-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Ly49C-Dependent Control of MCMV Infection by NK Cells Is -Regulated by MHC Class I Molecules
We previously identified a viral murine cytomegalovirus (MCMV) strain whose variant m157 immunoevasin can bind the inhibitory Ly49C NK cell receptor in addition to activating Ly49H receptor in B6 mice. Here we show that simultaneous engagement of the two receptors by m157 hampers NK cell activation. Most Ly49H+ NK cells lack Ly49C in B6 mice, as a result, NK cell population efficiently controls MCMV infection; however, the anti-viral response is reduced in transgenic mice where all NK cells express Ly49C. MHC I masks Ly49C through cis-interactions, which restricts its inhibitory function. Indeed, B6 Ly49C Tg, β2m ko mice, that are deficient for MHC I, are highly susceptible to low dose of MCMV. Our findings indicate that both the NK cell repertoire and MHC I molecules control susceptibility vs resistance to viral infections.
Published in the journal: . PLoS Pathog 10(5): e32767. doi:10.1371/journal.ppat.1004161
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004161Summary
We previously identified a viral murine cytomegalovirus (MCMV) strain whose variant m157 immunoevasin can bind the inhibitory Ly49C NK cell receptor in addition to activating Ly49H receptor in B6 mice. Here we show that simultaneous engagement of the two receptors by m157 hampers NK cell activation. Most Ly49H+ NK cells lack Ly49C in B6 mice, as a result, NK cell population efficiently controls MCMV infection; however, the anti-viral response is reduced in transgenic mice where all NK cells express Ly49C. MHC I masks Ly49C through cis-interactions, which restricts its inhibitory function. Indeed, B6 Ly49C Tg, β2m ko mice, that are deficient for MHC I, are highly susceptible to low dose of MCMV. Our findings indicate that both the NK cell repertoire and MHC I molecules control susceptibility vs resistance to viral infections.
Introduction
In humans, cytomegalovirus (CMV) is a pathogen responsible for causing significant mortality in immunocompromised patients [1] and in individuals lacking Natural Killer (NK) cells [2]. Mouse cytomegalovirus (MCMV) is a natural pathogen of mice. The similarities in structure and biology between human and mouse CMV make the latter a widely utilized model for human infection [3]. The study of MCMV has provided valuable insights into how the immune system responds to infection, and has helped to define the immune evasion mechanisms used by CMV to ensure that viral replication proceeds. NK cells play a crucial role in the early control of MCMV infection in resistant mouse strains; they limit viral replication and mortality during acute infection. The ability of NK cells to control viral infection is tightly regulated by their activating and inhibitory receptors [4]. Activating NK cell receptors include activating forms of killer cell immunoglobulin-like receptors (KIRs) in humans, and Ly49 receptors in mice. Both humans and mice express CD94/NKG2C which recognizes MHC class I molecules, and NKG2D which can be triggered by stress-induced ligands. NK cells also possess inhibitory receptors specific for MHC class I that permit discrimination of normal healthy cells from diseased ones, such as virus-infected cells, that display reduced MHC class I expression. These receptors include KIR in humans and members of the Ly49 family in mice, and LIR-1 and CD94/NKG2A in both species (reviewed in [5]). Inbred strains of mice express distinct NK cell receptor repertoires; NK cell receptors are encoded within a polygenic cluster in which each receptor gene is subject to polymorphism between the mouse strains; this variability results in resistance or susceptibility to specific viral infections.
Ly49H is the activating receptor responsible for resistance to MCMV infection in C57BL/6 (B6) mice [6]–[8]. Ly49H binds specifically to the m157 viral protein encoded by laboratory MCMV strains (Smith and K181) and triggers cytotoxicity and cytokine production [9], [10]. Arase et al showed that m157 binds to the inhibitory Ly49I receptor in 129/J mice, but not in B6 mice, while 129/J mice lack Ly49H [9]; this repertoire results in susceptibility to MCMV infection in the 129/J strain. In laboratory settings, immunological pressure through Ly49H was evidenced by the rapid selection of viral mutants producing m157 variants that escape recognition by this receptor [11]. Sequence analysis of m157 in a panel of MCMV isolates collected from a wild mouse population showed that only two isolates were identical to the laboratory MCMV strains (Smith and K181) [11], [12]. In addition, unlike the laboratory strains many of the viral isolates with m157 variants were able to replicate to high titers in resistant B6 mice [11]. We previously identified an MCMV strain (G1F) that was isolated from mice trapped in the wild; its m157 sequence shares over 93% homology with Smith and K181 strains but the protein displays a unusual binding profile to Ly49 receptors [13]. In addition to Ly49H, m157G1F can bind Ly49C in B6 and BALB/c mice [13], [14].
Inhibitory Ly49 receptors are thought to play a crucial role during NK cell education. Mechanisms of NK cell education are still unclear and different models co-exist. The current consensus states that NK cells expressing inhibitory receptors specific for self MHC class I molecules are fully educated (“licensed” [15] or “not disarmed” [16]) and have a greater response potential than NK cell subsets that lack such receptors. However, recent studies showed that Ly49C− NK cells are fully functional in B6 mice and indeed they dominate the Ly49H-dependent response to MCMV infection [17]. These results indicate that inhibition triggered by Ly49C binding to H-2 Kb overrides the responsive advantage gained by “licensing” and suggest that the inhibition mediated by Ly49C binding to H-2Kb regulates Ly49H-dependent NK cell activation. These data emphasize the need for a better understanding of the regulation of NK cells and how this impacts on NK cell function in the context of viral infection.
The high variability of the m157 sequence is not without similarities with human CMV, whose genome contains highly polymorphic loci that encode proteins (immunoevasins) with the potential to affect virulence through immune evasion [18]. The complex interactions of viral proteins with the host immune system are critical for determining a viral strain infectivity and pathogenicity. We undertook to study whether a viral immunoevasin able to bind multiple NK cell receptors can modulate the anti-viral immune response and to define the regulatory mechanisms. Our recent findings that m157G1F binds to two NK cell receptors with opposite functions in B6 mice, provide a unique opportunity to study, in a natural infection model, how the integration of competing signals determines tolerance versus killing and ultimately the outcome of an important viral infection in vivo.
Results
The m157G1F variant binds both Ly49H and Ly49C
We previously demonstrated the ability of m157 from the MCMVG1F strain (m157G1F) to bind both Ly49H and Ly49C in B6 mice [13]. Here, we analyzed the binding properties of m157G1F and aimed to determine whether it can bind the two receptors simultaneously. We transfected BWZ.36 cells to express either Ly49HB6 (BWZ-Ly49H) or Ly49CB6 (BWZ-Ly49C) and clones expressing the receptors at similar levels were selected to test m157G1F-Fc binding, by flow cytometry. Firstly, we measured the binding obtained at increasing concentrations of the m157G1F-Fc. Titration curves were similar for Ly49H and Ly49C expressing cells (Fig. 1A). No binding was detected to the parental BWZ.36 cells (data not shown). These results suggest that m157G1F binds to Ly49H and Ly49C with similar affinities. Next, we compared the kinetics of binding of m157G1F to these two receptors. Ly49H or Ly49C-expressing cells were incubated with saturating concentrations of m157G1F-Fc, as determined above, for 30 sec to 40 min. Fluorescence obtained after 40 min provided the maximal intensity value (100%), while the background to be subtracted was measured in the absence of fusion protein; the percentages of binding achieved over increasing incubation periods were calculated accordingly. Binding of m157G1F to Ly49C was found to occur slightly more quickly than to Ly49H, with 50% binding reached after 5 and 7 min, respectively (Fig. 1B). Thirdly, we measured the stability of the interactions. For this purpose, we used the optimal binding conditions previously determined, and analyzed dissociation rates. Excess amounts of anti-m157 antibody (6H121) prevented re-association of the fusion proteins to the receptors after they detached. We found that 50% of m157G1F that was initially bound to Ly49C had dissociated after approximately 20 min while over 50% was still attached to Ly49H after 90 min (Fig. 1C). These results suggest that m157G1F dissociates more quickly from Ly49C than Ly49H. A comparative binding analysis of m157K181 and m157G1F to Ly49H was also conducted as a control; our results showed similar binding characteristics of the two m157 variants to Ly49H (Fig. S1). This data confirmed our previous findings [13].
Fig. 1. Analysis of m157G1F binding to Ly49H and Ly49C expressing cells. 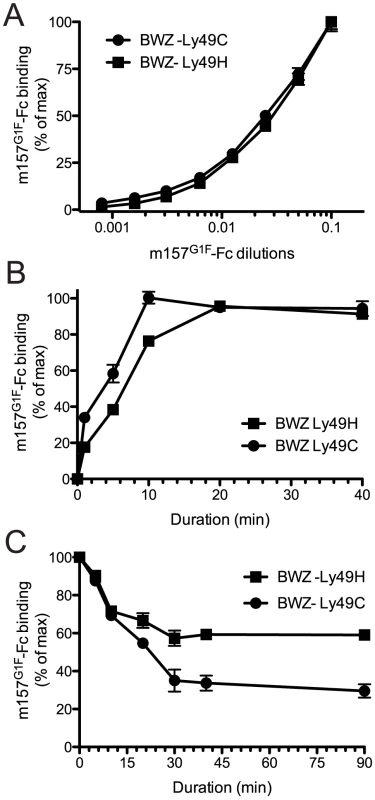
(A) BWZ-Ly49H and BWZ-Ly49C cells were incubated with increasing concentrations of m157G1F-Fc for 40 min at 4°C and binding was measured by flow cytometry. The m157G1F-Fc concentration that achieved the highest mean fluorescence intensity (MFI) was used as reference (100%) and values obtained at lower concentrations were normalized according to the formula: % of binding = ((sample MFI – background MFI)/(maximal MFI – background MFI))×100). Results show MFI of triplicate values +/− SEM and are from one experiment representative of six. (B) BWZ-Ly49H and BWZ-Ly49C cells were incubated with saturating concentrations of m157G1F-Fc for periods ranging from 1 to 40 min at 4°C and the resulting binding was analyzed by flow cytometry. The MFI obtained at 40 min was used as reference (100%) and values obtained after shorter incubation times were normalized according to the formula: % of binding = ((timed MFI – background MFI)/(MFI at 40 min – background MFI))×100). Results show mean of triplicate values +/− SEM and are from one experiment representative of six. (C) Ly49H and Ly49C-expressing BWZ cells were labeled with saturating concentrations of m157G1F-Fc at 4°C; the cells were then washed and incubated at 37°C in the presence of excess concentrations of anti-m157 blocking antibody (6H121) to prevent re-binding of detached m157-Fc. A measure of the MFI prior to the addition of 6H121 provided maximal binding values (100%). Binding decay was measured at times from 5 to 90 min of incubation at 37°C. Binding decay values were calculated using the formula: % of binding = ((timed MFI – background MFI)/(MFI at 0 min – background MFI))×100). Results show mean of triplicate values +/− SEM are from one experiment representative of four. Overall, these results suggest that m157G1F binds to Ly49H and Ly49C with similar affinities but interactions with Ly49H are more sustained.
Binding of m157G1F to Ly49C is limited by cis-interactions with MHC class I molecules
We previously showed that m157G1F binds to NK cells that are stained with the 5E6 mAb (i.e. NK cells expressing Ly49C and/or I) in B6 mice [13]. Here, we further analyzed m157G1F binding to NK cell subsets expressing Ly49H and/or Ly49C. Splenic NK cells were collected from the following mouse strains: B6 (H-2b), Cmv1r (H-2d background expressing B6 alleles in the NK cell receptor gene locus) [19] and B6 β2m ko (defective for MHC class I expression). Purified NK cells were stained with Ly49C-specific 4LO3311 and Ly49H-specific 3D10 antibodies in order to discriminate the respective NK cell subsets. We used the 4LO3311 antibody that is specific for Ly49C rather than 5E6, which also recognizes Ly49I [20]. 4LO3311 failed to stain B6 NK cells while it stained weakly Cmv1r NK cells; in contrast, NK cells from B6 β2m ko mice showed strong staining with 4LO3311 (Fig. 2A, left panels). In B6 mice, self-ligands for Ly49C are MHC class I molecules H-2 Kb; Ly49C can engage H-2 Kb on other cells (trans-interaction) as well as H-2 Kb expressed on the same NK cell membrane plan (cis-interaction) [21]. Cis-binding of the Ly49A inhibitory receptor with MHC class I has been shown to restrict the interactions with molecules presented in trans [22]. Likewise, the staining patterns detected on NK cells isolated from the three mouse strains analyzed were consistent with masking of Ly49C due to cis-interactions with H-2 Kb. In order to determine whether the differential staining illustrated in Fig. 2A were indeed due to cis-masking, we treated NK cells with an acid buffer; this treatment releases the β2m from MHC class I heavy chain, and disrupts cis-interactions [21], [22]. 4LO3311 bound to Ly49C on acid stripped B6 NK cells, while a more intense staining was achieved on Cmv1r NK cells; binding to B6 β2m ko NK cells remained unchanged (Fig. 2A, right panels). These results indicated that Ly49C is masked due to cis-interactions with MHC class I molecules in B6 mice. The existence of a partial masking in Cmv1r suggests that Ly49C also binds to H-2d MHC class I in cis but with a lower affinity than to H-2b molecules.
Fig. 2. Binding of m157G1F to NK cells isolated from various mouse strains. 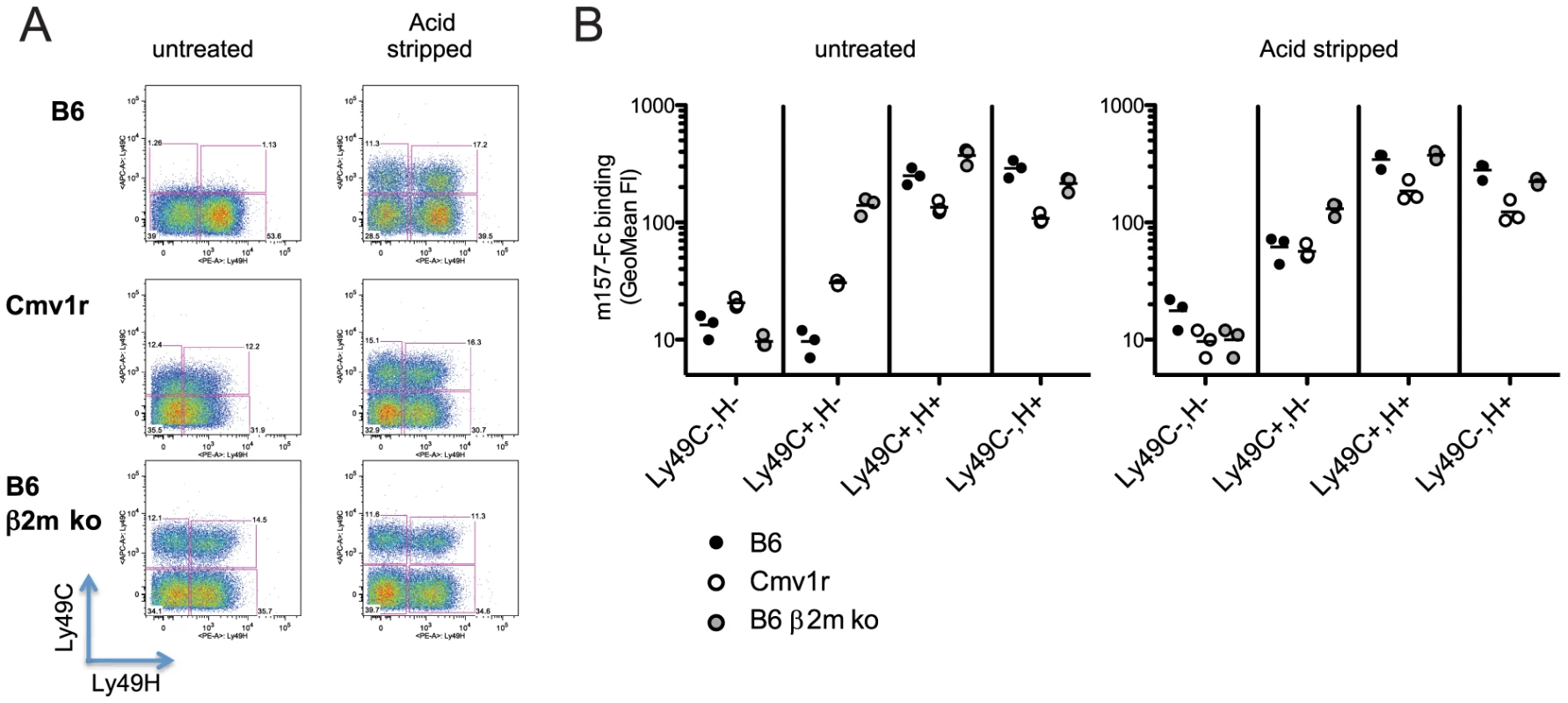
Splenic NK cells were enriched from naïve B6, Cmv1r or B6 β2m ko mice. NK cells were either untreated or acid stripped. (A) NK cells were incubated with fluorochrome-conjugated anti-Ly49C and Ly49H antibodies and analyzed by flow cytometry to discriminate the corresponding cell subsets. (B) NK cells were incubated with m157G1F-Fc and then with fluorochrome-conjugated antibodies to discriminate Ly49H and Ly49C NK cells subsets. The four following subsets were analyzed for m157G1F binding: 1- Ly49C−,H−, 2- Ly49C+,H−, 3- Ly49C+,H+, 4- Ly49C−,H+. Horizontal bars represent the mean values of m157G1F binding measured from three individual mice (each represented as a circle). Data are from one experiment representative of three performed. Next, we analyzed the binding of m157G1F to NK cell subsets expressing Ly49H and/or Ly49C (Fig. 2B). Using B6 β2m ko cells where Ly49C is free of cis-masking, m157G1F binding was detected to both Ly49H and Ly49C-expressing NK cell subsets. In B6, m157G1F binding was achieved only in the NK cell subsets expressing Ly49H, while in Cmv1r, m157G1F displayed weak binding to the Ly49C+, Ly49H− (Ly49C+,H−) NK cell subset. Acid stripping improved m157G1F binding to the Ly49C+,H− subset from B6 and Cmv1r and also resulted in enhanced binding to the subset co-expressing both receptors. The absence of m157G1F binding to untreated Ly49H− NK cells, despite a fraction of these cells expressing Ly49C, indicated that m157G1F cannot bind Ly49C when interactions in cis made it unavailable. As a control, we also incubated untreated and acid treated NK cells with the secondary antibody and with streptavidin in the absence of the first m157-Fc incubation step. We did not detect any fluorescence above the background, indicating the specificity of our multi-step staining in both untreated and acid treated NK cells (Fig. S2).
Consistent with the data we obtained using cell lines expressing NK cell receptors (Fig. 1), these results indicate that m157G1F can bind to NK cells expressing Ly49C and/or Ly49H. However, the ability to engage Ly49C is limited by MHC class I molecules (H-2b>H-2d) due to cis interactions.
Ly49C can limit Ly49H-dependent activation by m157
We previously demonstrated that m157G1F induces intracellular signals upon binding to both Ly49H and Ly49C [14]. Here, we analyzed whether m157G1F differentially activates NK cells that express Ly49H only or coexpress Ly49H and Ly49C. B6 NK cells were exposed to target cells expressing m157G1F (RMA m157G1F) or to parental RMA cells and analyzed for degranulation and production of IFN-γ (Fig. 3A). Upon exposure to RMA m157G1F, Ly49H+,C− NK cells degranulated (∼40% LAMP1+) and produced IFN-γ (∼15% IFN-γ+). NK cell activation was Ly49H-dependent as it was abolished in the presence of blocking 3D10 antibodies. However, co-expression of Ly49C did not alter the response. We hypothesized that cis-masking of Ly49C by H-2 Kb was responsible for the absence of inhibition. We therefore examined NK cells isolated from B6 β2m ko mice, where Ly49C receptors are not masked in cis (Fig. 2A). We used RMAS cells (MHC class I deficient) expressing m157G1F (RMAS m157G1F) as targets instead of RMA m157G1F cells in order to prevent competition between m157 and Kb for binding to Ly49C, thus, ensuring that inhibition through Ly49C would only be due to m157 binding. RMAS m157G1F cells strongly activated Ly49H+,C− NK cells but not the Ly49H+,C+ subset. As above, activation was Ly49H-dependent. Blocking of Ly49C enabled Ly49H+,C+ NK cell activation, albeit not as strongly as the Ly49H+,C− subset (Fig. 3B).
Fig. 3. m157G1F–Ly49C binding regulates Ly49H-dependent NK cell functions ex vivo. 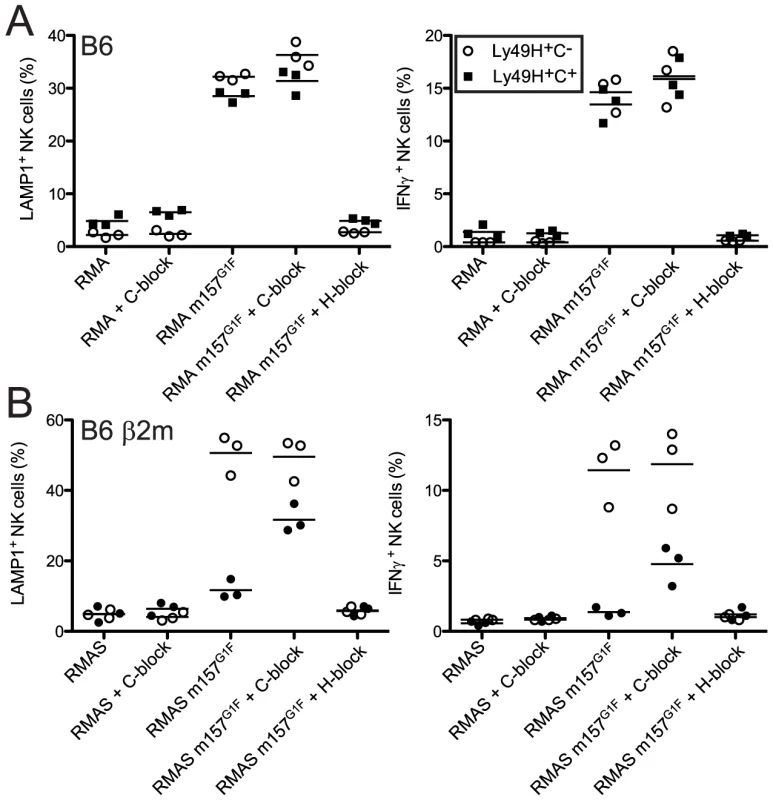
Splenic NK cells were enriched from naïve B6 wt (A) or B6 β2m ko (B) mice and exposed to parental or m157G1F expressing RMA (A) or RMAS cells (B). Excess concentrations of anti-Ly49C (4LO3311) or Ly49H (3D10) were added where indicated to block the corresponding receptors. At the end of the coculture, cells were stained with fluorochrome-conjugated antibodies to discriminate NK cell subsets (Ly49H+,C− (open circles) and the Ly49H+,C+ (filled squares)) and detect degranulation (LAMP-1, left panels) and IFN-γ production (right panels). Horizontal bars represent the mean values from three individual mice (each represented as a symbol). These data are from one experiment representative of three performed. Therefore, NK cell exposure to m157G1F-expressing cells triggers optimal Ly49H-dependent NK cell functions when Ly49C is not co-engaged. These results demonstrate that Ly49C can restrict Ly49H-dependent activation upon engagement of m157, but this inhibition is itself regulated in cis by MHC class I molecules.
Ly49C dampens NK cell activation by inhibiting cytoskeleton polymerization
Next we evaluated whether m157 binding to Ly49C affects the stability of cell conjugates formed between NK cells and their targets thereby preventing subsequent killing. B6 NK cells were expanded in IL2-containing medium and four subsets were sorted based on Ly49C and Ly49H expression (1. Ly49H−,C−, 2. Ly49H−,C+, 3. Ly49H+,C+ and 4. Ly49H+,C−). After further expansion, to eliminate residual fluorescence resulting from the sorting step, NK cells and target cells were labeled using intracellular dyes, CFSE and CMTMR respectively. NK cells were then exposed to various target cells and the formation of stable cell conjugates was assessed. Because B6 NK cells interactions with RMA cells do not result in killing, RMA cells were used as negative controls. RMA m157G1F were excluded as targets because they express Kb that can bind Ly49C and it would have been impossible to determine whether the effects of Ly49C on conjugate stability were due to engagement of m157 or Kb. Instead, we used MHC class I-deficient RMAS m157G1F cells. RMAS cells were also tested; they are killed by B6 NK cells due to “missing self recognition”, as described by [23], which is Ly49H-independent. Each of the four NK cell subsets indicated in Fig. 4A was exposed to target cells for 5 and 20 min and then the percentage of conjugated NK cells was analyzed by flow cytometry. In all four subsets, the fraction of NK cells conjugated with RMA cells remained below 20% (Fig. 4A, left panel). Using RMAS as targets, conjugation levels with NK cells expressing Ly49C increased to 40%, while they remained around 20% with the Ly49C− NK cells (Fig. 4A, middle panel). In B6 mice, Ly49C is involved in NK cell education [15], its expression resulting in a higher reactivity. This is consistent with the increased conjugate rates obtained when Ly49C+ NK cell subsets were exposed to RMAS cells. Interestingly, increased formation of conjugates by Ly49H−,C+ NK cells with RMAS (up to 40%, Fig. 4 A middle panel) were abolished when this subset was exposed to RMAS m157G1F (Fig. 4A right panel). This suggests that binding of m157G1F to Ly49C compensated for the absence of Kb and prevented “missing-self recognition”. Incubation of Ly49H+ NK cells with RMAS m157G1F resulted in high conjugation rates regardless of whether Ly49C was co-expressed (Fig. 4A, right panel). We hypothesized that the similar frequencies of cell conjugates detected in Ly49H+,C+ and Ly49H+,C− B6 NK cells with RMAS m157G1F (which suggested an absence of inhibitory effect mediated by Ly49C) was due to cis-masking by MHC class I molecules. Therefore, we isolated NK cells from B6 β2m ko mice to address this hypothesis. Although β2m ko mice are deficient for MHC class I expression, their NK cells undergo an education process that enables them to respond to MCMV infection as efficiently as wild type mouse cells (our unpublished results and [24]), while they are tolerant toward cells displaying altered MHC I expression. As described above, we sorted four NK cell subsets and exposed them to RMAS and RMAS m157G1F cells. The percentages of NK cells conjugated to RMAS cells remained around 20% for the four subsets. This result was consistent with NK cell tolerance toward MHC class I-devoid RMAS cells (Fig. 4B, left panel) in β2m ko mice. Incubation of RMAS m157G1F with the subsets expressing Ly49H resulted in increased conjugates, although to a lesser extent than seen with B6 NK cells (Fig. 4A and B). In addition, the Ly49H+,C+ NK cell subset demonstrated a reproducibly lower conjugation rate with RMAS m157G1F than the subset expressing Ly49H only (Fig. 4B, right panel).
Fig. 4. m157G1F hampers NK cell-target conjugate formation and cytoskeleton polymerisation. 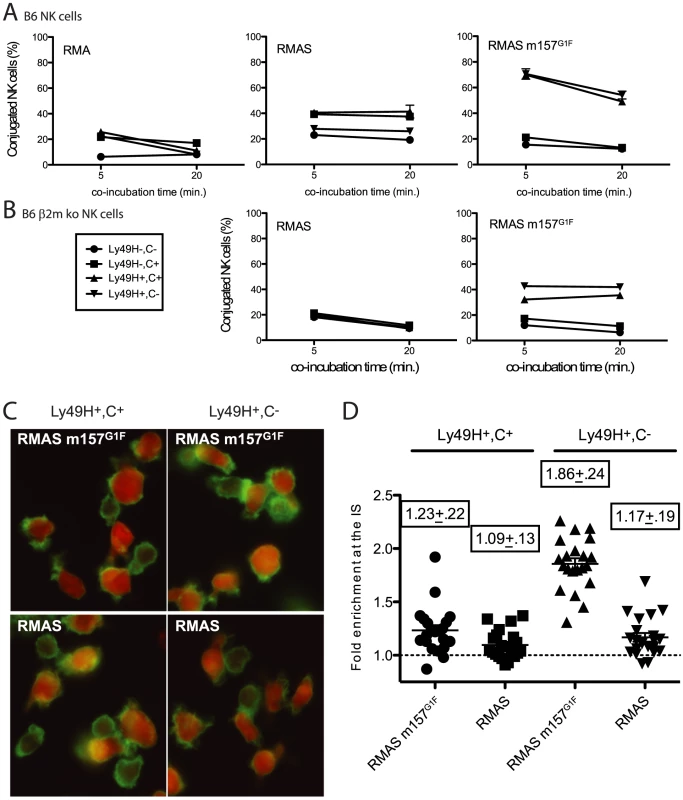
IL-2 expanded NK cells isolated from B6 wt (A) and B6 β2m ko mice (B) were sorted into four subsets according to Ly49H and Ly49C expression (1- Ly49H−,C−; 2- Ly49H−,C+; 3- Ly49H+,C+; and 4- Ly49H+,C−. They were further expanded in IL-2 containing medium to restore expression of antibody-free receptors before being tested for conjugate formation with the following target cells: RMA, RMAS or RMAS m157G1F. NK cells were labelled with CMTMR and targets with CFSE, then mixed and incubated for 5 or 20 min at 37°C. Percentages of conjugated NK cells were determined by flow cytometry. (C) Sorted B6 β2m ko NK cells (Ly49H+,C+: left panels; Ly49H+,C−: right panels) were exposed to CMTMR-labelled RMAS (bottom panels) or RMASm157G1F (top panels) for 15 min at 37°C. Polymerised cytoskeleton actin was detected using phalloidin-FITC and fluorescence microscopy analysis. (D) Enrichment of cytoskeleton F-actin at the cell contact area was quantified in the samples described in C using ImageJ software. Horizontal bars represent the mean of individual enrichment values (each symbol illustrates an individual conjugate). Mean values +/− SD for each condition tested are indicated above the symbols. These data are from one experiment representative of three performed. These results suggest that “missing-self recognition” and Ly49H-dependent signals synergize, resulting in more conjugated NK cells (70% when NK cells were from B6 wt mice, Fig. 4A), while the interactions exclusively due to missing self-recognition (Fig. 4A middle panel) or to Ly49H engagement (Fig. 4B right panel) resulted in more modest conjugation rates (40%). Thus, m157G1F binding to Ly49C disrupts the cell conjugates formed in the context of “missing self recognition”, but a simultaneous engagement of Ly49H can override this inhibition.
Interactions between NK cells and target cells that trigger killing events require the formation of stable cell conjugates [25]. During this process, cytoskeleton polymerization orients the cytotoxic granules toward the immunological synapse. Polymerization of the cytoskeleton actin into F-actin can be detected by microscopy using fluorescent phalloidin. B6 β2m ko NK cells were sorted according to their Ly49H and Ly49C expression as above, then were incubated with CMTMR-labeled RMAS or RMAS m157G1F cells and tested for actin polymerization. NK cell subsets expressing Ly49H but not Ly49C displayed a strong polarization of their cytoskeleton toward RMAS m157G1F (fig. 4C, upper right panel) whereas NK cells co-expressing Ly49H and Ly49C did not (fig. 4C, upper left panel). This process was m157-dependent as incubation with parental RMAS cells did not trigger actin polymerization (fig. 4C, lower panels). Quantification of F-actin within the immunological synapse further supported this result, confirming cytoskeleton accumulation toward m157G1F expressing cells within the Ly49H+,C− NK cell subset (fig. 4D). Thus, m157 binding to Ly49C reduces the level of cytoskeleton actin polymerization and accumulation toward the target cell.
Replication of MCMV m157G1F in Ly49H+ mice
We previously showed that the MCMV K181 laboratory strain in which m157 was substituted by m157G1F (MCMV m157G1F), replicates in B6 mice at a similar rate to the MCMV K181 wt virus [13]. In B6 mice, Ly49C is mostly unavailable for binding m157 due to cis-interaction with H-2 Kb molecules, as illustrated in Fig. 2. We analyzed MCMV m157G1F replication in a H-2d background using Cmv1r mice (Ly49H+) where cis-interactions are weak (Fig. 2). Mice were infected with either MCMV K181 wt, MCMV m157G1F or with a recombinant virus with m157 deleted (MCMV Δm157) and measured viral titers in the spleen, liver, lungs and salivary glands at various times post infection. Viral loads we measured in Cmv1r mice were similar to those observed previously in B6 mice (Fig. 5 and [13]). Only MCMV Δm157, which escapes NK cell surveillance, replicated at high titers while MCMV K181 wt and MCMV m157G1F were similarly controlled. A possible explanation is that, although weak in Cmv1r, cis-masking of Ly49C restrained sufficiently its ability to engage m157G1F, and so impeded its inhibitory effect over Ly49H-dependent activation. In addition, as most Ly49H+ NK cells do not co-express Ly49C in Cmv1r mice (as in B6) and therefore cannot be inhibited by m157, we proposed that this subset ensured virus elimination and compensated for a likely impaired response of the Ly49H+,C+ subset. To test this hypothesis, we generated a Ly49C transgenic mouse strain (B6 Ly49C Tg) in which all NK cells express this inhibitory receptor.
Fig. 5. Replication kinetics of MCMVm157G1F in Cmv1r mice. 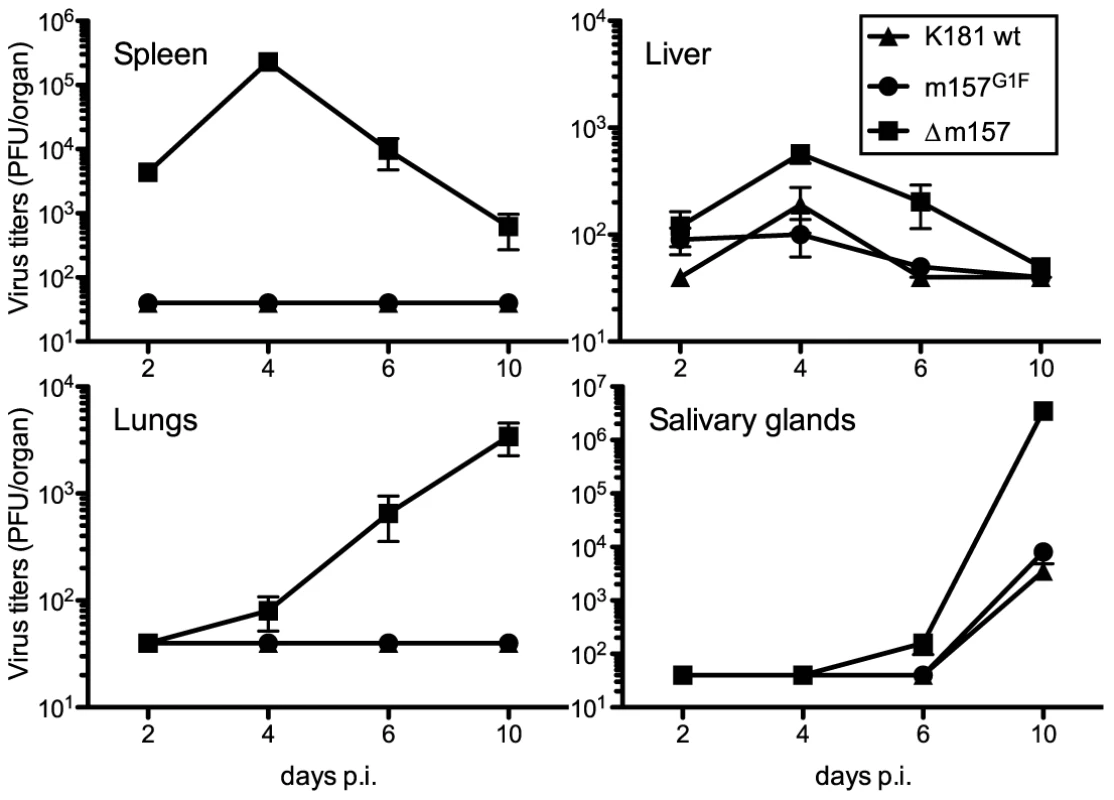
Cmv1r mice were infected with 1×104 PFU of MCMV K181 wt (triangles), MCMVm157G1F (circles) or MCMVΔm157 (squares) viruses and the organs indicated were harvested at days 2, 4, 6 and 10 p.i. Viral titers were determined from organ homogenates by standard plaque assay. Bars represent average values in each organ collected from 3 mice per group +/− SEM. Data are from one experiment representative of two performed. Replication of MCMV m157G1F in B6 Ly49C Tg mice
We generated the B6 Ly49C Tg mouse strain as indicated in the Materials and Methods section. Analysis of blood-borne NK cells indicated than over 95% expressed Ly49C (Fig. 6A). Ly49C was also found in over 50% of T cells and in most NKT cells; it was also expressed in a small fraction of B cells (<20%, data not shown), but in none of the other leukocyte populations (data not shown). The transgenic mice and negative littermates had splenic and bone marrow compartments of similar size (Fig. S3). Wild-type and transgenic spleens contained comparable frequencies of B cells, NKT cells and DCs; the T cell fraction was slightly reduced in the transgenic mice, while monocytes/macrophages, neutrophils and eosinophils were slightly increased (Fig. S3). Most importantly, the size of the Ly49H+ NK cell subset was unchanged (Fig. S4). A fraction of Ly49C receptors was found to be masked by cis interactions (Fig. S4).
Fig. 6. Infection of B6 Ly49C Tg mice by MCMVm157G1F. 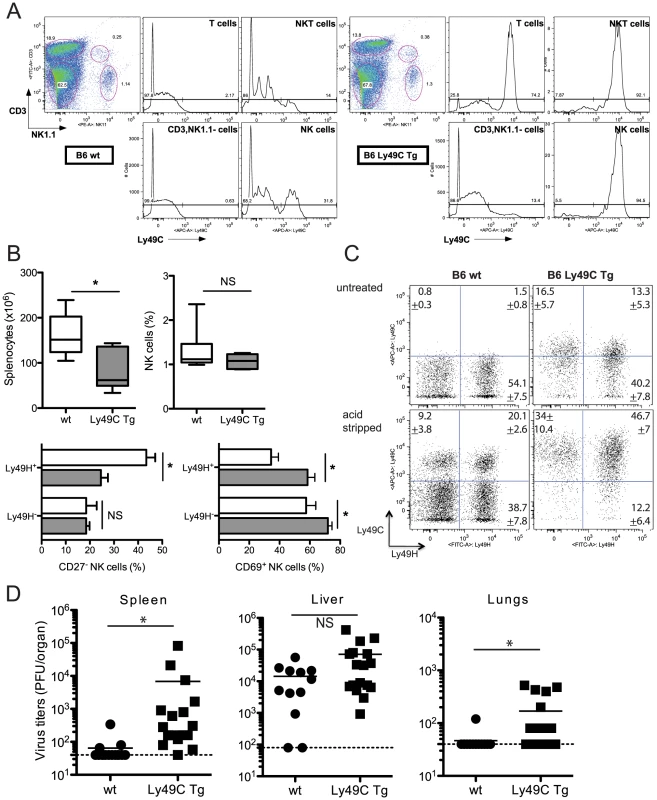
(A) Blood borne lymphocytes from Ly49C Tg B6 mice (right panels) or negative littermates (left panels) were exposed to acidic treatment and stained with fluorochrome conjugated antibodies for flow cytometric analysis. The histograms illustrate Ly49C expression in NK cells, NKT cells, T cells and CD3−, NK11− leukocytes. (B,C,D) Ly49C Tg and wt B6 mice were infected with 5×104 PFU of MCMVm157G1F. (B.C) Four days p.i. splenic NK cells were analysed by flow cytometry. Box and Whiskers graphs represent the range of individual mouse values spanning from 5 to 95 percentiles; they comprise 11 mice per group combining three independent experiments (B). (C) Dot plot analysis of untreated or acid stripped splenic NK cells. Values indicate percentages +/− SEM for each of the four cell subsets calculated from 11 mice per group that were analysed in three independent experiments. (D) Alternatively, spleen, liver and lungs were collected at d4 p.i. to measure virus titers by standard plaque assay. Horizontal bars represent the mean of individual values; each mouse is illustrated as a symbol (n = 12 B6 wt and n = 16 B6 Ly49C Tg). Differences between the groups were compared using the Mann-Whitney U test: *: p<0.05, NS: not significant). Ly49C Tg and negative littermates were infected with MCMV m157G1F and viral titers and NK cell activation tested after 4 days. The spleens in transgenic mice were not as enlarged as in wt mice upon infection, consistent with previous studies in Ly49H-devoid mouse strains [26], [27], although the percentage of splenic NK cells was similar (Fig. 6B). The frequency of Ly49H+ NK cells in wt and transgenic mice were also identical (Fig. 6C). CD27 dissects peripheral NK cells into two major subsets: naïve NK cells express CD27, while this marker is downregulated in activated and more mature NK cells [28]. We found that the frequency of the CD27− cells within Ly49H+ NK cells was higher in B6 wt than in Tg mice, while it was similar in Ly49H− NK cells (Fig. 6B). This suggests that Ly49H dependent activation was reduced in Ly49C Tg mice. We also analyzed the expression of the activation marker CD69 in Ly49H positive or negative NK cells. We found that CD69 was more increased in Tg than in wt mice, most particularly in Ly49H− NK cells (Fig. 6B). This sustained activation did not require Ly49H and was most likely due to higher cytokine levels associated with uncontrolled viral proliferation. Viral loads were measured in the spleen, liver and lungs after 4 days. A high dose of virus (5×104 PFU) was used so that viral titers found in wt mouse tissues would be just above the detection limit. Ly49C Tg mice had increased viral titers in the spleen and lungs; there was a trend for increased hepatic loads although the difference was not statistically significant (Fig. 6D). Thus expression of Ly49C on all NK cells results in an inhibitory effect of the immunoevasin m157 leading to higher viral replication in vivo.
Ly49C cis-masking was not as complete in B6 Ly49C Tg as in wt mice (Fig. 6C); we predicted that if more Ly49C receptors were rendered available in the absence of cis-interactions, the inhibitory effect on NK cells would be more intense and would lead to even more severe viral replication.
Replication of MCMV m157G1F in B6 Ly49C Tg, β2m ko mice
We crossed B6 Ly49C Tg mice with B6 β2m ko mice and obtained B6 Ly49C Tg, β2m ko double mutant mice. These were used to test in vivo the anti-viral NK cell response in the absence of Ly49C cis-masking. As in B6 Ly49C Tg mice, expression of the Ly49C transgene in the double mutant mice was limited to NK cells, NKT cells, most T cells (Fig. 7A) and a small fraction of B cells (data not shown). Ly49C was not detected elsewhere, and had little impact on the leukocyte population sizes (Fig. S5). B6 Ly49C Tg, β2m ko mice were infected along with B6 β2m ko and B6 Ly49C Tg using 5×104 PFU MCMVm157G1F, as previously. B6 Ly49C Tg, β2m ko mice were highly susceptible to viral infection with 3 out of 4 of these mice succumbing to infection by day 3. Infection with a lower inoculum (5×103 PFU) was performed and the viral titers were measured in the spleen, liver and lungs after 4 days (Fig. 7B). Splenic viral loads were higher in B6 Ly49C Tg, β2m ko mice than in B6 Ly49C Tg mice. Viral replication was completely controlled in B6 β2m ko mice, indicating that the absence of cytotoxic CD8 T cells due to the lack of MHC class I was not responsible for the impaired immune response at this early stage of acute infection.
Fig. 7. Infection of B6 Ly49C Tg β2m ko mice by MCMVm157G1F. 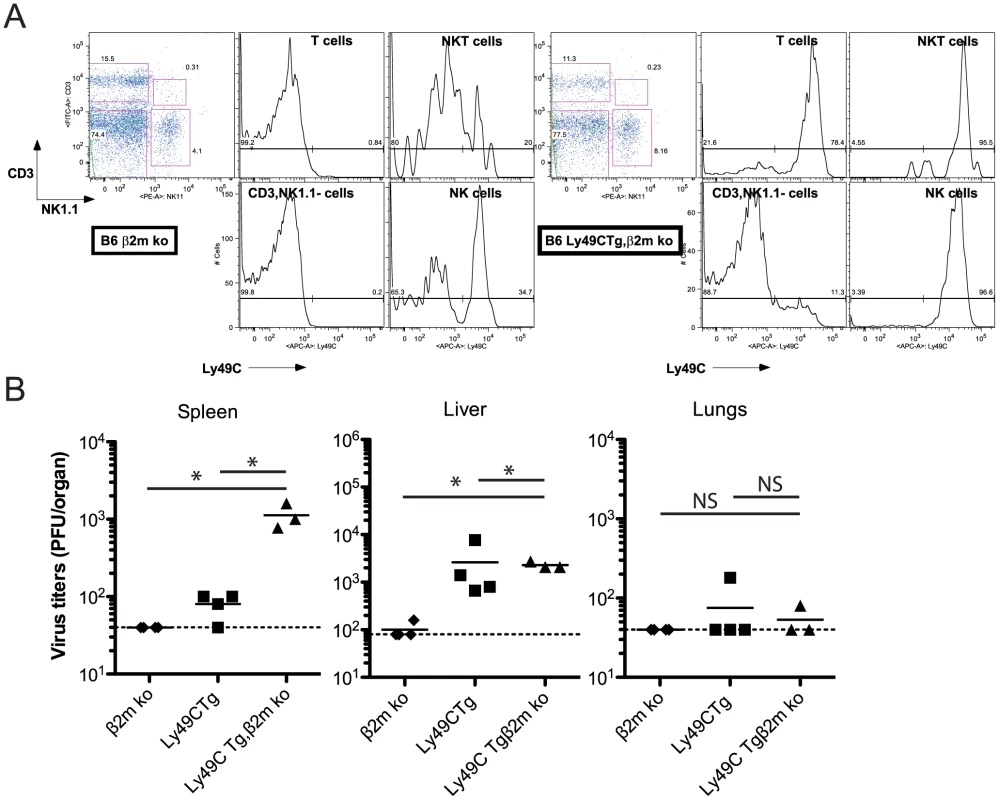
(A) Blood borne lymphocytes from Ly49C Tg, β2m ko B6 mice (right panels) or B6 β2m ko mice (left panels) were stained with fluorochrome conjugated antibodies for flow cytometric analysis. Histograms illustrate Ly49C expression in NK cells, NKT cells, T cells and CD3−, NK11− leukocytes. (B) B6 Ly49C Tg, β2m ko and B6 β2m ko mice were infected with 5×103 PFU of MCMVm157G1F. Four days p.i. the spleen, liver and lungs were collected to measure virus titers by standard plaque assay. Horizontal bars represent the mean of individual values; each mouse is illustrated as a symbol. Horizontal bars represent the mean of individual values, each mouse is illustrated as a symbol; groups are constituted of 6–8 mice. Differences between the groups were compared using Mann-Whitney U test: *: p<0.05, NS: not significant). We concluded that inhibitory Ly49C receptor is most efficient at negatively regulating NK cell functions when it are not engaged through cis-interactions and that targeting of this receptor by the m157 immunoevasin severely impairs the Ly49H-dependent response.
Inhibition of Ly49H-independent NK cell activation via m157G1F-Ly49C binding
Our results illustrated in Fig. 4A,B indicated that m157G1F binding to Ly49C could modulate “missing self recognition”. We investigated this possibility further in TC1 congenic mice; this strain has a B6 background, but has BALB/c alleles in the Ly49 cluster of the NKC and thus lacks Ly49H, as described in [19]. Missing-self recognition dependent killing in the B6 background is mainly mediated by Ly49C+ NK cells [16]. We hypothesized that RMAS m157G1F target cell killing by TC1 NK cells would be impaired due to Ly49C-mediated inhibition, even though the targets were devoid of MHC class I molecules.
TC1 NK cells were expanded in IL-2 containing medium and tested against RMAS and RMAS m157G1F in an in vitro killing assay. As expected, NK cells killed RMAS targets. By contrast, they spared RMAS m157G1F cells (Fig. 8A, left panel). Addition of excess concentrations of Ly49C-blocking antibody enabled NK cells to kill RMAS m157G1F targets (Fig. 8A, right panel). These results suggest that engagement of Ly49C by m157G1F induces inhibitory signals that compensate for the absence of MHC class I, and thereby prevents NK cell cytotoxicity. We then tested in vivo whether m157G1F could affect the control of MCMV infection mediated by NK cells in a Ly49H-independent manner. Some MCMV genes down-regulate MHC class I expression in infected cells [29]. Previous studies showed that levels of expression of MHC class I can affect NK cell responses during MCMV infection [30], [31]. We hypothesized that NK cell killing of infected cells with reduced MHC class I expression would be prevented by m157G1F binding to Ly49C. We infected Ly49H-devoid mice with MCMV m157G1F or with MCMV Δm157 and compared the replication of the two viruses; infected mice displayed similarly high titers in the spleen, liver and lungs after 4 days (Fig. 8B). We hypothesized that strong inflammatory conditions would promote a better anti-viral NK cell response. Administration of alpha-galactosyl ceramide (αGC) at the time of infection improves NK cell mediated control of MCMV in Ly49H-deficient mice; this effect is NKT cell independent [32]. Next, we administered αGC in combination with MCMVm157G1F or Δm157 and measured viral titers 4 days later. Treatment using αGC resulted in reduced replication of Δm157 virus in all organs (Fig. 8B). TC1 mice treated with αGC had similar viral titers following infection irrespective of which of the two viruses was used (Fig. 8B). We predicted that the absence of an effect associated with m157G1F expression was due to cis-masking of Ly49C. Therefore, we repeated the experiment in CT6 mice (H-2d, Ly49H−) [19], where cis-interactions are weak, as shown in Fig. 2. In this H-2d background, although Ly49C is not responsible for self recognition, m157G1F can bind the inhibitory receptor [13] and induce inhibitory signals. Infection with the Δm157 virus combined with αGC treatment was almost completely controlled, while the protection conferred by αGC was only partial after infection with the m157G1F virus. We also tested B6 β2m ko mice whose NK cell are tolerant to cells with reduced MHC class I expression and where Ly49C availability is not hampered by cis-masking. Only the Δm157 virus was tested in this mouse strain as the m157G1F virus would have triggered a dominant Ly49H-dependent response. Contrary to those measured in TC1 mice, identical Δm157 virus titers were measured with and without αGC treatment (Fig. 8B). These results are consistent with a Ly49C-mediated inhibition of a Ly49H-independent NK cell response; it is achieved upon binding of the m157 immunoevasin and is regulated through cis-interactions.
Fig. 8. m157G1F regulates Ly49H-independent NK cell activation. 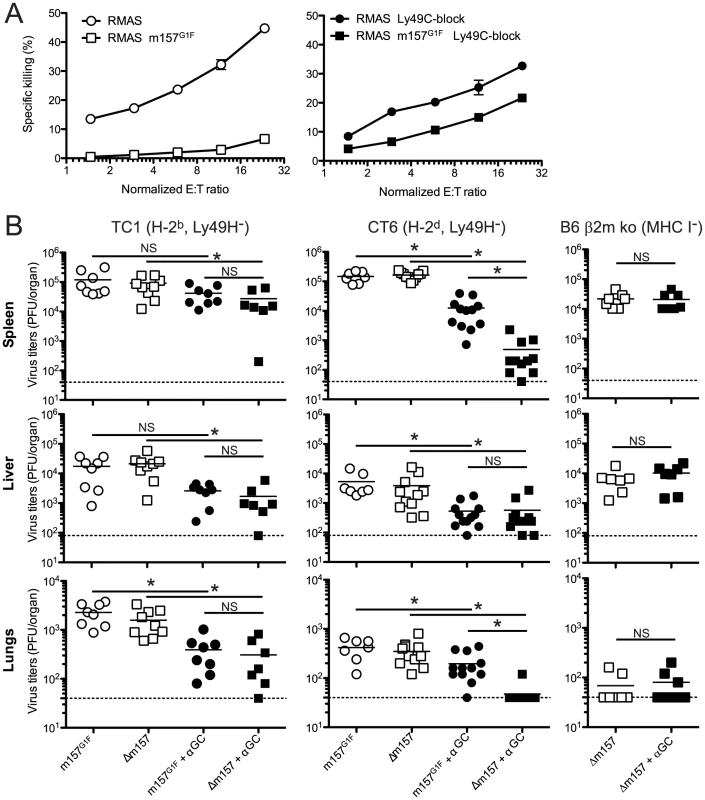
(A) Splenic NK cells were isolated from TC1 mice (H-2b, Ly49H−) and expanded in IL-2 containing medium. NK cells were left unblocked (left panel) or were incubated with anti-Ly49C (4LO3311) blocking antibody (right panel) and co-cultured for 4 h with 51Cr-loaded RMAS or RMAS m157G1F target cells at the indicated effector∶target ratios (E∶T). Each sample was tested in triplicate. The data illustrated represent means +/− SD and are from one experiment representative of four performed. (B) TC1 (H-2b, Ly49H−; left panels), CT6 (H-2d, Ly49H−; middle panels) or B6 β2m ko mice (no MHC I, Ly49H+; right panels) were infected with 1×104 PFU of MCMVm157G1F or MCMVΔm157; the viruses were administered alone or in combination with 2 µg alpha-galactosylCeramide (αGC). Four days p.i., spleen, liver and lungs were collected and viral titers within these organs were measured by standard plaque assay. Bars represent mean values from 7–12 mice/group pooled from 3 independent experiments, the dotted lines represent the limit of detection. Differences between the groups were compared using the Mann-Whitney U test: *: p<0.05, NS: not significant). Altogether, our results demonstrate that engagement of the inhibitory Ly49C receptor by the m157 immunoevasin inhibits NK cell activation leading to failure to control MCMV infection, and that Ly49C inhibitory function is modulated by MHC I molecules through cis-masking.
Discussion
We previously showed, and confirmed in this study, that m157 from the K181 and the G1F MCMV strains bind Ly49H with similar affinities [13]. Here, we analyzed the kinetics of binding of m157G1F to Ly49H or Ly49C. We found that the kinetics of association of m157G1F to Ly49C was slightly quicker than to Ly49H. In addition, the binding profiles suggested similar affinities for the two receptors, consistent with the results obtained by surface plasmon resonance analysis [14]. Kinetics of dissociation of m157 indicated that interactions with Ly49H were more sustained. This could suggest that after some degree of initial inhibition, Ly49H activation would proceed unchallenged when NK cells coexpress the two receptors. However, since Ly49C is not internalized upon engagement, unlike Ly49H, it is likely that the Ly49C receptors that have dissociated from m157 will establish new interactions and induce inhibitory signals as long as the NK cell-target pairs remain conjugated. Nevertheless, these different dissociation rates could lead to competition in a mixed population where some NK cells express exclusively Ly49H or Ly49C that would ultimately favor Ly49H+ engagement and thus lead to an efficient anti-viral NK cell response. This could explain why infection with either MCMV K181 or MCMV m157G1F resulted in similar viral replication in B6 [13] and in Cmv1r mice as shown here.
Analysis of m157 binding to NK cells revealed another degree of complexity in the parameters at play due to NK cell-borne MHC class I molecules able to interact in cis with Ly49C [21]. These cis-interactions impeded detection of Ly49C+ NK cells using the anti-Ly49C antibody 4LO3311 and prevented m157G1F binding. This lack of binding was reversed by acid treatment. Identical binding patterns obtained with the 4LO3311 antibody and m157 are consistent with the findings that both bind the stalk domain of Ly49C [14], [20]. Ly49C binding to Kb in trans requires a back-folded configuration, while binding in cis requires an extended configuration [33]. Interestingly, our recent findings show that m157G1F binds the Ly49C stalk region when the receptor adopts an extended configuration [14]. As the residues involved in Ly49C interactions with Kb in cis and m157 are distinct [14], one would predict that Ly49C may simultaneously bind both m157 and Kb. However, our data indicate that these two interactions are mutually exclusive, as no m157 binding was detected when Ly49C was engaged by MHC class I in cis, and no inhibition of NK cell functions was achieved. Hence, Ly49C-dependent inhibition of NK cell response upon infection with MCMVm157G1F was not as severe when the receptors could be masked in cis (infection of Ly49C Tg mice vs Ly49C Tg, β2m ko mice). It is possible that structural constraints in the Ly49C stalk domain that are incompatible with binding of m157 result from cis-binding to Kb.
Inhibitory Ly49 receptors, such as Ly49C, contain intracellular immunoreceptor tyrosine-based inhibitory motifs (ITIM). When phosphorylated, ITIMs recruit phosphatases that subsequently suppress phosphorylation-based activation signaling [34]. Trans-binding to inhibitory Ly49 induces ITIM phosphorylation, however, it is likely that binding in cis does not generate such a tonic inhibitory signal. Rather, cis-interactions restrict the pool of Ly49 receptors that are available for functional interaction with ligands on target cells [33]. Our functional assays highlighted an important role for cis interaction in regulating anti-viral NK cell responses. Using B6 NK cells where cis-interactions largely prevent binding of m157 to Ly49C, we detected an un-inhibited Ly49H-dependent NK cell activation characterized by IFN-γ production and degranulation. Conversely, activation of the Ly49H+,C+ NK cell subset was almost completely abolished in B6 β2m ko NK cells where Ly49C/MHC cis-interactions are absent. These functional results correlated with the binding patterns of the m157-Fc and supported a role for MHC class I molecules in controlling Ly49C inhibitory functions in cis. To determine whether Ly49C could control the stability of cell conjugates formed through Ly49H engagement, we measured the formation of conjugates when Ly49H+ NK cells were exposed to m157-expressing targets and analyzed F-actin polymerization and polarization towards the targets; these events indicate NK cell activation and lead to formation of the cytolytic synapse that is required to mediate killing [35], [36]. Fewer conjugates were formed and cytoskeleton polymerization did not occur in B6 β2m ko NK cells expressing Ly49C. Thus, engagement of Ly49C in trans leads to inhibitory signals that destabilize cell conjugation and impairs cytoskeleton reorganization.
Our initial pathogenesis studies showed equivalent viral replication in B6 [13] and Cmv1r mice infected with MCMV expressing m157K181 or m157G1F, despite the fact that m157G1F binds both inhibitory and activating receptors on NK cells, while m157K181 binds only activating receptors. We produced a transgenic mouse in which all NK cells expressed Ly49C. Expression of the activating Ly49H receptor remained unchanged; however, these mice had an increased number of NK cells in the spleen. Despite the presence of more NK cells expressing Ly49H, infection of B6 Ly49C Tg mice resulted in higher viral loads compared to those seen in B6. Analysis of NK cells in infected B6 Ly49C Tg mice showed that only 30% displayed detectable Ly49C, consistent with the regulatory role played in cis by MHC class I molecules. To determine the effect of cis-masking on Ly49C-induced NK cell inhibition, we bred B6 Ly49C Tg with B6 β2m ko mice and generated B6 Ly49C Tg, β2m ko double mutant mice. These mice proved to be highly susceptible to MCMV infection and when infected with the usual dose of 5×104 PFU of virus succumbed to infection within 3 days. This is consistent with a LD50 of 6.4×104 PFU seen in BALB/c mice (that are devoid of Ly49H receptor) infected with the MCMV Smith strain [37] and suggests that Ly49C can completely obliterate Ly49H function in the absence of cis-masking. These results confirm the regulation exerted by cis interactions on NK cells functions in vivo.
Beside their inhibitory function in mature NK cells, inhibitory Ly49 receptors play a crucial role during NK cell education. NK cells acquire functional competence (are licensed) upon Ly49 interactions with MHC class I molecules [15]. Educated cells elicit stronger responses than NK cell subsets devoid of self-specific receptors [15], [16]. In B6 mice, licensing requires engagement of the inhibitory receptor Ly49C and signaling via the intracellular ITIM. Ly49C+ NK cells from B6 wt but not β2m ko mice produce IFN-γ in response to NK1.1 stimulation [15]. However, another study indicated that Ly49H+ NK cells robustly proliferate and produce IFN-γ in both B6 β2m ko and wt mice in response to MCMV infection [24]. We addressed NK cell responsiveness in situations where Ly49C plays a role (B6 origin) or not (B6 β2m ko origin) in NK cell education and confirmed that the NK cell response to MCMV does not require education through Ly49C. Furthermore, it was shown that unlicensed NK cells (Ly49C−) dominate the response to MCMV infection [17]. In B6 mice, Ly49H-dependent NK cell activation was shown to be stronger in the Ly49C/I− subset, while Ly49C/I+ NK cells responded better to MHC class I deficient targets. Our experiments were conducted is similar settings to those described by Orr et al. albeit our target cells expressed m157G1F and not m157Smith; we obtained similar IFN-γ production and degranulation in Ly49C+,H+ and Ly49C−,H+ NK cells from B6 mice. However, using B6 β2m ko NK cells, the Ly49C-expressing subset elicited a limited response, which was increased upon blocking of Ly49C using antibodies, even though it remained weaker than with the Ly49H+,C− NK cells. These results suggest that the Ly49C+ subset had a lower intrinsic activity. The mechanisms responsible for the hyporesponsiveness of the Ly49C+ subset remain to be clarified.
Our data illustrated in figure 8A show a decreased killing of the untransfected RMA-S target cells when excess concentration of Ly49C blocking antibody was added to TC1 NK cells in vitro; this result was surprising because the blocking treatment should not interfere with binding to Ly49C of any ligand present on RMA-S in these settings. Instead, we propose that the decreased killing of RMA-S cells was due to a differential clusterization of the Ly49C receptor in untreated and anti-Ly49C treated NK cells. Indeed, it has been shown that restraints due to cis interactions limit the redistribution of Ly49A to the synapse, whereas unbound Ly49A dampens NK cell activation [38], [39]. Similarly, Ly49C receptors engaged in cis by MHC I molecules are likely to be contained in micro-clusters. Addition of excess concentration of blocking Ab may disrupt cis interactions, resulting in a re-localisation of the freed Ly49C to the immune synapse next to activating receptors where they can interfere with activating signaling pathways. Indeed, ITIM-bearing MHC I specific receptors such as Ly49C require activating receptors to be in the vicinity to perform their inhibitory function [40].
Interestingly, Ly49C engagement by m157G1F had an inhibitory effect on a Ly49H-independent pathway, most likely due to “missing-self recognition” (Fig. 4 and 8). This activation pathway plays a role in NK cell mediated control of MCMV infection [30]. To counter this, MCMV encodes a number of proteins (m04, m06, m152) that interfere with MHC expression [41]–[43]. In mouse backgrounds where Ly49C is involved in NK cell education, such as B6 mice, Ly49C+ NK cells are expected to represent a major subset that responds to infected cells with reduced MHC class I expression. We found that MHC class I-deficient target cells expressing m157 (RMAS m157G1F) avoided killing by B6 NK cells in vitro. Similarly, in vivo, mice lacking Ly49H controlled MCMVΔm157 infection whereas the m157G1F-expressing virus escaped immune surveillance. We propose that infected cells displaying reduced MHC class I expression were ignored by NK cells as a result of m157G1F binding to Ly49C. The response pattern we described here is not without analogies to that reported by Jonjic and colleagues [30] who showed that in BALB/c but not in BALB H-2b mice, NK cells control an MCMV mutant lacking the m04 immunoevasin. NK cell activation was likely due to missing-self recognition, as NK cells in BALB/c β2m ko mice failed to keep viral replication in check. Babic proposed that m04 stabilizes MHC class I and improves binding to Ly49A to a lesser extent to Ly49C. In the absence of m04 inhibitory signals were reduced, which allowed a better elimination of the virus. This resembles our results obtained with MCMVm157G1F: inhibition was induced by m157 through Ly49C, whereas a Δm157 virus (like Δm04 virus) failed to induce inhibitory signals. We verified the m04 gene sequence in the MCMVm157G1F recombinant virus and did not find any alterations (data not shown), which confirmed that the effects we noted were due to m157.
Our study provides a novel understanding of the parameters that direct how efficiently NK cells deal with viral infections. The variety of receptors encoded within the NKC, combined to a high level of sequence polymorphism between mouse strains, are critical determinants of whether NK cells can recognize and eliminate virus-infected cells. In addition, the host H-2 haplotype and MHC sequence polymorphism determine the possibility of interactions in cis that regulate NK cell receptor functions. Our in vivo results highlight why the stochastic expression of distinct NK cell receptors, which result in various NK cell subsets, is so important to host survival. In the ongoing race between the host and CMV, cis-binding of inhibitory NK cell receptors by MHC class I may have provided the host with a mechanism of countering viral immune evasion. Held and colleagues showed the role of cis-interactions between Ly49A and H-2 Dd in education and in regulating NK cell functions (reviewed in [33]). This mechanism could provide a strong evolutionary gain to mice that possess a suitable NK cell repertoire and specific sets of MHC class I molecules. We should also consider the possibility that an evolution-driven selection of individuals having developed Ly49 able to establish cis-interactions with MHC might have resulted from viruses producing immunoevasins able to inhibit NK cells through their self-specific receptors. A recent study demonstrated that the human inhibitory NK cell receptor LIR-1 interacts in cis with HLA-G; this affects the receptor accessibility for HCMV immunoevasin UL18 [44]. These results highlight the relevance of our work and its possible translation into human settings. Our findings provide novel insights into the mechanisms that dictate susceptibility to viral infections, such as HCMV, and thus may also indicate new immunotherapeutic approaches for regulating NK cell responses in settings of infection as well as possibly cancer and transplantation.
Materials and Methods
Ethics statement
All animal experiments were performed after approval by the Animal Experimentation and Ethics Committee of the University of Western Australia (AEC approval # RA/3/300/76 and RA/3/100/1079) and in accordance with the guidelines of the National Health and Medical Research Council of Australia (Australian code of the care and use of animals for scientific purposes 8th edition, 2013; ISBN: 1864965975).
Animals
Inbred C57BL/6 (B6) mice were obtained from the Animal Resources Centre (Perth, WA, Australia). BALB.B6-CT6 (H-2d, Ly49H−), BALB.B6-Cmv1r (H-2d, Ly49H+) and B6.BALB-TC1 (H-2b, Ly49H−) congenic mice and B6 Ly49C Tg, B6 Ly49C Tg, β2m ko and B6 β2m ko recombinant mice were bred at the University of Western Australia Animal Care Services (Perth, WA, Australia). Female mice aged 7–12 weeks were used for all experiments, except for virus stock preparation where 3 week-old mice were used. All animal experiments were performed in pathogen-free conditions.
Generation of Ly49C transgenic B6 mice
Splenic NK cells isolated from a B6 mouse were expanded in IL-2 for 12 d. Ly49C+ NK cells were sorted and mRNA was isolated using pureLink RNA minikit. A 907 bp long cDNA was amplified by PCR using the following primers Forward primer: CTC CAC CAG CAT CAC TCC G and Reverse primer: CAA GAA ACG AAT AAG GAT CAA CTC. A 850 bp product harboring Sal1 and BamH1 ends was generated by amplifying the ly49cB6 cDNA using the following primers Forward primer: TAT ATG TCG ACC TCC ACC AGC ATC ACT CCG and Reverse primer: TAT ATG GAT CCT TAA TCA GGG AAT TTA TCC. The ly49cB6 insert was inserted between the Sal1 and BamH1 restriction sites into the modified transgene cassette described in [45] and generously provided by Prof. W. Held (Ludwig Center for Cancer research, Lausanne, CH). The cassette was microinjected in fertilized C57BL/6 eggs (WEHI transgenic mouse facilities, Melbourne-Bundoora, VIC). Five founders were obtained and backcrossed to C57BL/6 mice. Appropriate offspring were identified by FACs analysis for Ly49C expression and checked by PCR; selected mice from a same dam were inter-crossed, four transgenic lines were generated among which one that displayed a homogenous Ly49C expression on over 95% NK cells was maintained.
Cells and cell lines
BWZ.36, RMA and RMAS cells were grown in RPMI 1640 containing 5% FCS (Invitrogen), glutamine, sodium pyruvate, penicillin, gentamicin and 2-ME. COS-7 cells and Platinum-E cells were grown in DMEM (Invitrogen) containing 10% FCS, glutamine, sodium pyruvate, penicillin and gentamicin. Primary mouse embryonic fibroblasts (MEFs) were produced by trypsin dispersion of 15–17 day-old embryos from ARC/S mice as previously described [46]. MEFs and M210B4 cells were maintained in MEM 10% NCS (Invitrogen). Immediately prior to infection the culture medium was replaced with media supplemented with 2% FCS. For in vitro NK cell expansion, splenocytes were passed through a nylon wool column and then cultured in DMEM containing 10% FCS, glutamine, non essential amino acids sodium pyruvate, penicillin and gentamicin (all from Invitrogen) and 250 ng/ml recombinant human IL-2 (Cell Sciences, Canton, MA, USA).
Production of m157-Fc soluble fusion proteins
The CP197 construct containing the Smith m157-Fc behind the CD150L sequence cloned in the CDM8 vector [9] was kindly provided by Prof. Lewis Lanier (University of California, San Francisco, CA USA). Soluble m157-Fc fusion proteins for the MCMV G1F isolate was constructed as described in [13].
Virus stocks
The MCMV laboratory strains used were Smith (originally obtained from E.S. Mocarski, Stanford University) and K181-Perth (K181). Origin of wild-derived isolate G1F has been previously reported in [13]. Tissue culture virus stocks were produced by propagation in M210B4 cells and titers determined by standard plaque assay as previously described [47]. Virulent salivary gland viral (SGV) stocks were prepared by infecting 3-week-old female BALB/c mice i.p. with 1×103 PFU of tissue-cultured passaged virus, and then preparing homogenates of salivary glands at 17 days post infection. Secondary SGV stocks were prepared by infecting 3-week-old female BALB/c mice with 1×103 PFU of the primary SGV stock. Viral DNA was produced from infected MEFs as previously described [48].
Construction of m157 gene-swap MCMV viruses
In order to produce m157 gene swap viruses we used a ‘BAC recombineering’ approach essentially as previously described [49]. The K181 BAC pARK25 DNA [50] kindly provided by A. Redwood (University of Western Australia). Generation of substitution mutants in which the K181 m157 gene has been replaced with the G1F m157 (MCMVm157G1F) sequences and deletion mutant in which the m157 gene has been deleted (MCMVΔm157) have been described in [13].
Generation of BWZ transfectants
BWZ HD12 cells were generously provided by Prof. Wayne Yokoyama (Washington University, St. Louis, MO). BWZ HD12 reporter cells were transduced with the pMX-s-IRES-Ly49CB6 construct to generate BWZ HD12C cells as indicated in [13]. Clones expressing a range of levels of Ly49HB6 and Ly49CB6 were selected following limit dilution cloning, including clones which expressed essentially equal levels of these two receptors. For construction of Ly49H/C chimeric cells which express the Ly49HB6 transmembrane and cytoplasmic domains fused to the extracellular domains of Ly49CB6, Ly49HB6 was PCR amplified using forward primer (A): 5′-TATATCTCGAGATGAGTGAGCAGGAGGTCAC and reverse primer 5′-TCATTGATTTCTTGTTTGTGTTGA. Ly49C B6 was amplified using forward primer 5′-CAAACAAGAAATCAATGAAACTCT and reverse primer (B): 5′-TATATGCGGCCGCTTAATCAGGGAATTTATCC. PCR products Ly49HB6 plus Ly49CB6 were sewed together by PCR using primers A and B above. The resultant PCR product was digested with XhoI and NotI and cloned into pMX-s-IG plasmid. The Plat-E packaging cell line was transfected with the plasmids and resulting retroviral supernatant used to transduce the BWZ-DAP12 reporter cells. Expression of Ly49C was confirmed by FACS.
Antibodies
The antibodies used for analysis for flow cytometry were directed against the following proteins: CD3ε (145.2C.11), CD4 (RM4-5), CD8α (53.6.7), CD11b (M1/70), CD19 (1D3), CD44 (1M7), CD49b (DX5), CD69 (H1-2F3), CD107a (1D4B), H-2 IA/E (M5/114.15.2), IFN-γ (XMG1.2), mouse IgG1 (A851), Ly6C (AL21) and Fluorochrome-conjugated Streptavidin were obtained from BD BioScience (San Diego, CA). Anti-CD27 (LG7F9) and Siglec H (440c) were obtained from eBioScience (San Diego, CA). Anti-CD11c (N418) was obtained from BioLegend (San Diego, CA). Anti-human IgG (Fc part)-Biotin by Jackson ImmunoResearch Laboratories (West Grove, PA). Anti-Ly49H Ab 3D10 producing hybridoma was kindly provided by Prof. W.M. Yokoyama (Washington University, St. Louis, MO) and anti-Ly49C Ab 4LO3311 producing hybridoma was kindly provided by Prof. S. Lemieux (INRS, Laval, Canada). Purified anti-Ly49C and Ly49H antibodies were prepared in-house and conjugated to fluorochrome using antibody-labeling kit according to the manufacturer's recommendations (Molecular Probes).
Staining for flow cytometry analysis
Staining with m157G1F-Fc fusion proteins was done as follows: cell lines or NK cells were incubated with m157G1F-Fc for 40 min, or shorter time when indicated, followed by anti human IgG (Fc part)-Biotin for 30 min and by a third step with Streptavidin-APC-Cy7 for 15 min. Non specific binding was provided in the absence of the initial incubation with the fusion protein. Live cells were gated based on exclusion of propidium iodide (PI, Sigma). Detection of Ly49C in B6 and Cmv1r NK cells was performed after release of the cis-interactions achieved by a mild acid treatment. Cells were washed twice in PBS and resuspended for 4 min at ambient temperature in Citrate buffer (0.133 M Citric acid, 0.066 M Na2HPO4, pH 3.3) at a density of 10.106 cells/ml. The acid treatment was stopped by adding an excess of PBS 5% FCS; the cells were then stained for flow cytometry analysis. Acid treatment did not affect cell viability as determined by PI exclusion. For the detection of intracellular IFN-γ and of degranulation (LAMP1 expression), cells were cultured at 37°C in presence of monensin (Sigma-Aldrich, St Louis, MO). For LAMP-1 staining, anti-CD107a antibody (1D4B, BD Bioscience) was added into the culture medium as described in [51]. After the culture, cells were stained with fluorochrome conjugated antibodies directed to surface markers, the cells were then fixed with Cytofix/Cytoperm and permeabilized with Perm/Wash according to the manufacturer's recommendations (BD Bioscience). Permeabilized cells were incubated with anti-IFN-γ and anti-Ly49H antibodies for 40 min at room temperature. Intracellular detection of Ly49H was required to detect cells that had internalized this receptor upon exposure to m157-expressing target cells. Antibody-labelled cells were analyzed using a BD FACSCanto (BD Biosciences).
Cell sorting
For in vitro assays, splenic cells were isolated and NK cells were expanded for 3–4 days in IL-2 containing medium; they were briefly treated in a mild acidic buffer, as above, and were stained with anti-NK1.1 (PK136), CD3ε (145.2C.11), Ly49H (3D10), Ly49C (4LO3311) fluorochrome-conjugated antibodies for cell sorting using a FACs Aria II cell sorter (BD Biosciences). Sorted NK cells were then placed back in IL-2 containing medium and expanded another 3 d to allow elimination of receptor-bound antibodies.
Determination of virus growth in vivo
The recombinant viruses MCMVm157G1F, MCMVΔm157 and parental K181 MCMV virus were used to infect intra peritoneally BALB.B6-Cmv1r mice at the dose of 1×104 PFU and viral titers in the target organs (spleen, liver, lungs and salivary glands) were determined at designated times post inoculation (p.i.). The mice were sacrificed and organs were collected, homogenized in cold MEM 2% Neonatal Calf serum (NCS, Gibco) and centrifuged at 3000 rpm for 15 min at 4°C. The supernatants were stored at −80°C and viral titers were quantified by standard Plaque assay on M210B4 cells by standard plaque assay as described [47].
Ly49H-independent NK cell response in vivo
B6.BALB TC1, BALB.B6-CT6 mice were infected intraperitoneally with 1×104 PFU MCMVm157G1F or MCMVΔm157; B6 β2m ko were infected with 1×104 PFU MCMVΔm157. Infection was done in combination, or not, of an intraperitoneal injection of 2 µg α-galactosylceramide (αGC) (KRN7000, Kirin Brewer Co, Ltd, Japan). Mice were sacrificed 4 days p.i., the spleen, liver and lungs were collected and the viral titers within these organs was determined by standard plaque assay as indicated above.
Conjugate assays
Sorted NK cells were labeled with 5 µM 5-(and -6)-(((4-chloromethyl) benzoyl) amino) tetramethyl-rhodamine orange (CMTMR, Molecular Probes, Carlsbad, CA) and target cells were labeled with 0.5 µM 5 - (and 6-) carboxyfluorescein diacetate succinimidyl ester (CFSE, Molecular Probes) according to the manufacturer's recommendations. 105 NK cells were mixed with 2×105 targets in a final volume of 100 µl of DMEM 5% FCS and were centrifuged for 2 min at 50 g. After incubation at 37°C for 5 or 20 min, the conjugates were gently resuspended in 100 µl ice-cold 4% paraformaldehyde. Percentages of conjugated NK cells were then determined on a BD FACs Canto flow cytometer.
Phalloidin staining
72 to 96 hours after sorting, Ly49H+,C+ and Ly49H+,C− NK cells from B6 β2m ko mice were exposed to CMTMR labelled RMAS (top panels) or RMASm157G1F. 1×105 sorted NK cells were mixed with 2×105 CMTMR labeled targets in a final volume of 100 µl in DMEM 5% FCS and were centrifuged for 2 min at 50 g. After incubation at 37°C for 15 min, conjugates were gently resuspended and allowed to adhere on poly(L-lysine) coated microscopic slides (Lomb Scientific, Taron Point, NSW) for 3 min at 37°C and then were fixed with 4% paraformaldehyde for 10 min. After washing twice in PBS, slides were extracted with 0.1% Triton X-100 in PBS for 5 min at room temperature and were washed twice again in PBS. The slides were then stained with 1 unit of Phalloidin-FITC (Molecular Probes) diluted in PBS 1% BSA for 30 min at 37°C. After two washes in PBS, slides were mounted using Fluoromount (Sigma Aldrich) and analyzed on a fluorescence microscope. Polymerization of cytoskeleton actin as detected by phalloidin was quantified by using ImageJ software (National Institutes of Health; http://rsbweb.nih.gov/ij/); F-actin mean density was measured at the immune (IS) synapse and within the rest of a conjugated NK cell in order to determine the enrichment at the IS. A value close to 1 indicated that no polarization of the cytoskeleton toward the target cell was detected.
Determination of NK cell cytotoxicity in in vitro assay
IL-2 activated NK cells were tested for cytotoxicity by standard 51Cr release assays [52]. Briefly, target cells were labeled with 100 µCi 51Cr (Perkin Elmer, Rowville, VIC Australia); radioactivity-labelled target cells were mixed with NK cells in triplicate at various E∶T ratios and incubated for 4 h at 37°C. Target cells incubated in Triton 1% provided the total radioactive content (maximal release) while targets cultured alone provided the spontaneous radioactive release values. Specific killing was determined by measuring the radioactivity released in cell-free supernatant according to the following formula: Specific killing = ((sample value – spontaneous release)/(maximal release – spontaneous release)×100). Radioactivity released in the culture medium upon target cell lysis was counted in cell-free culture supernatant using a Wallac Wizard γ-counter (Turku, Finland).
Statistical analysis
For statistical analysis, the nonparametric Mann-Whitney test was performed using the statistical software package GraphPad Prism (La Jolla, CA, USA).
Supporting Information
Zdroje
1. BironCA, ByronKS, SullivanJL (1989) Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med 320 : 1731–1735.
2. EidenschenkC, DunneJ, JouanguyE, FourlinnieC, GineauL, et al. (2006) A novel primary immunodeficiency with specific natural-killer cell deficiency maps to the centromeric region of chromosome 8. Am J Hum Genet 78 : 721–727.
3. Alford CA, Britt WJ (1996) Cytomegalovirus; Fields BN, editor. New York: Raven Press. 629–660 p.
4. PyzikM, KielczewskaA, VidalSM (2008) NK cell receptors and their MHC class I ligands in host response to cytomegalovirus: insights from the mouse genome. Semin Immunol 20 : 331–342.
5. VivierE, TomaselloE, BaratinM, WalzerT, UgoliniS (2008) Functions of natural killer cells. Nat Immunol 9 : 503–510.
6. BrownMG, DokunAO, HeuselJW, SmithHR, BeckmanDL, et al. (2001) Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292 : 934–937.
7. DanielsKA, DevoraG, LaiWC, O'DonnellCL, BennettM, et al. (2001) Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med 194 : 29–44.
8. LeeSH, GirardS, MacinaD, BusaM, ZaferA, et al. (2001) Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet 28 : 42–45.
9. AraseH, MocarskiES, CampbellAE, HillAB, LanierLL (2002) Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296 : 1323–1326.
10. SmithHR, HeuselJW, MehtaIK, KimS, DornerBG, et al. (2002) Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A 99 : 8826–8831.
11. VoigtV, ForbesCA, TonkinJN, Degli-EspostiMA, SmithHR, et al. (2003) Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci U S A 100 : 13483–13488.
12. BoothTW, ScalzoAA, CarrelloC, LyonsPA, FarrellHE, et al. (1993) Molecular and biological characterization of new strains of murine cytomegalovirus isolated from wild mice. Arch Virol 132 : 209–220.
13. CorbettAJ, CoudertJD, ForbesCA, ScalzoAA (2011) Functional Consequences of Natural Sequence Variation of Murine Cytomegalovirus m157 for Ly49 Receptor Specificity and NK Cell Activation. J Immunol 186 : 1713–1722.
14. BerryR, NgN, SaundersPM, VivianJP, LinJ, et al. (2013) Targeting of a natural killer cell receptor family by a viral immunoevasin. Nat Immunol 14 : 699–705.
15. KimS, Poursine-LaurentJ, TruscottSM, LybargerL, SongYJ, et al. (2005) Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436 : 709–713.
16. FernandezNC, TreinerE, VanceRE, JamiesonAM, LemieuxS, et al. (2005) A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 105 : 4416–4423.
17. OrrMT, MurphyWJ, LanierLL (2010) ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol 11 : 321–327.
18. PignatelliS, Dal MonteP, RossiniG, LandiniMP (2004) Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol 14 : 383–410.
19. ScalzoAA, BrownMG, ChuDT, HeuselJW, YokoyamaWM, et al. (1999) Development of intra-natural killer complex (NKC) recombinant and congenic mouse strains for mapping and functional analysis of NK cell regulatory loci. Immunogenetics 49 : 238–241.
20. BrennanJ, LemieuxS, FreemanJD, MagerDL, TakeiF (1996) Heterogeneity among Ly-49C natural killer (NK) cells: characterization of highly related receptors with differing functions and expression patterns. J Exp Med 184 : 2085–2090.
21. ScarpellinoL, OeschgerF, GuillaumeP, CoudertJD, LevyF, et al. (2007) Interactions of Ly49 Family Receptors with MHC Class I Ligands in trans and cis. J Immunol 178 : 1277–1284.
22. DouceyMA, ScarpellinoL, ZimmerJ, GuillaumeP, LuescherIF, et al. (2004) Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat Immunol 5 : 328–336.
23. KarreK, LjunggrenHG, PiontekG, KiesslingR (1986) Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319 : 675–678.
24. SunJC, LanierLL (2008) Cutting edge: viral infection breaks NK cell tolerance to “missing self”. J Immunol 181 : 7453–7457.
25. BrycesonYT, LongEO (2008) Line of attack: NK cell specificity and integration of signals. Curr Opin Immunol 20 : 344–352.
26. AndrewsDM, ScalzoAA, YokoyamaWM, SmythMJ, Degli-EspostiMA (2003) Functional interactions between dendritic cells and NK cells during viral infection. Nat Immunol 4 : 175–181.
27. Fodil-CornuN, LeeSH, BelangerS, MakrigiannisAP, BironCA, et al. (2008) Ly49h-deficient C57BL/6 mice: a new mouse cytomegalovirus-susceptible model remains resistant to unrelated pathogens controlled by the NK gene complex. J Immunol 181 : 6394–6405.
28. HayakawaY, SmythMJ (2006) CD27 Dissects Mature NK Cells into Two Subsets with Distinct Responsiveness and Migratory Capacity. J Immunol 176 : 1517–1524.
29. HengelH, ReuschU, GutermannA, ZieglerH, JonjicS, et al. (1999) Cytomegaloviral control of MHC class I function in the mouse. Immunol Rev 168 : 167–176.
30. BabicM, PyzikM, ZafirovaB, MitrovicM, ButoracV, et al. (2010) Cytomegalovirus immunoevasin reveals the physiological role of “missing self” recognition in natural killer cell dependent virus control in vivo. J Exp Med 207 : 2663–2673.
31. PyzikM, CharbonneauB, Gendron-PontbriandEM, BabicM, KrmpoticA, et al. (2011) Distinct MHC class I-dependent NK cell-activating receptors control cytomegalovirus infection in different mouse strains. J Exp Med 208 : 1105–1117.
32. van DommelenSL, TabariasHA, SmythMJ, Degli-EspostiMA (2003) Activation of natural killer (NK) T cells during murine cytomegalovirus infection enhances the antiviral response mediated by NK cells. J Virol 77 : 1877–1884.
33. HeldW, MariuzzaRA (2008) Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat Rev Immunol 8 : 269–278.
34. VeilletteA, LatourS, DavidsonD (2002) Negative regulation of immunoreceptor signaling. Annu Rev Immunol 20 : 669–707.
35. RadosevicK, van LeeuwenMT, Segers-NoltenIM, FigdorCG, de GroothBG, et al. (1994) Changes in actin organization during the cytotoxic process. Cytometry 15 : 320–326.
36. KrzewskiK, ChenX, StromingerJL (2008) WIP is essential for lytic granule polarization and NK cell cytotoxicity. Proc Natl Acad Sci U S A 105 : 2568–2573.
37. ScalzoAA, FitzgeraldNA, SimmonsA, La VistaAB, ShellamGR (1990) Cmv-1, a genetic locus that controls murine cytomegalovirus replication in the spleen. J Exp Med 171 : 1469–1483.
38. BackJ, ChalifourA, ScarpellinoL, HeldW (2007) Stable masking by H-2Dd cis ligand limits Ly49A relocalization to the site of NK cell/target cell contact. Proc Natl Acad Sci U S A 104 : 3978–3983.
39. ChalifourA, ScarpellinoL, BackJ, BrodinP, DevevreE, et al. (2009) A Role for cis Interaction between the Inhibitory Ly49A receptor and MHC class I for natural killer cell education. Immunity 30 : 337–347.
40. StebbinsCC, WatzlC, BilladeauDD, LeibsonPJ, BurshtynDN, et al. (2003) Vav1 dephosphorylation by the tyrosine phosphatase SHP-1 as a mechanism for inhibition of cellular cytotoxicity. Mol Cell Biol 23 : 6291–6299.
41. KleijnenMF, HuppaJB, LucinP, MukherjeeS, FarrellH, et al. (1997) A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J 16 : 685–694.
42. ReuschU, MuranyiW, LucinP, BurgertHG, HengelH, et al. (1999) A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J 18 : 1081–1091.
43. ZieglerH, ThaleR, LucinP, MuranyiW, FlohrT, et al. (1997) A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6 : 57–66.
44. LiNL, FuL, UchtenhagenH, AchourA, BurshtynDN (2013) Cis association of leukocyte Ig-like receptor 1 with MHC class I modulates accessibility to antibodies and HCMV UL18. Eur J Immunol 43 : 1042–1052.
45. PircherH, MakTW, LangR, BallhausenW, RuediE, et al. (1989) T cell tolerance to Mlsa encoded antigens in T cell receptor V beta 8.1 chain transgenic mice. EMBO J 8 : 719–727.
46. ChalmerJE, MackenzieJS, StanleyNF (1977) Resistance to murine cytomegalovirus linked to the major histocompatibility complex of the mouse. J Gen Virol 37 : 107–114.
47. FarrellHE, ShellamGR (1989) Immunoblot analysis of the antibody response to murine cytomegalovirus in genetically resistant and susceptible mice. J Gen Virol 70(Pt 10): 2573–2586.
48. XuJ, LyonsPA, CarterMD, BoothTW, Davis-PoynterNJ, et al. (1996) Assessment of antigenicity and genetic variation of glycoprotein B of murine cytomegalovirus. J Gen Virol 77(Pt 1): 49–59.
49. WarmingS, CostantinoN, CourtDL, JenkinsNA, CopelandNG (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33: e36.
50. RedwoodAJ, MesserleM, HarveyNL, HardyCM, KoszinowskiUH, et al. (2005) Use of a murine cytomegalovirus K181-derived bacterial artificial chromosome as a vaccine vector for immunocontraception. J Virol 79 : 2998–3008.
51. AlterG, MalenfantJM, AltfeldM (2004) CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods 294 : 15–22.
52. BrunnerKT, MauelJ, CerottiniJC, ChapuisB (1968) Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology 14 : 181–196.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Combined Systems Approaches Reveal Highly Plastic Responses to Antimicrobial Peptide Challenge inČlánek Two Novel Human Cytomegalovirus NK Cell Evasion Functions Target MICA for Lysosomal Degradation
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 5- Stillova choroba: vzácné a závažné systémové onemocnění
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Surveillance for Emerging Biodiversity Diseases of Wildlife
- The Emerging Role of Urease as a General Microbial Virulence Factor
- PARV4: An Emerging Tetraparvovirus
- Epigenetic Changes Modulate Schistosome Egg Formation and Are a Novel Target for Reducing Transmission of Schistosomiasis
- The Human Adenovirus E4-ORF1 Protein Subverts Discs Large 1 to Mediate Membrane Recruitment and Dysregulation of Phosphatidylinositol 3-Kinase
- A Multifactorial Role for Malaria in Endemic Burkitt's Lymphoma Pathogenesis
- Structural Basis for the Ubiquitin-Linkage Specificity and deISGylating Activity of SARS-CoV Papain-Like Protease
- Cathepsin-L Can Resist Lysis by Human Serum in
- Epstein-Barr Virus Down-Regulates Tumor Suppressor Expression
- BCA2/Rabring7 Targets HIV-1 Gag for Lysosomal Degradation in a Tetherin-Independent Manner
- The Evolutionarily Conserved Mediator Subunit MDT-15/MED15 Links Protective Innate Immune Responses and Xenobiotic Detoxification
- Suppressor of Cytokine Signaling 4 (SOCS4) Protects against Severe Cytokine Storm and Enhances Viral Clearance during Influenza Infection
- T Cell Inactivation by Poxviral B22 Family Proteins Increases Viral Virulence
- Dynamics of HIV Latency and Reactivation in a Primary CD4+ T Cell Model
- HIV and HCV Activate the Inflammasome in Monocytes and Macrophages via Endosomal Toll-Like Receptors without Induction of Type 1 Interferon
- Virus and Autoantigen-Specific CD4+ T Cells Are Key Effectors in a SCID Mouse Model of EBV-Associated Post-Transplant Lymphoproliferative Disorders
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis
- Squalene Synthase As a Target for Chagas Disease Therapeutics
- The Contribution of Viral Genotype to Plasma Viral Set-Point in HIV Infection
- Combined Systems Approaches Reveal Highly Plastic Responses to Antimicrobial Peptide Challenge in
- Anthrax Lethal Factor as an Immune Target in Humans and Transgenic Mice and the Impact of HLA Polymorphism on CD4 T Cell Immunity
- Ly49C-Dependent Control of MCMV Infection by NK Cells Is -Regulated by MHC Class I Molecules
- Two Novel Human Cytomegalovirus NK Cell Evasion Functions Target MICA for Lysosomal Degradation
- A Large Family of Antivirulence Regulators Modulates the Effects of Transcriptional Activators in Gram-negative Pathogenic Bacteria
- Broad-Spectrum Anti-biofilm Peptide That Targets a Cellular Stress Response
- Malaria Parasite Infection Compromises Control of Concurrent Systemic Non-typhoidal Infection via IL-10-Mediated Alteration of Myeloid Cell Function
- A Role for in Higher Order Structure and Complement Binding of the Capsule
- Hip1 Modulates Macrophage Responses through Proteolysis of GroEL2
- CD8 T Cells from a Novel T Cell Receptor Transgenic Mouse Induce Liver-Stage Immunity That Can Be Boosted by Blood-Stage Infection in Rodent Malaria
- Phosphorylation of KasB Regulates Virulence and Acid-Fastness in
- HIV-Infected Individuals with Low CD4/CD8 Ratio despite Effective Antiretroviral Therapy Exhibit Altered T Cell Subsets, Heightened CD8+ T Cell Activation, and Increased Risk of Non-AIDS Morbidity and Mortality
- A Novel Mechanism Inducing Genome Instability in Kaposi's Sarcoma-Associated Herpesvirus Infected Cells
- Structural and Biochemical Characterization Reveals LysGH15 as an Unprecedented “EF-Hand-Like” Calcium-Binding Phage Lysin
- Hepatitis C Virus Cell-Cell Transmission and Resistance to Direct-Acting Antiviral Agents
- Different Modes of Retrovirus Restriction by Human APOBEC3A and APOBEC3G
- TNFα and IFNγ but Not Perforin Are Critical for CD8 T Cell-Mediated Protection against Pulmonary Infection
- Large Scale RNAi Reveals the Requirement of Nuclear Envelope Breakdown for Nuclear Import of Human Papillomaviruses
- The Cytoplasmic Domain of Varicella-Zoster Virus Glycoprotein H Regulates Syncytia Formation and Skin Pathogenesis
- A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase
- Are We There Yet? The Smallpox Research Agenda Using Variola Virus
- High-Efficiency Targeted Editing of Large Viral Genomes by RNA-Guided Nucleases
- Dynamic Functional Modulation of CD4 T Cell Recall Responses Is Dependent on the Inflammatory Environment of the Secondary Stimulus
- Bacterial Superantigens Promote Acute Nasopharyngeal Infection by in a Human MHC Class II-Dependent Manner
- Follicular Helper T Cells Promote Liver Pathology in Mice during Infection
- A Nasal Epithelial Receptor for WTA Governs Adhesion to Epithelial Cells and Modulates Nasal Colonization
- Unexpected Role for IL-17 in Protective Immunity against Hypervirulent HN878 Infection
- Human Cytomegalovirus Fcγ Binding Proteins gp34 and gp68 Antagonize Fcγ Receptors I, II and III
- Expansion of Murine Gammaherpesvirus Latently Infected B Cells Requires T Follicular Help
- Venus Kinase Receptors Control Reproduction in the Platyhelminth Parasite
- Molecular Signatures of Hemagglutinin Stem-Directed Heterosubtypic Human Neutralizing Antibodies against Influenza A Viruses
- The Downregulation of GFI1 by the EZH2-NDY1/KDM2B-JARID2 Axis and by Human Cytomegalovirus (HCMV) Associated Factors Allows the Activation of the HCMV Major IE Promoter and the Transition to Productive Infection
- Inactivation of Fructose-1,6-Bisphosphate Aldolase Prevents Optimal Co-catabolism of Glycolytic and Gluconeogenic Carbon Substrates in
- New Insights into Rotavirus Entry Machinery: Stabilization of Rotavirus Spike Conformation Is Independent of Trypsin Cleavage
- Prophenoloxidase Activation Is Required for Survival to Microbial Infections in
- SslE Elicits Functional Antibodies That Impair Mucinase Activity and Colonization by Both Intestinal and Extraintestinal Strains
- Timed Action of IL-27 Protects from Immunopathology while Preserving Defense in Influenza
- HIV-1 Envelope gp41 Broadly Neutralizing Antibodies: Hurdles for Vaccine Development
- The PhoP-Dependent ncRNA Mcr7 Modulates the TAT Secretion System in
- Cellular Superspreaders: An Epidemiological Perspective on HIV Infection inside the Body
- The Inflammasome Pyrin Contributes to Pertussis Toxin-Induced IL-1β Synthesis, Neutrophil Intravascular Crawling and Autoimmune Encephalomyelitis
- Papillomavirus Genomes Associate with BRD4 to Replicate at Fragile Sites in the Host Genome
- Integrative Functional Genomics of Hepatitis C Virus Infection Identifies Host Dependencies in Complete Viral Replication Cycle
- Co-assembly of Viral Envelope Glycoproteins Regulates Their Polarized Sorting in Neurons
- Targeting Membrane-Bound Viral RNA Synthesis Reveals Potent Inhibition of Diverse Coronaviruses Including the Middle East Respiratory Syndrome Virus
- Dual-Site Phosphorylation of the Control of Virulence Regulator Impacts Group A Streptococcal Global Gene Expression and Pathogenesis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Venus Kinase Receptors Control Reproduction in the Platyhelminth Parasite
- Dual-Site Phosphorylation of the Control of Virulence Regulator Impacts Group A Streptococcal Global Gene Expression and Pathogenesis
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis
- High-Efficiency Targeted Editing of Large Viral Genomes by RNA-Guided Nucleases
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



