-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Telomere Length Affects the Frequency and Mechanism of Antigenic Variation in
Trypanosoma brucei is a master of antigenic variation and immune response evasion. Utilizing a genomic repertoire of more than 1000 Variant Surface Glycoprotein-encoding genes (VSGs), T. brucei can change its protein coat by “switching” from the expression of one VSG to another. Each active VSG is monoallelically expressed from only one of approximately 15 subtelomeric sites. Switching VSG expression occurs by three predominant mechanisms, arguably the most significant of which is the non-reciprocal exchange of VSG containing DNA by duplicative gene conversion (GC). How T. brucei orchestrates its complex switching mechanisms remains to be elucidated. Recent work has demonstrated that an exogenous DNA break in the active site could initiate a GC based switch, yet the source of the switch-initiating DNA lesion under natural conditions is still unknown. Here we investigated the hypothesis that telomere length directly affects VSG switching. We demonstrate that telomerase deficient strains with short telomeres switch more frequently than genetically identical strains with long telomeres and that, when the telomere is short, switching preferentially occurs by GC. Our data supports the hypothesis that a short telomere at the active VSG expression site results in an increase in subtelomeric DNA breaks, which can initiate GC based switching. In addition to their significance for T. brucei and telomere biology, the findings presented here have implications for the many diverse pathogens that organize their antigenic genes in subtelomeric regions.
Published in the journal: . PLoS Pathog 8(8): e32767. doi:10.1371/journal.ppat.1002900
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002900Summary
Trypanosoma brucei is a master of antigenic variation and immune response evasion. Utilizing a genomic repertoire of more than 1000 Variant Surface Glycoprotein-encoding genes (VSGs), T. brucei can change its protein coat by “switching” from the expression of one VSG to another. Each active VSG is monoallelically expressed from only one of approximately 15 subtelomeric sites. Switching VSG expression occurs by three predominant mechanisms, arguably the most significant of which is the non-reciprocal exchange of VSG containing DNA by duplicative gene conversion (GC). How T. brucei orchestrates its complex switching mechanisms remains to be elucidated. Recent work has demonstrated that an exogenous DNA break in the active site could initiate a GC based switch, yet the source of the switch-initiating DNA lesion under natural conditions is still unknown. Here we investigated the hypothesis that telomere length directly affects VSG switching. We demonstrate that telomerase deficient strains with short telomeres switch more frequently than genetically identical strains with long telomeres and that, when the telomere is short, switching preferentially occurs by GC. Our data supports the hypothesis that a short telomere at the active VSG expression site results in an increase in subtelomeric DNA breaks, which can initiate GC based switching. In addition to their significance for T. brucei and telomere biology, the findings presented here have implications for the many diverse pathogens that organize their antigenic genes in subtelomeric regions.
Introduction
Trypanosoma brucei is an extracellular human pathogen with an unparalleled capacity to evade host humoral immunity. The causative agent of African sleeping sickness in humans and nagana in cattle, T. brucei is transmitted into the bloodstream of its host by a tsetse vector and can grow to densities as high as 109 cells per milliliter of blood. The parasitemia is cyclically diminished to nearly undetectable levels, to be followed by another wave of immense growth [1]. These rounds of parasitemia reflect the battle between the host immune system and the pathogen's elegant mechanisms of immune evasion.
At the forefront of this battle is the T. brucei cell surface, which is primarily composed of about 107 copies of a single, densely packed Variant Surface Glycoprotein (VSG) [2]. VSG is highly immunogenic, yet T. brucei escapes immune recognition by switching the monoallelic expression of one VSG-encoding gene to another [1], [3]. This process of surface antigen variation is made possible by a genomic repertoire of more than 1000 highly divergent VSG-encoding genes and pseudogenes [4], [5], [6]. Furthermore, existing VSGs can recombine to form novel mosaic VSGs, making the depth of the repertoire potentially limitless [7].
VSGs are encoded throughout the T. brucei genome, which consists of 11 megabase chromosomes, numerous intermediate chromosomes, and ∼100 minichromosomes [6], [8]. Although most VSGs are found in VSG arrays within megabase chromosomes or singly on minichromosomes, they can only be transcribed by bloodstream-form parasites from one of the ∼15 Bloodstream Expression Sites (BES) at a time. Proper expression of VSG on the cell surface is essential for survival [1]. Each BES is composed of a collection of Expression Site Associated Genes (ESAGs), a long repetitive element (70-bp repeats), and a terminal VSG encoding gene, which are transcribed from one upstream promoter and spliced into separate mRNAs before translation [4], [9]. All known BESs are located within 60 kb from the end of the chromosome positioning the expressed VSG within >2 kb of repetitive telomeric DNA [4]. The organization of surface antigen encoding genes in subtelomeric regions is a common theme among pathogens that employ antigenic variation to evade host defenses [10], [11].
There are three predominant mechanisms by which T. brucei VSG switching can occur: In Situ (IS) transcriptional VSG switching — the inactivation of one BES coupled with activation of transcription from a new BES [12], [13], [14], [15], Reciprocal Telomeric Exchange (TE) — a homologous recombination event between two chromosome ends resulting in the balanced transfer of a new VSG to the active BES and the previously active VSG to a silent BES [16], [17], and Duplicative Gene Conversion (GC) — a non-reciprocal transfer of a VSG containing DNA to the active BES that results in loss of the previously active VSG from the genome [18], [19], [20], [21]. GC is predicted to account for the majority of VSG switching under natural conditions because it is the only mechanism that permits the expression of non-telomerically-encoded VSG genes. How each of these mechanisms is orchestrated is largely unknown.
Although the mechanistic basis of each type of VSG switch is unknown, it had long been predicted that GC would be initiated by a DNA break. Recent studies have shown that the artificial induction of a DNA double-stranded break (DSB) proximal to the BES repetitive region upstream of the active VSG increases the frequency of VSG switching by as much as 250-fold, recapitulating the rate estimated in natural isolates [22], [23]. As predicted, VSG switching under these conditions occurred mainly by GC [22]. Furthermore it was shown that DNA breaks accumulate in the repetitive region of the active BES [22]. However, the natural source of DNA breaks that precipitate GC remains a mystery.
A proposed source of GC initiating DSBs is related to the proximity of the BES encoded VSGs to the telomere (usually within >2 Kb) [4], [24]. The actively transcribed BES frequently experiences large stochastic terminal deletions, which are hypothesized to result from the very high levels of transcription at the end of the chromosome [25], [26]. Thus it has been suggested that when the telomere of the chromosome harboring the active BES is short the DNA breaks precipitating telomeric deletions would occur upstream of the VSG, resulting in an antigenic switch [24]. This claim was further correlated to the fact that strains of T. brucei that have been recently isolated from nature, who switch at a rate of approximately 10−2–10−3, have shorter telomeres than laboratory-adapted strains, whose rate of switching can be are 100–10,000-times lower [27], [28], [29], [30]. In all, this suggested an inverse correlation between telomere length and the amount antigenic switching in T. brucei [24], yet there were no data in direct support of this hypothesis.
T. brucei strains with the protein component of telomerase deleted (TERT−/−) are unable to repair telomeric breaks (such as those that occur frequently at the actively transcribed BES) and undergo progressive shortening of all the telomeres in the genome by 3–6 bp/Population Doubling (PD) [31]. Previously it was shown that when the telomere of a TERT−/− isolate is short, the actively expressed VSG is lost over the course of several weeks and replaced by a new VSG gene, which suggested an increase in VSG switching [32]. However, because of the time span in which those experiments took place, that study could not differentiate between the equivalent possibilities of an increase in switching versus the death of short-telomere clones being replaced by a subpopulation of switchers arising at the normal in vitro frequency [32].
In this study we directly tested the proposed correlation between VSG switching and telomere length. Using updated techniques, we compared the frequency of VSG switching between strains with wild-type length or shortened telomeres at the active BES. Large populations of switched clones were analyzed to identify their mechanism of switching and determine if those with short telomeres switch by way of a preferred mechanism. The findings presented here provide experimental support for the hypothesis that telomere length directly affects the frequency of VSG switching, and answer long-standing questions about the relationship between VSG switching, telomere length, and gene conversion.
Results
Telomere length affects antigenic switching frequency in vitro
To address the effect of telomere length on T. brucei antigenic switching, we first isolated strains with various telomere lengths at their active BES (BES1 expressing VSG427-2 [221]). The telomere of BES1 in a population wild-type isolates (WT) can range from ∼10–15 kb (FIG. 1. A). TERT−/− clones with short (∼1.5 kb), medium (∼5.0 kb), and long (>10 kb) BES1 telomeres were isolated and characterized by Southern blot (FIG. 1. B). The active BES telomere in a TERT−/− strain is not only prone to progressive shortening (3–6 bp/PD) but also massive truncations [31]. Thus, medium - and long-telomere clones can only be handled for a minimal number of passages before they shorten (as evidenced by the smear under the primary band in FIG. 1. B – “Long”). In contrast, critically short telomeres are stabilized by an unknown, telomerase-independent, mechanism that appears to be unique to T. brucei [33]. Thus resulting in the short-telomere clone used here (FIG. 1. B – “Short”), which can be stably maintained at a length of ∼1.5 kb for numerous passages.
Fig. 1. Correlating telomere length and VSG switching. 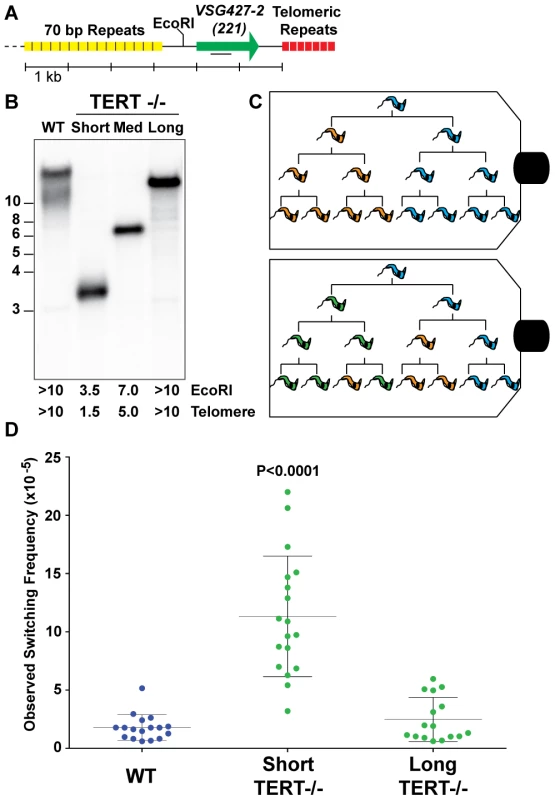
(A) Map displays proximity of the active VSG to the telomere, the position of the EcoRI site, and the position of DNA probe used in Southern blotting. (B) Lengths of wild-type (WT) and TERT−/− mutant telomeres shown by Southern blot analysis probed with VSG427-2 (221). (C) Illustration of two possible outcomes of fluctuation analysis resulting from a single trypanosome cell. (D) Observed Switching Frequency (OSF) of WT (n = 17), TERT−/− Short (n = 18), and TERT−/− Long telomere (n = 16) strains are shown. Each dot represents the OSF value arising from a single cell clone following fluctuation analysis for an equivalent number of population doublings. The P value applies to the comparison of TERT−/− short-telomere to either WT or TERT−/− long-telomere data sets. Populations of trypanosomes, as with any organism, are heterogeneous and this can affect both the expressed VSG and telomere length. Furthermore, accurate determination of the frequency of VSG switching requires that the populations being compared undergo a comparable number of population doublings (PD) during the experiment. Therefore, WT, TERT−/− short - and long-telomere clones with similar growth rates (FIG. S1) were grown from single-cells to ∼5×107 in vitro, thereby performing a modified Luria-Delbrück fluctuation analysis [34] (FIG. 1C). The VSG switching frequency of the resulting populations of trypanosomes was determined using the previously published magnetic-activated cell sorting (MACS) depletion of the initiating VSG followed by flow cytometry quantification [22].
TERT−/− short clone populations switched their expressed VSG at a significantly (P<0.0001) higher frequency (11.3×10−5±4.6) than both WT (1.8×10−5±1.1) and TERT−/− long-telomere clones (2.5×10−5±1.9), which were not significantly different from each other (P = 0.3136) (FIG. 1D). The TERT−/− long-telomere clones serve as a proxy for TERT complementation in this study because a previous study showed that ectopic TERT expression results in rapid elongation of the active site telomere (∼160 bp/PD) [33], which prevents the analysis of a short-telomere TERT complemented clone by this method. The switching frequencies of the TERT−/− short-telomere clones covered a broad range of values (3.2–22×10−5), which correspond to a 6 - to 36-fold increase in switching compared to WT. These data might be explained by a stochastic increase in subtelomeric DNA breakage that occurs when the telomere of the active site is short. Alternatively, in the absence of TERT complementation data, the increase in switching could arise from a combinatorial effect of having a short telomere in the context of a telomerase mutant. In either case these data support the notion that subtelomeric breaks promote antigenic switching.
The output of the VSG switching assay represents the proportion of the input population that is no longer expressing the starting VSG type, but this value does not account for replication of progeny, the number of VSG switching events intervening before the final measurement, or potential genotypic diversity of phenotypically identical cells in the resulting population (FIG. 1C). We therefore adopted the nomenclature Observed Switching Frequency (henceforth referred to as switching frequency or OSF) to indicate the limitations of this measured value (FIG. 1D).
Expressed VSG diversity following TERT−/− short-telomere switching
Hypothetically, populations that switch more often will contain a greater diversity of expressed VSGs. Thus we predicted that populations with higher OSF values would contain a larger diversity of VSG transcripts. To investigate this prediction, the MACS eluates from 12 of the 18 TERT−/− short-telomere switched populations (FIG. 1D) were grown to a sufficient extent for RNA extraction to identify the expressed VSGs, which were then compared with the determined OSF for that population (FIG. 2A, y-axis values correspond directly with FIG. 1D TERT−/− short data). This type of analysis is only possible due to the extensive nucleotide sequence diversity of VSG-encoding genes, which allows each VSG to be accurately distinguished from another [35]. Although there was a subtle trend of populations with OSF>10 to express a greater diversity of VSGs (∼3–6) than those with an OSF<10 (1–2 VSGs) (FIG. 2B), we did not observe a linear relationship between the switching frequency of these populations and the depth of their VSG diversity (FIG. 2). This is highlighted by the fact that the highest OSF (22×10−5) of this set contained only one expressed VSG (VSG427-3 [224]) (FIG. 2A & 2B).
Fig. 2. VSGs expressed in TERT−/− short-telomere switched populations. 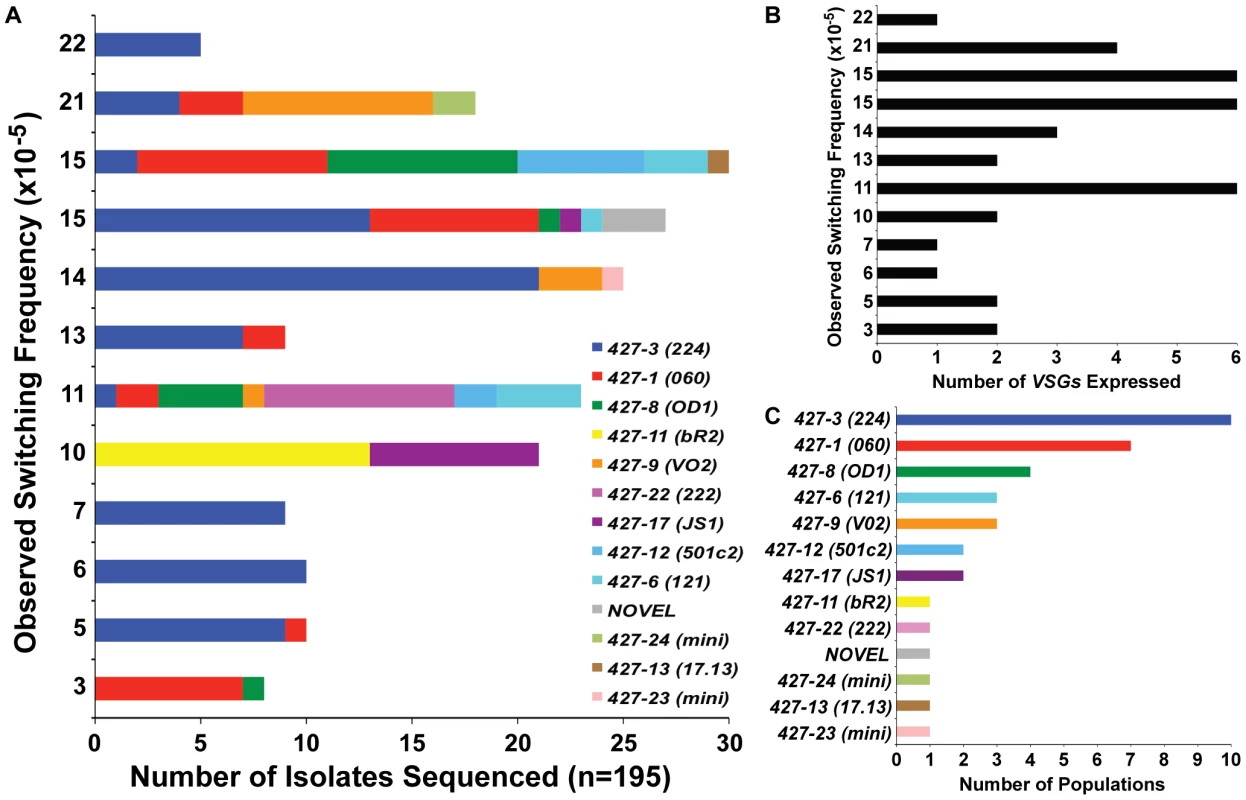
(A) Depiction of the diversity of VSGs present in specific populations of TERT−/− short-telomere switchers. X-axis shows the number of times each VSG was identified by bacterial colony sequencing versus the OSF value (same as those shown in FIG. 1D) on the y-axis. (B) Correlation of the total number of VSGs expressed in each population (x-axis) versus the OSF of that population (y-axis). (C) The number of populations (out of 12 total) (x-axis) in which each expressed VSG (y-axis) was identified. It is worth noting that the diversity of VSGs identified in TERT−/− short-telomere VSG switching assays was similar to those seen in other studies [1]. VSG427-3 (224) was the most commonly observed VSG, which was present in 10 of the 12 populations analyzed, and the sole VSG expressed in 3 of those populations (FIG. 2A & 2C). The second most commonly expressed was VSG427-1 (060) in 7 populations, followed by VSG427-8 (OD1) in 4 populations (FIG. 2C). These data support previous in vivo and in vitro studies suggesting that certain VSGs are favored donors and that VSG switching follows a semi-predictable order [36]. In addition to the predictably expressed VSGs, we isolated two VSGs that were recently demonstrated to reside on minichromosomes (427-23 & 427-24) [22] and a VSG that had not previously been annotated (FIG. 3, “NOVEL”).
Fig. 3. Genetic diversity of VSG427-3 (224) switchers. 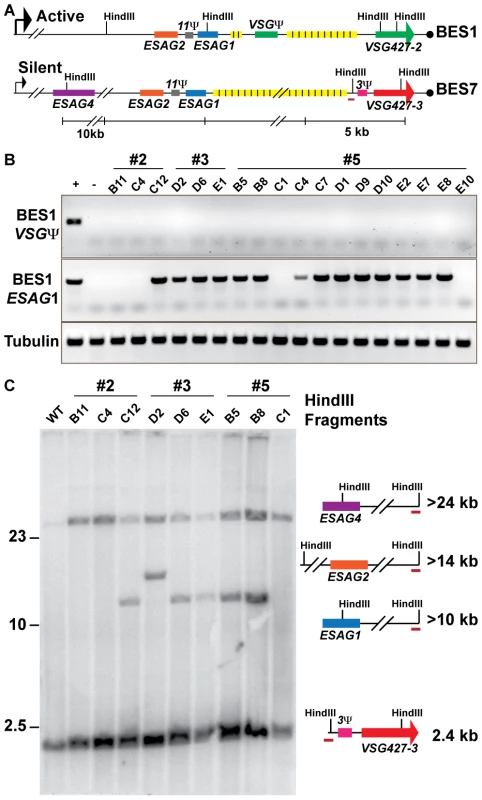
(A) Comparative maps of BES1 (active) and BES7 (silent; source of VSG427-3) with critical experimental features illustrated: HindIII sites, genes (ESAG1 & VSG Ψ), and BES7-unique probe (red line). (B) PCR products from 18 VSG427-3 switcher originating from 3 VSG427-2 starting clones (#2, #3, & #5) are shown for BES1 VSG Ψ, BES1 ESAG1, and Tubulin control in comparison to positive (WT) and negative (BES1 deletion strain courtesy of Kim & Cross). The control strain demonstrates the presence or absence of features in the genome. (C) FIGE Southern blot analysis of VSG427-3 switchers from HindIII digested gDNA probed with BES7-unique probe (BES7 red underline). Illustrations and predicted sizes of restriction fragments shown to the right of the blot image. Phenotypic diversity under-represents genetic diversity of switched isolates
The lack of a clear linear relationship between the switching frequency and the depth VSG diversity (FIG. 2) could arise from two alternative biological situations: extensive propagation of an early switch event (FIG. 1C, top) or multiple switch events that result in expression of the same VSG (FIG. 1C, bottom). As noted, the population of TERT−/− short telomere with the highest OSF (22×10−5) contained only VSG427-3 (224) (FIG. 2A). Did this population arise from one or multiple VSG switch events? To address this question, we isolated TERT−/− short-telomere trypanosomes that had switched from VSG427-2 (expressed from BES1) to VSG427-3 (originating from BES7) from three separate fluctuation analysis experiments. The genotypic differences among isolates from the same starting clone were then compared by PCR and Southern blot analysis (FIG. 3).
Eighteen switchers expressing VSG427-3 were isolated from three populations (3 clones from two populations [#2 & #3] and 12 from the third [#5]). All 18 clones had switched to VSG427-3 by GC (expressed from BES1), as evidenced by the loss of the VSG pseudogene from BES1 (FIG. 3A; BES1 map VSGΨ green box). Using PCR primers unique to ESAG1 [4] in BES1, we determined that VSG427-3 switchers from populations #2 and #5 contained a mixture of clones that had either lost or retained BES1 ESAG1 during GC (2 of 3 in population 2 and 2 of 12 in population 5 had lost ESAG1). Loss of BES1 ESAG1 indicates that resolution of GC occurred upstream of ESAG1. Therefore, clones expressing VSG427-3 in these populations arose from at least two distinct VSG switching events.
To further analyze the genotypes of the VSG427-3 switchers, we used a unique region in BES7 to probe a HindIII-digested large-fragment separation gel and Southern blot (FIG. 3A). There are two HindIII sites in BES1 between the repeat region and ESAG4 that distinguish it from BES7 (FIG. 3A). Thus, GC resolution downstream of ESAG1 results in the formation of a >10 kb fragment, upstream of ESAG1 but downstram of ESAG2 results in a >14 kb fragment, and upstream of ESAG2 but downstream of ESAG4 results in a fragment >24 kb (FIG. 3C). Because BES7 is intact in the genome regardless of switching, all strains produce the >24 kb and ∼2.4 kb fragments. Exact prediction of product sizes for fragments resolved beyond the BES7 repeat region (FIG. 3A, yellow boxes) is not possible due to missing sequence data [4].
Southern blot data largely agreed with PCR data (FIG. 3B), such that the VSG427-3 switchers that retained BES1 ESAG1 by PCR produced the restriction fragment associated with resection downstream of ESAG1 (>10 kb fragment present for #2 = 2/3, #3 = 2/3, & #5 = 3/12 [only partial data shown]). However, one isolate from clone 3 retained ESAG1 but produced a fragment that was predicted to result in the loss of ESAG1 (#3 D2, >14 kb). This combination of data could arise from a recombination event of unpredicted complexity. These results further demonstrate that each population of phenotypically identical VSG427-3 switchers analyzed are composed of at least 2 genotypes, and therefore must arise from multiple genetic events. Fine mapping or sequencing the point of resection for these strains could show further switching diversity, but is not possible due to the >90% sequence similarity between BES1 and BES7 [4]. Therefore, the OSF values displayed in figure 1 under-represent genetic switching frequency by not accounting for the number of VSGs expressed in the population (FIG. 2) and the genetic diversity of the switched population (FIG. 3).
TERT−/− short-telomere clones preferentially switch by duplicative gene conversion
To determine if telomere length affects the mechanism of switching as well as frequency, populations arising from single cells clones of wild-type and TERT−/− short-telomere strains were MACs sorted, their OSF determined (FIG. 4B – using the same methodology used to produced the data in FIG. 1 with new starting populations), and plated to limiting dilution. The secondary clones were then analyzed for the expression of VSG427-2 (221) by high-throughput screening (HTS) flow cytometry, and all cultures not expressing VSG427-2 were deemed “switched clones.” From 12 populations of WT, we screened 1617 secondary clones, identifying 189 WT switched clones (12% of the post-MACS screened population), whereas screening of 255 secondary clones from 6 populations of TERT−/− short-telomere clones resulted in a similar number (188) of switched clones to be further analyzed (74% of the post-MACS screened population). The mechanism of switching for all secondary clones was then determined, based on three criteria: (1) resistance or sensitivity to a BES1 promoter-proximal antibiotic marker (WT marked with hygromycin [HYG] & TERT−/− marked with blasticidin [BSD]), (2) the presence or absence of VSG427-2 (221) in the genome, and (3) the presence or absence of the promoter-proximal resistance marker in the genome. Thus the switch type can be counted in the following way: IS events correspond to MarkerS, 427-2(221)+, Marker+; TE events are MarkerR, 427-2(221)+, Marker+; GC results in MarkerR, 427-2(221)−, Marker+; ES GC (Expression Site Gene Conversion), a subtype of GC in which the entire active BES is replaced by the donor BES, are MarkerS, 427-2(221)−, Marker−; and UD, for Undetermined, when switched clones did not fit the other criteria (FIG. 4A & TABLE S3). These data were also used to establish a minimal number of independent switch events. By creating a matrix based on the 12 known combinations of phenotype and genotype analyzed (VSG427-3 [224] expression as determined flow cytometry [data not shown]), switch type [IS, TE, GC, ES GC or UD], & BES1 VSG Ψ+/−) against the source populations (12 for WT & 6 for Short), we determined that the 189 WT switched clones arose from at least 47 independent switchers and the 188 Short switched clones from at least 25 (TABLE S3). The higher number for WT is an artifact of the increased complexity of its matrix (i.e. more originating populations). Determining the mechanism of switching in TERT−/− long-telomere clones was not possible because of their propensity to break at the active BES, which results in the rapid formation of a heterogenic population of telomere lengths.
Fig. 4. Mechanisms of VSG switching in WT and short-telomere isolates. 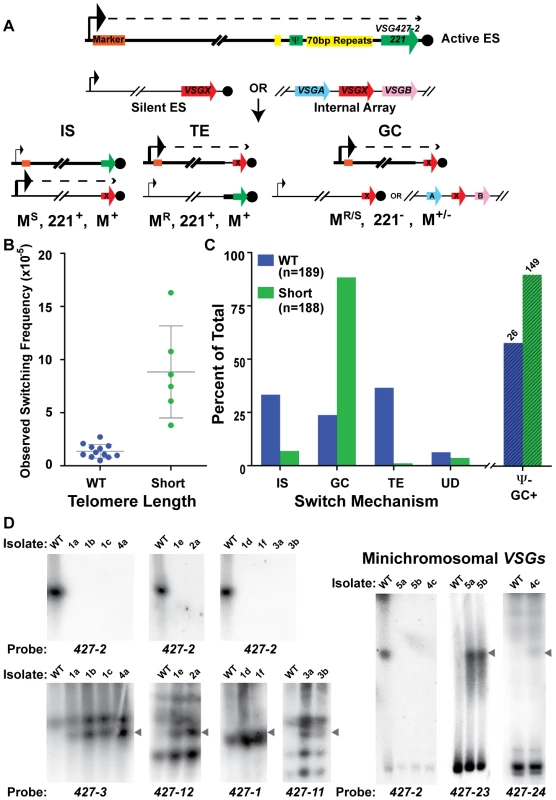
(A) Cartoon depicting the critical details of the three predominant mechanisms of VSG switching- IS (In situ), TE (telomere exchange), and GC (duplicative gene conversion). Large arrowhead indicates active promoter; small arrowhead depicts the silent promoter; dashed line indicates transcription; orange box shows the position of the antibiotic selection marker. VSG427-2 nomenclature is used interchangeably with its colloquial name VSG221 (B) OSF of the populations of WT (blue, n = 12 populations) and TERT−/− short-telomere (green, n = 6 populations) switchers. (C) Percent of WT (blue, n = 189) and TERT−/− short-telomere (green, n = 188) VSG switched isolates versus switching mechanism: IS (in situ), GC (all gene conversion = GC+ES−GC), TE (telomere exchange), or UD (undetermined). Percent loss of BES1 VSG pseudogene from GC (“Ψ− GC+”) switched isolates (WT n = 26 & Short n = 149) is shown beyond the broken line (WT, lined blue bar & short-telomere, lined green bar). (D) RAGE separated chromosomes blotted and probed with VSG specific probes for a subset of TERT−/− short-telomere switched clones. WT VSG switched clones produced similar average levels of IS (33%), GC (24%), and TE (37%). This was in contrast to the TERT−/− short-telomere VSG switchers, which showed a clear preference for GC-based switching (88%), only a small amount of IS (7%) and negligible TE (1%) (FIG. 4C [“GC” shown is the sum of both GC and ES GC] & TABLE S1, S2, S3). Switching by GC for a subset of TERT−/− short-telomere clones was further confirmed by pulsed-field gel separation of chromosomes followed by Southern blot analysis using specific VSG probes, which showed both the loss of the initiating VSG (427-2) and the duplication of the newly expressed VSG to the active BES (FIG. 4D). These data, in conjunction with published data that induction of a DNA break in the active site initiates a GC based switch [22], suggest that a short telomere at the active site can increase switch initiating subtelomeric DNA breaks. Further mathematical analysis of switched clone data suggested that the heightened level of GC could account for the overall increase in switching frequency observed when the telomere is short (mathematical validation shown in Experimental Procedures & TABLE S3). The same analytical process showed that the frequency of IS switching (FIS = average OSF×% IS, for both WT and TERT−/− short-telomere clones) was not significantly different between WT and TERT−/− short-telomere clones (TABLE S3). To further analyze the nature of the GC events in WT and TERT−/− short, all isolates identified as GC were assayed for the presence of BES1 VSG pseudogene (PSD or Ψ) in the genome, a parameter that indicates the location of GC resolution. BES1 VSG PSD was lost from TERT−/− short-telomere switchers (90%) ∼30% more often than WT (58%) (FIG. 4C, lined bars “Ψ − GC+”). This indicates that the resolution of GC events in TERT−/− switchers often occurs farther upstream than in WT VSG switchers and suggests that the initiating lesion itself may occur farther upstream when the telomere is short.
The body of isolated switcher data was further analyzed to determine if individual populations, OSF values, or isolation dates, affected the propensity to switch by a given mechanism (TABLE S3, FIG. S2 & S3). Each initiating clone results in a population of secondary switched clones with a somewhat variable percentage of each switching mechanism. Generally, population variations did not affect the results, but there were two WT populations (#2-5 & #2-6) with a large number of switchers (34 & 25, respectively) that had a high percentage of TE (76% & 80%, respectively) (FIG. S2 & TABLE S3), which gave the average WT TE value of 37%. If reanalyzed, the median values for WT were IS = 37%, GC = 20%, and TE = 19%, thus reducing the overall significance of TE. Analysis of the percent switch mechanism in comparison with the switching frequency for each population showed a very subtle trend for populations with higher OSF values to contain a higher level of diversity in the mechanisms represented, which was similarly true for both WT and short-telomere isolates (TABLE S3). Due to different growth rates following limiting dilution, VSG switch isolates were selected 7, 9, or 11 days after initial plating. Analysis of the VSG switch mechanism by isolation date for WT and TERT−/− short-telomere clones displayed a trend of increased IS and decreased GC with time (this pattern was more apparent in WT), while TE did not (FIG. S3). This suggests that isolates that switch by IS initially grow more slowly than those that switch by GC; the basis of this difference is unknown. None of the alternative analysis of the data in this section detract from the central result, namely that TERT−/− short-telomere clones switch preferentially by GC.
Discussion
Telomeres are structures of DNA and protein that protect the ends of chromosomes from DNA loss and damage. Yet, somewhat counterintuitively, T. brucei is only one of the many pathogens whose critical antigenic diversity genes are organized in potentially fragile subtelomeric regions [10], [37]. In 2007 Dreesen et al. proposed a model in which telomere length (at the active BES) had an inverse correlation with the frequency of VSG switching in T. brucei. The model also predicted that switching at short telomeres would occur by GC, resulting from an increase in BES internal DSBs when the telomere is short [24]. The 2007 proposal was based on a correlation between two main bodies of data: 1) the telomeres of laboratory-adapted strains are longer than those of strains recently isolated from nature [30] & 2) populations of TERT−/− short-telomere strains progressively loose the initially expressed VSG, which suggested, but did not demonstrate an increase in the frequency of VSG switching [33]. Although this model gained popular support, its predictions were unsubstantiated.
Here we have used updated techniques to rigorously test the proposed correlation between telomere length and VSG switching. Our data demonstrate, for the first time, that antigenic switching in T. brucei increases in a TERT−/− mutant when the telomere of the active BES is short (in direct contrast to TERT−/− strains with long a telomere whose switching is like WT) (FIG. 1). Furthermore, we show that the increase in switching can be accounted for by a significant increase in GC (in comparison with other measured switching mechanisms), which is likely due to an increase in DNA breaks in the region upstream of the VSG in the active/short-telomere BES (evidenced by an increased loss of VSG pseudogene [Ψ]) (FIG. 4).
There are two equally valid mechanistic interpretations of these data. The first is that telomere shortening per se leads to an increase in the frequency of DNA breakage in locations that surround the VSG, thus precipitating a switching event, as hypothesized originally by Cross and colleagues [24]. Alternatively, the phenotype we observe might be due to a telomere-capping defect, which would be the result of the combined effect of shortened telomeres in the context of a telomerase-null mutant. Indeed, such a capping defect has been shown to increase gene conversion in the budding yeast Kluyveromyces lactis [38]. To distinguish between these two mechanistic possibilities, we have made concerted efforts to complement telomerase expression in TERT−/− short-telomere T. brucei so that their switching frequency could be compared to un-complemented TERT−/− short-telomere strains. However, we have been unable to set up a strictly regulated inducible system to reconstitute TERT expression while retaining a short telomere at the active expression site (even at exceedingly low expression levels). This is due to the inherent leakiness of inducible systems available for T. brucei, together with the impressive efficiency of catalytically active telomerase to rapidly elongate a short telomere at the active expression site [30], [31]. Also, the impact of telomerase mutations, if any, on t-loop formation and telomere capping in T. brucei is not known at this time. Thus, at the moment, we cannot distinguish between the possibility that the phenotype we observe in the TERT−/− short-telomere strains is due to a direct effect of telomere shortening, as originally proposed, or a capping defect (combined with telomere shortening) due to lack of telomerase function, along the lines of K. lactis [38]. Nevertheless, the general appropriation of conserved mechanisms of chromosome end protection (breakage, lengthening and capping) for the purposes of increased antigenic variation is an intriguing possibility as an aspect of antigenic variation in T. brucei.
T. brucei strains that have been recently isolated from nature are distinct from their laboratory propagated cousins in that they switch more frequently (approximately 10−2–10−3) [27], have shorter telomeres [30], and preferentially switch by GC [39]. Thus, it would appear that by producing a T. brucei strain with an artificially shortened telomere we have, at least partially, recapitulated these characteristics of a natural isolate. The 10 - to 100-fold increase OSF measured in the TERT−/− short-telomere strain does not cover the approximately 100 - to 10,000-fold difference in the rate of switching published between recent isolates (approximately 10−2–10−3) and laboratory-adapted strains in vitro (approximately 10−5–10−6) [28], [29]. Comparative analysis of switch rate approximations has been a central challenge in the field due to significant variability among the methodologies used and the very small sets of VSG switchers (often less than 10) from which these rate approximations were often derived [27], [28], [29]. The OSF value is simply a measurement of the proportion of cells in the population that are no longer coated in the parental VSG, which does not account for the phenotypic diversity of VSGs in the population (which was included in previous approximate rate derivations) (considered in FIG. 2) or the genotypic diversity (for which no method has ever accounted) (considered in FIG. 3). The findings presented here suggest that the shorter telomeres of natural isolates could contribute to their comparatively high level of VSG switching.
The question that persists is, how naturally occurring populations keep their telomeres short? In addition, T. brucei telomeres grow by 6–8 bp/PD during growth under laboratory conditions (a process that has only been observed in this organism) [25], [30]. What is distinct between the natural T. brucei lifecycle and their laboratory propagation that could account for the observed differences in telomere lengths? Although telomerase regulation in T. brucei is not understood, in other organisms telomerase is activity is regulated by the cell cycle such that telomeres only lengthen in the transition from S phase to G2 [40], [41], [42]. If this were also the case for trypanosomes, the organisms increased in vivo growth rate (which is more than 2 times higher than in vitro) could result in a reduced duration of telomerase activity and thus progressively shorter telomeres. An alternative possibility is that a regulated component of telomere structure or stability, or possibly telomerase itself, is affected when trypanosomes are not permitted to undergo their natural life cycle, which includes passage through the tsetse. In support of this hypothesis, laboratory adapted T. brucei can recover its in vivo switch rate following passage through the tsetse [29]. Although little is known about the regulation of telomerase and other telomere-associated proteins in trypanosomes, perhaps this is a missing connection between the switching behavior of natural isolates and extensively adapted T. brucei strains.
Materials and Methods
Trypanosoma brucei
Cell lines were generated from Lister427 bloodstream-form trypanosomes derived from the ‘single marker’ line [43], with a hygromycin resistance marker at the BES1 promoter [22] (“wild-type” [WT]), or homozygous telomerase (Gene ID: 3664223 & protein accession: XP_829083) deletion mutant with blasticidin resistance marker at the BES1 promoter (TERT−/−) [31], [32]. TERT−/− short and long active-site telomere BES clones were isolated from single cell cultures of TERT−/− and telomere lengths were determined by Southern Blot analysis (below) [31]. Trypanosomes were cultured in vitro in HMI-9 medium at 37°C [44].
Southern blot analysis
DNA restriction fragments were separated by standard agarose gel electrophoresis (1–15 kb), Field Inversion Gel Electrophoresis (FIGE) [45] (1–25 kb, BIORAD “Program 1”), Rotating Agarose Gel Electrophoresis [46] (“Classical Program” for separation of megabase, intermediate, and minichromosomes), using published methods. Southern blots were produced using established methods of capillary blotting by neutral transfer (GE Scientific). DNA probes were made by PCR amplification using previously published primer sequences [4], [22], 32P radiolabeled using Prime-It II Random Labeling Kit (Stratagene), and purified over G-50 microcolumns (GE Healthcare). Blots were probe-hybridized, washed, and visualized by phosphorimaging as described (GE Healthcare).
OSF determination by MACS and flow cytometry-based fluctuation analysis
Strains expressing VSG427-2 were single cell cloned by limiting dilution and expanded to a total of ∼5×107 cells prior to MACS isolation and flow cytometry-based quantification of VSG switching frequency as previously described [22]. The VSG switching frequency, here termed the “Observed Switching Frequency (OSF)”, is a direct measure of the proportion of living (measured by propidium iodide staining) trypanosomes in a population that no longer express VSG427-2 on their surface, compared to the total input population, both of which are normalized to a control sample (CountBright Beads purchased from and used according to Invitrogen instructions).
Identification of expressed VSGs
Following MACS depletion of VSG472-2 expressing cells, half of the effluent was removed prior to OSF determination by flow cytomertry and grown in 50 mL HMI-9 with pen-strep until confluent. RNA was extracted, cDNA synthesized, VSG RT-PCR amplified and subcloned (pGEM-T Easy, Promega) using established methods (tryps.rockefeller.edu/trypsru2_protocols_index.html, “VSG cloning for mRNA”), prior to sequencing and expressed VSG determination by NCBI BLAST.
Isolation and analysis of VSG427-3 (224) switchers
Following MACS depletion of VSG427-2 expressing cells (from single cell cultures grown for the specific purpose of VSG427-3 switcher isolation and not OSF determination), the effluent was labeled with anti-224 antibody and bound to a second MACS column. Following standard MACS binding and wash steps the bound trypanosomes (anticipated VSG427-3+ switchers) were plunged from the column, cloned and screened for VSG427-3 expression by flow cytometry. VSG427-3 expressing clones were grown for genomic DNA isolation [46], which was analyzed by PCR using BES1 VSG pseudogene primers (pseudo5-F: 5′ - GCGCCGAATTTAATGCAATATGCAACG & pseudo5-R: 5′ - GCAGGCCGTCTTTTGAGTTGTAGTAAG) & BES1 ESAG1 primers (ESAG1 Fw1 : 5′-GAGCAAACTGATAGGTTGGAAAAG & ESAG1 Rv1 5′-GCACTGGCGGCCACTCCATTGTC) and HindIII FIGE Southern blot analysis using a BES7 specific probe (FIG. 4A – red bar) produced by PCR using unique primers (BES7 UR1 Fw: 5′-GCAACTAACTACTGTAATTCCC & BES7 UR Rv: 5′-GCTACTAATGTGTTTCAATATGCG).
Determination of VSG switch mechanism
Following expansion of VSG427-2 (221) expressing WT and TERT−/− short-telomere cells from single cells to ∼5×107 total cells and MACS depletion of VSG472-2 expressing cells as described above [22], the effluent was split into two samples. Half of the resulting cells were used to determine the OSF and the other half was cloned by limiting dilution in 96-well tissue-culture plates. Clone growth was observed for two weeks following initial plating. At 7, 9, and 11 (WT only) days after plating, confluent cultures were split into new 96-well plates and allowed to become confluent. Final identification of VSG427-2− switchers was performed by high-throughput-screening (HTS) flow cytometry using anti-VSG221 antibody on an LSRII with HTS adaptor for 96 well plate analysis (BD Biosciences). VSG427-2 negative clones were then analyzed for BES1 promoter activity by antibiotic selection of the BES1 promoter proximal marker (WT: hygromycin & TERT−/−: blasticidin) and the Southern dot blots for the presence of VSG427-2 (221), the promoter proximal antibiotic resistance gene and BES1 VSG pseudogene in the genome (using radiolabeled probes to VSG221, hygromycin, blasticidin, pseudogene). Southern dot blot was adapted for trypanosomes from manufacturers protocols (GE Healthcare) by adding ∼1×106 cells/well to the membrane, cell lysis with denaturation buffer, neutralization, and fixation with UV prior to radiolabeled probe hybridization, washing, and visualization as described above for Southern blot analysis. Primers for PCR production of Southern dot blot probes: 221 Prb Fw (5′ - GTAACAGCTACTGCAACAGCGAGC)& 221 Prb Rv(5′ - GCTTCTTCAACAAGCTTGGTAACG), HYGRO Prb Fw (5′-GCTCTCGATGAGCTGATGCTTTGG) & HYGRO Prb Rv (5′-GATAGAGTTGGTCAAGACCAATGC), BES1 PSEUDO Fw (5′ - CATTAAATTCAAGCGTCTAGACCGCAGC) & BES1 PSUEDO Rv (5′ - GCGCGTTGTTCCGTATCTGCTGAGC)
Mathematical analysis of GC contribution to OSF
If ΣFShort≈ΣFWT+ΔFGC, then GC accounts for the increase in short-telomere OSF. Where FMech = Average OSF×% Mechanism and ΔFGC = FGC,Short−FGC,WT. First the F for each mechanism for both WT and Short were determined, the sum of which is equal to the average OSF for each strain (ΣFWT = 1.38 & ΣFShort = 8.84). Then the ΔFGC was determined and used to show that ΣFWT+ΔFGC = 8.86 which is fundamentally identically to the ΣFShort (8.84), thus making ΣFShort≈ΣFWT+ΔFGC a valid statement (TABLE S3).
Supporting Information
Zdroje
1. BarryJD, McCullochR (2001) Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol 49 : 1–70.
2. CrossGA (1975) Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 71 : 393–417.
3. PaysE (2005) Regulation of antigen gene expression in Trypanosoma brucei. Trends Parasitol 21 : 517–520.
4. Hertz-FowlerC, FigueiredoLM, QuailMA, BeckerM, JacksonA, et al. (2008) Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One 3: e3527.
5. Van der PloegLH, ValerioD, De LangeT, BernardsA, BorstP, et al. (1982) An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res 10 : 5905–5923.
6. BerrimanM, GhedinE, Hertz-FowlerC, BlandinG, RenauldH, et al. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309 : 416–422.
7. MarcelloL, BarryJD (2007) Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Res 17 : 1344–1352.
8. WicksteadB, ErsfeldK, GullK (2004) The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res 14 : 1014–1024.
9. BerrimanM, HallN, SheaderK, BringaudF, TiwariB, et al. (2002) The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol Biochem Parasitol 122 : 131–140.
10. DeitschKW, LukehartSA, StringerJR (2009) Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol 7 : 493–503.
11. HornD, BarryJD (2005) The central roles of telomeres and subtelomeres in antigenic variation in African trypanosomes. Chromosome Res 13 : 525–533.
12. JohnsonPJ, BorstP (1986) Mapping of VSG genes on large expression-site chromosomes of Trypanosoma brucei separated by pulsed-field gradient electrophoresis. Gene 43 : 213–220.
13. ZomerdijkJC, OuelletteM, ten AsbroekAL, KieftR, BommerAM, et al. (1990) The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J 9 : 2791–2801.
14. BorstP, UlbertS (2001) Control of VSG gene expression sites. Mol Biochem Parasitol 114 : 17–27.
15. HornD, CrossGA (1997) Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol Biochem Parasitol 84 : 189–201.
16. PaysE, GuyauxM, AertsD, Van MeirvenneN, SteinertM (1985) Telomeric reciprocal recombination as a possible mechanism for antigenic variation in trypanosomes. Nature 316 : 562–564.
17. RudenkoG, McCullochR, Dirks-MulderA, BorstP (1996) Telomere exchange can be an important mechanism of variant surface glycoprotein gene switching in Trypanosoma brucei. Mol Biochem Parasitol 80 : 65–75.
18. De LangeT, KooterJM, MichelsPA, BorstP (1983) Telomere conversion in trypanosomes. Nucleic Acids Res 11 : 8149–8165.
19. HoeijmakersJH, FraschAC, BernardsA, BorstP, CrossGA (1980) Novel expression-linked copies of the genes for variant surface antigens in trypanosomes. Nature 284 : 78–80.
20. MylerP, NelsonRG, AgabianN, StuartK (1984) Two mechanisms of expression of a predominant variant antigen gene of Trypanosoma brucei. Nature 309 : 282–284.
21. PaysE, DelauwMF, Van AsselS, LaurentM, VervoortT, et al. (1983) Modifications of a Trypanosoma b. brucei antigen gene repertoire by different DNA recombinational mechanisms. Cell 35 : 721–731.
22. BoothroydCE, DreesenO, LeonovaT, LyKI, FigueiredoLM, et al. (2009) A yeast-endonuclease-generated DNA break induces antigenic switching in Trypanosoma brucei. Nature 459 : 278–281.
23. GloverL, McCullochR, HornD (2008) Sequence homology and microhomology dominate chromosomal double-strand break repair in African trypanosomes. Nucleic Acids Res 36 : 2608–2618.
24. DreesenO, LiB, CrossGA (2007) Telomere structure and function in trypanosomes: a proposal. Nat Rev Microbiol 5 : 70–75.
25. BernardsA, MichelsPA, LinckeCR, BorstP (1983) Growth of chromosome ends in multiplying trypanosomes. Nature 303 : 592–597.
26. PaysE, LaurentM, DelinteK, Van MeirvenneN, SteinertM (1983) Differential size variations between transcriptionally active and inactive telomeres of Trypanosoma brucei. Nucleic Acids Res 11 : 8137–8147.
27. LamontGS, TuckerRS, CrossGA (1986) Analysis of antigen switching rates in Trypanosoma brucei. Parasitology 92(Pt 2):355–367.
28. MylerPJ, AllenAL, AgabianN, StuartK (1985) Antigenic variation in clones of Trypanosoma brucei grown in immune-deficient mice. Infect Immun 47 : 684–690.
29. TurnerCM (1997) The rate of antigenic variation in fly-transmitted and syringe-passaged infections of Trypanosoma brucei. FEMS Microbiol Lett 153 : 227–231.
30. DreesenO, CrossGA (2008) Telomere length in Trypanosoma brucei. Experimental Parasitol 118 : 103–110.
31. DreesenO, LiB, CrossGA (2005) Telomere structure and shortening in telomerase-deficient Trypanosoma brucei. Nucleic Acids Res 33 : 4536–4543.
32. DreesenO, CrossGA (2006) Consequences of telomere shortening at an active VSG expression site in telomerase-deficient Trypanosoma brucei. Eukaryot cell 5 : 2114–2119.
33. DreesenO, CrossGA (2006) Telomerase-independent stabilization of short telomeres in Trypanosoma brucei. Mol Cell Biol 26 : 4911–4919.
34. LuriaSE, DelbruckM (1943) Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics 28 : 491–511.
35. FieldMC, BoothroydJC (1996) Sequence divergence in a family of variant surface glycoprotein genes from trypanosomes: coding region hypervariability and downstream recombinogenic repeats. J Mol Evol 42 : 500–511.
36. MorrisonLJ, MajiwaP, ReadAF, BarryJD (2005) Probabilistic order in antigenic variation of Trypanosoma brucei. Int J Parasitol 35 : 961–972.
37. DeitschKW, MoxonER, WellemsTE (1997) Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol Mol Biol Rev 61 : 281–293.
38. McEachernMJ, IyerS (2001) Short telomeres in yeast are highly recombinogenic. Mol Cell 7 : 695–704.
39. RobinsonNP, BurmanN, MelvilleSE, BarryJD (1999) Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol Cell Biol 19 : 5839–5846.
40. HugN, LingnerJ (2006) Telomere length homeostasis. Chromosoma 115 : 413–425.
41. LiS (2011) Cell-cycle-dependent telomere elongation by telomerase in budding yeast. Biosci Rep 31 : 169–177.
42. LiS, MakovetsS, MatsuguchiT, BlethrowJD, ShokatKM, et al. (2009) Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136 : 50–61.
43. WirtzE, LealS, OchattC, CrossGA (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99 : 89–101.
44. HirumiH, HirumiK (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol 75 : 985–989.
45. DawkinsHJ, FerrierDJ, SpencerTL (1987) Field inversion gel electrophoresis (FIGE) in vertical slabs as an improved method for large DNA separation. Nucleic Acids Res 15 : 3634–3635.
46. Munoz-JordanJL, CrossGA, de LangeT, GriffithJD (2001) t-loops at trypanosome telomeres. EMBO J 20 : 579–588.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Exon Level Transcriptomic Profiling of HIV-1-Infected CD4 T Cells Reveals Virus-Induced Genes and Host Environment Favorable for Viral Replication
- Five Mechanisms of Manipulation by Bacterial Effectors: A Ubiquitous Theme
- Nonhuman Primate Models for HIV Cure Research
- The Ebola Virus Glycoprotein Contributes to but Is Not Sufficient for Virulence
- Trichomonosis, a Common Curable STI, and Prostate Carcinogenesis—A Proposed Molecular Mechanism
- Host Defense and Tolerance: Unique Challenges in the Placenta
- CPAF: A Chlamydial Protease in Search of an Authentic Substrate
- Small Protease Sensitive Oligomers of PrP in Distinct Human Prions Determine Conversion Rate of PrP
- Invariant NKT Cells: Regulation and Function during Viral Infection
- Human Monoclonal Antibody HCV1 Effectively Prevents and Treats HCV Infection in Chimpanzees
- Chemokine Receptor Ccr1 Drives Neutrophil-Mediated Kidney Immunopathology and Mortality in Invasive Candidiasis
- Phagocyte Responses to Protozoan Infection and How Meets the Challenge
- Telomere Length Affects the Frequency and Mechanism of Antigenic Variation in
- A Biofilm-Induced Pathway for Matrix Glucan Delivery: Implications for Drug Resistance
- The HSV-1 Exonuclease, UL12, Stimulates Recombination by a Single Strand Annealing Mechanism
- Inhibition of Fatty Acid Synthase (Fas2) Induces Mitochondrial Cell Death in Serum
- Interferon-alpha Subtype 11 Activates NK Cells and Enables Control of Retroviral Infection
- Transposon-mediated Chromosomal Integration of Transgenes in the Parasitic Nematode and Establishment of Stable Transgenic Lines
- Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease
- Upregulation of Retinal Dehydrogenase 2 in Alternatively Activated Macrophages during Retinoid-dependent Type-2 Immunity to Helminth Infection in Mice
- Measles Immune Suppression: Lessons from the Macaque Model
- Fungi and the Rise of Mammals
- Bacterial Cell Surface Heterogeneity: A Pathogen's Disguise
- Cytoplasmic Entry Induces Fetal Wastage by Disrupting Maternal Foxp3 Regulatory T Cell-Sustained Fetal Tolerance
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Invariant NKT Cells: Regulation and Function during Viral Infection
- Host Defense and Tolerance: Unique Challenges in the Placenta
- Nonhuman Primate Models for HIV Cure Research
- Exon Level Transcriptomic Profiling of HIV-1-Infected CD4 T Cells Reveals Virus-Induced Genes and Host Environment Favorable for Viral Replication
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



