-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: A prospective study in children
Anette-Gabriele Ziegler and colleagues report their novel genetic risk score for identifying infants with a high risk of developing type 1 diabetes.
Published in the journal: . PLoS Med 15(4): e32767. doi:10.1371/journal.pmed.1002548
Category: Research Article
doi: https://doi.org/10.1371/journal.pmed.1002548Summary
Anette-Gabriele Ziegler and colleagues report their novel genetic risk score for identifying infants with a high risk of developing type 1 diabetes.
Introduction
Precision medicine typically relies on our ability to identify individuals with precise genetic elements that define a disease. These elements may be used not only to select optimal treatment modalities, but also to identify individuals who may benefit from preventative interventions. In pediatric disease, current studies seeking to elucidate disease etiology, as well as clinical trials aimed at prevention, rely on identifying and enrolling infants with increased risk [1–7]. The risk for diseases such as allergy, type 1 diabetes, and celiac disease is often assessed in terms of family history [1–3,7], which, at best, identifies 10% of children who subsequently develop the condition [7,8].
In type 1 diabetes, genotypes in the human leukocyte antigen (HLA) DRB1, DQA1, and DQB1 loci are sometimes used to identify at-risk infants from the general population [2,9,10]. Risk is 5% in children with the 2 highest-risk HLA genotypes (DR3 and DR4-DQ8 or homozygous for DR4-DQ8), and 40% of cases of childhood type 1 diabetes have 1 of these 2 genotypes [11]. Although the HLA loci are the strongest genetic risk markers for type 1 diabetes, many other regions of the genome also confer susceptibility to type 1 diabetes [12]. Therefore, it is conceivable that risk stratification could be improved if risk is calculated according to genetic information derived from multiple genetic susceptibility regions [13,14].
We previously applied logistic regression to the Type 1 Diabetes Genetics Consortium (T1DGC) case–control dataset and developed a weighted genetic score derived from HLA and 40 type 1 diabetes susceptibility loci (Winkler score) [15]. Independently, a genetic score derived from HLA plus 25 susceptibility loci was developed in the UK using Wellcome Trust Case Control Consortium (WTCCC) data (Oram score) [16]. These studies suggested that the scores might improve our ability to predict and diagnose type 1 diabetes. Hence, genetic scores could become a new paradigm for stratifying type 1 diabetes risk and for recruitment into primary prevention trials, and provide a proof of principle for other diseases with multiple known genetic susceptibility markers. With this in mind, the 2 consortia joined efforts to determine how the 2 genetic scores and a merged score performed in a prospective study.
The Environmental Determinants of Diabetes in the Young (TEDDY) study, a multicenter cohort study set in Germany, Finland, Sweden, and the US, has intensively followed several thousand HLA-selected children from birth for the development of islet autoantibodies and of diabetes [17]. The presence of 2 or more islet autoantibodies (multiple islet autoantibodies) in genetically at-risk children defines a pre-symptomatic stage of type 1 diabetes where progression to type 1 diabetes is around 80% over 10 years [18,19]. TEDDY offers the unique opportunity to test the multiple-locus genetic scores in a prospectively studied cohort of children who have high-risk HLA genotypes in the absence of family history of type 1 diabetes [2,17]. The objective of our analysis was to determine whether the genetic scores could identify infants from the general population who had at least a 10% risk for type 1 diabetes, a risk threshold that has been used for primary prevention trials and that has previously only been achievable in infants with a family history of type 1 diabetes [3].
Methods
Case–control cohort
We reasoned that our target risk of 10% could only be achieved by applying our multi-locus genetic scores in individuals who had the highest-risk HLA genotypes. We obtained data for controls from the UK Biobank (https://www.ukbiobank.ac.uk/) [20] and data for controls and cases from the WTCCC [21], and calculated the Winkler and Oram scores in 4,371 non-diabetic individuals who were heterozygous for HLA DR3-DQA1*0501-DQB1*0201 and DR4-DQA1*030X-DQB1*0302 (HLA DR3/DR4-DQ8) or who were homozygous for HLA DR4-DQA1*030X-DQB1*0302 (HLA DR4-DQ8/DR4-DQ8) (controls) and 781 patients with type 1 diabetes who had 1 of these 2 genotypes (cases). UK Biobank participants were aged 40 to 69 years, and the WTCCC patients were all aged <50 years when sampled.
TEDDY cohort
TEDDY is a prospective cohort study conducted at 3 centers in the US (Colorado, Georgia/Florida, and Washington) and 3 centers in Europe (Finland, Germany, and Sweden) [2,17]. Between 1 September 2004 and 28 February 2010, a total of 421,047 newborn children were screened for high-risk HLA genotypes for type 1 diabetes [22]. HLA genotype screening was conducted as previously described [22]. The families of children with type 1 diabetes risk HLA genotypes were invited to participate in the follow-up study in which blood samples were obtained every 3 months for the first 4 years and biannually thereafter for the analysis of islet autoantibodies (glutamic acid decarboxylase antibody [GADA], insulinoma antigen-2 antibody [IA-2A], and insulin autoantibodies [IAAs]). The HLA genotypes were confirmed by the central HLA Reference Laboratory at Roche Molecular Systems (Oakland, CA) for enrolled participants. The present report includes TEDDY children with the HLA DR3/DR4-DQ8 or the HLA DR4-DQ8/DR4-DQ8 genotype, without a first-degree relative with type 1 diabetes, if at least 1 blood sample was obtained after birth (Fig 1). This included 4,543 participants (2,278 [50.1%] girls). At analysis (follow-up to 31 May 2016), the median age of these children was 6.7 years (interquartile range, 2.5 to 8.6 years). Written informed consent was obtained for all study participants from a parent or primary caretaker for genetic screening and to participate in the prospective follow-up. The study was approved by local institutional review boards and is monitored by an external advisory board established by the US National Institutes of Health.
Fig. 1. Flow diagram of the TEDDY study participants included in this analysis. 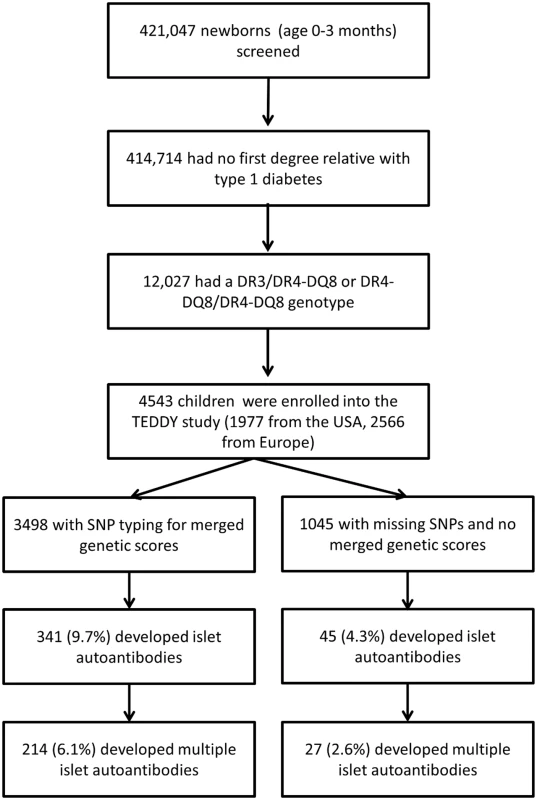
TEDDY study outcomes
Islet autoantibodies (IAAs, GADA, and IA-2A) were measured by radiobinding assays every 3 months for the first 4 years and biannually thereafter. In the US, autoantibodies were measured at the Barbara Davis Center for Childhood Diabetes at the University of Colorado Denver reference laboratory. In Europe, autoantibodies were measured at the University of Bristol, the UK reference laboratory. All radiobinding assays were performed as previously described [2,23]. Samples positive for islet autoantibodies were retested at the second reference laboratory for confirmation. The outcome of islet autoantibody positivity was defined as a positive result at both reference laboratories (confirmed) and the presence of islet autoantibodies (GADA, IA-2A, or IAAs) on 2 or more consecutive visits (persistent). The date of seroconversion to islet autoantibodies (time to first autoantibody) was defined as the date of drawing the first of the 2 consecutive positive samples. The presence of persistent multiple islet autoantibodies was defined as the presence of at least 2 persistent and confirmed islet autoantibodies. The date of persistent multiple islet autoantibodies was defined as the date of drawing the first sample for which the second persistent and confirmed islet autoantibody was detected.
Children with positive islet autoantibodies that were due to maternal IgG transmission were not considered to be positive for that autoantibody unless the child had a negative sample before the first positive sample or the autoantibody persisted beyond 18 months of age [2].
Diabetes was diagnosed according to American Diabetes Association criteria [24].
Single nucleotide polymorphism typing
In the TEDDY study, single nucleotide polymorphisms (SNPs) of immune-related genes were genotyped using the Illumina ImmunoChip [25]. For SNPs rs11755527 (BACH2) and rs689 (INS), which were not available on the immunochip, the SNPs rs3757247 (BACH2) and rs1004446 (INS) were used (S1 Table). No proxy SNPs were available for rs917997 (IL18RAP).
Genetic scores
Genetic scores were determined as described by Winkler et al. [15], without including the intercept value from the logistic regression, and as described by Oram et al. [16]. The Winkler score was originally derived from the Type 1 Diabetes Genetics Consortium case–control dataset, and the Oram score was originally based on the odds ratios available on ImmunoBase (http://www.t1dbase.org/). The genetic score of each individual was derived from weighted values given to the HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype plus a weighted value assigned to each susceptible allele of non-HLA SNPs for the Winkler score and HLA class I and non-HLA SNPs for the Oram score (S1 Table). A total of 39/40 non-HLA class II SNPs used in the Winkler score and 26/28 non-HLA class II SNPs used in the Oram score were available to calculate the genetic score in the TEDDY children, while 35/40 and 26/28 SNPs were available for the case–control cohort. For both scores, the HLA DR-DQ genotype weights were added to the weighted risks for each SNP according to the child’s number of risk alleles (0, 1, or 2) for each SNP (S1 Table). Additionally, since the Winkler and Oram scores were derived from partially overlapping genetic loci and each had distinct features, a merged genetic score was derived using the information for all available SNPs contained in the Winkler and Oram scores and was calculated for the TEDDY children (S1 Table). For simplicity, when SNPs overlapped in the Winkler and Oram scores, the mean weight of each SNP in the Winkler and Oram scores was used in the merged score, and for SNPs that were unique in the Winkler or the Oram score, the weight used in the original score was used for the merged score. Exceptions were for 2 SNPs (rs2069763 and rs3825932) that had a negative weight in the Winkler score but a positive weight in the Oram score, where the original Oram score weight was used to calculate the merged score.
Statistical analyses
An analysis plan was submitted to the TEDDY data coordinating center and approved by the TEDDY steering committee prior to compiling and analyzing the data (S1 Appendix). The merged score was added to this once both the Winkler and Oram scores were found to stratify risk. The Cox analysis and specificity analysis prescribed in the analysis plan were no longer considered to be sufficiently informative to include in the final analysis. The analysis was extended to include type 1 diabetes risk during revision of the manuscript.
For TEDDY children, the cumulative risks of developing islet autoantibodies, multiple islet autoantibodies, and diabetes were estimated using the Kaplan–Meier method and were compared between risk groups using the log-rank test. The risks of islet autoantibodies, multiple islet autoantibodies, and diabetes were calculated for increasing thresholds of the Winkler, Oram, and merged genetic scores. Analyses were also performed after stratification by HLA genotype, geographic location (US, Europe), and sex. The sensitivity of the genetic scores was assessed by calculating the proportion of children who developed islet autoantibodies, multiple islet autoantibodies, and diabetes whose genetic score was above the threshold value. Spearman’s correlation coefficient was used to assess whether the autoantibody risk by age 6 years or diabetes risk by age 10 years—and sensitivity for cases that developed by age 6 years or by age 10 years—changed with increasing score thresholds. The proportion of children in the general population who would be expected to have a genetic score above the threshold was calculated based on the frequency of children with the HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype (2.9%) identified in the screening phase of the TEDDY study [22].
For the case–control dataset, we calculated the proportions of non-diabetic controls and cases of type 1 diabetes whose genetic score exceeded the thresholds, with score increments of 0.1. The sensitivity of the genetic scores was assessed by calculating the proportion of cases within the cohort who had a score above the threshold. The empirical risk was calculated as the ratio of the proportion of cases to the proportion of controls above the threshold multiplied by the assumed background risk of 5% for individuals with the DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype [11].
The distribution of genetic scores was compared among groups defined by islet autoantibody outcome, geographic location (US, Europe), or sex using the Mann–Whitney U test.
All analyses were performed using R 3.3.2 software (R Foundation for Statistical Computing, Vienna, Austria), IBM SPSS version 22.0 (IBM, Armonk, NY), and SAS 9.4 (SAS Institute, Cary, NC).
The datasets generated and analyzed during the current study are available in the NIDDK Central Repository at https://www.niddkrepository.org/studies/teddy. TEDDY immunochip (SNP) data that support the findings of this study have been deposited in NCBI’s Database of Genotypes and Phenotypes (dbGaP) with the primary accession code phs001037.v1.p1.
Results
Genetic scores in the case–control population
The Winkler and Oram genetic scores in the WTCCC HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 cases were increased as compared to the UK Biobank HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 controls (P < 0.001; S1 Fig). Using the Winkler score, the calculated actual risk reached 10% above a threshold of 11.72, corresponding to a sensitivity of 58.7% (95% CI 55.2%–62.2%) for the patients who had the HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype. Using the Oram score, an actual risk of 10% was reached above a score threshold of 11.67, corresponding to a sensitivity of 36.6% (95% CI 33.2%–40.0%; S1 Fig).
Having verified both scores for type 1 diabetes risk stratification in the case–control dataset, we reasoned that a composite score that included all the features from the Winkler and Oram scores would be justified. We therefore developed a merged genetic score that represented the average weighted values of loci, genotypes, and alleles common to the Winkler and Oram scores, and the original weighted values for loci and alleles that were unique to 1 of the scores (S1 Table). Using the prospectively followed TEDDY cohort, we then asked how well the Winkler, Oram, and merged scores could stratify the risk for pre-symptomatic type 1 diabetes.
Baseline risk for islet autoantibodies and diabetes in TEDDY children with HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype without family history of type 1 diabetes
Seroconversion to islet autoantibodies occurred in 386 children (8.5%) (166 [43.0%] girls), and 4,157 children (91.5%) remained islet autoantibody negative (2,112 [50.8%] girls). Of the 386 children with islet autoantibodies, 241 children (62.4%) developed multiple islet autoantibodies (102 [42.3%] girls; 81 [33.6%] from US). A total of 107 (2.3%) developed diabetes by age 10 years (47 [43.9%] girls). The cumulative risk for developing islet autoantibodies was 9.2% (95% CI 8.2%–10.1%; Fig 2A), of developing multiple islet autoantibodies (pre-symptomatic type 1 diabetes) by age 6 years was 5.8% (95% CI 5.0%–6.6%; Fig 2B), and of developing diabetes by age 10 years was 3.7% (95% CI 3.0%–4.4%; Fig 2C).
Fig. 2. Cumulative risks of 1 or more islet autoantibody, multiple islet autoantibody, and type 1 diabetes in TEDDY children with the HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype. 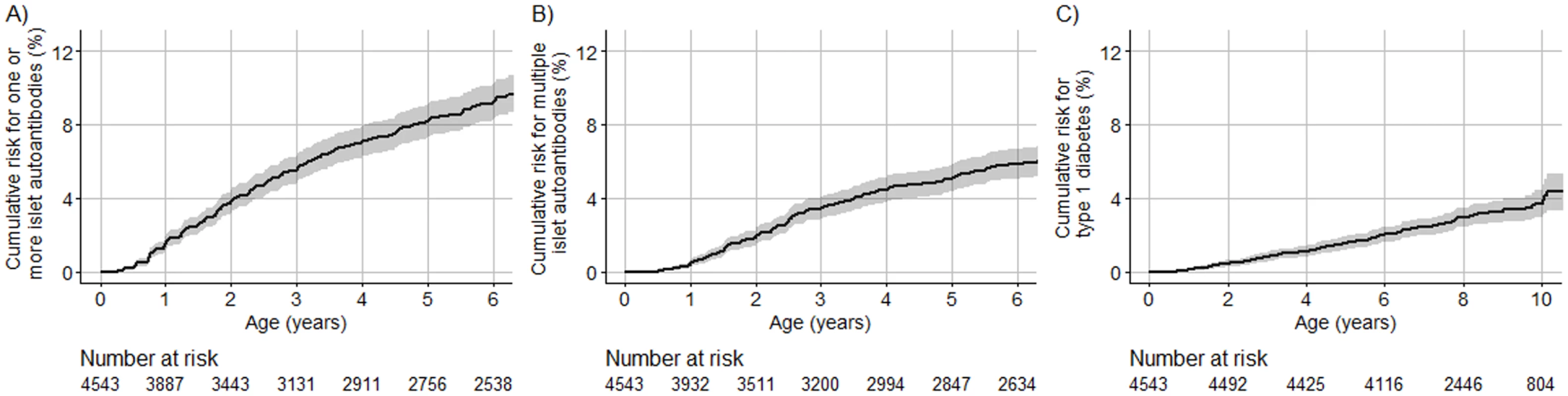
The cumulative risk for 1 or more islet autoantibodies (A), multiple islet autoantibodies (B), and type 1 diabetes (C) for TEDDY children (y-axis) is shown relative to the age of the children (x-axis) and was calculated using the Kaplan–Meier method. The shaded area represents the 95% confidence interval of the cumulative risk. The numbers at risk indicate the number of children included in the analysis at each age. Genetic scores in TEDDY children
We examined whether Winkler, Oram, and merged genetic scores were increased in children who developed islet autoantibodies. The genetic scores were calculated in 3,498 (1,471 US) children who had material for additional genetic analysis. The median follow-up in these children was 7.39 years. For each of the Winkler, Oram, and merged scores, the score was greater in children who developed islet autoantibodies by 6 years of age as compared to children who remained islet autoantibody negative (P < 0.001; Fig 3A and S2 Fig). The median merged score was 14.3 (IQR, 13.6–14.9) in children who developed islet autoantibodies versus 13.7 (IQR, 13.1–14.4) in children who remained islet autoantibody negative. The genetic scores were also slightly greater in European children (median merged score, 13.8; IQR, 13.1–14.5) than in US children (13.7; IQR, 13.1–14.4; P = 0.003; Fig 3B and S2 Fig). The frequencies of minor alleles differed between the US and European children for 7 of 43 SNPs (Bonferroni-corrected P of 0.05/43 = 0.0012; S2 Table). Scores were not different between boys and girls (P = 0.69; Fig 3C and S2 Fig).
Fig. 3. Merged genetic score in TEDDY children according to their islet autoantibody outcome, geographic location, and sex. 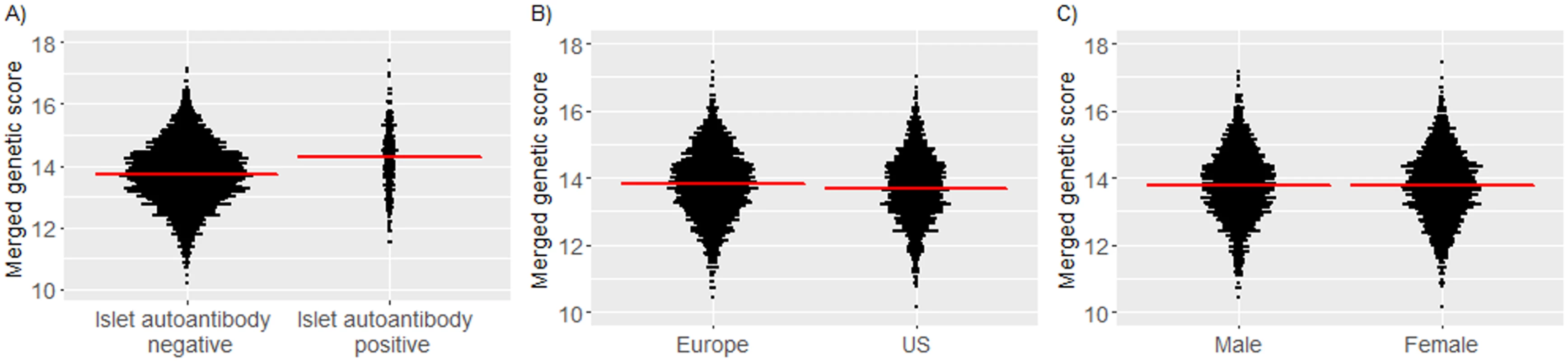
Islet autoantibody outcome (A); geographic location (B); sex (C). Red horizontal lines indicate the median genetic score value in each group. Risk for islet autoantibodies and diabetes according to the genetic scores
We next asked if and how much the genetic scores could stratify risk in TEDDY children without a family history of type 1 diabetes. To address this, the cumulative risk for developing islet autoantibodies and for diabetes was compared between HLA DR3/DR4-DQ8 and DR4-DQ8/DR4-DQ8 children who were in the upper quartile, middle 2 quartiles, and lower quartile of the merged genetic score (Fig 4). The cumulative risk for developing islet autoantibodies by 6 years of age was 16.0% (95% CI 13.3%–18.6%) among children with a merged genetic score of >14.4, representing the upper quartile, compared with 6.9% (95% CI 5.9%–8.0%) in children with a score of ≤14.4 (P < 0.001). The cumulative risk for developing multiple islet autoantibodies by 6 years of age was 11.0% (95% CI 8.7%–13.3%) in children with a score of >14.4, compared with 4.1% (95% CI 3.3%–4.9%) in children with a score of ≤14.4 (P < 0.001). The cumulative risk for developing diabetes by age 10 years was 7.6% (95% CI 5.3%–9.9%) in children with a score of >14.4, compared with 2.7% (95% CI 1.9%–3.6%) in children with a score of ≤14.4 (P < 0.001). The risks were also stratified by the Winkler and Oram scores (P < 0.001; S3 Fig). However, the merged genetic score performed better than both the Winkler and Oram scores in identifying the HLA DR3/DR4-DQ8 and HLA DR4-DQ8/DR4-DQ8 children who developed multiple islet autoantibodies (S4 Fig).
Fig. 4. Cumulative risks of 1 or more islet autoantibody, multiple islet autoantibody, and type 1 diabetes development in TEDDY children with the HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype stratified by their merged score. 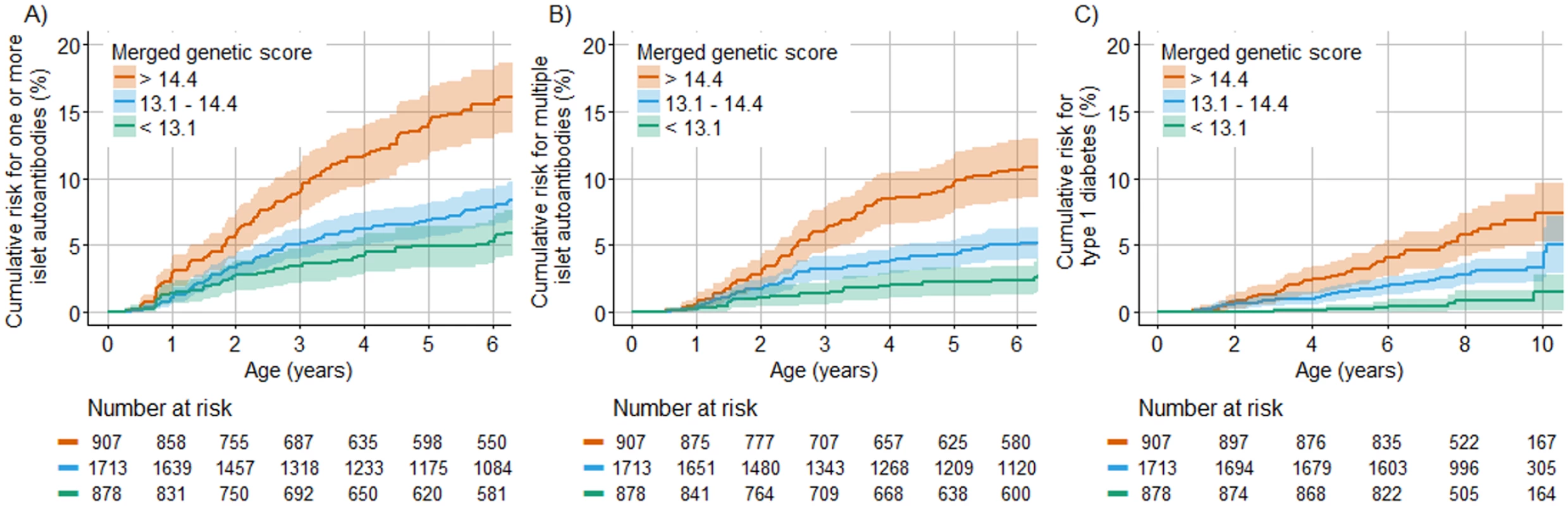
The cumulative risk of developing 1 or more islet autoantibodies (A), multiple islet autoantibodies (B), and type 1 diabetes (C) (y-axis) is shown relative to age in years (x-axis) and was calculated using the Kaplan–Meier method. Curves are shown for children with genetic scores in the upper (orange line), lower (green line), and 2 middle (blue line) quartiles. The shaded areas represent the 95% confidence interval of the cumulative risk. The numbers at risk indicate the number of children included in the analysis at each age. The merged genetic score stratified the risk for islet and multiple islet autoantibodies and for diabetes both in children who had the HLA DR3/DR4-DQ8 genotype and in children who had the HLA DR4-DQ8/DR4-DQ8 genotype (S5 Fig). The risks of islet autoantibodies, multiple islet autoantibodies, and diabetes in children with a merged genetic score of >14.4 were not significantly different between US and European children (P = 0.16, P = 0.97, and P = 0.96, respectively; S6 Fig), but autoantibody risks were higher in boys than in girls (P = 0.001 for 1 or more islet autoantibodies and P = 0.01 for multiple islet autoantibodies; S6 Fig).
Sensitivity versus risk for islet autoantibodies and diabetes according to the merged genetic score
Islet autoantibody and diabetes risk appeared incremental with increasing merged genetic score. Since the efficiency of a test is measured by both sensitivity and positive predictive value (risk), we examined the relationship between sensitivity and risk at increasing thresholds for the genetic score (Fig 5; S3–S5 Tables). As predicted, the cumulative risk for developing islet autoantibodies or multiple islet autoantibodies by age 6 years and diabetes by age 10 years increased (P < 0.001) and the sensitivity decreased (P < 0.001) with each increment in the genetic score threshold by the 5th percentile of the cohort. The risk for multiple islet autoantibodies reached a maximum of 13.2% (95% CI 9.2%–17.1%) above a threshold of >15.1, which identified 38 of the 173 children who developed multiple islet autoantibodies by age 6 years, corresponding to a sensitivity of 22.0% (95% CI 16.4%–28.7%). Above a threshold of 14.4, representing the upper 25% of merged genetic score values and where risk for multiple islet autoantibodies was 11.0% (95% CI 8.7%–13.3%), 82 of the 173 children who developed multiple islet autoantibodies by age 6 years were identified (sensitivity, 47.4%; 95% CI 40.1%–54.8%). In a population where the prevalence of HLA DR3/DR4-DQ8 and HLA DR4-DQ8/DR4-DQ8 genotypes is similar to that of the TEDDY cohort (2.9%), using the threshold of 14.4 would identify <1% of all newborns without a prior family history of type 1 diabetes.
Fig. 5. Cumulative risks and the proportion of cases identified for 1 or more islet autoantibodies, multiple islet autoantibodies, and type 1 diabetes in TEDDY children with the HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype according to increasing thresholds of the merged genetic score. 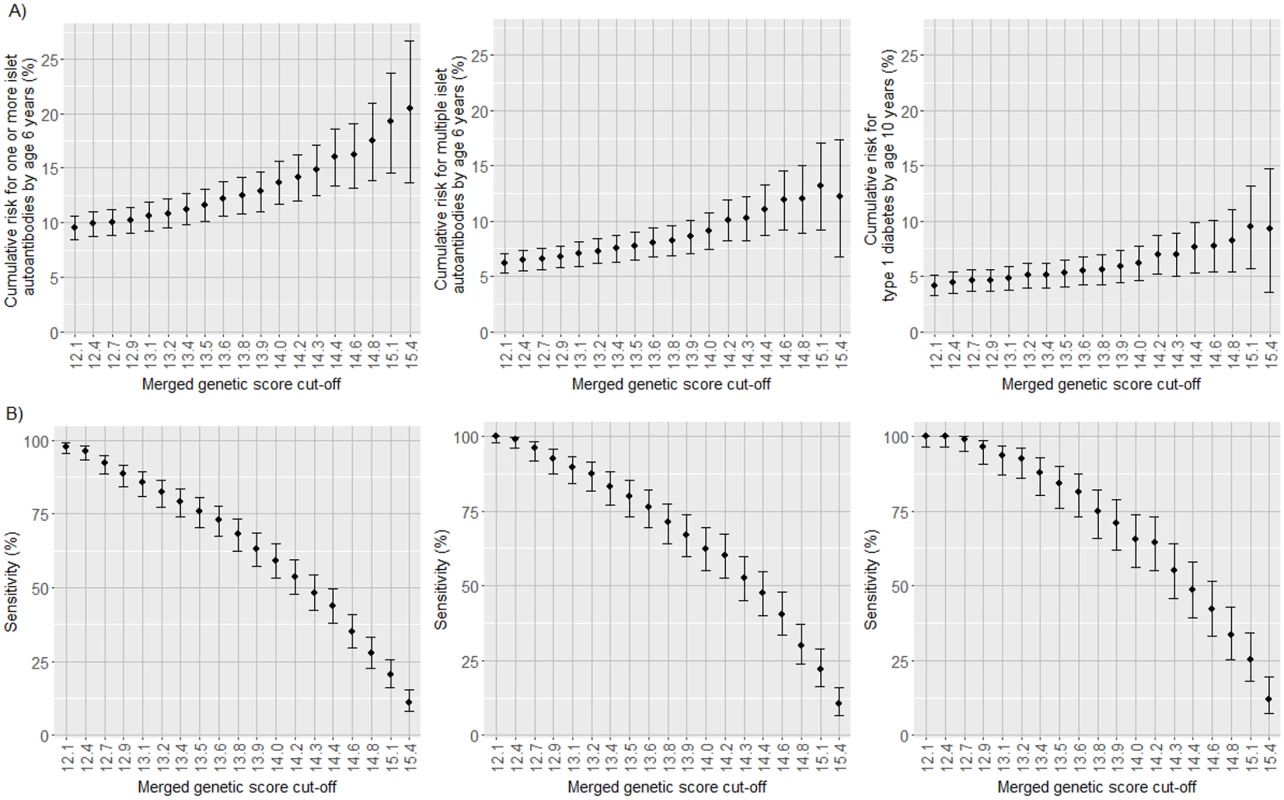
Cumulative risk for developing islet autoantibodies by age 6 years and diabetes by age 10 years (A) and the proportion of cases positive for islet autoantibodies by age 6 years and diabetes by age 10 years (sensitivity; B) in TEDDY children with the HLA DR3/DR4-DQ8 or DR4-DQ8/DR4-DQ8 genotype stratified by their merged genetic score. The risk and sensitivity are shown for each increment in the genetic score by the 5th percentile of scores in the TEDDY children, ranging from >12.1 (the 5th percentile of children) to >15.4 (the 95th percentile of children). The risk and sensitivity are shown for the development of 1 or more islet autoantibodies (left panels), multiple islet autoantibodies (middle panels), and type 1 diabetes (right panels). Error bars indicate 95% confidence intervals. Discussion
Genetic scores derived from logistic regression of numerous loci associated with type 1 diabetes susceptibility were able to stratify the risk for pre-symptomatic and clinical type 1 diabetes in a prospective cohort of children with high-risk HLA genotypes but no family history of type 1 diabetes. The risks of developing islet autoantibodies, multiple islet autoantibodies, and diabetes increased with each increment in the genetic score. A genetic score that would identify <1% of all newborn infants was associated with a risk for developing multiple islet autoantibodies of >10% by 6 years of age. This compares to a background population risk of around 0.4% [26]. These findings provide a paradigm for identifying infants whose risk for developing type 1 diabetes is more than 25 times that of the general population, twice that of infants identified by the highest-risk HLA genotypes alone, and higher than that of children with a first-degree relative with type 1 diabetes.
The study was performed using a large number of children who were prospectively followed for the development of islet autoantibodies from infancy. The findings were consistent between US and European children and for 2 independently derived genetic scores that were used to develop the merged genetic score. We note, however, that the risk scores were generated using the outcome of clinical, or stage 3, type 1 diabetes. It has been previously noted in multiple studies, including TEDDY, that many, but not all of the genes associated with type 1 diabetes confer risk for islet autoantibodies [25,27]. For this reason, the performance of a genetic score for identifying pre-symptomatic type 1 diabetes might improve if genes and weights for islet autoantibody susceptibility are incorporated into the score. Genetic score performance may also be improved if more accurate estimates of risk weight for homozygous versus heterozygous alleles were available. Of note, the score does not include all type 1 diabetes susceptibility genes and does not contain weights for several HLA class I alleles that confer susceptibility for type 1 diabetes [28–30]. Finally, the current genetic scores were derived from cohorts of mostly individuals of European descent, and it is likely that the genetic scores may not be suitable for all races or ethnic groups.
The study was performed to extend the opportunities for early identification of individuals at increased risk for disease. Previous primary prevention trials in type 1 diabetes involved HLA typing of infants with a family history of type 1 diabetes [3,4,31]. The enrollment of participants into these trials took several years, and the proportion of all cases of childhood type 1 diabetes that were represented by the inclusion criteria was less than 5%, limiting the generalizability of trial outcomes. Screening that is limited to HLA typing of the general population can identify individuals with 3% to 5% risk, which may be insufficient for enrollment into primary prevention studies in which infants are exposed to treatment. Indeed, the TRIGR study used a combination of HLA typing and family history in order to identify infants whose risk for type 1 diabetes was 10% [3]. Therefore, we set a risk target of 10%, which was achieved in our study when we used the development of multiple islet autoantibodies as a marker for pre-symptomatic type 1 diabetes. The risk threshold was reached when the Winkler, Oram, or merged genetic score was used in children with the 2 highest-risk HLA genotypes, DR3/DR4-DQ8 and DR4-DQ8/DR4-DQ8, which can be detected by typing 3 SNPs. In a European population, these 2 genotypes were present in around 40% of all cases of childhood type 1 diabetes [11]. The merged genetic score threshold of >14.4 identified almost 50% of children with these genotypes who developed multiple islet autoantibodies or diabetes. Therefore, we surmise that our risk score threshold would identify up to 20% of children without family history of type 1 diabetes who will develop the disease. Moreover, the screening strategy is relatively inexpensive, and is now being used in the Primary Oral Insulin Trial (https://clinicaltrials.gov/ct2/show/NCT03364868), where DNA extraction from blood spots and typing are performed for less than US$8 per sample. Extending the strategy to individuals with other HLA genotypes is possible, but the other genotypes are less frequent in type 1 diabetes and are associated with a lower risk than that conferred by the DR3/DR4-DQ8 and DR4-DQ8/DR4-DQ8 genotypes. Therefore, the inclusion of other genotypes is unlikely to further improve risk stratification.
In conclusion, a genetic score based on 3 SNPs for HLA class II genotyping and 41 SNPs in other genes identified <1% of newborn children who, in the absence of a family history of type 1 diabetes, had a >10% risk for developing multiple islet autoantibodies by 6 years of age. This greatly extends the possibilities of enrolling participants into clinical trials aimed at evaluating type 1 diabetes prevention strategies that could be applied in infancy and before the development of autoimmunity [32].
Supporting Information
Zdroje
1. Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, et al. Randomized trial of peanut consumption in infants at risk for peanut allergy. N Engl J Med. 2015;372 : 803–13. doi: 10.1056/NEJMoa1414850 25705822
2. Krischer JP, Lynch KF, Schatz DA, Ilonen J, Lernmark Å, Hagopian WA, et al. The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia. 2015;58 : 980–7. doi: 10.1007/s00125-015-3514-y 25660258
3. Knip M, Åkerblom HK, Becker D, Dosch HM, Dupre J, Fraser W, et al. Hydrolyzed infant formula and early β-cell autoimmunity: a randomized clinical trial. JAMA. 2014;311 : 2279–87. doi: 10.1001/jama.2014.5610 24915259
4. Bonifacio E, Ziegler AG, Klingensmith G, Schober E, Bingley PJ, Rottenkolber M, et al. Effects of high dose oral insulin on immune responses in children at high risk for type 1 diabetes: the Pre-POINT randomized clinical trial. JAMA. 2015;313 : 1541–9. doi: 10.1001/jama.2015.2928 25898052
5. Vriezinga SL, Auricchio R, Bravi E, Castillejo G, Chmielewska A, Crespo Escobar P, et al. Randomized feeding intervention in infants at high risk for celiac disease. N Engl J Med. 2014;371(14):1304–15. doi: 10.1056/NEJMoa1404172 25271603
6. Lionetti E, Castellaneta S, Francavilla R, Pulvirenti A, Tonutti E, Amarri S, et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N Engl J Med. 2014;371(14):1295–303. doi: 10.1056/NEJMoa1400697 25271602
7. Liu E, Lee HS, Aronsson CA, Hagopian WA, Koletzko S, Rewers MJ, et al. Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med. 2014;371(1):42–9. doi: 10.1056/NEJMoa1313977 24988556
8. Bonifacio E. Predicting type 1 diabetes using biomarkers. Diabetes Care. 2015;38(6):989–96. doi: 10.2337/dc15-0101 25998291
9. Näntö-Salonen K, Kupila A, Simell S, Siljander H, Salonsaari T, Hekkala A, et al. Nasal insulin to prevent type 1 diabetes in children with HLA genotypes and autoantibodies conferring increased risk of disease: a double-blind, randomised controlled trial. Lancet. 2008;372(9651):1746–55. doi: 10.1016/S0140-6736(08)61309-4 18814906
10. Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, et al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY). Diabetologia. 1996;39(7):807–12. 8817105
11. Lambert AP, Gillespie KM, Thomson G, Cordell HJ, Todd JA, Gale EA, et al. Absolute risk of childhood-onset type 1 diabetes defined by human leukocyte antigen class II genotype: a population-based study in the United Kingdom. J Clin Endocrinol Metab. 2004;89 : 4037–43. doi: 10.1210/jc.2003-032084 15292346
12. Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41(6):703–7. doi: 10.1038/ng.381 19430480
13. Winkler C, Krumsiek J, Lempainen J, Achenbach P, Grallert H, Giannopoulou E, et al. A strategy for combining minor genetic susceptibility genes to improve prediction of disease in type 1 diabetes. Genes Immun. 2012;13(7):549–55. doi: 10.1038/gene.2012.36 22932816
14. Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet. 2009;5(7):e1000540. doi: 10.1371/journal.pgen.1000540 19584936
15. Winkler C, Krumsiek J, Buettner F, Angermüller C, Giannopoulou EZ, Theis FJ, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia. 2014;57 : 2521–9. doi: 10.1007/s00125-014-3362-1 25186292
16. Oram RA, Patel K, Hill A, Shields B, McDonald TJ, Jones A, et al. A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care. 2016;39(3):337–44. doi: 10.2337/dc15-1111 26577414
17. TEDDY Study Group. The environmental determinants of diabetes in the young (TEDDY) study: study design. Pediatr Diabetes. 2007;8 : 286–98. doi: 10.1111/j.1399-5448.2007.00269.x 17850472
18. Insel RA, Dunne JL, Atkinson MA, Chiang JL, Dabelea D, Gottlieb PA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care. 2015;38 : 1964–74. doi: 10.2337/dc15-1419 26404926
19. Ziegler AG, Rewers M, Simell O, Simell T, Lempainen J, Steck A, et al. Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. JAMA. 2013;309(23):2473–9. doi: 10.1001/jama.2013.6285 23780460
20. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779 25826379
21. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447 : 661–78. doi: 10.1038/nature05911 17554300
22. Hagopian WA, Erlich H, Lernmark A, Rewers M, Ziegler AG, Simell O, et al. The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421000 infants. Pediatr Diabetes. 2011;12(8):733–43. doi: 10.1111/j.1399-5448.2011.00774.x 21564455
23. Bonifacio E, Yu L, Williams AK, Bingley PJ, Marcovina SM, Adler K, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for national institute of diabetes and digestive and kidney diseases consortia. J Clin Endocrinol Metab. 2010;95(7):3360–7. doi: 10.1210/jc.2010-0293 20444913
24. American Diabetes Association (2014) Standards of medical care in diabetes—2014. Diabetes Care. 2014;37(Suppl 1):S14–80.
25. Törn C, Hadley D, Lee HS, Hagopian W, Lernmark Å, Simell O, et al. Role of type 1 diabetes-associated SNPs on risk of autoantibody positivity in the TEDDY study. Diabetes. 2015;64(5):1818–29. doi: 10.2337/db14-1497 25422107
26. Raab J, Haupt F, Scholz M, Matzke C, Warncke K, Lange K, et al. Capillary blood islet autoantibody screening for identifying pre-type 1 diabetes in the general population: design and initial results of the Fr1da study. BMJ Open. 2016;6(5):e011144. doi: 10.1136/bmjopen-2016-011144 27194320
27. Rich SS, Concannon P. Role of type 1 diabetes-associated SNPs on autoantibody positivity in the Type 1 Diabetes Genetics Consortium: overview. Diabetes Care. 2015;38(Suppl 2):S1–3.
28. Evangelou M, Smyth DJ, Fortune MD, Burren OS, Walker NM, Guo H, et al. A method for gene-based pathway analysis using genome wide association study summary statistics reveals nine new type 1 diabetes associations. Genet Epidemiol. 2014;38(8):661–70. doi: 10.1002/gepi.21853 25371288
29. Lenz TL, Deutsch AJ, Han B, Hu X, Okada Y, Eyre S, et al. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat Genet. 2015;47(9):1085–90. doi: 10.1038/ng.3379 26258845
30. Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450 : 887–92. doi: 10.1038/nature06406 18004301
31. Hummel S, Pflüger M, Hummel M, Bonifacio E, Ziegler AG. Primary dietary intervention study to reduce the risk of islet autoimmunity in children at increased risk for type 1 diabetes: the BABYDIET study. Diabetes Care. 2011;34(6):1301–5. doi: 10.2337/dc10-2456 21515839
32. Ziegler AG, Danne T, Dunger DB, Berner R, Puff R, Kiess W, et al. Primary prevention of beta-cell autoimmunity and type 1 diabetes—the Global Platform for the Prevention of Autoimmune Diabetes (GPPAD) perspectives. Mol Metab. 2016;5(4):255–62. doi: 10.1016/j.molmet.2016.02.003 27069865
33. Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32 : 5381–97. doi: 10.1002/sim.5958 24027076
34. Guinney J, Wang T, Laajala TD, Winner KK, Bare JC, Neto EC, et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol. 2017;18 : 132–42. doi: 10.1016/S1470-2045(16)30560-5 27864015
35. Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90 : 773–95.
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2018 Číslo 4- Není statin jako statin aneb praktický přehled rozdílů jednotlivých molekul
- Magnosolv a jeho využití v neurologii
- Moje zkušenosti s Magnosolvem podávaným pacientům jako profylaxe migrény a u pacientů s diagnostikovanou spazmofilní tetanií i při normomagnezémii - MUDr. Dana Pecharová, neurolog
- S prof. Vladimírem Paličkou o racionální suplementaci kalcia a vitaminu D v každodenní praxi
- Biomarker NT-proBNP má v praxi široké využití. Usnadněte si jeho vyšetření POCT analyzátorem Afias 1
-
Všechny články tohoto čísla
- Preprints in medical research: Progress and principles
- From surviving to thriving: What evidence is needed to move early child-development interventions to scale?
- Preprints: An underutilized mechanism to accelerate outbreak science
- Attacks on medical workers in Syria: Implications for conflict research
- Determining the scope of attacks on health in four governorates of Syria in 2016: Results of a field surveillance program
- Estimating the health and economic effects of the proposed US Food and Drug Administration voluntary sodium reformulation: Microsimulation cost-effectiveness analysis
- Maternal age and offspring developmental vulnerability at age five: A population-based cohort study of Australian children
- Two-year impact of community-based health screening and parenting groups on child development in Zambia: Follow-up to a cluster-randomized controlled trial
- Universal versus conditional day 3 follow-up for children with non-severe unclassified fever at the community level in Ethiopia: A cluster-randomised non-inferiority trial
- Universal versus conditional day 3 follow-up for children with non-severe unclassified fever at the community level in the Democratic Republic of the Congo: A cluster-randomized, community-based non-inferiority trial
- Breastfeeding during infancy and neurocognitive function in adolescence: 16-year follow-up of the PROBIT cluster-randomized trial
- Impacts 2 years after a scalable early childhood development intervention to increase psychosocial stimulation in the home: A follow-up of a cluster randomised controlled trial in Colombia
- Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: A prospective study in children
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Breastfeeding during infancy and neurocognitive function in adolescence: 16-year follow-up of the PROBIT cluster-randomized trial
- Preprints in medical research: Progress and principles
- Determining the scope of attacks on health in four governorates of Syria in 2016: Results of a field surveillance program
- Estimating the health and economic effects of the proposed US Food and Drug Administration voluntary sodium reformulation: Microsimulation cost-effectiveness analysis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



