-
Medical journals
- Career
Evaluation pattern within tumor microenvironment and consequent gene expression in oral cancer
Authors: A. Vasheghani Farahani; M. Kavousi; F. Jamshidian
Authors‘ workplace: Department of Biology, Faculty of Biological Sciences, East Tehran Branch (Ghiamdasht), Islamic Azad University, Tehran, Iran
Published in: Klin Onkol 2024; 37(1): 34-39
Category: Original Articles
doi: https://doi.org/10.48095/ccko202434Overview
Background: Oral squamous cell carcinoma (OSCC) is one of the most common cancers in the head and neck squamous cell cancer group. The increasing frequency of oral carcinomas and their late-stage appearance is a major worldwide health concern. MicroRNAs (miRNAs) appear to play an important role in cancer growth and progression, according to growing data, whereas no information is available regarding miR-7113-3p and miR-6721-5p involvement in OSCC. In this article, the expression of MAP2K1, miR-7113-3p, and miR-6721-5p was examined for possible biological functions in the advancement of oral squamous cell carcinoma. Material and methods: We used quantitative real-time PCR (to examine the mRNA expression of MAP2K1, miR-7113-3p, and miR-6721-5p in fresh frozen OSCC tissues and adjacent normal fresh frozen tissues from 30 patients, and we investigated their relationship with clinical parameters. Results: MAP2K1 expression was found to be dramatically increased in tumor tissues than in normal tissues, whereas miR7113-3p and miR-6721-5p expression was significantly decreased. Furthermore, a statistical correlation of P = 0.04 was also observed between increased MAP2K1 expression and perineural invasion. Additionally, we noted that the downregulation of miR-7113-3p appears to correlate positively with overexpression of MAP2K1 (P = 0.0218), and a negative correlation was observed between downregulation of miR-6721-5p and overexpression of MAP2K1 (P = 0.7771). Conclusion: Based on these findings, miR-7113-3p and miR-6721-5p might be prospective biomarkers for OSCC patients, and could be utilized to detect OSCC at an early stage for future diagnosis. MAP2K1 overexpression has been linked to the development of OSCC and perineural invasion.
Keywords:
OSCC – MAP2K1 target gene – miR-7113-3p – miR-6721-5p – quantitative real-time PCR
Material and methods
Cell collection
The study utilized 30 pairs of tumor and adjacent normal cell line collection. The samples were available at the Tumor Bank of Cancer Institute approved by an orthodontic specialist and a pathologist. We immediately preserved fresh tissue samples in liquid nitrogen and stored them at −80°C until RNA extraction.
RNA extraction and quantitative real-time PCR
After following the manufacturer’s instructions, TRIZOL reagent was used to extract RNA (Invitrogen, Sigma, USA). Electrophoresis in 1.5% agarose gel and a Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) were used to confirm the quality and quantity of extracted RNAs, respectively (the light absorption ratio of 260–280 nm in pure RNA is around 1.9–2.0 and it has a 28S to 18S bond strength of 2 : 1). Total extracted RNA was reverse transcribed using BioFACT’s cDNA Synthesis kit to synthesize complementary DNA (cDNA) (Daejeon, South Korea), according to the manufacturer’s protocol. Additionally, cDNA for miRNAs was synthesized using appropriate stem-loop RT primers and the MiR-Amp kit (Pars Genome, Iran). The SYBR Green RT-PCR Kit (BioFact, Daejeon, South Korea) was used to conduct the quantitative real-time PCR analysis on a Roche ExicyclerTM 96 thermocycler. The following thermal cycling profile was used for quantitative real-time PCR on miRNAs and MAP2K1: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s for denaturation, 60 °C for 30 s for annealing, and 72 °C for 20 s for elongation. By employing ACTB as a housekeeping gene, the expression of MAP2K1 was normalized. In addition, the expression of miR-7173-3p and miR-6721-5p was standardized using U6 as an endogenous control. Following completion of the preceding stages, the received information was checked for Melting curve and the obtained diagrams were examined for dimer formation. The findings of the melting curve of these samples revealed that the miRNAs product was proprietary and had their own TM, as well as a single peak, thus confirming the correctness of the primers and the accuracy of real-time PCR. Finally, the CT number was calculated using the provided data. Primers were designed using Oligo Analyzer and the Primer3plus program, evaluated for optimal properties through the BLAST program, and synthesized by BIONEER (Daejeon, South Korea). A summary of the primer sequences can be found in Tab. 1.
1. Real-time quantitative polymerase chain reaction primers. 
1. Downregulated expressions of miR-7113-3p (A) and miR-6721-5p (B) in oral squamous cell carcinoma. Data are shown as means ± SD of three separate experiments. The transcript levels were normalized to U6 expression. (P < 0.050; N = 30). 
Statistical analysis
The results were provided as the mean ± standard error of the mean of three identical experiments carried out in triplicate. GraphPad Prism software 9.0.0 (GraphPad Software, Inc., San Diego, CA, USA) and SPSS software were used to analyze the data (version 21.0; SPSS, Inc., Chicago, IL, USA). To determine the normal distribution of sample data, the one-sample Kolmogorov-Smirnov test was performed. The independent-sample Kruskal-Wallis tests were used to evaluate the association between MAP2K1 levels and clinicopathological features in OSCC patients. Furthermore, the one-way analysis of variance (ANOVA) was employed to compare the levels of MAP2K1 expression in different tumor sizes and clinical stages. The correlation between miR-7113-3p, miR-6721-5p, and MAP2K1 expression was investigated applying Pearson’s correlation and regression analysis. Gene expression differences were calculated using Genex6 software. In order to analyze the relationship between the levels of variables and disease probability, the odds ratio method was employed. This parameter was calculated using logistic regression in SPSS software. Finally, the diagnostic value was evaluated using the receiver operating characteristic (ROC) curve. A P-value ≤ 0.050 was regarded as statistically significant.
Results
miR-7113-3p and miR-6721-5p expression was downregulated in OSCC
The expression patterns of miR-7113-3p and miR-6721-5p were examined in 30 paired OSCC tissues and adjacent normal oral tissues using quantitative real-time PCR. MiR-7113-3p and miR-6721-5p expression levels were both reduced, (4.24-folds and 1.85-folds, respectively) in OSCC tissues compared to normal tissues (P = 0.00000 and P = 0.00001, respectively) (Graph 1).
MAP2K1 gene expression was upregulated in OSCC
In this investigation, quantitative real-time PCR was used to evaluate the expression of MAP2K1 as a possible target for miR-7113-3p and miR-6721-5p in 30 paired OSCC tissues and adjacent normal oral tissues. Mirwalk and miRDB algorithms were used to discover potential co-targets of miR-7113-3p and miR-6721-5p in OSCC. Following that, online bioinformatics databases confirmed that MAP2K1 might be an acceptable direct target for the corresponding miRNAs. MAP2K1 expression was observed to be considerably higher (3.087-folds) in tumor tissues compared to adjacent normal oral tissues (P = 0.00000) (Graph 2).
2. 
Quantitative real time polymerase chain reaction analysis of MAP2K1 expression in oral squamous cell cancer tissues and adjacent normal tissues (N = 30). The transcript levels were normalized to ACTB expression. The data are presented as means ± SD (P < 0.050). 2. Clinicopathological characteristics and MAP2K1 expression. 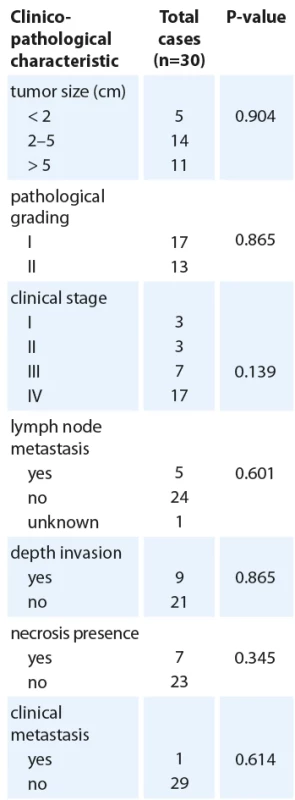
Correlation between MAP2K1 expression and miR-7113-3p, miR-6721-5p in OSCC patients
The Pearson’s correlation analysis was used to examine the connection between miR-7113-3p and miR-6721-5p levels and MAP2K1 expression in OSCC. We discovered an inverse and significant correlation between miR-7113-3p downregulation and MAP2K1 target gene overexpression in OSCC (r = −0.295, P = 0.021). A direct and nonsignificant correlation was also identified between miR-6721-5p downregulation and MAP2K1 overexpression (r = 0.037, P = 0.777) (Graph 3).
Potential diagnostic values of MAP2K1 in OSCC
Based on ROC curve analysis, MAP2K1 was evaluated for its potential to diagnose OSCC. The area under the curve (AUC) of MAP2K1 was 0.9466 (95% CI = 0.8934–0.9999; P = 0.00000). The best cutting point is indicated by the threshold. This cutting point’s sensitivities and specificities are also provided. To choose the best cut point, a value of J or the Youden index is employed (J = 0.8333). The optimal MAP2K1 cutting point is Dct = 6.8125, with a sensitivity of 0.8667 and a specificity of 0.9667 (Graph 4).
Potential diagnostic values of miR-7113-3p and miR-6721-5p in OSCC
Furthermore, the potential diagnostic value of miR-7113-3p and miR-6721-5p for OSCC was assessed by ROC curve analysis. According to the following tables, the value of AUC for miR-7113-3p is 0.9666 (95% CI = 0.9284–1; P = 0.00000), and for miR-6721-5p, the AUC is equal to 0.8261 (95% CI = 0.7155–0.9367; P = 0.00000) (Graph 5, 6).
Association between MAP2K1 expression and clinicopathological features
The association between MAP2K1 expression levels and some other clinicopathological parameters was investigated in Tab. 2 to gain a better awareness of its possible function in the development of oral cancer. It was found that MAP2K1 expression was remarkably associated with tumor PNI (P = 0.041). According to the presence of PNI in 30 patients, 37% were positive (N = 11), and 63% were negative (N = 19). MAP2K1 expression was increased in all patients, although considering the small number of patients with PNI, the presence of PNI was significantly associated with MAP2K1 upregulation (P < 0.05) (Tab. 2).
Discussion
HNSCC is a serious public health issue globally, with a high fatality rate. The most frequent kind of HNSCC is OSCC, which remains a concern for head and neck specialists despite major advances in diagnostic techniques and treatments [15]. Oral cancer is a multifactorial disease caused by a combination of genetic abnormalities and environmental factors, the most important of which are tobacco and alcohol use [16]. Epigenetic alterations, such as DNA methylation, histone modifications, and non-coding RNA modifications (miRNAs), have been shown to play an important regulatory role in the development and progression of oral cancer [17]. MiRNAs seem to be essential in the epigenetic regulation of cellular processes such as cell cycle regulation, differentiation, apoptosis, and migration. MiRNA dysregulation leads to tumor-related events throughout cancer development [18]. In this way, miRNAs can control gene expression that is involved in cancer biology by acting as oncogenes or tumor suppressors [19].
In numerous recent studies, it has been shown that miRNAs expression is altered in oral squamous cell carcinoma, and some miRNAs are shown to function as tumor suppressors or tumor promoters during tumorigenesis. Tumor suppressor miRNAs like miR-26-a, miR-99a-5p, miR-375, and miR-139-5p were discovered to be downregulated in oral cancer and inhibit oncogenes, whereas oncomiRs like miR-21, miR-31, miR-93, miR-211, and miR-373 were observed to be up-regulated in oral cancer and inhibit tumor suppressors [20]. Furthermore, it has been demonstrated in a study examining the expression of numerous miRNAs that miR-31 may be an ideal candidate for clinical application in oral cancer due to its high sensitivity in tissue, saliva, and plasma [21].
There have been some important candidate miRNAs implicated in progression of oral cancer as earlier studies demonstrated. Downregulation of miR-125a, miR-184, and miR-16 as well as upregulation of miR-96 were noted in both oral tumors and surgical margins, suggesting combinatorial regulation of these miRNAs and target transcription factors contributes to oral tumorigenesis and is useful in detecting minimal residual disease after surgery [22].
While miR-7113-3p and miR-6721-5p have frequently been reported to contribute in a variety of cancers, no study has evaluated their expression in OSCC. For instance, miR-7113 was upregulated by AnAc in MDA-MB-231 cells and targets host gene NDUFS8 to cause breast cancer [23]. According to Guo’s research, hsa-miR-7113-3p participates in the LINC00973-miRNA-mRNA cRNA network, which is enhanced in non-small-cell lung cancer tissues [24]. According to the findings, the circ 0034467_ miR-6721-5p - SLC19A1 regulatory network may serve as a key regulator in prostate cancer [25]. Additionally, one study demonstrated that miR-6721-5p was downregulated by HOXC6, another gene related to cancer progression [26]. Based on these results, miR-7113-3p and miR-6721-5p could represent potential biomarkers in OSCC and different cancers by exerting oncogenic or tumor - suppressive functions. It is, however, necessary to conduct more research to verify these findings.
The current study aimed to discover new diagnostic or prognostic biomarkers for OSCC. MiR-7113-3p and miR-6721-5p are significantly downregulated in OSCC tissues compared to normal tissues, according to our analyses. The role of MAP2K1 in tumorigenesis and cancer progression has been noted previously as a candidate for further studies. Activated MAP2K1 promotes cancer cell proliferation and confers drug resistance. The results of Zhe Jin’s study suggested that blocking MAP2K1 and miR-330-3p also inhibited the ability of HepG2 cells to migrate. In this study, miR-330-3p suppressed migration of liver cancer cells by interacting with MAP2K1 [27]. In addition, You et al. observed MAP2K1 overexpression in non-small cell lung cancer, and discovered that miR-449a regulated MAP2K1 expression by directly targeting its 3’UTR [28]. MAP2K1 mutations have been identified at a lower frequency in several cancers, including lung adenocarcinoma, melanoma and gastric cancer. About 1% of HNSCC cases exhibit MAP2K1 mutations, the same as lung cancer [29]. MAP2K1 has been shown to regulate tumorigenic development in OSCC. It is primarily responsible for cancer proliferation, chemoresistance, invasion, and migration in oral cancer [30]. Further studies identified that MAP2K1 activation increased CD44 expression and promoter activity, whereas CD44 attenuation reduced both in vitro migration and in vivo oral tumor formation [31]. According to another study, MAP2K1 activation frequently occurs in oral malignancies and is linked to tumor cell proliferation, migration, and invasion by regulating antiapoptotic and proliferative pathways [32]. These findings confirmed what we had discovered.
Based on bioinformatics analysis, we identified MAP2K1 as a direct target of miR-7113-3p and miR-6721-5p. We discovered a significant increase in MAP2K1 gene expression in tumor tissues, particularly in comparison to adjacent normal tissues from OSCC patients, which supports previously reported results. Moreover, miR-7113-3p and miR--6721-5p expression levels were significantly decreased. In the current study, we correlated the expression level of miR-7113-3p and miR-6721-5p to MAP2K1 mRNA and we observed a significant inverse correlation between miR-7113-3p downregulation and MAP2K1 target gene overexpression in OSCC (r = −0.295, P = 0.021). There was also a non-significant association between miR-6721-5p downregulation and MAP2K1 overexpression (r = 0.037, P = 0.777). PNI is a form of tumor progression in which cancer cells encroach along nerves [33]. PNI is well known to be associated with a poor outcome in cancers of the colorectal, pancreas, and salivary glands. PNI has been reported to occur in 2–82 % of oral squamous cell carcinoma. There is also a correlation between PNI and prognostic factors [34]. According to the present study, there was a significant association between MAP2K1 overexpression and PNI status in OSCC tumors (P = 0.041) and no remarkable association was found between vascular and depth invasions with MAP2K1 overexpression (P = 0.627 and P = 0.865 respectively). Additionally, we observed increased MAP2K1 expression in tumors in late stages (grade II), but no significant correlation was found (P = 0.139). According to the results of the present study, the overexpression of MAP2K1 is not correlated with necrosis presence (P = 0.345), clinical metastasis (P = 0.614), tumor size (P = 0.904), pathological grading (P = 0.865), smoking status (P = 0.443) and family history (P = 0.456). To confirm these results, better understand the connection between the MAP2K1 gene and miR-7113-3p and miR-6721-5p expression in oral cancer malignancy, and modify the aggressive behavior of oral cancer cells in clinical trials, additional research on the expression of the MAP2K1 protein is required. Sample size, repetition cycles and multiple analyses to endorse the result were the limitations of our study.
Conclusion
The results of this study revealed the first evidence of evaluation of miR-7113-3p and miR-6721-5p expression in OSCC and showed increased expression of the MAP2K1 gene and decreased expression of miR-7113-3p and miR-6721-5p in tumor tissues, compared to normal adjacent tissues. As potential diagnostic and prognostic biomarkers for OSCC patients, miR-7113-3p and miR-6721-5p have the potential to become powerful biomarkers in the near future, and they may even contribute to the early diagnosis and prognosis of this disease.
Dr. Faranak Jamshidian
Department of Biology
Faculty of Biological Sciences
East Tehran Branch (Ghiamdasht)
Islamic Azad University
P.O. Box: 33955/16,
Tehran, Iran
e-mail:faranak.jamshidian@gmail.comSubmitted/Obdrženo: 1. 2. 2023
Accepted/Přijato: 21. 9. 2023
Sources
1. Zhao W, Cui Y, Liu L et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ 2020; 27(3): 919–933. doi: 10.1038/ s41418-019-0423-5.
2. Roi A, Roi CI, Negruțiu ML et al. The challenges of OSCC diagnosis: salivary cytokines as potential biomarkers. J Clin Med 2020; 9(9): 2866. doi: 10.3390/ jcm9092866.
3. Ries J, Baran C, Wehrhan F et al. The altered expression levels of miR-186, miR-494 and miR-3651 in OSCC tissue vary from those of the whole blood of OSCC patients. Cancer Biomark 2019; 24(1): 19–30. doi: 10.3233/ CBM-180032.
4. Chang K-W, Liu C-J, Chu T-H et al. Association between high miR-211 microRNA expression and the poor prognosis of oral carcinoma. J Dent Res 2008; 87(11): 1063–1068. doi: 10.1177/ 154405910808701116.
5. Peng Q, Deng Z, Pan H et al. Mitogen-activated protein kinase signaling pathway in oral cancer. Oncol Lett 2018; 15(2): 1379–1388. doi: 10.3892/ ol.2017.7491.
6. Lawal B, Lee C-Y, Mokgautsi N et al. mTOR/ EGFR/ iNOS/ MAP2K1/ FGFR/ TGFB1 are druggable candidates for N-(2, 4-difluorophenyl)-2’, 4’ - difluoro-4-hydroxybiphenyl-3-carboxamide (NSC765598), with consequent anticancer implications. Front Oncol 2021; 11 : 656738. doi: 10.3389/ fonc.2021.656738.
7. Baghaei F, Abdollahi A, Mohammadpour H et al. PTEN and miR-26b: promising prognostic biomarkers in initiation and progression of oral squamous cell carcinoma. J Oral Pathol Med 2019; 48(1): 31–35. doi: 10.1111/ jop.12794.
8. Shan C, Chen X, Cai H et al. The emerging roles of autophagy-related microRNAs in cancer. Int J Biol Sci 2021; 17(1): 134–150. doi: 10.7150/ ijbs.50773.
9. Acunzo M, Romano G, Wernicke D et al. MicroRNA and cancer – a brief overview. Adv Biol Regul 2015; 57 : 1–9. doi: 10.1016/ j.jbior.2014.09.013.
10. Yoshizawa JM, Wong DT. Salivary microRNAs and oral cancer detection. Methods Mol Biol 2013; 936 : 313–324. doi: 10.1007/ 978-1-62703-083-0_24.
11. Bam M, Yang X, Busbee BP et al. Increased H3K4me3 methylation and decreased miR-7113-5p expression lead to enhanced Wnt/ b-catenin signaling in immune cells from PTSD patients leading to inflammatory phenotype. Mol Med 2020; 26(1): 110. doi: 10.1186/ s10 020-020-00238-3.
12. Santoro G, Lapucci C, Giannoccaro M et al. Abnormal circulating maternal miRNA expression is associated with a low (< 4%) cell-free DNA fetal fraction. Diagnostics 2021; 11(11): 2108. doi: 10.3390/ diagnostics11112108.
13. http:/ / mirwalk.uni-hd.de.
14. http:/ / mirdb.org).
15. Capote-Moreno A, Brabyn P, Muñoz-Guerra M et al. Oral squamous cell carcinoma: epidemiological study and risk factor assessment based on a 39-year series. Int J Oral Maxillofac Surg 2020; 49(12): 1525–1534. doi: 10.1016/ j.ijom.2020.03.009.
16. Rivera C. Essentials of oral cancer. Int J Clin Exp Pathol 2015; 8(9): 11884–11894.
17. Irimie AI, Ciocan C, Gulei D et al. Current insights into oral cancer epigenetics. Int J Mol Sci 2018; 19(3): 670. doi: 10.3390/ ijms19030670.
18. Rapado-González Ó, López-López R, López-Cedrún JL et al. Cell-free microRNAs as potential oral cancer biomarkers: from diagnosis to therapy. Cells 2019; 8(12): 1653. doi: 10.3390/ cells8121653.
19. Barbato S, Solaini G, Fabbri M. MicroRNAs in oncogenesis and tumor suppression. Int Rev Cell Mol Biol 2017; 333 : 229–268. doi: 10.1016/ bs.ircmb.2017.05.
001.20. D’Souza W, Kumar A. MicroRNAs in oral cancer: moving from bench to bed as next generation medicine. Oral Oncol 2020; 111 : 104916. doi: 10.1016/ j.oraloncology.2020.104916.
21. Fang C, Li Y. Prospective applications of microRNAs in oral cancer. Oncol Lett 2019; 18(4): 3974–3984. doi: 10.3892/ ol.2019.10751.
22. Maleki N, Karami F, Heyati S et al. MiR-329-containing small extracellular vesicles derived from breast tumor cells promote endothelial cell angiogenesis through activating KDM1A and VEGF. Ukr Biochem J 2021; 93(4): 37–44 doi: 10.15407/ ubj93.04.037.
23. Schultz DJ, Muluhngwi P, Alizadeh-Rad N et al. Genome-wide miRNA response to anacardic acid in breast cancer cells. PloS One 2017; 12(9): e0184471. doi: 10.1371/ journal.pone.0184471.
24. Guo Q, Li D, Luo X et al. The regulatory network and potential role of LINC00973-miRNA-mRNA ceRNA in the progression of non-small-cell lung cancer. Front Immunol 2021; 12 : 684807. doi: 10.3389/ fimmu.2021.684
807.25. Xia Q, Ding T, Zhang G et al. Circular RNA expression profiling identifies prostate cancer-specific circRNAs in prostate cancer. Cell Physiol Biochem 2018; 50(5):
1903–1915. doi: 10.1159/ 000494870.26. Jeong S, Kim S-A, Ahn S-G. HOXC6-mediated miR-188-5p expression induces cell migration through the inhibition of the tumor suppressor FOXN2. Int J Mol Sci 2021; 23(1): 9. doi: 10.3390/ ijms23010009.
27. Jin Z, Jia B, Tan L et al. MiR‑330‑3p suppresses liver cancer cell migration by targeting MAP2K1. Oncol Lett 2019; 18(1): 314–320. doi: 10.3892/ ol.2019.10280.
28. You J, Zhang Y, Li Y et al. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res 2015; 5(9): 2730–2744.
29. Jain AP, Patel K, Pinto S et al. MAP2K1 is a potential therapeutic target in erlotinib resistant head and neck squamous cell carcinoma. Sci Rep 2019; 9(1): 18793. doi: 10.1038/ s41598-019-55208-5.
30. Lien M-Y, Chang A-C, Tsai H-C et al. Monocyte chemoattractant protein 1 promotes VEGF-A expression in OSCC by activating ILK and MEK1/ 2 signaling and downregulating miR-29c. Front Oncol 2020; 10 : 592415. doi: 10.3389/ fonc.2020.592415.
31. Judd NP, Winkler AE, Murillo-Sauca O et al. ERK1/ 2 regulation of CD44 modulates oral cancer aggressiveness. Cancer Res 2012; 72(1): 365–374. doi: 10.1158/ 0008-5472.CAN-11-1831.
32. Lin C, Tu C, Ma Y et al. Curcumin analog EF24 induces apoptosis and downregulates the mitogen activated protein kinase/ extracellular signal-regulated signaling pathway in oral squamous cell carcinoma. Mol Med Rep 2017; 16(4): 4927–4933. doi: 10.3892/ mmr.2017.7189.
33. Zhang M, Zhu Z-L, Gao X-L et al. Functions of chemokines in the perineural invasion of tumors. Int J Oncol 2018; 52(5): 1369–1379. doi: 10.3892/ ijo.2018.
4311.34. Matsushita Y, Yanamoto S, Takahashi H et al. A clinicopathological study of perineural invasion and vascular invasion in oral tongue squamous cell carcinoma. Int J Oral Maxillofac Surg 2015; 44(5): 543–548. doi: 10.1016/ j.ijom.2015.01.018.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2024 Issue 1-
All articles in this issue
- Evropský plán boje proti rakovině a Mise rakovina – co nám přináší?
- Cardiac conduction system as a new organ at risk in radiotherapy
- Gut microbiome and pancreatic cancer
- Molecular basis of multiple myeloma
- Evaluation pattern within tumor microenvironment and consequent gene expression in oral cancer
- Analysis of the effect of baseline detection and early clearance of ct-DNA, on survival outcomes among patients with advanced EGFR-mutant non-small cell lung cancer
- Immunohistochemical analysis of CD9, CD29 and epithelial to mesenchymal transition in triple-negative breast cancer
- Klinická zkušenost s kabozantinibem u pacientů s metastatickým karcinomem ledviny
- Treatment of tobacco dependence in cancer patients - Recommendations of the Section of Supportive Treatment and Care and the Section of Preventive Oncology of the Czech Cancer Society of the Czech Medical Association of J. E. Purkyně, Working Group for the Prevention and Treatment of Tobacco Dependence of the Czech Medical Association of J. E. Purkyně, and the Society for Treatment of Tobacco Dependence
- Pokročilé léčebné strategie metastatického kolorektálního karcinomu a karcinomu pankreatu
- MU Dr. Libor Havel (1967–2023)
- Doc. Ing. Čestmír Altaner, DrSc. oslávil vzácne životné jubileum – 90 rokov
- Poděkování recenzentům
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Molecular basis of multiple myeloma
- Gut microbiome and pancreatic cancer
- Analysis of the effect of baseline detection and early clearance of ct-DNA, on survival outcomes among patients with advanced EGFR-mutant non-small cell lung cancer
- Cardiac conduction system as a new organ at risk in radiotherapy
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

