arbitrary units, respectively. The ion transfer capillary temperature was set at 350°C with typical ion gauge pressure of 0.90×10−5. Full scan spectra were acquired from 100–1500 amu at unit mass resolution with maximum injection time set to 200 ms in one micro scan. Acquisition was performed in both positive/negative switching modes. A sucrose tune file in negative/positive modes at high LC flow rate was used during all the LC/ESI-MS acquisitions on the LTQ mass spectrometer.
Metabolite annotation: Annotation of the MS and MS/MS spectra was done using commercial NIST05/Wiley Registry; METLIN, Mass-Bank, Human Metabolome Database, Lipid Maps, and in-house built mass spectral libraries. Annotation was further validated with de novo structure identification. The unique elemental formula was searched against the CAS database using the strategy of Explore Substances – Chemical Structure (American Chemical Society, Washington, DC) for known compounds, or input into the MolGen 3.5,20 generating all of the possible structural isomers corresponding to the elemental formula. The chemical structures were saved and imported to Mass Frontier 5.0 (HighChem Ltd., Bratislava, Slovakia) for MS/MS fragmentation modeling analysis. The Mass Frontier Fragments and Mechanisms module is an expert system providing information about basic fragmentation and rearrangement processes based on literature, starting from a user-supplied chemical structure. The theoretical fragments generated by Mass Frontier were compared to those acquired from LC/MS. Parent compounds that had the best match of MS/MS fragmentation pattern were considered as the molecular structures of the potential biomarkers. For validation purpose, the proposed molecules were searched against chemical structure and property databases or search engines such as publicly available PubChem, Chemical Structure Lookup Service (CSLS), CRC Dictionary of Natural Products (DNP), ChemSpider, and proprietary Beilstein Database using MDL Crossfire Commander and Chemical Abstracts Database (CAS) using SciFinder Scholar.
Raw data processing: Prior to data processing, original Xcalibur LC/MS files were converted into netCDF format using the XConverter (Thermo Fisher), then converted into WIFF format usingAnalyst QS for use in MarkerView software (v1.2, Applied Biosystems, Foster City, CA, USA). For HILIC LC/MS data, peak finding options were set as follows: subtraction offset, 10 scans; subtraction multiplication factor, 1.3; noise threshold, 3; minimum spectral peak width, 5 amu; minimum retention time peak width, 5 scans; and maximum retention time width, 1000 scans. Peak alignment options were set as follows: retention time tolerance, 0.5 min; mass tolerance, 0.8 amu; and maximum number of peaks, 5000. Peaks found in fewer than 3 of the samples were discarded using filter setting. Peak area integration was performed using raw data. Datasets were normalized using Euclidean norm by scaling each sample-vector to unit vector norm, which can be interpreted geometrically as a projection of the samples x to a hyper-sphere with the length of this sample vector scaled to one. Peaks were, then, normalized to the total absolute area of all detected metabolites in each sample. Datasets were routinely processed in MarkerView for principal component analysis. The data that constituted retention time, mass-to-charge ratio, and peak areas of detected and aligned peaks were exported from MarkerView into into MetaboAnalyst and MetPA for further analysis.
Quantitative determination of total bile acids
Colon contents were weight, homogenized in 1 ml PBS and analyzed using a colorimetric Total Bile Acids Assay Kit (Bio-Quant). Briefly, conversion of bile acids and Thio-NADH to 3-keto steroids and Thio-NADH by 3-α.hydroxysteroid dehydrogenase was followed by measuring change of absorbance at 405 nm. Bile acid concentrations were determined using a conjugated cholic acids standard using a protocol provided by the manufacturer.
Statistical analysis
Competitive indices were converted logarithmically (log10) prior to statistical analysis using a one-tailed parametric test (Student's t tests). Concentrations of total bile acids were analyzed using a one-tailed Student's t tests. A P value of <0.05 was considered to be significant.
Fold differences in metabolites between mock-infected mice and S. Typhimurium infected mice were converted logarithmically (log2) and a Student's t was used to determine whether differences were statistically significant. False discovery rates were estimated using the q value method. A P value of <0.05 and q value of <0.1 were considered a significant change.
Supporting Information
Zdroje
1. RabschW, TschapeH, BaumlerAJ (2001) Non-typhoidal salmonellosis: emerging problems. Microb Infect 3 : 237–247.
2. GalánJE, CurtissRIII (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA 86 : 6383–6387.
3. OchmanH, SonciniFC, SolomonF, GroismanEA (1996) Identification of a pathogenicity island for Salmonella survival in host cells. Proc Natl Acad Sci USA 93 : 7800–7804.
4. TsolisRM, AdamsLG, FichtTA, BaumlerAJ (1999) Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67 : 4879–4885.
5. StecherB, RobbianiR, WalkerAW, WestendorfAM, BarthelM, et al. (2007) Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5 : 2177–2189.
6. BarmanM, UnoldD, ShifleyK, AmirE, HungK, et al. (2008) Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76 : 907–915.
7. WinterSE, ThiennimitrP, WinterMG, ButlerBP, HusebyDL, et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 : 426–429.
8. ThiennimitrP, WinterSE, WinterMG, XavierMN, TolstikovV, et al. (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA 108 : 17480–17485.
9. RaffatelluM, GeorgeMD, AkiyamaY, HornsbyMJ, NuccioSP, et al. (2009) Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microb 5 : 476–486.
10. StelterC, KappeliR, KonigC, KrahA, HardtWD, et al. (2011) Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PloS One 6: e20749.
11. LiuJZ, JellbauerS, PoeAJ, TonV, PesciaroliM, et al. (2012) Zinc sequestration by the neutrophil protein calprotectin enhances salmonella growth in the inflamed gut. Cell Host Microb 11 : 227–239.
12. BatchelorRA, HaraguchiGE, HullRA, HullSI (1991) Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol 173 : 5699–5704.
13. BatchelorRA, AlifanoP, BiffaliE, HullSI, HullRA (1992) Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol 174 : 5228–5236.
14. MurrayGL, AttridgeSR, MoronaR (2003) Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol 47 : 1395–1406.
15. DanielsC, MoronaR (1999) Analysis of Shigella flexneri wzz (Rol) function by mutagenesis and cross-linking: wzz is able to oligomerize. Mol Microbiol 34 : 181–194.
16. HolzerSU, SchlumbergerMC, JackelD, HenselM (2009) Effect of the O-antigen length of lipopolysaccharide on the functions of Type III secretion systems in Salmonella enterica. Infect Immun 77 : 5458–5470.
17. HapfelmeierS, EhrbarK, StecherB, BarthelM, KremerM, et al. (2004) Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72 : 795–809.
18. IlgK, EndtK, MisselwitzB, StecherB, AebiM, et al. (2009) O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect Immun 77 : 2568–2575.
19. MurrayGL, AttridgeSR, MoronaR (2006) Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol 188 : 2735–2739.
20. MurrayGL, AttridgeSR, MoronaR (2005) Inducible serum resistance in Salmonella typhimurium is dependent on wzz(fepE)-regulated very long O antigen chains. Microb Infect 7 : 1296–1304.
21. ParkhillJ, DouganG, JamesKD, ThomsonNR, PickardD, et al. (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413 : 848–852.
22. LawleyTD, BouleyDM, HoyYE, GerkeC, RelmanDA, et al. (2008) Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun 76 : 403–416.
23. LeavittJW (1992) “Typhoid Mary” strikes back. Bacteriological theory and practice in early twentieth-century public health. Isis 83 : 608–629.
24. PutnamP (1927) The trend of typhoid fever mortality in the united states. Am J Hyg 7 : 762–781.
25. SantosRL, ZhangS, TsolisRM, KingsleyRA, AdamsLG, et al. (2001) Animal models of Salmonella infections: enteritis versus typhoid fever. Microb Infect 3 : 1335–1344.
26. CoburnB, LiY, OwenD, VallanceBA, FinlayBB (2005) Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73 : 3219–3227.
27. BarthelM, HapfelmeierS, Quintanilla-MartinezL, KremerM, RohdeM, et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71 : 2839–2858.
28. NorrisFA, WilsonMP, WallisTS, GalyovEE, MajerusPW (1998) SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci U S A 95 : 14057–14059.
29. GunnJS, LimKB, KruegerJ, KimK, GuoL, et al. (1998) PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27 : 1171–1182.
30. GodinezI, RaffatelluM, ChuH, PaixaoTA, HanedaT, et al. (2009) Interleukin-23 orchestrates mucosal responses to Salmonella enterica serotype Typhimurium in the intestine. Infect Immun 77 : 387–398.
31. FischbachMA, LinH, ZhouL, YuY, AbergelRJ, et al. (2006) The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc Natl Acad Sci U S A 103 : 16502–16507.
32. AntunesLC, ArenaET, MenendezA, HanJ, FerreiraRB, et al. (2011) Impact of salmonella infection on host hormone metabolism revealed by metabolomics. Infect Immun 79 : 1759–1769.
33. EjderhamnJ, RafterJJ, StrandvikB (1991) Faecal bile acid excretion in children with inflammatory bowel disease. Gut 32 : 1346–1351.
34. ArakiY, AndohA, TsujikawaT, FujiyamaY, BambaT (2001) Alterations in intestinal microflora, faecal bile acids and short chain fatty acids in dextran sulphate sodium-induced experimental acute colitis in rats. Europ J Gastroenterol Hepatol 13 : 107–112.
35. CrawfordRW, GibsonDL, KayWW, GunnJS (2008) Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immun 76 : 5341–5349.
36. CrawfordRW, Rosales-ReyesR, Ramirez-Aguilar MdeL, Chapa-AzuelaO, Alpuche-ArandaC, et al. (2010) Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc Natl Acad Sci U S A 107 : 4353–4358.
37. HernandezSB, CotaI, DucretA, AusselL, CasadesusJ (2012) Adaptation and preadaptation of Salmonella enterica to Bile. PLoS Genet 8: e1002459.
38. TsaiCM, FraschCE (1982) A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Analyt Biochem 119 : 115–119.
39. UrayamaS, ZouW, BrooksK, TolstikovV (2010) Comprehensive mass spectrometry based metabolic profiling of blood plasma reveals potent discriminatory classifiers of pancreatic cancer. Rap Commun Mass Spectrom 24 : 613–620.
40. SimonR, PrieferU, PuhlerA (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1 : 784–791.
41. StojiljkovicI, BaumlerAJ, HeffronF (1995) Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177 : 1357–1366.
42. MillerVL, MekalanosJJ (1988) A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170 : 2575–2583.
43. KinderSA, BadgerJL, BryantGO, PepeJC, MillerVL (1993) Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R-M+ mutant. Gene 136 : 271–275.
44. BäumlerAJ, TsolisRM, van der VeldenAWM, StojiljkovicI, AnicS, et al. (1996) Identification of a new iron regulated locus of Salmonella typhi. Gene 193 : 207–213.
45. WangRF, KushnerSR (1991) Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100 : 195–199.
46. HanedaT, WinterSE, ButlerBP, WilsonRP, TukelC, et al. (2009) The capsule-encoding viaB locus reduces intestinal inflammation by a Salmonella pathogenicity island 1-independent mechanism. Infect Immun 77 : 2932–2942.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 9- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- A Population Genomics Perspective on the Emergence and Adaptation of New Plant Pathogens in Agro-Ecosystems
- A Novel Rhabdovirus Associated with Acute Hemorrhagic Fever in Central Africa
- Ifitm3 Limits the Severity of Acute Influenza in Mice
- Copper at the Front Line of the Host-Pathogen Battle
- The Human Cytomegalovirus Assembly Compartment: A Masterpiece of Viral Manipulation of Cellular Processes That Facilitates Assembly and Egress
- Insights from Genomics into Bacterial Pathogen Populations
- Slc15a4, a Gene Required for pDC Sensing of TLR Ligands, Is Required to Control Persistent Viral Infection
- Relacin, a Novel Antibacterial Agent Targeting the Stringent Response
- Global Assessment of Genomic Regions Required for Growth in
- The Battle over mTOR: An Emerging Theatre in Host–Pathogen Immunity
- Very Long O-antigen Chains Enhance Fitness during -induced Colitis by Increasing Bile Resistance
- and Beyond—Inositol Utilization and Its Implications for the Emergence of Fungal Virulence
- Antifungal Drug Discovery: Something Old and Something New
- Deep Sequencing of Antiviral T-Cell Responses to HCMV and EBV in Humans Reveals a Stable Repertoire That Is Maintained for Many Years
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Insights from Genomics into Bacterial Pathogen Populations
- Relacin, a Novel Antibacterial Agent Targeting the Stringent Response
- Ifitm3 Limits the Severity of Acute Influenza in Mice
- Antifungal Drug Discovery: Something Old and Something New
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



