-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
HIV-1 Transmission during Early Infection in Men Who Have Sex with Men: A Phylodynamic Analysis
Background:
Conventional epidemiological surveillance of infectious diseases is focused on characterization of incident infections and estimation of the number of prevalent infections. Advances in methods for the analysis of the population-level genetic variation of viruses can potentially provide information about donors, not just recipients, of infection. Genetic sequences from many viruses are increasingly abundant, especially HIV, which is routinely sequenced for surveillance of drug resistance mutations. We conducted a phylodynamic analysis of HIV genetic sequence data and surveillance data from a US population of men who have sex with men (MSM) and estimated incidence and transmission rates by stage of infection.Methods and Findings:
We analyzed 662 HIV-1 subtype B sequences collected between October 14, 2004, and February 24, 2012, from MSM in the Detroit metropolitan area, Michigan. These sequences were cross-referenced with a database of 30,200 patients diagnosed with HIV infection in the state of Michigan, which includes clinical information that is informative about the recency of infection at the time of diagnosis. These data were analyzed using recently developed population genetic methods that have enabled the estimation of transmission rates from the population-level genetic diversity of the virus. We found that genetic data are highly informative about HIV donors in ways that standard surveillance data are not. Genetic data are especially informative about the stage of infection of donors at the point of transmission. We estimate that 44.7% (95% CI, 42.2%–46.4%) of transmissions occur during the first year of infection.Conclusions:
In this study, almost half of transmissions occurred within the first year of HIV infection in MSM. Our conclusions may be sensitive to un-modeled intra-host evolutionary dynamics, un-modeled sexual risk behavior, and uncertainty in the stage of infected hosts at the time of sampling. The intensity of transmission during early infection may have significance for public health interventions based on early treatment of newly diagnosed individuals.
Please see later in the article for the Editors' Summary
Published in the journal: . PLoS Med 10(12): e32767. doi:10.1371/journal.pmed.1001568
Category: Research Article
doi: https://doi.org/10.1371/journal.pmed.1001568Summary
Background:
Conventional epidemiological surveillance of infectious diseases is focused on characterization of incident infections and estimation of the number of prevalent infections. Advances in methods for the analysis of the population-level genetic variation of viruses can potentially provide information about donors, not just recipients, of infection. Genetic sequences from many viruses are increasingly abundant, especially HIV, which is routinely sequenced for surveillance of drug resistance mutations. We conducted a phylodynamic analysis of HIV genetic sequence data and surveillance data from a US population of men who have sex with men (MSM) and estimated incidence and transmission rates by stage of infection.Methods and Findings:
We analyzed 662 HIV-1 subtype B sequences collected between October 14, 2004, and February 24, 2012, from MSM in the Detroit metropolitan area, Michigan. These sequences were cross-referenced with a database of 30,200 patients diagnosed with HIV infection in the state of Michigan, which includes clinical information that is informative about the recency of infection at the time of diagnosis. These data were analyzed using recently developed population genetic methods that have enabled the estimation of transmission rates from the population-level genetic diversity of the virus. We found that genetic data are highly informative about HIV donors in ways that standard surveillance data are not. Genetic data are especially informative about the stage of infection of donors at the point of transmission. We estimate that 44.7% (95% CI, 42.2%–46.4%) of transmissions occur during the first year of infection.Conclusions:
In this study, almost half of transmissions occurred within the first year of HIV infection in MSM. Our conclusions may be sensitive to un-modeled intra-host evolutionary dynamics, un-modeled sexual risk behavior, and uncertainty in the stage of infected hosts at the time of sampling. The intensity of transmission during early infection may have significance for public health interventions based on early treatment of newly diagnosed individuals.
Please see later in the article for the Editors' SummaryIntroduction
Variation in the timing of transmissions over the course of an infection can have large consequences for the design of HIV prevention programs [1]. For example, interventions focused on increasing treatment rates (treatment as prevention [TasP]) are only able to block transmissions that occur after diagnosis, entry into care, and consistent adherence to antiretroviral treatment. Consequently, TasP will preempt fewer transmissions as the proportion of transmissions from early infections rises. Highly elevated transmission rates early in infection are both biologically and sociologically plausible. The transmissibility of HIV per sexual encounter depends on the viral load within infected hosts [2],[3], which peaks during early HIV infection (EHI) and also rises during stage 3 (AIDS) [4]. Likewise, a recently infected individual is likely to have been infected during a period of high-risk behavior; if the high-risk behavior extends through EHI, then the interaction of elevated viral load and risky behavior can potentially elevate transmission rates during EHI.
Several mathematical analyses have argued that transmission rates during early infection drive the HIV-1 epidemic [5],[6]. That early infection transmissions drove the early epidemic is clear. But what drives transmission later in the epidemic remains controversial. A recent survey of estimates of the proportion of transmissions from EHI based on mathematical models of HIV transmission found very wide ranges, including scenarios where nearly all or almost no transmissions were coming from EHI [2]. The surveyed mathematical studies [2] attempted to fit diverse models of HIV transmission to indirect data such as diagnoses over time. The variation in parameter estimates could be due to real differences between the study populations, but it could also be due to a fundamental limitation of the data used to fit these kinds of models. Mathematical models of HIV transmission are essential for understanding HIV dynamics, but model-based analysis of diagnosis data alone has not yielded consistent estimates of the timing of HIV transmission over the course of infection.
Direct measurement of the timing of HIV transmission is possible in large population-based cohort studies that follow recently infected individuals and their uninfected partners [7],[8]. Careful monitoring for seroconversion of the uninfected partner and administration of questionnaires about sexual practices can paint an empirically driven picture of the timing of transmission events over the course of an infection. However, longitudinal partner studies are expensive and lack generalizability because risk behavior is highly variable over time and between risk groups.
Traditional surveillance data that are used to estimate incidence and prevalence of infection have little value for estimating the intensity of transmission during EHI, because outside of the very early epidemic period, a given incidence curve can be consistent with either high or low levels of EHI transmission. Methods have been developed to back calculate the incidence over time [9],[10] using information about the stage of infection of patients at the time of diagnosis, and estimates can be further refined by incorporating diverse data sources such as behavioral surveillance and seroprevalence surveys [11]. These methods are essential in evaluating the efficacy of prevention programs; however, the timing of transmission events cannot be identified from incidence data alone [12].
HIV genetic sequence data can potentially augment traditional surveillance data to estimate the timing of HIV transmissions. After the advent of highly effective antiretroviral therapy (HAART), increasing concerns about transmitted drug-resistant mutant strains of HIV, coupled with rapidly dropping prices for genetic sequencing, led to an abundance of HIV genetic sequence data from infected individuals in nearly all regions of the United States. There is substantial molecular epidemiological evidence that variation in transmission rates over the course of infection influences the genetic diversity of HIV [13]–[16]. For example, viral sequences isolated from patients who were recently infected tend to be phylogenetically clustered (more closely related to one another than expected by chance). Simple models of HIV transmission have been shown to reproduce these phylogenetic patterns [17], suggesting that the transmission rate from EHI could be identifiable from genetic data. The probability of observing a particular viral phylogeny depends not only on the historical dynamics of HIV in the population but also on the stage of each patient at the time of sampling. For example, a sample comprised of only EHI patients will yield a different phylogeny than one comprised of only AIDS patients, the former having many more short external branches [17].
Recent advances [18]–[21] in population genetic methods have enabled the fitting of formal epidemiological models to viral sequence data. We use these methods to estimate HIV incidence, HIV prevalence, and the timing of transmission using both genetic sequence data and conventional HIV surveillance data. These methods may detect intensified transmission during EHI and reduced transmission following diagnosis, and may illustrate how the fraction of transmissions attributable to EHI has varied over the course of the epidemic.
Methods
Ethics Statement
This research was reviewed by the Institutional Review Boards at the University of Michigan and the Michigan Department of Community Health (MDCH). Data used in this research were originally collected for HIV surveillance purposes. Data were anonymized by staff at the MDCH before being provided to investigators. Because this research falls under the original mandate for HIV surveillance and the data were de-identified, the study was classified as human subjects research but was exempt from further Institutional Review Board review.
Data
As previously described in [17], the MDCH curates a database of partial pol HIV-1 sequences collected as part of routine clinical care and surveillance of drug-resistant mutant strains. MDCH provided an anonymized database of 9,002 sequences linked to clinical, demographic, and behavioral covariates of the patients from whom the sequences were isolated. Sequences were collected from October 14, 2004, through February 24, 2012 (Figure S1). 2,808 of these sequences correspond to men who have sex with men (MSM) in the Detroit metropolitan area (DMA), Michigan. To be included in the analysis each record must (1) have an HIV-1 subtype B sequence, (2) have a sequence from a HAART-naive patient within 12 mo of initial diagnosis, and (3) be collected from a man who has sex with men, residing in the DMA. Additionally, to achieve an analytically tractable sample size, we restricted our analyses to records that (4) have a high-quality sequence of at least 1,200 nucleotides. 662 of 2,808 sequence records collected from DMA MSM met all inclusion criteria. Demographic and clinical attributes of the study sample and the population are described in Table 1 and in Data S1. Details of the sequence selection, alignment, and quality control are contained in Text S1.
Tab. 1. Comparison of demographic and clinical variables for DMA MSM with sequences and those included in the estimated phylogenies. 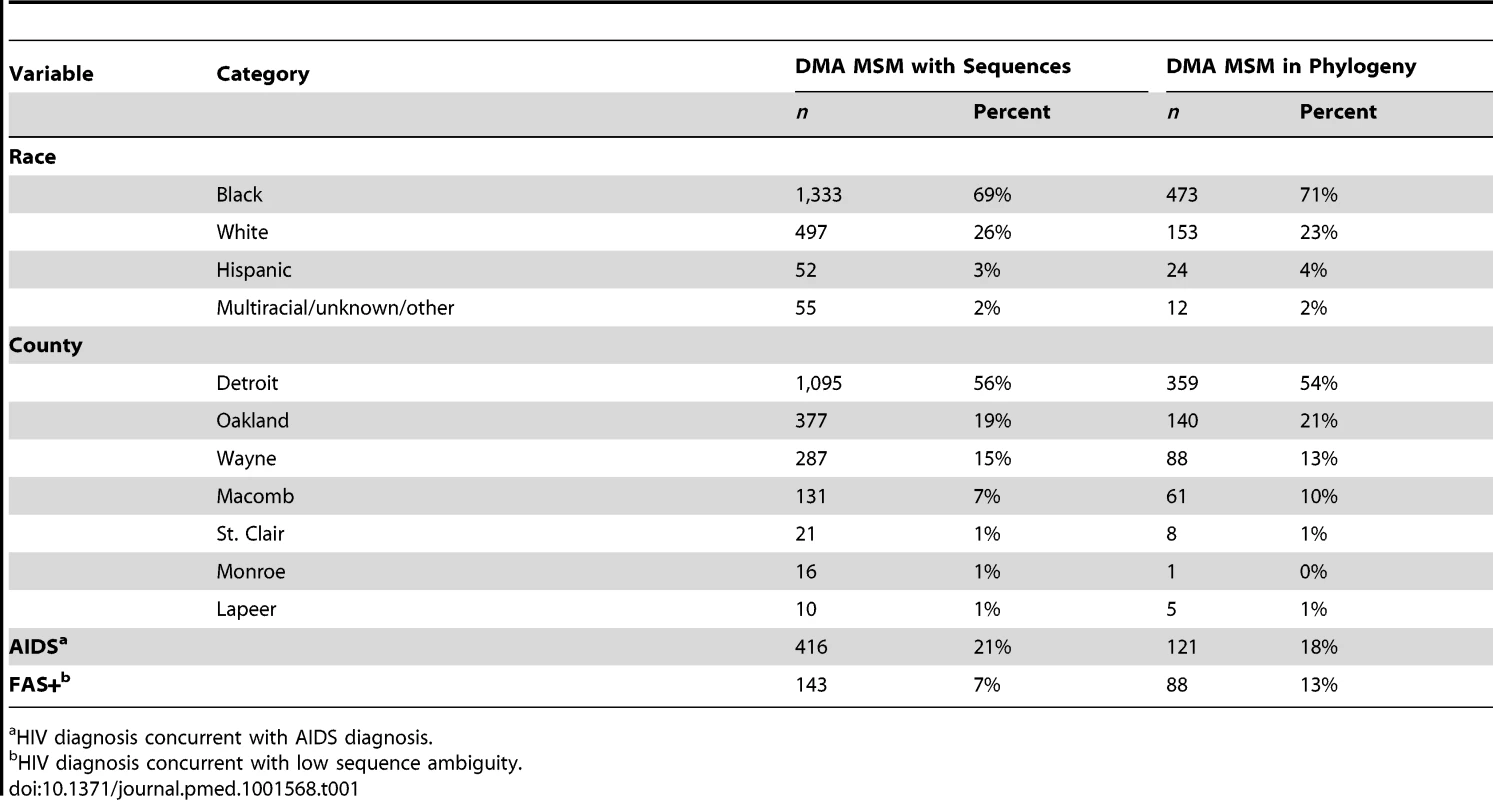
a HIV diagnosis concurrent with AIDS diagnosis. MDCH also provided anonymized surveillance data for 30,200 diagnoses reported in Michigan through March of 2012. 9,127 of these records corresponded to diagnoses from MSM in the DMA. These records contained CD4 cell counts, primary risk behavior, county of residence, and diagnosis dates of any AIDS-defining illnesses. The mean CD4 count upon diagnosis and number of AIDS/non-AIDS diagnoses were abstracted from these data and used for model-fitting and validation. Throughout this manuscript, “AIDS” refers to stage 3 HIV infection as defined in [4].
Phylogenetic Inference
The pattern and timing of coalescent events were inferred using relaxed clock phylogenetic methods [22] implemented in BEAST [23]. We used a log-normal relaxed clock model informed by the sequence data [22] that accounts for variable evolutionary rates within [24] and between lineages. The full parameters of the BEAST analysis, convergence diagnostics, and details of sequence alignment and quality control are reported in Text S1 [25]–[29].
To give insight into how frequently HIV lineages are introduced into DMA MSM from other geographic or risk behavior groups, we supplemented our data with 100 sequences from the Los Alamos National Laboratory HIV Sequence Database. Sequences with high similarity to at least one of 662 DMA MSM sequences were sampled. These sequences were included in subsequent phylogenetic and coalescent analysis.
To ease the computational burden of analyzing 662 sequences, phylogenetic analysis proceeded in two steps. First a neighbor joining tree was calculated using all sequences (TN93+Gamma model). Then the tree was divided into nine clades by selecting branches close to the root of the tree. Nine disjoint multiple sequence alignments were generated, corresponding to taxa in each clade, which were then independently analyzed with BEAST. Ten trees were sampled from the results of these nine BEAST analyses (90 phylogenies in total) in order to capture uncertainty in topology and branch lengths. These trees were used in subsequent coalescent analysis.
We found that the estimate of the height of the tree—the time to the most recent common ancestor (TMRCA) of the whole sample—was not well identified by the sequence data alone. This common issue arises when the height of the tree and the mean evolutionary rate cannot be fully resolved. To alleviate this problem, we constrained the height of the tree by using a uniform prior on the TMRCA between 1970 and 1982. In subsequent coalescent analyses, we sampled the posterior distribution of trees estimated by BEAST and then merged nine clades at a polytomous root. To render branch lengths comparable between different BEAST analyses, we calculated the mean substitution rate across a sample of trees from each of nine BEAST analyses and then adjusted the substitution rate within each tree to this mean value. Because phylogenetic relationships in the distant past carry little information about the epidemic close to the present, and in order to reduce the computational burden of fitting the population genetic model, the subsequent coalescent analysis used only the portion of the tree dating from 1990 onwards.
Transmission Model
The transmission model that we used to estimate the incidence, prevalence, and timing of transmission events is an extension of a model used by Bezemer et al. [12] and Hogan et al. [30]. There are three essential components to the model structure: the incidence rate, the diagnosis rate, and the natural history of infection. The model is illustrated in Figure 1 and described in detail in Text S2. The natural history of infection is modeled with a system of ordinary differential equations that tracks infected individuals as they progress through EHI, three chronic stages of infection, and AIDS. Diagnosed and treated individuals progress through infection at a reduced rate. The model incorporates empirical death rates from natural and AIDS-related causes. The model closely reproduces empirical observations regarding the time from infection to AIDS (Text S2).
Fig. 1. HIV transmission model and phylogeny. 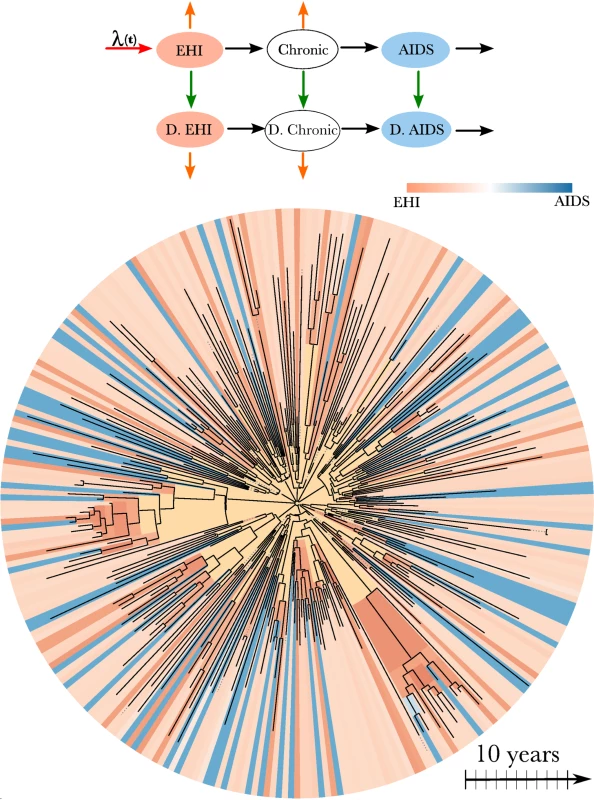
Top: A flow diagram describing the mathematical model fitted to surveillance time series and the HIV-1 phylogeny. Arrows of different colors represent the time-dependent rates at which transitions occur. Infected individuals progress from EHI to AIDS and may also become diagnosed (“D.”), as represented by black and green arrows. Orange arrows represent natural mortality. Incidence occurs at the rate λ(t) (red arrow). A more detailed diagram is shown in Figure S6. Bottom: HIV-1 phylogeny comprising virus samples from 662 patients and ancestral states estimated using the methods in [18]. The tree has been randomly downsampled to include 250 terminals for perspective. Colors at the terminals of the phylogeny represent the estimated stage of infection of the host at the time of sampling based on clinical data. Colors on the interior of the phylogeny represent the estimated stage of infection of the host harboring virus that is ancestral to the sample. Yellow corresponds to lineages that are likely to represent infections from outside of the DMA MSM risk group. We defined EHI to have a mean duration of 1 y. This duration was chosen instead of the more commonly used durations of 2 or 6 mo because the simulation studies described in Text S3 revealed that transmission rates for shorter periods were not identifiable given the available number and quality of HIV sequences.
Both the diagnosis rate and incidence rate were modeled using separate cubic B-splines [30]. Splines are a semi-parametric method for defining a very wide range of curves with relatively few parameters. Using splines is a flexible approach that can approximately capture patterns generated by heterogeneities in behavior and diagnosis patterns without explicitly modeling them.
We also modeled importation of lineages into the DMA MSM risk group by adding an additional compartment that represents infected hosts outside of the DMA MSM risk group. DMA MSM emigrate out of the risk group at a constant per capita rate, and immigration rates balance emigration rates by design, such that prevalence is unchanged by migration dynamics.
We also estimated the HIV incidence from the surveillance data using a back-calculation method [9]. The back-calculation estimator is derived from sample survey statistics and is based on the ratio of the number of total diagnoses to the subset determined to have been recently infected [31].
Parameter Estimation
The stage of infection at the time of sequencing of every patient with sampled sequence data was estimated using a naive Bayes classification method as described in “Estimating Stage of Infection” in Text S4. Covariates that were used for estimating stage of infection included the CD4 counts within 6 mo of diagnosis, whether the patient was diagnosed with AIDS within 2 mo of HIV diagnosis, and a measure of HIV sequence diversity. High sequence diversity is an indicator of a diverse intra-host viral population resulting from a long period of intra-host evolution and has previously been shown to be highly informative about the recency of infection [32]. The transmission model was fitted by maximum likelihood. The likelihood of the joint diagnosis time series data and of the genetic sequence data is given in Text S4.
Full derivations of the likelihood of the genetic data are given in [18],[20],[33]. These methods model each lineage in the viral phylogeny at each time point as corresponding to a single infected host. The model does not assume complete sampling and correctly accounts for the possibility that a lineage may pass through multiple unsampled hosts. Each node in the phylogeny is modeled as corresponding to a transmission event. These are reasonable approximations for many rapidly evolving RNA viruses including HIV [20],[34],[35] (see simulations in Text S3). The likelihood of the genetic data is computed by deriving the probability that each lineage in the phylogeny at each time point corresponds to an infected individual at a given stage of infection and diagnosis status. For example, the estimated stage of infection on the interior of the phylogeny is illustrated in Figure 1 for the HIV phylogeny of 662 patients. This figure shows that the state of an ancestral lineage at an internal node of the tree is likely to correspond to an EHI, owing to the large fraction of transmissions attributable to newly infected individuals.
The likelihood was numerically optimized using the simplex method via optim in R [36] to obtain maximum likelihood estimates (MLEs) of the transmission model parameters. Likelihood profiles were calculated for each transmission parameter. Credible intervals were calculated using an empirical Bayes approximation [37] (see “Model fitting” in Text S4). This approach uses a prior distribution that is calculated directly from the data. We constructed a multivariate uniform prior with bounds given by the 97.5% CIs calculated for each transmission parameter using the profile method.
To estimate parameters describing incidence, prevalence, and diagnosis rates through time, the model depicted in Figure 1 was fitted to diagnostic time series without using genetic data. To estimate the relative contribution of different stages of infection to total transmissions, the model was fitted to the genetic data while keeping fixed the parameters that describe incidence, prevalence, and diagnosis rates through time. This approach does not make full use of the genetic data, which may carry information about incidence and prevalence as well, but it is computationally efficient, since only a few parameters need to be estimated with the genetic data.
The robustness of inferences to phylogenetic error was assessed both by simulation techniques and by replicating parameter estimates across multiple independent estimated phylogenies. Uncertainty of the topology and branch lengths of estimated phylogenies (Figure S2) can lead to error in parameter estimates that is difficult to quantify. In “Simulations and Sensitivity Analysis” in Text S3, a simulation experiment is described that demonstrates that transmission rates in the first 6 mo of the infectious period cannot be inferred on the basis of the number and length of currently available sequences. Simulation experiments also show that it is feasible to estimate transmission rates within the first year (our definition of EHI) and to distinguish differing rates between chronic infection and late infection.
Results
Figure 2A and 2B illustrates the estimated fractions and total numbers of transmissions that originated from EHI and chronic infections. Transmissions by EHI dominated during the early epidemic (1980–1990), reflecting the larger prevalence of EHI as a proportion of the total number of infections and the relative intensity of transmission from this group [6]. The estimated fraction of transmissions attributable to EHI has stabilized since the early 1990s. Combined analysis of genetic and time series data yields an estimate of 44.7% (95% CI, 42.2%–46.4%) for the fraction of transmissions originating from EHI (approximately the first year of infection) at the beginning of 2007. This reflects about a 4.2-fold increase in transmission rates during EHI relative to the entire infectious period and an 8-fold increase in transmission rates relative to chronic infection. This credible interval and subsequent credible intervals are based on the fitting of a single model. These estimates do not reflect uncertainty due to model misspecification. Models with different parameterizations for the infectiousness of diagnosed individuals or different interaction effects could yield different estimates (Table S1).
Fig. 2. Estimated transmission patterns through time. 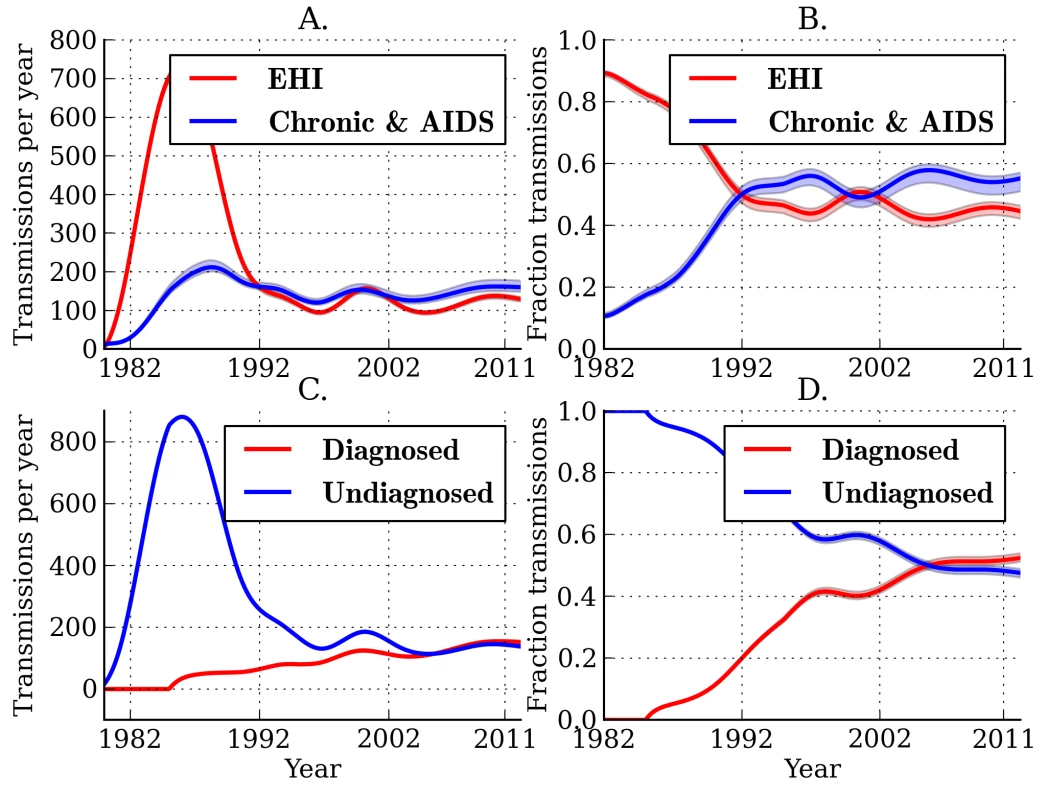
Lines show the MLE, and shaded regions show the 95% credible interval. (A) Estimated number of transmissions originating from individuals in different stages of infection: EHI, chronic infection, and AIDS. (B) The estimated fraction of transmissions attributable to EHI, chronic infection, and AIDS. (C) Estimated number of transmissions originating from diagnosed and undiagnosed individuals through time. (D) The estimated fraction of transmissions attributable to diagnosed and undiagnosed infections through time. Estimated credible intervals reflect the fit of a single model to the data and do not incorporate uncertainty due to model misspecification error. The true number of transmissions from diagnosed individuals depends on many factors, including the number of infected individuals who are diagnosed, the extent to which knowledge of infection reduces infectiousness, and the effectiveness of HAART at reducing transmission probabilities per sexual act. We estimate that transmissions from diagnosed individuals have trended upwards in recent years, which reflects that a steadily increasing proportion of infections are now diagnosed (Figure 2). We estimate that 52.4% (95% CI, 51.1%–53.9%) of transmissions originated from diagnosed individuals in 2007 (Figure 2C and 2D) [38]. We found that the proportion of transmissions from EHI mirrors the trend in diagnoses late in infection over time. Figure 3 shows the fraction of diagnoses concurrent with AIDS diagnosis and the estimated number of diagnoses during EHI. We categorized all diagnoses prior to the availability of the first HIV test (1985) as AIDS. AIDS diagnoses have fallen over time, while the number of diagnoses during EHI has risen dramatically since the mid-1990s from zero to an estimated 21.7% of current diagnoses. Estimated diagnosis rates are shown in Figure S3. The reported number of diagnoses that are concurrent with AIDS diagnosis is in close agreement with the model-estimated number of AIDS diagnoses in Figure 3. We found that diagnosis rates have risen consistently since the HIV test became available in 1985, and the time from infection to diagnosis has steadily decreased.
Fig. 3. HIV diagnoses and CD4 cell count through time. 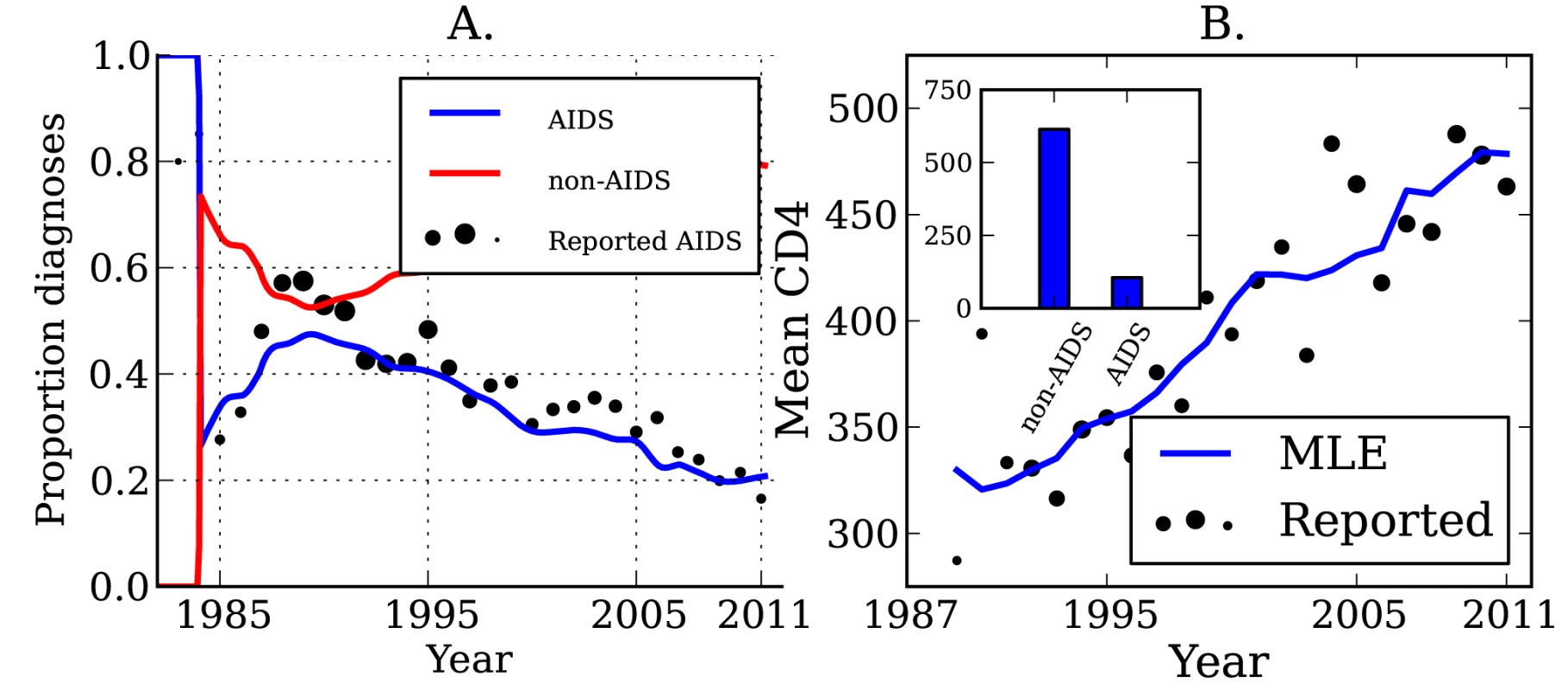
(A) The sample proportion (points) and estimated proportion (lines) of diagnoses that are concurrent with AIDS diagnosis over time, and the estimated proportion of diagnoses that are not concurrent with AIDS. The diameter of points is proportional to the number of diagnoses used to calculate the proportions. (B) The sample mean (points) and estimated mean (lines) CD4 cell count in newly diagnosed individuals over time. The mean is calculated from CD4 counts aggregated by year. The diameter of points is proportional to the number of CD4 counts used to calculate the mean. Inset: The mean CD4 cell count by stage of infection, which gives the best fit (least squares) to the observed trend in mean CD4 count over time. The mean CD4 cell count of patients at the time of diagnosis is indicative of the trend of increasing diagnosis rates. The average CD4 count for new diagnoses by year is shown in Figure 3. While CD4 counts are a very noisy and unreliable indicator of time since infection on an individual basis, aggregated CD4 counts follow an almost linear trend. The CD4 data were not used when estimating diagnosis rates, but are a useful check that the model is giving realistic estimates. Also shown in Figure 3 is the mean CD4 count for new diagnoses predicted by the MLE model fit, as well as the best-fitting CD4 counts by stage of infection as described in “CD4 and Model Validation” in Text S3.
Estimated incidence and cumulative diagnoses of HIV infection for DMA MSM are shown in Figure 4. As of the beginning of 2012, there have been a total of 9,127 HIV diagnoses in DMA MSM. We estimate that there have been a total of 12,139 infections in DMA MSM, of which 6,084 are still living; of these 6,084, we estimate that 5,233 have been diagnosed. The estimated current number of prevalent infections exceeds the number of living diagnosed individuals by 16.3%. Approximately one in seven infected individuals is unaware of his infection [39]. Estimates using the HIV model are very similar to those obtained from widely used back-calculation methods [10],[31] (Figure 4B), despite the fact that the HIV model uses much more data and more realistically models diagnosis rates and the natural history of infection.
Fig. 4. Estimated HIV diagnoses and HIV incidence through time. 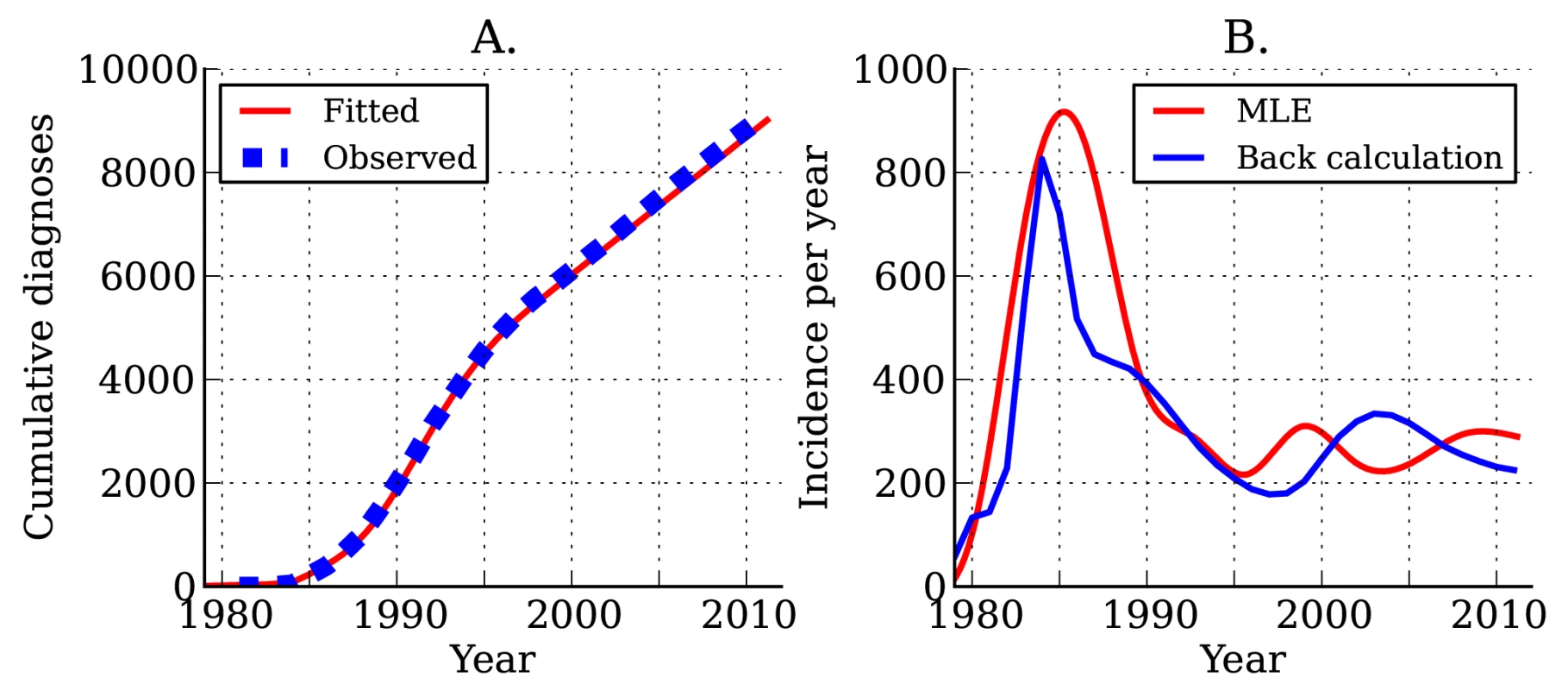
(A) Actual (blue) and estimated (red) cumulative HIV diagnoses in DMA MSM. (B) Estimated incidence of infection over time. The red line shows estimated incidence from surveillance time series data. The blue line shows estimated incidence using the back-calculation method. Following the initial rapid rises in the 1980s, we found that incidence (Figure 4B) and the number of undiagnosed infections (Figure S4) have been steady since the mid-1990s. The total number of individuals living with diagnosed infections has trended upwards since 1995 because of reduced mortality with effective treatment (Figure S4). We estimate that incidence at the beginning of 2007 stood at 256 new infections per year, and incidence at the beginning of 2012 stood at 290 new infections per year; however, there is a great deal of uncertainty in estimated incidence close to the present.
Infectiousness can vary with stage of infection and diagnosis status; however, it is not clear a priori which factors are necessary to include in a model to provide a satisfactory fit to the data. We conducted a data-driven comparison of several models that differed in how relative infectiousness was described. Results of this analysis are shown in Table S1. In the most general model, three parameters describe the relative infectiousness of diagnosed individuals, those with chronic infection, and those with AIDS. The simplest models include only one parameter describing the relative infectiousness of those with late infection (chronic and AIDS) or diagnosed individuals. Analyzing the fits of these models to the phylogenetic data provides strong support for the two-parameter model, which includes parameters only for infectiousness of chronic infection and AIDS. Thus, we do not detect reduced infectiousness with diagnosis; however, this finding may be related to sample selection. Only sequences from recently diagnosed individuals were considered in this analysis. It is possible that analysis of sequences from individuals who have been diagnosed for a long time may reveal reduced infectiousness with diagnosis and treatment; however, the present HIV model is not equipped to fit such data. The estimated fractions of transmissions from EHI and diagnosed individuals are consistent across model variants, with the exception of one model that fit the data poorly.
To give greater intuition into why variation in transmission rates is identifiable from genetic data, we present simulated trees in “Simulated Trees” in Text S3. Simulations were carried out under scenarios in which individuals with EHI transmit at greater rates than those with chronic infections or at equal rates. The times and states of patients in the simulated trees were chosen to match the real data. In addition to the results presented in [17], these simulations give a graphical representation of how EHI transmission influences HIV phylogenetic structure.
Discussion
We have estimated that almost half of transmissions occur within the first year of infection in the contemporary HIV epidemic among MSM in the DMA. This inference was made possible by recently developed population genetic methods [18],[20],[33] that enable characterization of the major sources of transmission. These findings may have significance for control strategies based on prophylactic use of antiretroviral medications. TasP [1],[40] is a strategy based on early administration of HAART following HIV diagnosis in an effort to suppress viral loads and reduce transmission probabilities. The impact that transmission during early infection will have on TasP strategies has been a subject of recent debate. Cohen et al. [41] present contrasting views regarding the potential impact of EHI transmission on TasP effectiveness. A simplistic interpretation of our results would hold that a lower fraction of transmissions will be prevented by TasP because a large fraction of transmissions are likely to occur before diagnosis. A more nuanced view presented by Cohen et al. [41] holds that high transmission during EHI and observed incidence trends are consistent with a low reproduction number, and therefore TasP may nevertheless have large population-level impacts even if it prevents few transmissions directly. Contact patterns and fluctuations in risk behavior can markedly raise the fraction of transmissions from early infection [42], and these factors were not modeled in this study. These same factors may yield lower reproduction numbers at any given endemic prevalence level, so that TasP would have to prevent only a small fraction of all transmissions in order to have very large population effects.
The robustness of our conclusions depends on the sensitivity of the conclusions to potential violations of several assumptions on which the population genetic model is based. The population genetic model accounts for the effects of incomplete sampling and the possibility that a lineage in the viral phylogeny may pass through more than one infected host. However, the approach relies on the assumption that a single lineage at a given time corresponds to a single infected host. This assumption would be problematic if multiple lineages circulate in a host and are independently transmitted. But a growing body of evidence suggests that new infections are established by a very small number of viral particles [34],[43],[44]. Virus derived from a single transmitting host at a single transmission event is likely to have limited diversity. In contrast, dual infection from distinct partners may present a greater challenge to attempts to reconstruct epidemiological dynamics from genetic data. Virus derived from distinct partners is likely to be more diverse, which in the presence of high levels of recombination makes estimation of an accurate phylogeny difficult. Recent studies [45],[46] have found low prevalence of HIV dual infections (including super - and coinfection [47]). There is some evidence based on African heterosexual cohorts that the incidence of super-infection is comparable to the incidence of infection generally [48]. However, bias due to dual infection will depend on when transmission occurs during the course of infection. If transmission occurs early, it is more likely to occur before a host is multiply infected. Additionally, if the super-infecting strain has low abundance within the host, it is unlikely to have a large influence on phylogenies estimated from consensus sequences. We removed all sequences with evidence of recombination, further reducing the possibility of bias from dual-infected hosts. In “Dual Infection” in Text S3, we provide a simple calculation that gives the approximate bias that can be expected if the prevalence of super-infection is 10%. In this scenario, the estimated HIV incidence would be biased downwards by at most 3.75%.
This analysis assumes that the internal nodes of the phylogeny represent transmission events. In reality, the viral lineage that is transmitted may have arisen in the host some time before transmission occurred [35]. The magnitude of the potential bias introduced by this assumption is an empirical question that we have addressed by simulation in “Simulations and Sensitivity Analysis” in Text S3. By incorporating an empirical distribution [49],[50] for the time of common ancestry within hosts into epidemic simulations, we have established that intra-host evolutionary dynamics are unlikely to introduce large bias into our estimates.
The methods that we presented in this paper are sensitive to uncertainty both in the phylogeny and in the stage of infection of patients at the time of sampling. Estimation of stage of infection is very imprecise and is limited by the available clinical and self-reported data. Improved antibody avidity assays promise to greatly improve our ability to determine when newly diagnosed individuals were infected. Serial sampling with deep sequencing of the virus within hosts is another promising strategy to estimate the time since infection [51]. Increasing the number, length, and quality of sequence data can greatly improve the quality of the phylogenetic reconstruction. In this study, we were limited to using only about a quarter of potentially informative sequences by the computational demands of both estimating a relaxed clock phylogeny and fitting complex models to the estimated phylogeny (see “Computation” in Text S4). The value of genetic data for epidemiological inference will increase as computational techniques are developed that allow for the incorporation of more sequences.
The accuracy of the phylogenetic reconstruction may also be affected by sampling from different stages of infection. In our analysis, the sample was skewed towards viral sequences from EHI or late infections. The mean substitution rate varies over the course of infection [24], adding an extra layer of complexity to the phylogenetic analysis. We found that the mean substitution rate in external branches of the phylogeny is significantly correlated (Pearson correlation −0.17, p<0.001) with the estimated stage of infection of the patient from whom the virus was sampled (Figure S5). This correlation suggests that the relaxed clock methods that we used to estimate the branch lengths in units of time was flexible enough to account for variable rates of evolution over the course of infection.
While our conclusions are sensitive to many different sources of error, we can evaluate the robustness of our conclusions by reestimating parameters with simulated data where the true parameter values are known. In Text S3, we describe several simulation experiments designed to test the robustness of our estimates to error in the phylogeny and to errors arising from stochastic population dynamics. These simulations demonstrate that it is feasible to estimate EHI transmission rates given the available data. This analysis can be repeated with different HIV transmission models, which may be appropriate when more is known about heterogeneous risk behaviors and sexual networks. Sexual network heterogeneity influences HIV phylogenetic structure [52],[53], which may make it possible to estimate features of the sexual network from phylogenetic data [54]. Our estimated credible intervals are based on the fit of a single model to the data, and models that more realistically account for individual-level heterogeneities may yield different estimates. Although we report estimates based on the model that best fit the data, our estimated credible intervals do not account for error due to model misspecification.
The analysis we have presented can be replicated for other cities and risk groups where drug-resistant mutant strain sequence databases are available and can be linked to clinical and behavioral covariates for each patient. Phylodynamic analysis of HIV can supplement routine surveillance, addressing the need to monitor sources of transmission and generating the evidence necessary to efficiently allocate resources and assess control program effectiveness.
Supporting Information
Zdroje
1. CohenM, ChenY, McCauleyM, GambleT, HosseinipourM, et al. (2011) Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med 365 : 493–505.
2. CohenM, ShawG, McMichaelA, HaynesB (2011) Acute HIV-1 infection. N Engl J Med 364 : 1943–1954.
3. PilcherC, TienH, EronJJr, VernazzaP, LeuS, et al. (2004) Brief but efficient: acute HIV infection and the sexual transmission of HIV. J Infect Dis 189 : 1785–1792.
4. SchneiderE, WhitmoreS, GlynnK, DominguezK, MitschA, et al. (2008) Revised surveillance case definitions for HIV infection among adults, adolescents, and children aged <18 months and for HIV infection and AIDS among children aged 18 months to <13 years—United States, 2008. MMWR Recomm Rep 57 : 1–12.
5. PowersK, GhaniA, MillerW, HoffmanI, PettiforA, et al. (2011) The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet 378 : 256–268.
6. JacquezJA, KoopmanJS, SimonCP, LonginiIMJr (1994) Role of the primary infection in epidemics of HIV infection in gay cohorts. J Acquir Immune Defic Syndr 7 : 1169–1184.
7. BaetenJ, OverbaughJ (2003) Measuring the infectiousness of persons with HIV-1: opportunities for preventing sexual HIV-1 transmission. Curr HIV Res 1 : 69–89.
8. GrayR, WawerM, BrookmeyerR, SewankamboN, SerwaddaD, et al. (2001) Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357 : 1149–1153.
9. KaronJ, SongR, BrookmeyerR, KaplanE, HallH (2008) Estimating HIV incidence in the United States from HIV/AIDS surveillance data and biomarker HIV test results. Stat Med 27 : 4617–4633.
10. PrejeanJ, SongR, HernandezA, ZiebellR, GreenT, et al. (2011) Estimated HIV incidence in the United States, 2006–2009. PLoS ONE 6: e17502 doi:10.1371/journal.pone.0017502
11. van VeenMG, PresanisAM, ContiS, XiridouM, StengaardAR, et al. (2011) National estimate of HIV prevalence in the Netherlands: comparison and applicability of different estimation tools. AIDS 25 : 229–237.
12. BezemerD, de WolfF, BoerlijstM, van SighemA, HollingsworthT, et al. (2010) 27 years of the HIV epidemic amongst men having sex with men in the Netherlands: an in depth mathematical model-based analysis. Epidemics 2 : 66–79.
13. PaoD, FisherM, HuéS, DeanG, MurphyG, et al. (2005) Transmission of HIV-1 during primary infection: relationship to sexual risk and sexually transmitted infections. AIDS 19 : 85–90.
14. YerlyS, JunierT, Gayet-AgeronA, AmariE, von WylV, et al. (2009) The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS 23 : 1415–1423.
15. CuevasM, Muñoz-NietoM, ThomsonM, DelgadoE, IribarrenJ, et al. (2009) HIV-1 transmission cluster with T215D revertant mutation among newly diagnosed patients from the Basque Country, Spain. J Acquir Immune Defic Syndr 51 : 99–103.
16. AldousJ, PondS, PoonA, JainS, QinH, et al. (2012) Characterizing HIV transmission networks across the United States. Clin Infect Dis 55 : 1135–1143.
17. VolzE, KoopmanJ, WardM, BrownA, FrostS (2012) Simple epidemiological dynamics explain phylogenetic clustering of HIV from patients with recent infection. PLoS Comput Biol 8: e1002552 doi:10.1371/journal.pcbi.1002552
18. VolzE (2012) Complex population dynamics and the coalescent under neutrality. Genetics 190 : 187–201.
19. FrostS, VolzE (2010) Viral phylodynamics and the search for an ‘effective number of infections’. Philos Trans R Soc Lond B Biol Sci 365 : 1879–1890.
20. VolzE, PondS, WardM, Leigh BrownA, FrostS (2009) Phylodynamics of infectious disease epidemics. Genetics 183 : 1421–1430.
21. StadlerT, KouyosR, von WylV, YerlyS, BöniJ, et al. (2012) Estimating the basic reproductive number from viral sequence data. Mol Biol Evol 29 : 347–357.
22. DrummondA, HoS, PhillipsM, RambautA (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88 doi:10.1371/journal.pbio.0040088
23. DrummondA, RambautA (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7 : 214.
24. LeeH, PerelsonA, ParkS, LeitnerT (2008) Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS Comput Biol 4: e1000240 doi:10.1371/journal.pcbi.1000240
25. Rambaut A (2011). Path-O-Gen [computer program]. Available: http://tree.bio.ed.ac.uk/software/pathogen/. Accessed 4 November 2013.
26. PondS, PosadaD, StawiskiE, ChappeyC, PoonA, et al. (2009) An evolutionary model-based algorithm for accurate phylogenetic breakpoint mapping and subtype prediction in HIV-1. PLoS Comput Biol 5: e1000581 doi:10.1371/journal.pcbi.1000581
27. BennettD, CamachoR, OteleaD, KuritzkesD, FleuryH, et al. (2009) Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE 4: e4724 doi:10.1371/journal.pone.0004724
28. Pond S, Muse S (2005) Hyphy: hypothesis testing using phylogenies. In: Nielsen R, editor. Statistical methods in molecular evolution. pp. 125–181.
29. Los Alamos National Laboratory (2011) Quality control: HIV-1 sequence quality analysis. Available: http://www.hiv.lanl.gov/content/sequence/QC/index.html. Accessed 4 November 2013.
30. HoganD, ZaslavskyA, HammittJ, SalomonJ (2010) Flexible epidemiological model for estimates and short-term projections in generalised HIV/AIDS epidemics. Sex Transm Infect 86: ii84–ii92.
31. YanP, ZhangF, WandH (2011) Using HIV diagnostic data to estimate HIV incidence: method and simulation. Stat Commun Infect Dis 3 : 1–28.
32. KouyosR, von WylV, YerlyS, BöniJ, RiederP, et al. (2011) Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis 52 : 532–539.
33. RasmussenD, RatmannO, KoelleK (2011) Inference for nonlinear epidemiological models using genealogies and time series. PLoS Comput Biol 7: e1002136 doi:10.1371/journal.pcbi.1002136
34. ZhuT, MoH, WangN, NamD, CaoY, et al. (1993) Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261 : 1179–1181.
35. LeitnerT, AlbertJ (1999) The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc Natl Acad Sci U S A 96 : 10752–10757.
36. R Core Team (2012) R: a language and environment for statistical computing. R Vienna: Foundation for Statistical Computing. Available: http://www.R-project.org/. Accessed 4 November 2013.
37. Efron B (2010) Large-scale inference: empirical Bayes methods for estimation, testing and prediction. Cambridge: Cambridge University Press.
38. MarksG, CrepazN, JanssenR (2006) Estimating sexual transmission of HIV from persons aware and unaware that they are infected with the virus in the USA. AIDS 20 : 1447–1450.
39. CampsmithM, RhodesP, HallH, GreenT (2010) Undiagnosed HIV prevalence among adults and adolescents in the United States at the end of 2006. J Acquir Immune Defic Syndr 53 : 619–624.
40. MontanerJS (2013) Treatment as prevention: toward an AIDS-free generation. Top Antivir Med 21 : 110–114.
41. CohenMS, DyeC, FraserC, MillerWC, PowersKA, et al. (2012) HIV treatment as prevention: debate and commentary—will early infection compromise treatment-as-prevention strategies? PLoS Med 9: e1001232 doi:10.1371/journal.pmed.1001232
42. ZhangX, ZhongL, Romero-SeversonE, AlamSJ, HenryCJ, et al. (2012) Episodic HIV risk behavior can greatly amplify HIV prevalence and the fraction of transmissions from acute HIV infection. Stat Commun Infect Dis 4 : 1041.
43. LiH, BarK, WangS, DeckerJ, ChenY, et al. (2010) High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog 6: e1000890 doi:10.1371/journal.ppat.1000890
44. FischerW, GanusovV, GiorgiE, HraberP, KeeleB, et al. (2010) Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS ONE 5: e12303 doi:10.1371/journal.pone.0012303
45. CourgnaudV, SengR, BecquartP, BoulahtoufA, RouziouxC, et al. (2007) HIV-1 co-infection prevalence in two cohorts of early HIV-1 seroconverters in France. AIDS 21 : 1055–1056.
46. van der KuylA, ZorgdragerF, JurriaansS, BackN, PrinsJ, et al. (2009) Incidence of human immunodeficiency virus type 1 dual infections in Amsterdam, The Netherlands, during 2003–2007. Clin Infect Dis 48 : 973–978.
47. SmithD, RichmanD, LittleS (2005) HIV superinfection. J Infect Dis 192 : 438–444.
48. ReddA, MullisC, SerwaddaD, KongX, MartensC, et al. (2012) The rates of HIV superinfection and primary HIV incidence in a general population in Rakai, Uganda. J Infect Dis 206 : 267–274.
49. ShankarappaR, MargolickJ, GangeS, RodrigoA, UpchurchD, et al. (1999) Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 73 : 10489–10502.
50. LemeyP, PondSLK, DrummondAJ, PybusOG, ShapiroB, et al. (2007) Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol 3: e29 doi:10.1371/journal.pcbi.0030029
51. PoonA, McGovernR, MoT, KnappD, BrennerB, et al. (2011) Dates of HIV infection can be estimated for seroprevalent patients by coalescent analysis of serial next-generation sequencing data. AIDS 25 : 2019–2026.
52. LeventhalGE, KouyosR, StadlerT, von WylV, YerlyS, et al. (2012) Inferring epidemic contact structure from phylogenetic trees. PLoS Comput Biol 8: e1002413 doi:10.1371/journal.pcbi.1002413
53. FrostSD, VolzEM (2013) Modelling tree shape and structure in viral phylodynamics. Philos Trans R Soc Lond B Biol Sci 368 20120208.
54. BrownAJL, LycettSJ, WeinertL, HughesGJ, FearnhillE, et al. (2011) Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 204 : 1463–1469.
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2013 Číslo 12- Není statin jako statin aneb praktický přehled rozdílů jednotlivých molekul
- Magnosolv a jeho využití v neurologii
- Moje zkušenosti s Magnosolvem podávaným pacientům jako profylaxe migrény a u pacientů s diagnostikovanou spazmofilní tetanií i při normomagnezémii - MUDr. Dana Pecharová, neurolog
- Biomarker NT-proBNP má v praxi široké využití. Usnadněte si jeho vyšetření POCT analyzátorem Afias 1
- Antikoagulační léčba u pacientů před operačními výkony
-
Všechny články tohoto čísla
- Editors' Wishes for an Illuminated Season and an Open New Year
- Artemisinin Combination Therapy: A Good Antimalarial, but Is the Dose Right?
- Early HIV Infection in the United States: A Virus's Eye View
- Financial Conflicts of Interest and Reporting Bias Regarding the Association between Sugar-Sweetened Beverages and Weight Gain: A Systematic Review of Systematic Reviews
- Investigating the Intersection of Policing and Public Health
- Improving Treatment of Children with Autism Spectrum Disorder in Low- and Middle-Income Countries: The Role of Non-specialist Care Providers
- Data Sharing in a Humanitarian Organization: The Experience of Médecins Sans Frontières
- Why Growing Retractions Are (Mostly) a Good Sign
- Challenges in Addressing Plagiarism in Education
- Experiences with Policing among People Who Inject Drugs in Bangkok, Thailand: A Qualitative Study
- Non-Specialist Psychosocial Interventions for Children and Adolescents with Intellectual Disability or Lower-Functioning Autism Spectrum Disorders: A Systematic Review
- Malaria and Severe Anemia: Thinking beyond
- Financing Essential HIV Services: A New Economic Agenda
- Timing and Completeness of Trial Results Posted at ClinicalTrials.gov and Published in Journals
- Circulating Mitochondrial DNA in Patients in the ICU as a Marker of Mortality: Derivation and Validation
- The Effect of Dosing Regimens on the Antimalarial Efficacy of Dihydroartemisinin-Piperaquine: A Pooled Analysis of Individual Patient Data
- Major Burden of Severe Anemia from Non-Falciparum Malaria Species in Southern Papua: A Hospital-Based Surveillance Study
- HIV-1 Transmission during Early Infection in Men Who Have Sex with Men: A Phylodynamic Analysis
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Artemisinin Combination Therapy: A Good Antimalarial, but Is the Dose Right?
- Circulating Mitochondrial DNA in Patients in the ICU as a Marker of Mortality: Derivation and Validation
- Timing and Completeness of Trial Results Posted at ClinicalTrials.gov and Published in Journals
- Malaria and Severe Anemia: Thinking beyond
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



