-
Medical journals
- Career
Analysis of the effect of baseline detection and early clearance of ct-DNA, on survival outcomes among patients with advanced EGFR-mutant non-small cell lung cancer
Authors: A. Joel 1; R. Abarna 2; R. T. Chacko 1; A. Singh 1; J. T. Georgy 1; A. O. John 1; D. B. Thumaty 1; S. Balukrishna 3; R. Isiah 3; S. Paavamani 3; T. A. Kodiatte 4; S. Rima 4; G. Rebekah 5; R. Pai 2
Authors‘ workplace: Department of Medical Oncology, Christian Medical College and Hospital, Vellore, India 1; Department of Molecular Pathology, Christian Medical College and Hospital, Vellore, India 2; Department of Radiotherapy, Christian Medical College and Hospital, Vellore, India 3; Department of Pathology, Christian Medical College and Hospital, Vellore, India 4; Department of Biostatistics, Christian Medical College and Hospital, Vellore, India 5
Published in: Klin Onkol 2024; 37(1): 40-49
Category: Original Articles
doi: https://doi.org/10.48095/ccko202440Overview
Background: To determine if circulating tumor DNA (ct-DNA) dynamics of epidermal growth factor receptor (EGFR) mutation in plasma can identify a subset of patients with EGFR-mutant (EGFR - m) non-small cell lung cancer (NSCLC) with inferior survival outcomes, we analyzed and compared survival outcomes among patients with and without baseline presence and early clearance of EGFR ct-DNA in plasma. Material and methods: For 66 patients newly diagnosed with EGFR - m NSCLC, plasma samples were collected at baseline and 1st response assessment at 12–24 weeks for extraction of ct-DNA. Estimation of ct-DNA (EGFR exons 18, 19, 20 and 21) was done using droplet digital polymerase chain reaction (dd-PCR) on the QX200 ddPCR system (BioRad, USA). Patients with detectable EGFR ct-DNA at baseline (sample 1), with either undetectable or persistent detectable ct-DNA in sample 2 were classified as clearers and non-clearers, respectively. Results: Fifty-three patients received 1st/ 2nd generation EGFR tyrosine kinase inhibitors (TKIs) and 13 received either 3rd generation TKI (osimertinib) or chemotherapy plus gefitinib. The baseline ct-DNA-positive group had more patients with extra thoracic disease (60.4 vs. 48.5%). For the entire cohort, there was no difference in median progression-free survival (PFS) among baseline ct-DNA-negative (13.57 months) vs. ct-DNA-positive patients (12.32 months) (HR 0.74). There was a significant improvement of PFS among early ct-DNA clearers vs. non-clearers (12.32 vs. 9.92 months; HR 0.57). For those treated with 1st/ 2nd generation EGFR TKIs, this improvement in median PFS among early ct-DNA clearers vs. non-clearers was more apparent (11.76 vs. 6.8 months; HR 0.34). Conclusions: Baseline detection of the presence of ct-DNA of EGFR mutation in plasma was not predictive of first-line PFS, but is associated with extra thoracic disease. Patients with EGFR mutation and persistence of ct-DNA at first follow-up have worse PFS and overall survival (OS) in comparison to those clearing the same in plasma, especially among those treated with 1st/ 2nd generation EGFR TKIs.
Keywords:
advanced non-small cell lung cancer – EGFR mutation – ct-DNA – mutant allele frequency – EGFR tyrosine kinase inhibitors
Introduction
The therapeutic landscape of first-line management of advanced non-small-cell lung cancer (NSCLC) with a driver mutation in the epidermal growth factor receptor (EGFR) now ranges from EGFR tyrosine kinase inhibitors (TKIs), a combination of chemotherapy with 1st generation EGFR TKIs, EGFR TKIs + vascular endothelial growth factor (VEGF) inhibitors (bevacizumab/ ramucirumab) to chemo-immunotherapy [1–3]. Patients treated with 1st/2nd generation EGFR TKIs eventually develop resistance and progress within 9–12 months of treatment, a large percentage of which is attributed to the acquired mutant T790M [2]. Though testing systems to detect T790M are readily available, a significant proportion of patients may not receive second or subsequent lines of systemic therapy in the real world; more so in low middle income countries (LMIC) [1,4,5]. It is also well established that first-line chemotherapy + TKI combinations show an overall survival (OS) benefit and are feasible in the LMIC setting, though this option is associated with three-weekly hospital visits, higher toxicity when compared to TKIs alone, and is also beset by the relative dearth of information regarding the patterns of resistance that emerge while on treatment [1,6].
The initial use of detection of the presence of EGFR mutation in the ct-
-DNA in plasma was limited to the detection of the presence of resistant mutations (mostly T790M). With the arrival of newer and more sensitive techniques like the digital droplet polymerase chain reaction (ddPCR), the detection of the presence of various EGFR mutants in ct--DNA in blood and their quantification in the form of mutant allele frequency (MAF), is feasible, with prior reports correlating baseline presence of ct-DNA in blood with poorer survival outcomes [7–11]. The ddPCR platform not only detects the presence of T790M mutation (both at baseline and progression) but also enables its quantification in the form of MAF; MAF-based longitudinal monitoring in the T790M mutant population has helped predict clinical outcomes in the second line setting [7,11–13].In addition, there is emerging information with respect to the correlation between the baseline presence and early clearance of ct-DNA of EGFR mutation in plasma to survival outcomes among patients treated with EGFR TKIs in the first-line setting [10,11,14–18] These data thus point towards the utility of early ct--DNA clearance as a prognostic indicator in the management of these patients.
Our study is an initial analysis of a longitudinal analysis of ct-DNA dynamics among patients with EGFR-mutant (EGFR - m) NSCLC treated in the first-line and aimed to determine whether baseline detection and early clearance of ct-DNA of EGFR mutation in plasma (using dd-PCR) correlated with survival outcomes.
Materials and methods
Study setting
This prospective single centre cohort study was carried out at a teaching hospital in South India. We enrolled all consecutive newly diagnosed cases of advanced EGFR - m NSCLC, who were treated in the first line between August 2018 and November 2020.
Patient selection
This is an initial report of a prospective study designed to assess the role of serial longitudinal monitoring of plasma ct-DNA among all patients with EGFR - m NSCLC, commencing first-line treatment with 1st/ 2nd generation EGFR TKI. However, after the emergence of practice changing data supporting the use of both osimertinib and chemotherapy + gefitinib combination as first-line treatments, we included these patients, too.
All patients with newly diagnosed, histologically proven EGFR-m (exon 18, 19, 20 or 21 mutation) NSCLC-adenocarcinoma (stage IIIB or IV), aged ≥ 18 years, with Eastern Cooperative Oncology Group (ECOG) performance status 0–2, planned for the treatment with EGFR TKIs or chemotherapy + EGFR TKI combination, were enrolled. Written informed consent was obtained from all patients. Patients who did not have a baseline blood sample, received palliative radiotherapy to the primary mass prior to the start of TKI or harbored a de novo T790M mutation detected during routine baseline evaluation were excluded. The study was approved by the Institutional Review Board (IRB) of the Christian Medical College, Vellore (IRB Min No 10452 dated 14/ 12/ 2016).
Sample collection
At baseline, EGFR mutational analysis was performed on formalin-fixed paraffin embedded tissues (FFPE) sections. Following confirmation of EGFR mutation in tissue, after obtaining informed consent, the first blood sample (Sample 1 - baseline) was collected prior to initiation of first-line treatment. The next blood sample (Sample 2) was collected at the next follow-up, between 3–4 months from initiation of first-line treatment. Since a large number of our patients travel from other states for treatment at our centre and since patients recruited in 2020 could not report at 3–4 months for their first reassessment due to the COVID pandemic, we included those reporting up to 6 months from diagnosis for assessment of the second sample (Sample 2) for analysis for early clearance of ct-DNA of EGFR mutation in plasma. Patients with a detectable EGFR ct-DNA in plasma at baseline, which became undetectable in the second sample were classified as clearers while those in whom the EGFR ct--DNA was persistently detectable in the second sample were classified as non-clearers.
Molecular testing for EGFR-technique
A) DNA extraction from tissue: Formalin fixed paraffin embedded (FFPE) blocks were examined and the area with maximum tumor cellularity was identified. Extraction was performed with 3–4 5-µm sections using the DNA FFPE tissue extraction kit (Qiagen India). The DNA was quantitated using the Nanodrop (NanoDrop technologies, USA), the 260/ 280 ratio was determined and used for EGFR mutational analysis by ddPCR.
B) Blood: Ten mL of peripheral venous blood was collected in PAXgene blood cf-DNA tube (QIAGEN, India). Briefly, the tube was centrifuged at 1,900 g for 15 min at room temperature, the plasma was removed into another conical tube and frozen at −800 °C until further use. Cell-free DNA (cf-DNA) was extracted from plasma using the Quick cf-DNA serum and plasma kit (Zymo research, USA) and the DNA was stored at −200 °C until further use.
C) Droplet digital PCR (ddPCR): Droplet digital PCR was performed on the QX200 ddPCR system (BioRad, USA) using the prime PCR assays for detection of mutants including E746-A750 deletion, L858R, and T790M (Bio-Rad, USA). The cycling conditions for PCR included an initial incubation at 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 55 °C for 60 s and final enzyme inactivation at 98 °C for 10 min. The digital PCR data was analyzed using the Quanta Soft analytical software package version 1.3.2.0 (BioRad, USA). The baseline tissue, baseline plasma sample and all subsequent plasma samples were analyzed by ddPCR. The data generated were expressed as copies of target/ microliter (copies/ μL). The concentration was calculated using the formula: −ln (Nneg/ N)/ Vdroplet, where Vdroplet = volume of droplet, Nneg = number of negative droplets, N = total number of droplets and E = observed fraction of droplets. During standardization, DNA from NCI-H1975 (harbouring T790M) was serially diluted in human reference genomic DNA (catalogue no. G1471, Promega) to achieve decreasing ratios (from 1 : 100 to 1 : 10,000) helping to determine the sensitivity of the assay. In all subsequent runs, the same human reference genomic DNA was also included as a negative control.
Clinical parameters
We collected information on patient demographics, smoking status, staging, disease extent, details of treatment including date of initiation and periodic follow-up. All patients with EGFR-m NSCLC were treated with 1st/ 2nd generation EGFR TKIs (gefitinib, erlotinib or afatinib), 3rd generation TKI (osimertinib) or chemotherapy (pemetrexed and carboplatin) + TKI (gefitinib) combination. Dose modifications were done as per standard clinical practice and first-line treatment was continued till either disease progression or unacceptable toxicity. Patients underwent reassessment with contrast enhanced CT of the thorax at their first follow-up visit, 3–4 months from the start of their first-line treatment. Details at progression were also noted, including the presence of T790M mutation. Following disease progression, data for OS analysis were still collected; by review of outpatient records or by telephonic follow-up.
Efficacy assessments
Our primary and secondary endpoints were progression free survival (PFS) and OS, respectively. These were further compared among patients with ct-DNA positive vs. negative at baseline and also among clearers vs. non-clearers. Response Evaluation Criteria in Solid Tumours (RECIST), version 1.1, were used to assess patients’ response to treatment.
Statistical analysis
The data were entered in Epi Info Version 7.2.3.1 and analyzed on Statistical Package for the Social Sciences (SPSS), version 21. Graphs were plotted using RStudio Version 1.3.1093; R version 4.0.0. Categorical variables were compared using the chi square and Fisher’s exact tests. Survival analysis was done using the Kaplan-Meier method, and comparison between groups using the Cox--Mantel log-rank test. PFS was calculated from the date of initiation of first-line treatment to the date of progression or death. OS was calculated from the date of diagnosis of advanced NSCLC to the date of the last follow-up or death.
1. STROBE diagram for study population. 
STROBE – strengthening the reporting of observational studies in epidemiology. Results
Patients
We recruited 66 patients in the study (Scheme 1). The demographics and clinical characteristics are summarised in Tab. 1. The median age at diagnosis was 57 years (range 31–85). Three patients (4.5%) had oligometastatic disease. De novo brain metastases were present in 7 (10.6%) patients. Extra thoracic disease involvement was seen in 36 (54.5%) patients and 6 (9.1%) patients had ≥ 2 sites of extra thoracic involvement. Forty-nine (74.2%) patients were treated with 1st generation EGFR TKI (gefitinib/ erlotinib), 4 (6%) with second generation TKI (afatinib), 12 (17.9%) with 3rd generation TKI (osimertinib) and 1 (1.5%) was treated with chemotherapy + TKI (carboplatin + pemetrexed) with gefitinib. The median follow-up was 13.8 months (range 1–31.44).
1. Baseline characteristics (N = 66). 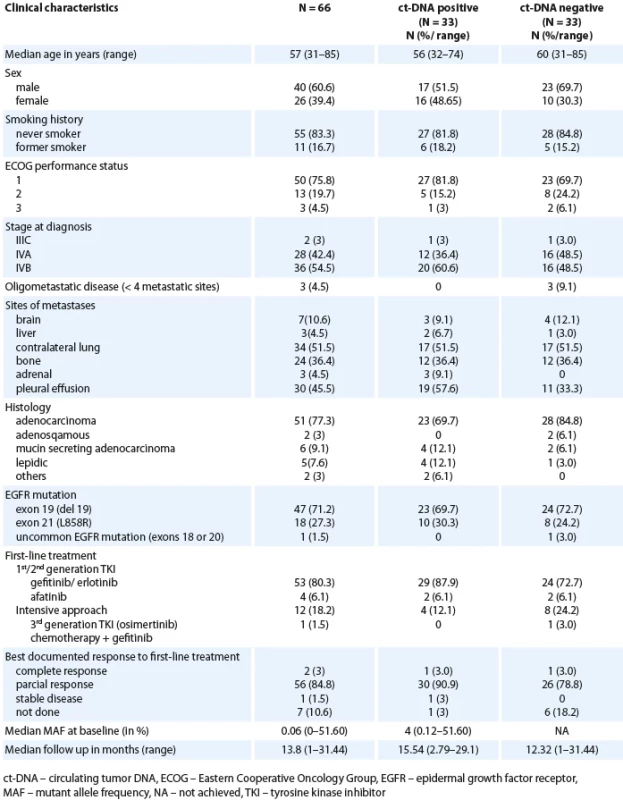
Baseline plasma ct-DNA detection of EGFR exon 19/ 21
At diagnosis, 33 (50%) patients had a detectable ct-DNA in plasma (based on the MAF cutoff 0.1%). This cutoff was used as this was the documented lower limit of detection for the dd-PCR assay. Thus, the concordance between the tissue and liquid biopsy for the detection of EGFR mutation on dd-PCR was 50% (33 out of 66).
Correlation of baseline ct-DNA detection in plasma with baseline characteristics, disease burden and response rates
This is highlighted in Tab. 1. All three patients with oligometastatic disease were ct-DNA negative at baseline. A higher proportion of patients in the baseline ct-DNA positive group had extra thoracic disease (60.4%) in contrast to those who were ct-DNA negative at baseline (48.5%). The overall response rates were 93.9% and 81.8% in the ct-DNA positive and ct-DNA negative cohorts, respectively. Response assessment was not available in 6 (18.2%) patients in the ct-DNA negative cohort. The proportion of non-smokers were equal in the two groups (baseline ct-DNA positive vs. ct--DNA negative).
ct-DNA clearance in second sample
The median time to the second sample (Sample 2) was 3.45 months (interquartile range – IQR 2.99–4.60) among 47 (60.7%) patients who gave the second plasma sample. Among 33 patients with initially detectable ct-DNA, 26 provided a second sample; 17 demonstrated clearance of the EGFR mutation at first follow-up (Scheme 1). With the MAF cutoff > 0.1%, 9 patients (34.6%) had persistent detectable ct-DNA in the plasma and were classified as non-clearers.
1. Progression free survival as per baseline ct-DNA. A) All patients (N = 66); B) patients on 1st/2nd generation TKI (N = 53). 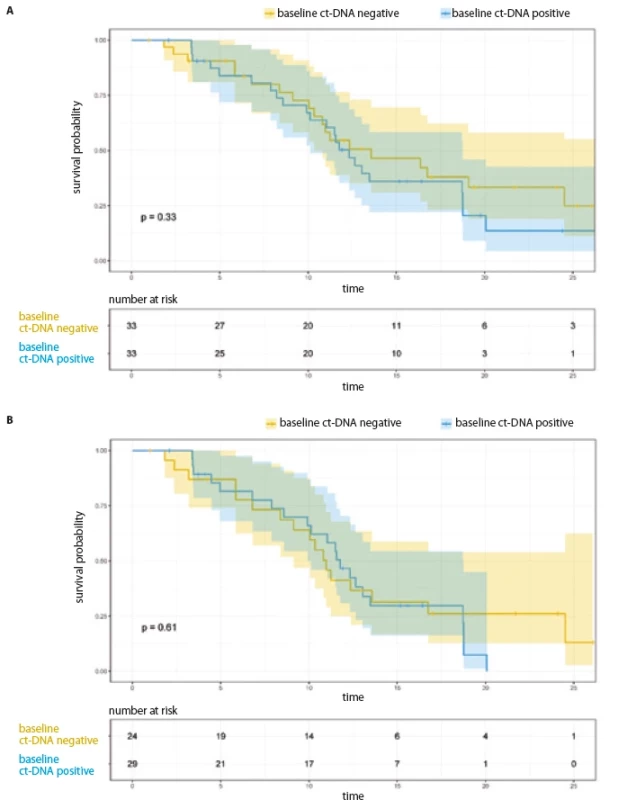
ct-DNA – circulating tumor DNA PFS analysis
The median PFS for the entire cohort was 12.65 months (95% CI 10.51–14.79) and 11.57 months (95% CI 9.92–13.21) for the 53 patients who were treated with 1st/ 2nd generation EGFR TKI. The median PFS among the 13 patients treated with osimertinib/ chemotherapy + TKI combination was 26.35 months (95% CI 15.77–36.93).
PFS analysis based on baseline EGFR ct-DNA in plasma and its early clearance at first follow-up (entire cohort, N = 66)
For the entire cohort, there was no significant difference in the PFS, based on baseline ct-DNA detection (Tab. 2, Fig. 1A). Among 33 patients with initially detectable ct-DNA, 26 provided a second sample; 17 demonstrated clearance of the EGFR mutation at first follow-up. On comparison of median PFS among the clearers (median 12.32 months; 95% CI 9.63–15.01) vs. non-clearers (median 9.92 months; 95% CI 3.32–16.52) (HR 0.57; 95% CI 0.22–1.44), there was a trend towards better PFS in the group who cleared the plasma ct-DNA at their first follow-up (Tab. 2, Fig. 2A).
2. Median PFS and OS as per baseline ct-DNA detection and ct-DNA clearance (all patients, N = 66). 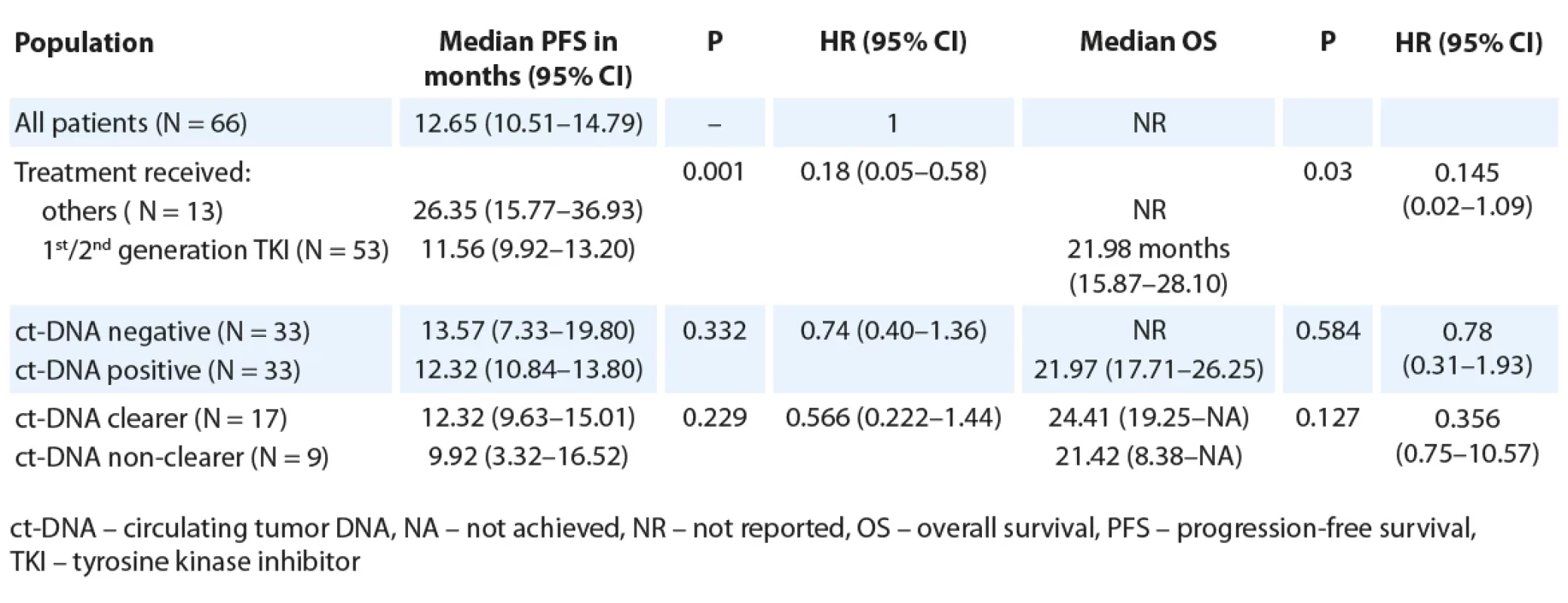
PFS analysis based on baseline EGFR ct-DNA in plasma and its early clearance at first follow-up (patients treated with 1st/ 2nd generation TKI, N = 53)
Restricting the PFS analysis to the 53 patients treated with 1st/ 2nd generation EGFR TKI, the median PFS was 11.57 months (95% CI 9.92–13.21). Twenty-nine patients with detectable ct-DNA at baseline were treated with 1st/ 2nd generation TKI. There was no significant difference in PFS with respect to baseline ct-DNA positivity (Fig. 1B, Tab. 3). Out of 29 patients treated with 1st/ 2nd generation TKI and with detectable ct-DNA at baseline, 22 patients gave a second sample. Here, among the 14 clearers, we noted a trend towards an improved PFS (12.04 months; 95% CI 10.12–NR), in comparison to the 8 non-clearers (PFS 6.80 months; 95% CI 4.46–NR) (HR 0.34; 95% CI 1.03–8.38) (Tab. 3, Fig. 2B).
3. Median PFS and OS as per baseline ct-DNA detection and ct-DNA clearance among patients treated with 1st/2nd generation EGFR TKI (N = 53). 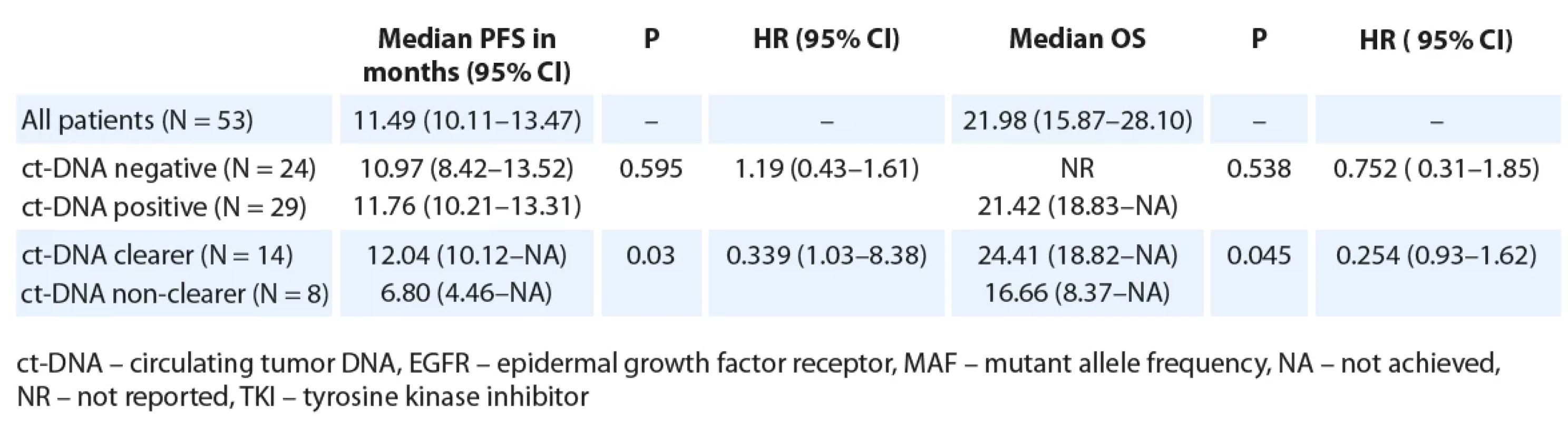
OS analysis
At data cutoff, 21 (31.8%) patients were still continuing first-line treatment. Eighteen (27.3%) were alive on subsequent lines of treatment and 8 (12.1%) were on 1st line TKI beyond radiological progression.
OS analysis based on baseline ct-DNA detection and clearance at first follow-up (entire cohort, N = 66)
The median OS of all patients was not reached. The median OS among patients treated with 1st/ 2nd generation EGFR TKIs, was lower (21.98 months; 95% CI 15.87–28.10) than those who were treated with either osimertinib or chemotherapy/ TKI combination (not reached; P = 0.03, HR 0.15; 95% CI 0.02–1.09). A similar trend towards an inferior OS was seen in patients with ct-DNA positivity at baseline (21.97 months; 95% CI 17.71–26.25) in contrast with those who were ct-DNA negative at baseline (median OS NR, P = 0.584, HR 0.78; 95% CI 0.31–1.93) (Tab. 2). There was no significant difference in OS among ct-DNA clearers (median: 24.41 months; 95% CI 14.78–34.04) vs. non-clearers (median 21.4 months; 95% CI 7.25–35.59; P = 0.110, HR 0.36 (0.09–1.34) (Tab. 2).
OS analysis based on baseline ct-DNA and clearance at first follow-up (patients treated with 1st/ 2nd generation TKI, N = 53)
For the patients treated with 1st/ 2nd generation EGFR TKI, the median OS was 21.98 months (95% CI 15.87–28.10). There was a trend towards an improved OS for patients who were ct-DNA negative at baseline (Tab. 3). Also, a trend towards improved median OS for ct-DNA clearers (24.4 months) vs. non-clearers (16.6 months) (P = 0.045, HR 0.25; 95% CI 0.06–1.07) was seen in this subgroup of patients (Tab. 3).
T790M incidence at progression and second-line treatment
There were no patients with de novo T790M mutation at diagnosis. Among 66 patients, 43 had a PFS event (either death or progressive disease) and 20 (46.5%) underwent mutation testing (either on liquid biopsy; N = 19, or tissue biopsy; N = 1) at progression. T790M mutation at progression was documented in 9 (16.9%) out of 53 patients on 1st/ 2nd generation TKI. Among these, 3 were detected during serial plasma samples; while on first-line therapy and the appearance of T790M in plasma preceded clinico-radiological progression by 12, 15 and 17 weeks respectively. One patient developed C797S mutation at progression on second-line osimertinib and progressed despite a 6-week empirical course of osimertinib 80 mg once daily in combination with gefitinib 250 mg once daily.
2. Progression free survival based on ct-DNA clearance. A) All patients (N = 26); B) patients on 1st/2nd generation TKI (N = 22). 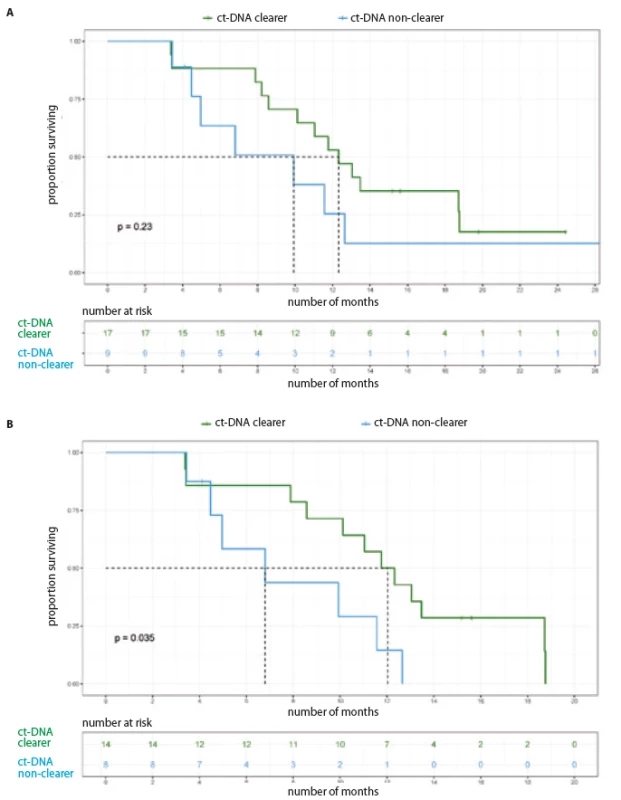
ct-DNA – circulating tumor DNA Discussion
The past half decade has witnessed heightened activity in terms of the increasing number of treatment options available for first-line treatment of EGFR - m NSCLC. However, in LMIC, despite a high prevalence of EGFR - m NSCLC, osimertinib, bevacizumab and ramucirumab in the first or subsequent lines of treatment are inaccessible to most patients. The type of mutation (exon 19 vs. exon 21 L858R) does not predict differential survival benefit with either TKIs (1st/ 2nd or 3rd generation) or chemotherapy/ TKI combination in the first-line setting [1,2,19]. While the world awaits the results of the first-line chemotherapy + osimertinib combination (FLAURA-2), it would be prudent in the LMIC setting (where majority of patients are still treated with 1st/ 2nd generation EGFR TKIs as the first line), to determine which subsets of these patients with EGFR - m NSCLC have inferior survival outcomes and hence may benefit from a more intensive first-line approach, i.e. 3rd generation TKI or chemotherapy/ TKI combination.
In our study, there was a concordance of 50% between liquid biopsy (using dd-PCR in plasma) and tissue testing of EGFR mutation at baseline. This is in contrast to other studies where percentages as high as 81% have been described, though a fair number have also reported lower concordance as seen in our study [8,14,19–21].
In our cohort, the presence of detectable ct-DNA in plasma at baseline correlated with increasing disease stage (IVB vs. others) and number of extra thoracic metastases. These are well established as poor prognostic factors for survival outcomes among patients who are treated with 1st/ 2nd generation EGFR TKIs [5,13,17].
This study failed to demonstrate that the presence of detectable ct-DNA in plasma at baseline is a negative prognostic factor which is predictive of inferior survival outcomes (OS and PFS) among our cohort of patients with EGFR - m NSCLC. This is in contrast to prior studies (using 1st/ 2nd generation TKIs and osimertinib) which have shown an improvement in PFS among patients who are baseline ct-DNA negative in comparison to those who are ct--DNA positive at baseline [10,15,17,18]. Further, the largest database in this regard, i.e. a recent meta-analysis by Phan et al. has shown that the presence of EGFR mutation in pre treatment plasma was associated with a shorter PFS and OS. [17] The exact reason behind this difference in outcome between our cohort and prior datasets is unclear and could be attributable to the smaller number of patients in our study.
Importantly, we demonstrated an improvement in PFS and OS among ct--DNA clearers vs. non-clearers at first follow-up, consistent with similar prior studies [10,11,14–18,20]. The meta-analysis by Phan et al. was also in keeping with this finding and showed worse PFS and OS in patients with persistent EGFR mutation in plasma post-treatment. This finding is similar across all studies irrespective of the testing platform and treatments used [10,14,15,17].
There is an immediate need to prospectively validate these findings in a larger cohort of patients who have a detectable ct-DNA at baseline. This may have clinical implications with regard to identification of a subset of patients with EGFR - m NSCLC who are at higher risk of early progression and may hence benefit from escalation or intensification of therapy; with either the addition of chemotherapy to 1st generation TKI or switching to osimertinib. Such an approach will then require prospective evaluation in the form of a randomized clinical trial comparing continuation of standard treatment vs. treatment escalation among patients who are persistently ct-DNA positive (ct-DNA non-clearers) at first follow-up. This has tremendous implications for treatment of EGFR - m NSCLC in the LMIC setting where a higher proportion of patients present with advanced disease, extra thoracic involvement, are more likely to be treated with 1st or 2nd generation EGFR TKIs and may not end up receiving later lines of treatment at progression.
Our rates of T790M mutation at progression are lower than previous cohorts due to two crucial factors: a) a significant proportion of our patients are still on 1st line treatment and b) fewer patients were able to undergo molecular testing upon progression due to the COVID-19 pandemic.
To our knowledge, ours is one of the very few studies of longitudinal monitoring and ct-DNA dynamics of EGFR - m NSCLC in the first-line setting, using the dd-PCR testing platform. The majority of data that exists is based on findings shown using allele specific PCR [8,10,14]. Using the ultra-sensitive dd-PCR platform allows for detection of low quantities of ct-DNA and its quantification by MAF and in turn correlation of various MAF cutoffs with survival outcomes [11]. We did not study this correlation between various MAF cutoff and survival outcomes in our cohort as the number of patients with ct-DNA persistence was only nine and we would not be able to draw meaningful conclusions due to the small number. Li et al. have studied this using a ‘targeted’ NGS panel and have also demonstrated similar findings correlating ct-DNA clearance and survival outcomes [16].
Our study is not without limitations. Though we initially planned to include only patients treated with 1st/ 2nd generation EGFR TKIs, we had to include patients who were started on first-line osimertinib or chemotherapy/ TKI combination during the latter half of the study; in keeping with practice changing studies which were published during our recruitment period [1,3]. Also, due to the COVID-19 pandemic, some of our patients were unable to travel to our centre for serial longitudinal mutational monitoring plasma samples, at the planned 3–4 monthly intervals. Moreover, despite its sensitivity, dd-PCR will not be able to detect the presence of any other concomitant driver mutations which could also influence responses to EGFR TKIs. In our patient cohort, the study of the various concomitant gene mutations (using next generation sequencing), among those with persistence of ct-DNA, could have provided more clarity towards the molecular basis behind the lack of response to standard treatments [21,22].
Baseline plasma detection of ct-DNA EGFR mutation positively correlates with a higher disease burden as evidenced by association with higher stage and extra thoracic metastatic involvement. Persistence of ct-DNA on plasma among patients with EGFR - m NSCLC at first follow-up is predictive of inferior survival outcomes (PFS and OS); especially among patients treated with 1st/ 2nd generation EGFR TKIs and allows us to identify patients with inferior survival outcomes and opens up an opportunity for intensification of their treatment.
Conclusion
Detectable ct-DNA in plasma at baseline correlates with disease burden (disease stage and number of extra thoracic metastasis). Patients with EGFR mutation and persistence of ct-DNA at first follow-up have worse PFS and OS in comparison to those clearing the same in plasma; more so among those treated with 1st/ 2nd generation EGFR TKIs.
Acknowledgements
This study was funded by a research grant from the Department of Science of Technology, Government of India (EMR/2016/005292). We acknowledge and thank Mrs Anandhi Jegan and Mrs Saraswathi from the Department of Medical Oncology, CMC Vellore for their assistance in patient follow-up.
Sources
1. Noronha V, Patil VM, Joshi A et al. Gefitinib versus gefitinib plus pemetrexed and carboplatin chemotherapy in EGFR-mutated lung cancer. J Clin Oncol 2020; 38(2):
124–136. doi: 10.1200/ JCO.19.01154.2. Zhao Y, Liu J, Cai X et al. Efficacy and safety of first line treatments for patients with advanced epidermal growth factor receptor mutated, non-small cell lung cancer: systematic review and network meta-analysis. BMJ 2019; 367: l5460. doi: 10.1136/ bmj.l5460.
3. Soria J-C, Ohe Y, Vansteenkiste J et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018; 378(2): 113–125. doi: 10.1056/ NEJMoa1713137.
4. Noronha V, Patil VM, Joshi A et al. Epidermal growth factor receptor positive lung cancer: the nontrial scenario. Indian J Cancer 2017; 54(1): 132–135. doi: 10.4103/ 0019-509X.219583.
5. Garg A, Batra U, Choudhary P et al. Clinical predictors of response to EGFR-tyrosine kinase inhibitors in EGFR-mutated non-small cell lung cancer: a real-world multicentric cohort analysis from India. Curr Probl Cancer 2020; 44(3): 100570. doi: 10.1016/ j.currproblcancer.2020.100570.
6. Hosomi Y, Morita S, Sugawara S et al. Gefitinib alone versus gefitinib plus chemotherapy for non-small-cell lung cancer with mutated epidermal growth factor receptor: NEJ009 study. J Clin Oncol 2020; 38(2): 115–123. doi: 10.1200/ JCO.19.01488.
7. Buder A, Hochmair MJ, Setinek U et al. EGFR mutation tracking predicts survival in advanced EGFR-mutated non-small cell lung cancer patients treated with osimertinib. Transl Lung Cancer Res 2020; 9(2): 239–245. doi: 10.21037/ tlcr.2020.03.02.
8. Mok T, Wu Y-L, Lee JS et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015; 21(14): 3196–3203. doi: 10.1158/ 1078-0432.CCR-14-2594.
9. O’Kane GM, Liu G, Stockley TL et al. The presence and variant allele fraction of EGFR mutations in ctDNA and development of resistance. Lung Cancer 2019; 131 : 86–89. doi: 10.1016/ j.lungcan.2019.03.019.
10. Gray JE, Okamoto I, Sriuranpong V et al. Tissue and plasma EGFR mutation analysis in the FLAURA trial: osimertinib versus comparator EGFR tyrosine kinase inhibitor as first-line treatment in patients with EGFR-mutated advanced non-small cell lung cancer. Clin Cancer Res 2019; 25(22): 6644–6652. doi: 10.1158/ 1078-0432.CCR-19-1126.
11. Provencio M, Serna-Blasco R, Franco F et al. Analysis of circulating tumour DNA to identify patients with epidermal growth factor receptor-positive non-small cell lung cancer who might benefit from sequential tyrosine kinase inhibitor treatment. Eur J Cancer 2021; 149 : 61–72. doi: 10.1016/ j.ejca.2021.02.031.
12. Su K-Y, Tseng J-S, Liao K-M et al. Mutational monitoring of EGFR T790M in cfDNA for clinical outcome prediction in EGFR-mutant lung adenocarcinoma. PloS One 2018; 13(11): e0207001. doi: 10.1371/ journal.pone.0207
001.13. Lettig L, Sahnane N, Pepe F et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl Lung Cancer Res 2019; 8(5): 584–592. doi: 10.21037/ tlcr.2019.09.18.
14. Behel V, Choughule A, Noronha V et al. Clinical utility of liquid biopsy (cell-free DNA) based EGFR mutation detection post treatment initiation as a disease monitoring tool in patients with advanced EGFR-mutant NSCLC. Clin Lung Cancer 2022; 23(5): 410–418. doi: 10.1016/
j.cllc.2022.04.002.15. Fukuhara T, Saito H, Furuya N et al. Evaluation of plasma EGFR mutation as an early predictor of response of erlotinib plus bevacizumab treatment in the NEJ026 study. EBioMedicine 2020; 57 : 102861. doi: 10.1016/ j.ebiom.2020.102861.
16. Li Y, Xu Z, Wang S et al. Disease monitoring of epidermal growth factor receptor (EGFR)-mutated non-small-
-cell lung cancer patients treated with tyrosine kinase inhibitors via EGFR status in circulating tumor DNA. Thorac Cancer 2022; 13(15): 2201–2209. doi: 10.1111/ 1759-7714.14545.17. Phan TT, Tran VT, Tran B-T et al. EGFR-plasma mutations in prognosis for non-small cell lung cancer treated with EGFR TKIs: a meta-analysis. Cancer Rep 2022; 5(8): e1544. doi: 10.1002/ cnr2.1544.
18. Ebert EBF, McCulloch T, Hansen KH et al. Clearing of circulating tumour DNA predicts clinical response to first line tyrosine kinase inhibitors in advanced epidermal growth factor receptor mutated non-small cell lung cancer. Lung Cancer 2020; 141 : 37–43. doi: 10.1016/ j.lungcan.2019.12.016.
19. Noronha V, Patil VM, Joshi A et al. Epidermal growth factor receptor positive lung cancer: the nontrial scenario. Indian J Cancer 2017; 54(1): 132–135. doi: 10.4103/ 0019-509X.219583.
20. Kuo C-Y, Lee M-H, Tsai M-J et al. The factors predicting concordant epidermal growth factor receptor (EGFR) mutation detected in liquid/ tissue biopsy and the related clinical outcomes in patients of advanced lung adenocarcinoma with EGFR mutations. J Clin Med 2019; 8(11): 1758. doi: 10.3390/ jcm8111758.
21. Zhang G, Cheng R, Niu Y et al. Efficacy differences of first-line EGFR-TKIs alone vs. in combination with chemotherapy in advanced lung adenocarcinoma patients with sensitive EGFR mutation and concomitant non-EGFR genetic alterations. Zhongguo Fei Ai Za Zhi 2022; 25(9):
651–657. doi: 10.21203/ rs.3.rs-254564/ v1.22. Hong S, Gao F, Fu S et al. Concomitant genetic alterations with response to treatment and epidermal growth factor receptor tyrosine kinase inhibitors in patients with EGFR-mutant advanced non-small cell lung cancer. JAMA Oncol 2018; 4(5): 739–742. doi: 10.1001/ jamaoncol.2018.0049.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2024 Issue 1-
All articles in this issue
- Evropský plán boje proti rakovině a Mise rakovina – co nám přináší?
- Cardiac conduction system as a new organ at risk in radiotherapy
- Gut microbiome and pancreatic cancer
- Molecular basis of multiple myeloma
- Evaluation pattern within tumor microenvironment and consequent gene expression in oral cancer
- Analysis of the effect of baseline detection and early clearance of ct-DNA, on survival outcomes among patients with advanced EGFR-mutant non-small cell lung cancer
- Immunohistochemical analysis of CD9, CD29 and epithelial to mesenchymal transition in triple-negative breast cancer
- Klinická zkušenost s kabozantinibem u pacientů s metastatickým karcinomem ledviny
- Treatment of tobacco dependence in cancer patients - Recommendations of the Section of Supportive Treatment and Care and the Section of Preventive Oncology of the Czech Cancer Society of the Czech Medical Association of J. E. Purkyně, Working Group for the Prevention and Treatment of Tobacco Dependence of the Czech Medical Association of J. E. Purkyně, and the Society for Treatment of Tobacco Dependence
- Pokročilé léčebné strategie metastatického kolorektálního karcinomu a karcinomu pankreatu
- MU Dr. Libor Havel (1967–2023)
- Doc. Ing. Čestmír Altaner, DrSc. oslávil vzácne životné jubileum – 90 rokov
- Poděkování recenzentům
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Molecular basis of multiple myeloma
- Gut microbiome and pancreatic cancer
- Analysis of the effect of baseline detection and early clearance of ct-DNA, on survival outcomes among patients with advanced EGFR-mutant non-small cell lung cancer
- Cardiac conduction system as a new organ at risk in radiotherapy
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

