-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
Aging is a general biological process among the living organisms which is affected by environmental stimuli but also genetically controlled. Genome instability is one of the aging hallmarks and has long been implicated as one of the main causal factors in aging. DNA double strand breaks (DSBs) are the most deleterious DNA damages that cause genome instability. To counteract DNA damage of DSBs and maintain high level of genome integrity, cells have evolved powerful repair systems such as homologous recombination (HR). HR is crucial for DNA repair and genome integrity maintenance, and is generally believed to be essential for assurance of cell longevity. Telomeres, the physical ends of eukaryotic linear chromosomes, are preferentially elongated by telomerase, a specialized reverse transcriptase, in most cases. However, due to the resemblance of telomeres to DSBs, HR can not be eliminated but rather readily takes place on telomeres, even in the presence of telomerase. Here we show that HR at yeast telomeres elicits genome instability and accelerates cellular aging. Inactivation of the evolutionarily conserved KEOPS complex subunit Cgi121 specifically inhibits telomere HR and results in extremely long lifespan, indicating a dark side of HR in longevity regulation.
Published in the journal: . PLoS Genet 11(3): e32767. doi:10.1371/journal.pgen.1005071
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005071Summary
Aging is a general biological process among the living organisms which is affected by environmental stimuli but also genetically controlled. Genome instability is one of the aging hallmarks and has long been implicated as one of the main causal factors in aging. DNA double strand breaks (DSBs) are the most deleterious DNA damages that cause genome instability. To counteract DNA damage of DSBs and maintain high level of genome integrity, cells have evolved powerful repair systems such as homologous recombination (HR). HR is crucial for DNA repair and genome integrity maintenance, and is generally believed to be essential for assurance of cell longevity. Telomeres, the physical ends of eukaryotic linear chromosomes, are preferentially elongated by telomerase, a specialized reverse transcriptase, in most cases. However, due to the resemblance of telomeres to DSBs, HR can not be eliminated but rather readily takes place on telomeres, even in the presence of telomerase. Here we show that HR at yeast telomeres elicits genome instability and accelerates cellular aging. Inactivation of the evolutionarily conserved KEOPS complex subunit Cgi121 specifically inhibits telomere HR and results in extremely long lifespan, indicating a dark side of HR in longevity regulation.
Introduction
Aging is generally defined as the time-dependent functional decline and increased mortality in most living organisms. Although aging appears to be a natural process, increasing evidence indicates that aging is genetically controlled. In order to elucidate how aging is influenced by intrinsic cellular traits, researchers have developed and employed various model organisms including yeast, worm, fly, fish, mouse and monkey to study the pathways that affect aging. The single-cell organism, budding yeast Saccharomyces cerevisiae represents a widely used tool for aging study [1,2,3]. A single yeast mother cell can only generate a limited number of daughter cells before its mitotic arrest [4]. This aging-associated phenotype is called replicative aging [5]. The organismal aging for multicellular species is likely (or at least partially) to be attributed to cellular aging in their corresponding organs and/or tissues.
The genome, which carries the genetic information of a cell, is continuously threatened by exogenous damages, as well as by endogenous threats such as DNA replication errors [6]. Genome instability is one of the aging hallmarks, and has long been implicated as one of the main causal factors in aging [7,8]. DNA damage (e.g. double strand break, DSB) is one of the major causes for genome instability. When the repair pathways are not efficient enough to cope with a given level of damage, cells may undergo cell cycle arrest, cellular senescence and cell death. For example, the Werner syndrome and Bloom syndrome, two typical progeroid syndromes, are respectively caused by defective helicases WRN and BLM, which are involved in DNA repair [9]. The cells from both syndromes show increased DNA damage accumulation [9]. Consistently, the deficiency in yeast Sgs1 helicase, the homologue of human WRN and BLM, also results in genome instability, such as enhancement of rDNA recombination and fragmentation of nucleolus, and leads to premature cellular aging [10]. To maintain genome stability, genome maintenance pathways have emerged during evolution, and function in longevity assurance. For example, homologous recombination (HR) and non-homologous end joining (NHEJ) pathways have been evolved to repair the most deleterious DNA damages, the DNA double strand breaks (DSBs). Accordingly, mutation of yeast DSB repair genes, such as RAD50, RAD51, RAD57 and RAD52, greatly reduces yeast replicative lifespan [11].
Telomeres are the physical ends of eukaryotic linear chromosomes, and are crucial for genome integrity and stability [12]. Although telomeres may look like DSBs as the chromosomal ends, they are distinguished by the specialized architecture, consisting of repetitive guanine-rich DNA bound by telomere-specific proteins. The yeast telomeric DNA consists of ∼350 bp of TG1–3/C1–3A repeats, and the G strand extends beyond its complementary strand to form a single-stranded overhang, called the G-overhang [12,13,14]. The telomerase complex, which consists of the catalytic subunit Est2, the template RNA moiety Tlc1, and two accessory subunits Est1 and Est3, is responsible for telomeric G-strand elongation, as well as telomere protection [15,16,17,18]. When telomerase is inactivated, telomeres keep shortening and most cells undergo critically short telomere-triggered cell cycle arrest, a process termed “replicative senescence” [16,19,20]. “Replicative senescence” is usually considered to be different from “replicative aging” as the former is largely attributed to critically short telomeres. Although telomeres are well protected and excluded from DSB repair at most of the time, yet there are several traits that make telomeres highly prone to be recombined. Firstly, all the telomeres are much alike in their repetitive sequences which could be favorable substrates for homologous recombination activities. Additionally, quite a few proteins (or protein complexes) involved in DNA repair pathways also bind and function at telomeres [14]. The yKu70/80 heterodimer, which is required for NHEJ [21,22], is indispensable for telomere protection, telomerase recruitment and telomeric heterochromatin maintenance [23,24,25,26,27,28]. Mre11/Rad50/Xrs2 heterotrimer, DNA helicase Sgs1 and endonuclease Sae2, which are critical for resection of the ends of DSBs in HR-mediated DSB repair, are involved in telomere 5’ end resection after DNA replication [29,30,31]. Moreover, the 3’ overhang generated by telomere end resection could be perceived as intermediates of DSB, as the strand invasion step requires the ssDNA [32]. Thus, considering all the traits of telomeres mentioned above, we propose that recombination activity in yeast might covet telomeres and interfere with telomerase to elicit genome instability under physiological conditions, and thereby affect cellular longevity.
The evolutionary conserved KEOPS complex which consists of five subunits, i.e. Cgi121, Bud32, Kae1, Gon7 and Pcc1 in yeast, was first identified as a telomere regulator [33,34]. Deletion of CGI121 or BUD32 reduces single-stranded telomeric DNA accumulated in cdc13-1 cells, and suppresses the temperature sensitivity of cdc13-1 mutant grown at 28°C [33], indicating that loss of Cgi121 or Bud32 limits the amount of ssDNA generated at uncapped telomeres. Moreover, deletion of any subunit of KEOPS complex results in defect in telomere recombination [35], suggesting that KEOPS complex promotes telomeric TG1–3 tracts recombination. In addition to telomere regulation, KEOPS complex also participates in tRNA modification (t6A) [36,37]. Interestingly, the Cgi121 subunit of the KEOPS complex is indispensable for both telomere length regulation and recombination, but not required for tRNA modification [33,37]. We therefore exploited the separation-of-function subunit Cgi121 to dissect the functions of KEOPS in telomere recombination from those in tRNA modification. Our data presented in the current work indicate that activation of telomere recombination accelerates cellular aging, and attenuation of telomere recombination, e.g. by inactivation of Cgi121, promotes cell longevity.
Results
Telomerase-null survivors have shorter replicative lifespan than telomerase-proficient cells
When telomerase is inactivated by deletion of a gene encoding telomerase subunit (Est1, Est2, Est3 or Tlc1), telomeres gradually shorten, and most cells undergo senescence after 75 to 100 generations due to the critical short telomeres [16,19,20]. Because critical short or deprotected telomeres are highly recombinogenic, a small percentage of the telomerase-null cell can overcome the crisis by using homologous recombination to maintain their telomeres. These so-called “survivors” either have telomeres with amplified subtelomeric Y’ sequence and short TG1–3 tracts (the Type I survivors) [38], or harbor long heterogeneous TG1–3 sequence (Type II survivors) [39].
In order to address the effect of telomere recombination on replicative aging, we examined lifespan of the survivor cells. After deletion of telomerase subunit TLC1, cells were serially passaged in solid or liquid medium to obtain Type I and Type II survivors respectively (Fig. 1A, left) [35]. Lifespan assay was performed with both types of survivors. The results showed that both the Type I and Type II survivors have much shorter lifespan than wild-type cells, and the lifespan of Type I survivors is extremely short (Fig. 1A, right). This result is consistent with our previous data that est2Δ Type II survivors have shorter lifespan [40].
Fig. 1. Inactivation of telomerase accelerates yeast replicative aging, and reintroduction of telomerase recovers lifespan. 
(A) Telomere structure of tlc1Δ Type I and Type II survivors were examined by telomere Southern blot (left) and the replicative lifespan of these strains were determined (right). Statistical significance was determined by a Wilcoxon rank sum test and significant differences were stated for p < 0.05. The statistical data of all the lifespan experiments in this study were shown in S2 Table. (B) After integration of TLC1 genes, tlc1Δ Type I or Type II strains were continuously passaged and cells at different time point were subjected to telomere Southern blot analyses. The numbers above each lane indicate the numbers of restreaks after TLC1 reintroduction. (C) and (D) Lifespan analysis of tlc1Δ Type I (C) or Type II (D) survivor cells at the 10th restreak after TLC1 reintroduction. Re-activation of telomerase in telomerase-null survivors inhibits telomere recombination and at least partially restores cellular lifespan
The shorter lifespan in telomerase-null survivors suggests that telomere recombination elicits genome instability in telomerase-null survivors to accelerate cellular aging. To test this hypothesis, we re-introduced TLC1 gene back into the survivors that were derived from tlc1Δ cells, by integrating an intact copy of this gene into the genome. After serial passages, telomere structures were examined. The Southern blotting results showed that replenishment of telomerase activity leads to elongation of terminal telomeric TG1–3 tracts in Type I cells and the telomere pattern is stably maintained without further Y’ amplification (Fig. 1B). On the other hand, reduction of the heterogeneity of the long telomeres was observed in Type II cells after replenishment of TLC1, and the telomere structure was gradually restored to a wild-type pattern (Fig. 1B). These results indicate that the recombination activity on telomeres is inhibited by telomerase. Accordingly, reactivation of telomerase partially and completely restores the lifespan of Type I and Type II cells, respectively (Fig. 1C and 1D). These results support the notion that telomere recombination results in genome instability which causes shorter replicative lifespan. Notably, re-introduction of TLC1 only partially restored lifespan of Type I survivors (Fig. 1C). This phenotype is likely attributed to the abnormal karyotypes resulting from telomere end-to-end fusions during Type I survivor generation [38,41]. This explanation is supported by the observation that the severe growth defect of Type I survivors was partially recovered after re-introduction of TLC1.
Y’ element rearrangement mediated by HR occurs in telomerase-positive cells
In telomerase-proficient cells telomere recombination is largely inhibited. However, previous studies have indicated that telomerase seems not to be able to completely eliminate telomere recombination. For example, recombination-mediated telomere rapid deletion (TRD) has been observed in telomerase positive cells [42]. In mouse cells lacking the amino-terminal basic domain of TRF2, t-loop-sized telomeric circles can be excised from leading strand telomeres via homologous recombination [43]. Additionally, homologous recombination occurs with the same frequency in human telomerase-positive and telomerase-negative ALT (alternative lengthening of telomeres) cells [44]. It is conceivable that homologous recombination is intermittently competing with telomerase to contribute to telomere elongation. Due to the repetitive nature of telomeric DNA sequence, telomere recombination products cannot be readily distinguished from those generated by telomerase.
In order to detect telomere recombination event(s) in telomerase-proficient cells, we performed a chromosome healing (de novo telomere formation) assay (see Experimental procedures). The system is modified from that developed by Gottschling’s lab [45]. Briefly, 81 bp of TG1–3 telomeric “seed” (TG81) is inserted into the left arm of chromosome VII at the ADH4 locus, flanked by a TRP1 marker gene and an HO endonuclease cutting site (Fig. 2A). Upon HO endonuclease induction by galactose, the HO site is cut and the TG1–3 sequence is exposed to the very end. The newly formed telomere of 81 bp is critically short and has to be repaired through telomerase or recombination pathways. As a control, an isogenic strain with no TG1–3 seed imbedded (TG0) was included in the experiment. HO-cut will generate a none-telomeric DNA double strand break, which must be repaired to maintain cell viability. In order to examine the repair efficiency of the end generated by HO-cut, we cultured the TG81 or TG0 cells in galactose-containing solid medium, and colonies in which the short telomeres (in TG81 strains) or the DSBs (TG0) have presumably been repaired were counted. The repair efficiency was defined by the number of colonies formed on the galactose plate (cut) divided by that of the same strain on the glucose plate (uncut). The de novo generated short telomere can be efficiently repaired with an efficiency of ∼100% in the TG81 strain (Fig. 2B). Notably, the efficiency was reduced to ∼10% by deletion of TLC1 (Fig. 2B), suggesting a crucial role of telomerase in the elongation of this new short telomere. Deletion of RAD50, RAD51 or RAD52 resulted in no or slight reduction in the repair efficiency (Fig. 2B), supporting a predominant role of telomerase rather than recombination in the repairing process. In contrast, the none-telomeric control TG0 strain has very low repair efficiency (∼0.36%) (Fig. 2B). Deletion of TLC1 in the TG0 strain has little effect on the repair efficiency (Fig. 2B), consistent with the notion that the regular DSB generated by HO-cut can hardly be repaired by telomerase. These data indicate that HO-induced double strand break in TG81 strain can generate a bona fide new telomere, which can be readily elongated by telomerase while recombination could still make a minor contribution to its repair.
Fig. 2. Subtelomeric Y’ element recombination occurs in the presence of telomerase. 
(A) Schematic representation of the strategy to detect Y’ recombination in the presence of telomerase (not to scale). 81 bp of TG1–3 seed (in purple) is inserted at the ADH4 gene locus on chromosome VII, flanked by a TRP1 marker gene (in green) and an HO endonuclease cutting site. The LYS2 gene was placed between the natural telomere of VII-L and the ADH4 locus to serve as a genetic marker to monitor HO cutting. After HO induction by galactose, a short telomere with 81 bp of TG1–3 sequence is generated de novo through HR activity in two distinct regions of the donor telomere: one in TG1–3 tracts (in orange) between the STR and the Y’ element (Type I) and the other in the terminal TG1–3 tracts (Type II). The telomerase-mediated elongation is omitted in this diagram. The resulting Type I recombination products can be detected by PCR amplification using primers specific for TRP1 (indicated by a green arrow) and Y’ consensus sequences (indicated by gray arrow) followed by Southern blot with probes hybridized to TG1–3 repeats (indicated by dashes). (B) Cell viability assay for chromosome healing. Proportional yeast cells were plated onto galactose (cut) or glucose-containing medium (uncut). The numbers of colonies formed on the plates were counted and repair efficiency was calculated by dividing the number of colonies on “cut” plates by that on “uncut” plates. The error bars indicates the standard deviations. *p < 0.05 and **p < 0.01. (C) Southern blot detection of Y’ recombination. After HO induction by galactose in liquid culture for 24 h, the isogenic strains (labeled on the top) were subjected to genomic DNA extraction (cut). The uninduced strains were also included in the assay as “uncut” controls. PCR was then performed using primers indicated in A. The PCR products were then subjected to Southern blot with probes recognizing TG1–3 sequence (Y’ rec). As the loading control, the proportional genomic DNA was digested with EcoRI endonuclease to generate a DNA fragment of 1 kb containing the POL1 gene sequence and subjected to Southern blot using a probe specific for POL1 sequence (ctrl). The DNA signals were quantified by software (Multi Gauge). The number below each lane indicates the Y’ recombination efficiency, which is defined by dividing the intensity of signal of Y’ recombination with that of the internal control. The efficiency of all the samples were compared to that of TG81 cut sample which was set as “1”. (D) A representative sequence of one of the three clones showing the sequence of Y’ recombination products. Part of the TRP1 promoter (in green), 78 bp of TG1–3 seed sequence (in purple), 9 bp of recombined TG1–3 sequence (in orange) and part of the Y’ sequence from telomere VIL (Tel06L-YP, in gray) are shown. The underlined sequences are primers for PCR as indicated in (A). The full sequence for the PCR product is shown in S1 Fig. Next, we used this system to detect whether telomere recombination takes place in telomerase-positive cells. Although it is technically difficult to distinguish the telomeric tracts added by terminal TG1–3 recombination (Type II recombination) from those by telomerase, the addition of Y’ element to the end of this telomere (Type I recombination) can be detected by Southern blot following PCR amplification with primers specific for TRP1 region and Y’ consensus sequence (Fig. 2A). In this set of experiments, cells were harvested after induction with galactose for 24 h, and genomic DNA was extracted. PCR amplifications were performed using the genomic DNA as templates. The PCR products were subjected to Southern blot with a TG1–3 probe. Meanwhile, proportional amount of genomic DNA was hybridized to a POL1 probe as the internal control. DNA signals in the Southern blot results were quantified and the level of Y’ recombination was normalized to the corresponding internal control. The Y’ recombination efficiency of all the samples were compared with that of TG81 cut sample which was defined as “1”. We successfully detected the telomere recombination events in TG81 but not TG0 strain by Southern blot (Fig. 2C). To validate that the PCR-amplified fragments contained Y’-sequence, we cloned and sequenced some of the PCR products. The representative sequences of three clones are shown (Fig. 2D and S1 Fig.). As expected, the sequences of the PCR products contain part of the TRP1 promoter sequence (in green color), variable lengths (87 to 271 bp) of TG1–3 repeats (purple for TG seed and orange for telomere sequence from the donor chromosome) and the proximal parts of Y’ elements (in gray). Notably, sequences of three clones vary at both the length of internal TG1–3 tracts and the origins of Y’ elements. Two of the three clones captured the Y’ element from the left telomere of chromosome VI, and the third one copied the Y’ element from the right telomere of chromosome VIII. These data indicate that HO-induced short telomeres can be repaired by HR in the presence of telomerase, most likely through break-induced replication (BIR) [46].
We also checked the role of some recombination regulators in telomere recombination in TG81 strain in the presence of telomerase. Interestingly, deletion of RAD50 results in significantly reduced level of such recombination (Fig. 2C). RAD51-null cells have unaffected Y’ recombination, and deletion of RAD52 modestly reduces such recombination (Fig. 2C). Paradoxically, it is generally believed that Rad51 and Rad50 are required for the formation of Type I and Type II survivors respectively, and Rad52 is thought to be essential for virtually all homologous recombination activity [32,47]. It remains elusive why Rad51 is dispensable, or Rad52 plays a minor role for the Y’ telomere recombination in the presence of telomerase. This kind of recombination events may occur in a way similar to that of BIR, as it was reported that a rad51Δ strain still allows BIR to proceed [48], and the Rad51-independent BIR pathway is largely dependent on another set of recombination genes including RAD50 and RAD59 [46]. Nevertheless, these results support the argument that HR takes place in telomerase-positive cells.
Deletion of CGI121 compromises telomere recombination
Because telomere recombination affects cellular lifespan, we propose that inhibition of telomere recombination will be beneficial to cell longevity. Our previous genetic screenings have shown that the evolutionarily conserved KEOPS complex was required for telomeric TG1–3 tracts recombination [35]. Additionally KEOPS complex is likely to be involved in generation of telomeric ssDNA because deletion of either CGI121 or BUD32 reduces telomeric ssDNA level in cdc13-1 mutant and suppresses the temperature sensitivity [33]. We therefore wanted to establish the functional relevance of KEOPS complex between telomere recombination and cellular longevity regulation. We focused our efforts on Cgi121 due to several concerns. (1) Deletion of any of the other four subunits confers severe growth defect [33,34,35,49,50], while deletion of CGI121 only has minor effect on cell growth. (2) Structural studies indicate that lack of Cgi121 doesn’t affect the interactions between other subunits [49]. (3) More importantly, Cgi121 is not required for tRNA modification, but indispensable for both telomere length regulation and recombination [33,35,37], providing us a separation-of-function tool to conduct genetic analyses.
To validate that Cgi121 promotes telomere recombination, we firstly examined telomere recombination efficiency of cgi121Δ mutant in the presence of telomerase as in Fig. 2C. The result showed that the Y’ recombination efficiency was modestly reduced by deletion of CGI121 in telomerase-positive cells (Fig. 3A). Then we performed telomere sequencing to examine the role of Cgi121 in telomeric TG1–3 recombination in telomerase-negative tlc1Δ and tlc1Δ cgi121Δ cells. We constructed heterozygous diploid cells with one copy of telomerase RNA gene TLC1 and CGI121 deleted (BY4743 TLC1/ tlc1Δ CGI121/cgi121Δ). After sporulation and tetrad dissection, spores with different genotypes (tlc1Δ and tlc1Δ cgi121Δ) were identified. Spores from the same tetrad were used for further analysis as they had the same initial telomere lengths. Cells were cultured for 50 generations after tetrad dissection and genomic DNA was extracted. Telomere PCR was then performed as previously described to amplify the TG1–3 sequence of telomere IL [51,52]. The PCR products were then cloned to T vector for sequencing analyses. About 100 clones of each genotype were obtained and analyzed. The results showed that 20.59% of telomeres (21 out of 102) were elongated through recombination in tlc1Δ cells (Fig. 3B). In contrast, only 11.54% of telomeres (12 out of 104) were repaired through recombination in tlc1Δ cgi121Δ cells (Fig. 3B). These results confirmed that Cgi121 plays a positive regulatory role in telomere recombination in both telomere-positive and -negative cells [35].
Fig. 3. Deletion of KEOPS subunit gene CGI121 compromises telomere recombination and inhibits cellular aging. 
(A) Y’ recombination of cgi121Δ cells. The assay was performed as in Fig. 2C. (B) Telomere sequencing results of tlc1Δ and tlc1Δ cgi121Δ cells. Spores with genotype of tlc1Δ and tlc1Δ cgi121Δ were obtained from the same tetrad and grown for 50 generations before genomic DNA extraction. PCR of telomere IL was performed and the PCR products were ligated to pMD18-T vector and subjected to sequencing. Each column represents one sequenced telomere. The constant parts of the telomere sequences were indicated in blue and the divergent parts in pink (representing the recombined telomere sequences). About 100 clones of each strain were analyzed. (C) Lifespan assay of BY4742 cgi121Δ mutant strain. (D) Telomeric Southern blot and lifespan assay of tlc1Δ cgi121Δ pre-senescing cells. Spores with different genotypes and the same mating type α were grown for 50 generations and then subjected to telomere Southern blot (left) and lifespan analysis (right). (E) After super-elongation of telomeres, telomere length and lifespan of spores with different genotypes and mating type α were determined as in (D). Inactivation of Cgi121 promotes cell longevity
Since Cgi121 promotes telomere recombination (Fig. 3A and 3B), and inhibition of telomere recombination restores cellular lifespan (Fig. 1C and 1D), we reasoned that deletion of CGI121 would suppress the recombination activity at telomeres, and thereby extend the replicative lifespan. Following this thought, we deleted CGI121 in a long lived yeast strain BY4742 (Mat α) which is commonly used in aging research and examined lifespan of the mutant. Deletion of CGI121 slowed down aging process strikingly, both the mean and maximum lifespan of cgi121Δ cells increased about 50% (Fig. 3C). The long live phenotype was also observed in the isogenic BY4741 cgi121Δ strain of Mat a mating type (S2A Fig.), indicating that Cgi121 affects lifespan independently of the mating type. Thus, we identified Cgi121 as a novel longevity regulator.
As we have mentioned above, in wild-type cells telomeres are maintained mainly by telomerase while telomere recombination occasionally occurs and brings the risk of genome instability. Deletion of CGI121 in these wild-type cells may inhibit telomere recombination and promote genome stability and cell longevity. If this is the case in the presence of telomerase, the activated telomere recombination in the absence of telomerase could also be compromised by deletion of CGI121, and extension of lifespan in telomerase–null cells would be expected.
To examine the role of Cgi121 on lifespan of telomerase-null pre-senescing cells, we obtained spores from heterozygous diploid cells (BY4743 TLC1/tlc1Δ CGI121/cgi121Δ) by tetrad dissection. Spores with Mat α (the same mating type as that of BY4742) were selected to perform the following lifespan assays. According to previously published data in our lab, cells immediately dissected have lifespan similar to that of wild-type cells as telomere recombination is not activated yet [40]. Thus, in our experiment, spores were grown for 50 generations after dissection so that telomeres are modestly shortened and telomere recombination level is elevated (Fig. 3D, left). These cells were then subjected to lifespan assay. As expected, tlc1Δ senescing cells show shortened lifespan, and deletion of CGI121 extends lifespan of tlc1Δ mutant (Fig. 3D, right). Consistently, this phenotype is also observed in est2Δ mutant (S2B Fig.). We noticed that lifespan of tlc1Δ cgi121Δ double mutant was not restored to the level seen in the cgi121Δ single mutant (Fig. 3D, right). That’s likely attributed to the continuous telomere shortening in the double mutant as the lifespan assay progressing. The emerging critically short telomeres can trigger cell cycle arrest and senescence. Therefore, the double mutant doesn’t have a full replicative capacity as the cgi121Δ single mutant.
To avoid the interference of critically short telomere(s) on lifespan, we generated heterozygous diploid cells with over-elongated telomeres by introducing a plasmid harboring a Cdc13-Est2 fusion protein [53]. Cells were serially passaged and telomeres were examined by telomeric Southern blot. Expression of the fusion gene conferred super-long telomeres of about 1 kb to the diploid cells (S2C Fig.). Then the plasmid encoding Cdc13-Est2 fusion protein was popped-out by negative selection and tetrad dissection was performed to obtain spores with different genotypes. The super-long telomeres in the dissected spores are about 800 bp (Fig. 3E, left), a length that prevents critically short telomeres from emerging during the lifespan assay. Over-elongating telomere has no effect on lifespan of wild-type cells (S2D Fig.). The tlc1Δ mutant with long telomeres shows similar lifespan to that of wild-type cells (Fig. 3E, right), probably because the long telomeres in this mutant result in similar telomere recombination state to that of wild-type strain. Deletion of CGI121 extends lifespan of tlc1Δ mutant significantly and the double mutant has lifespan similar to that of cgi121Δ single mutant (Fig. 3E, right). These data further support our hypothesis that inhibition of telomere recombination by deletion of CGI121 promotes cellular longevity.
Deletion of CGI121 promotes longevity in yku80-4 mutant
It remains elusive how Cgi121 promotes telomere recombination to affect cell longevity. One possibility is through regulating generation of telomeric ssDNA which is essential for initiation of recombination events. Previous report suggests that Cgi121 functions in generation of telomeric ssDNA, as deletion of CGI121 inhibits accumulation of telomeric ssDNA in the temperature sensitive cdc13-1 mutant [33]. In wild-type cells, the level of telomeric ssDNA is relatively low. In yku80Δ cells, telomeres become deprotected and telomeric ssDNA is accumulated [23,25], and accordingly the replicative lifespan is shortened (S3 Fig.) [54]. However, the shortened lifespan of yku80Δ mutant is not restored to the length of wild-type cells by deletion of CGI121 (S3 Fig.). Considering that yKu80 plays multiple roles in DNA damage repair, as well as telomere maintenance, we then used a separate-of-function yku80tel allele, yku80-4, which retains the ability of DNA end-joining and telomerase activity regulation, but displays severe defects in telomere protection [55]. The yku80-4 mutant was constructed by integrating a plasmid bearing a yku80-4 allele into the genome of yku80Δ strain. In parallel, the vector plasmid or the plasmid harboring a wild-type copy of YKU80 was integrated to yku80Δ mutant respectively. As the yku80Δ null mutant, the yku80-4 mutant also shows significantly shortened lifespan (Fig. 4A), probably due to accumulated ssDNA [55]. Strikingly, the lifespan of yku80-4 cgi121Δ double mutant is fully restored and extended to a level equivalent to that of the cgi121Δ single mutant (Fig. 4B). These data support the conclusion that Cgi121 may facilitate ssDNA generation at telomeres [33], and therefore accelerate cellular aging.
Fig. 4. Inactivation of CGI121 extends lifespan of yku80-4 cells. 
(A) Lifespan assay of the yku80 mutants. (B) Lifespan analysis of yku80-4 and yku80-4 cgi121Δ mutants. Cgi121 and Fob1 independently regulate cellular lifespan
In budding yeast, the rDNA consists of ∼150 copies of 9.1 kb rRNA genes, and is highly recombinogenic [56,57,58]. rDNA instability is promoted by Fob1-dependent DNA replication fork stalling which may cause DSBs within the rDNA [59,60,61]. Elimination of FOB1 gene reduces the rate of rDNA recombination [61], and significantly extends cellular lifespan [62]. In contrast, deletion of SIR2 gene disrupts the heterochromatin structure of rDNA loci and rDNA recombination level is elevated which confers shortened lifespan [54,63,64,65]. To investigate whether the effect of Cgi121 on cell longevity is attributed to rDNA recombination, we performed a marker loss assay to analyze the rDNA recombination rate [40]. As controls, sir2Δ cells show significantly elevated rDNA recombination level while fob1Δ cells have very low level of rDNA recombination (Fig. 5A). The rDNA recombination rate in cgi121Δ mutant was comparable to that in wild-type cells (Fig. 5A), indicating that Cgi121 is not involved in rDNA recombination. Consistently, the long lifespan of cgi121Δ cells could be further extended by deleting FOB1 (Fig. 5B). These results demonstrate that rDNA recombination and telomere recombination affect cellular lifespan in different pathways, and inhibition of both recombination activities have additive effect on cell longevity. Interestingly, deletion of Cgi121 affects neither homologous recombination activity at other genomic loci (S4A Fig.) nor the NHEJ efficiency (S4B Fig.), and the gross chromosomal rearrangement (GCR) rate is modestly elevated in cgi121Δ mutant (S4C Fig.). Thus, we conclude that the effect of Cgi121 in longevity regulation is attributed specifically to its role in telomere recombination.
Fig. 5. Cgi121 does not affect rDNA recombination and functions in parallel with Fob1 in aging regulation. 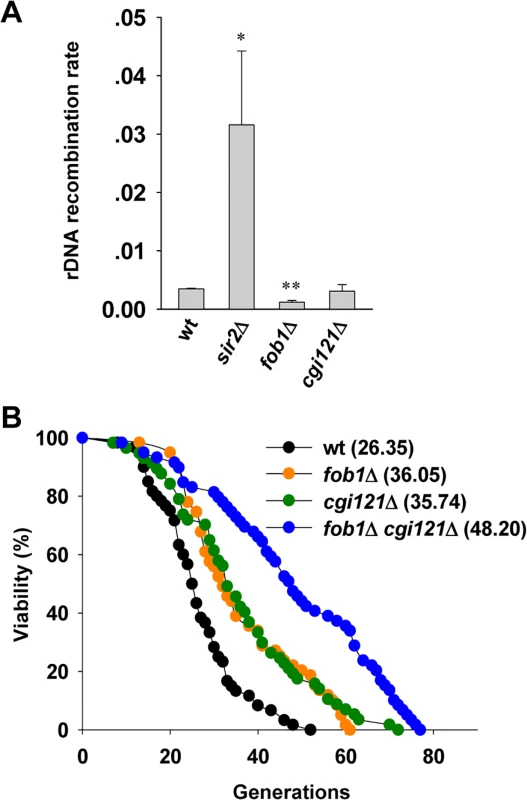
(A) Examination of rDNA recombination rate in cgi121Δ, fob1Δ and sir2Δ mutants. The error bars indicates the standard deviations. *p < 0.05 and **p < 0.01. (B) Lifespan assay of cgi121Δ, fob1Δ and fob1Δ cgi121Δ mutants. Extension of lifespan in cgi121Δ cells requires TOR1
Calorie restriction (CR) slows aging and increases life span in many organisms [66,67]. The life span extension by CR in yeast is mediated by the coordinated activity of three nutrient-responsive kinases: TOR (target of rapamycin), Sch9, and protein kinase A (PKA) [68,69,70,71]. To better understand the longevity regulation by Cgi121, we examined the lifespan of cgi121Δ cells under CR condition. CR treatment was achieved by reducing the glucose concentration in the growth medium from 2% to 0.05% [72]. In this assay, CR treatment extends lifespan of wild-type cells while the long lifespan of cgi121Δ cells could not be maintained under CR condition (Figs. 6A and S5A). Consistently, deletion of TOR1 which genetically mimics CR [69] shortens the mean and maximum life span of cgi121Δ cells (Fig. 6B). The phenotype of lifespan shortening by CR in cgi121Δ mutant is quite unexpected, but similar to those observed in W303AR cells, which has been commonly used in yeast aging research [73], as well as in long-lived Osh6 overexpression cells [74].
Fig. 6. CR shortens lifespan of cgi121Δ mutant. 
(A) Lifespan assay of cgi121Δ mutant under CR condition (reducing glucose concentration in the medium from 2% to 0.05%). (B) Lifespan assay of tor1Δ and tor1Δ cgi121Δ mutants. The observation that the longevity of cgi121Δ cells requires TOR activity leads us to speculate that Tor1 might be involved in telomere recombination. To test this possibility, we constructed heterozygous diploid cells with one copy of TOR1 and TLC1 deleted, and then obtained spores with different genotypes by tetrad dissection. These spores were cultured and serially passaged in solid or liquid medium to see whether deletion of TOR1 affects either type of telomere recombination. When cultured on solid medium, Type I survivors were readily obtained in tor1Δ tlc1Δ cells. 9 out of the 10 randomly selected clones were Type I survivors and the other clone was Type II (S5B Fig.). The emerging frequency of Type I survivors (90%) is highly consistent with previous reports [38,39], suggesting that Y’ recombination is unaffected by deletion of TOR1. On the other hand, when cultured in liquid medium, tlc1Δ cells showed a typical senescence phenotype, and deletion of TOR1 had no effect on senescence rate according to the growth curve (S5C Fig.). Southern blot analysis revealed that Type II survivors arised in both clones of either tlc1Δ or tor1Δ tlc1Δ cultures (S5D Fig.), arguing that TOR1 does not affect TG recombination. Taken together, these data suggest that Tor1 doesn’t affect telomere recombination.
Discussion
Homologous recombination (HR) is generally a universal biological process across the living organisms. It not only serves to eliminate deleterious chromosome lesions (such as DSBs and interstrand crosslinks), but also is critical for the stabilization of stalled replication forks and chromosome segregation in meiosis. Therefore HR is indispensable for general maintenance of genome integrity and stability. Deletion of the genes in RAD52 epistasis group inhibits telomere recombination, but also results in inability of repairing deleterious lesions inevitably occurring at other chromosomal loci than telomeres. Thus the overall impact of inactivation of RAD52 epistasis genes on lifespan is negative, leading to genome instability and lifespan shortening [11]. In this work, we surprisingly found that HR activities at telomeres can elicit genome instability and pose a negative effect on cellular longevity. Deletion of the KEOPS subunit gene CGI121 specifically inhibits telomere recombination and significantly slows down replicative aging (Fig. 3). Cgi121 appears to be only required for HR at telomeres (Fig. 3A and 3B), but not for regular DSB repair by HR at other genomic loci including the rDNA (Figs. 5A and S4A), nor for other DNA repair pathways like NHEJ or GCR (S4B and S4C Fig.). Such a separation-of-function property of Cgi121 provides us a specific tool to assess the effect of telomere recombination in cellular longevity.
Telomerase and HR activities function competitively in telomere maintenance and longevity regulation
Although telomerase-mediated telomere replication is the major pathway that elongates telomeres in most eukaryotes, there are eukaryotes that do not have telomerase, but solely use recombination to maintain telomeres. From the evolutional point of view, HR may represent the earliest telomere maintenance mechanism, which precedes the evolution of telomerase-dependent maintenance of chromosomal termini [75]. In budding yeast, both telomerase and recombination can efficiently function to replicate telomeres, though the former is more preferred. In telomerase-proficient cells, the engagement of recombination in telomere elongation can have both beneficial and detrimental effects (Fig. 7A). On one hand, the recombination activity seems to be complementary to telomerase activity in telomere elongation (Fig. 2). This idea is also supported by the study in the Kluyveromyces lactis stn1-M1 mutant, in which recombination takes a dominant role in telomere elongation in spite of proficient telomerase activity [76]. Thus it is not surprising, but rather logical, to see that HR is able to function as a back-up system for telomere replication when telomerase pathway fails. On the other hand however, telomeres possess multiple characteristics of DSBs and recruit HR activity to undergo “repair” process. The HR-mediated repair activity tangles or competes with telomerase activity on telomeres (Fig. 2), leading to false-alarms which make the cells undergo aging. Thus, the balance of both activities results in metastable telomeres and normal lifespan (Fig. 7A).
Fig. 7. A model of telomere recombination on aging regulation. 
(A) In wild-type cells, telomerase and HR competes at telomeres to affect cell longevity. (B) In cells that have no or less HR activity (e.g. cgi121Δ mutant), telomerase is not disturbed by HR activity. Telomeres are more stable, and results in extended lifespan. (C) In telomerase-deficient cells (e.g. tlc1Δ cells), HR activity is the sole force to maintain telomeres, leading to unstable telomeres and shortened lifespan. In cells with no or less HR activities at telomeres (Fig. 7B), telomerase is not (or less) “harassed” by the HR activity, and the more stable telomeres result in longer lifespan (Fig. 7B). The KEOPS complex subunit Cgi121 is required for efficient telomere recombination (Fig. 3A), probably by functioning in telomeric ssDNA generation (Fig. 4B) [33]. Deletion of CGI121 specifically inhibits telomere recombination and confers extended lifespan (Fig. 3).
In telomerase-negative yeast cells, HR becomes the only means to maintain telomere length, and the unstable telomeres elicit genome instability. The trade-off of ensuring the viability of a population of cells through telomere recombination is to partially sacrifice the longevity of the mother cells (Fig. 7C). This notion is supported by several lines of evidence. The yeast cells that solely relied on HR (telomerase-null survivors) exhibited shorter lifespan than those that use telomerase to replicate telomeres (wild-type cells) (Fig. 1A) [40]. Additionally, replenishment of telomerase in telomerase-null yeast cells efficiently inhibits telomere recombination (Fig. 1B), and at least partially restores lifespan (Fig. 1C and 1D). Furthermore, attenuation of telomere recombination by deletion of CGI121 significantly increases lifespan in telomerase-negative cells (Figs. 3D, 3E and S2B). Therefore, we prefer the model that the illegitimate recombination activity involuntarily competes and interferes with telomerase activity to cause genome instability at telomeres, and results in acceleration of replicative aging (Fig. 7). It is possible that both genome stability and cell longevity are driving forces for the evolution of HR-to-telomerase in telomere maintenance mechanism.
HR activities at different genomic loci have additive effects on aging
HR is prone to take place at the genomic loci that share similar or the same DNA sequences. In yeast genome, both telomere regions and rDNA loci have repetitive sequence, and they are hot spots for intra and/or inter-chromosomal HR. At chromosomal ends, telomere recombination occurs in the presence of telomerase (Fig. 2). Deletion of CGI121 specifically inhibits telomere recombination and extends cell longevity both in telomerase-positive and -negative cells (Fig. 3). At rDNA loci, the Fob1-dependent replication fork stall causes replication stress and rDNA instability, and triggers recombination-mediated pop-out of rDNA circles [59,60,61]. Deletion of FOB1 reduces rDNA recombination [61], and extends cellular lifespan [62]. In spite of the differences between telomere and rDNA recombination, inhibition of recombination at both sites seen in the fob1Δ cgi121Δ double mutant cells has additive effect on slowing down of aging (Fig. 5B). This evidence further supports our model that the unregulated and/or illegitimate HR events occurring at certain genomic loci such as telomeres and rDNA elicit genome instability, and thereby pose a negative impact on cell longevity. It might be therapeutically significant to find a means to promote cell longevity by blocking recombination-mediated repair of telomeres.
The KEOPS complex and aging regulation
In addition to Cgi121, the KEOPS complex contains four other subunits including a protein kinase (Bud32) and an ATPase (Kae1) [33,34]. It remains elusive whether the other subunits also regulate longevity. Due to the severe growth defect of the other four KEOPS mutants, lifespan assay could not be performed. When a second copy of the KEOPS genes respectively integrated into the genome, the mRNA levels of these genes were elevated respectively according to the qRT-PCR results, suggesting that the five subunits were overexpressed respectively (S6A Fig.). However, the lifespan of the cells overexpressing any of the five subunits was unchanged (S6B and S6C Fig.). We speculate that the subunits of KEOPS complex do not function individually, but rather as a whole complex to regulate aging. Considering the high conservation of KEOPS complex in evolution, it is also intriguing to investigate whether the counterpart of the KEOPS in higher organisms functions in the same way as in yeast in longevity regulation.
Materials and Methods
Yeast strains
All the yeast strains used in this study were listed in S1 Table. Strains used in lifespan assay were all in BY4742 background unless stated otherwise. The de novo telomere addition system was modified from that reported by Gottschling’s lab [45]. The yku80-4 and related strains were constructed by integrating an MscI-linearized plasmid pRS303 bearing a copy of yku80-4 or YKU80 gene, or simply the vector plasmid into the his3Δ1 locus in the genome of yku80Δ mutant. Strains overexpressing KEOPS subunits were constructed by integrating an MscI-linearized plasmid pRS303 bearing sequences including ORF, endogenous promoter and terminator of the target genes into the his3Δ1 locus in the genome. Yeast strains were constructed either by transformation with a lithium acetate procedure or genetic cross (mating and tetrad dissection). The plasmids for gene deletion were constructed based on the pRS series [77].
Replicative lifespan assay
Lifespan assay was performed as described previously [40]. Yeast strains were pre-grown overnight on solid YPD plates at 30°C. Cells were then streaked onto fresh YPD plates and grew for about 2 hours. Single cells were randomly selected and arrayed to the plates using a micromanipulator (Singer MSM). After 2 hours (about 1–2 divisions), virgin daughter cells were isolated as buds from mother cells and subjected to lifespan analysis. Daughter cells were then removed by gentle agitation with a dissecting needle and tabulated every 1–2 cell divisions until all the cells stopped dividing. Each experiment was performed with 50–60 cells for each strain. Statistical significance was determined by a Wilcoxon rank sum test using Stata 8 software and significant differences were stated for p < 0.05. The statistical data of the replicative lifespan experiments in this study were shown in S2 Table.
Telomere Southern blot
Genomic DNA was extracted and digested with XhoI and then subjected to telomere Southern blot as described previously using the TG1–3 probe [52].
Cell viability assay for chromosome healing
Yeast cells were inoculated in yeast complete medium lacking both uracil and lysine (YCU-K-), containing 2.5% of raffinose (Sigma). The TG81 rad52Δ or TG81 rad50/rad51/rad52Δ cells were inoculated in YC medium lacking lysine (YCK-). Proportional cells were plated onto YC medium lacking uracil, containing glucose (2%) or galactose (3%) (Sigma). The repair efficiency was defined as the number of colonies on galactose plates (cut) divided by that on glucose plates (uncut). The data were summarized from four independent experimental duplicates and the error bars indicates the standard deviations. Statistic significances were calculated by Student t-test (*p < 0.05 and **p < 0.01).
Y’ recombination detection
Yeast cells were inoculated in YCU-K- medium plus 2.5% of raffinose, then diluted into YCU- plus 2.5% of raffinose and cultured to logarithmic phase. For TG81 rad52Δ or TG81 rad50/rad51/rad52Δ strain, cells were inoculated in YCK- medium and diluted into complete YC medium. Galactose was then added to a final concentration of 3%, and cells were cultured for an additional 24 h. Cells were harvested and genomic DNA was extracted following PCR amplification with primers specific for TRP1 promoter and consensus sequence of all the Y’ elements. The PCR products were subjected to Southern blot using a probe specific for TG1–3 repeats as used in other telomeric Southern blot in this study. Proportional genomic DNA was digested with EcoRI endonuclease to generate a POL1 DNA fragment of about 1 kb which was detected by a probe specific for POL1 gene and serves as internal control. Meanwhile, aliquots of the PCR products were cloned to pMD18-T vector (TAKARA) and subjected to sequencing.
Telomere sequencing
Spores derived from diploid strain BY4743 TLC1/tlc1Δ CGI121/cgi121Δ were cultured for about 50 generations after dissection. Genomic DNA was extracted and subjected to telomere PCR as described previously [51,52]. PCR products were cloned to pMD18-T vector (TAKARA) and then subjected to sequencing.
rDNA recombination rate
rDNA recombination rate is assessed by the rate of loss of the URA3 reporter gene inserted at rDNA loci [40]. Cells grown in log phase were plated to YC medium with or without 0.15% 5-FOA. rDNA recombination rate is determined by dividing the number of colonies grown on 5-FOA-containing YC plates by that on YC plate without 5-FOA. The error bars indicates the standard deviations of data from three independent experiments. Significant differences were stated for *p < 0.05 and **p < 0.01.
Supporting Information
Zdroje
1. Guarente L, Kenyon C (2000) Genetic pathways that regulate ageing in model organisms. Nature 408 : 255–262. 11089983
2. Longo VD, Shadel GS, Kaeberlein M, Kennedy B (2012) Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16 : 18–31. doi: 10.1016/j.cmet.2012.06.002 22768836
3. Kaeberlein M (2010) Lessons on longevity from budding yeast. Nature 464 : 513–519. doi: 10.1038/nature08981 20336133
4. Barton AA (1950) Some aspects of cell division in saccharomyces cerevisiae. J Gen Microbiol 4 : 84–86. 15415559
5. Mortimer RK, Johnston JR (1959) Life span of individual yeast cells. Nature 183 : 1751–1752. 13666896
6. Hoeijmakers JH (2009) DNA damage, aging, and cancer. N Engl J Med 361 : 1475–1485. doi: 10.1056/NEJMra0804615 19812404
7. Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The hallmarks of aging. Cell 153 : 1194–1217. doi: 10.1016/j.cell.2013.05.039 23746838
8. McMurray MA, Gottschling DE (2004) Aging and genetic instability in yeast. Curr Opin Microbiol 7 : 673–679. 15556042
9. Burtner CR, Kennedy BK (2010) Progeria syndromes and ageing: what is the connection? Nat Rev Mol Cell Biol 11 : 567–578. doi: 10.1038/nrm2944 20651707
10. Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91 : 1033–1042. 9428525
11. Park PU, Defossez PA, Guarente L (1999) Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol Cell Biol 19 : 3848–3856. 10207108
12. McEachern MJ, Krauskopf A, Blackburn EH (2000) Telomeres and their control. Annu Rev Genet 34 : 331–358. 11092831
13. Wellinger RJ, Wolf AJ, Zakian VA (1993) Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72 : 51–60. 8422682
14. Wellinger RJ, Zakian VA (2012) Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191 : 1073–1105. doi: 10.1534/genetics.111.137851 22879408
15. Lingner J, Cech TR, Hughes TR, Lundblad V (1997) Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci U S A 94 : 11190–11195. 9326584
16. Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266 : 404–409. 7545955
17. Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43 : 405–413. 3907856
18. Tong XJ, Li QJ, Duan YM, Liu NN, Zhang ML, et al. (2011) Est1 protects telomeres and inhibits subtelomeric y'-element recombination. Mol Cell Biol 31 : 1263–1274. doi: 10.1128/MCB.00831-10 21220516
19. Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57 : 633–643. 2655926
20. Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144 : 1399–1412. 8978029
21. Boulton SJ, Jackson SP (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res 24 : 4639–4648. 8972848
22. Milne GT, Jin S, Shannon KB, Weaver DT (1996) Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol 16 : 4189–4198. 8754818
23. Polotnianka RM, Li J, Lustig AJ (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol 8 : 831–834. 9663392
24. Boulton SJ, Jackson SP (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J 17 : 1819–1828. 9501103
25. Gravel S, Larrivee M, Labrecque P, Wellinger RJ (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science 280 : 741–744. 9563951
26. Fisher TS, Taggart AK, Zakian VA (2004) Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11 : 1198–1205. 15531893
27. Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17 : 2384–2395. 12975323
28. DuBois ML, Haimberger ZW, McIntosh MW, Gottschling DE (2002) A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161 : 995–1013. 12136006
29. Nakada D, Hirano Y, Sugimoto K (2004) Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol Cell Biol 24 : 10016–10025. 15509802
30. Mimitou EP, Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455 : 770–774. doi: 10.1038/nature07312 18806779
31. Nakada D, Matsumoto K, Sugimoto K (2003) ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 17 : 1957–1962. 12923051
32. McEachern MJ, Haber JE (2006) Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75 : 111–135. 16756487
33. Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, et al. (2006) A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 124 : 1155–1168. 16564010
34. Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, et al. (2006) Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J 25 : 3576–3585. 16874308
35. Hu Y, Tang HB, Liu NN, Tong XJ, Dang W, et al. (2013) Telomerase-null survivor screening identifies novel telomere recombination regulators. PLoS Genet 9: e1003208. doi: 10.1371/journal.pgen.1003208 23390378
36. El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, et al. (2011) A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 30 : 882–893. doi: 10.1038/emboj.2010.363 21285948
37. Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, et al. (2011) The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 30 : 873–881. doi: 10.1038/emboj.2010.343 21183954
38. Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1 - senescence. Cell 73 : 347–360. 8477448
39. Teng SC, Zakian VA (1999) Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19 : 8083–8093. 10567534
40. Chen XF, Meng FL, Zhou JQ (2009) Telomere recombination accelerates cellular aging in Saccharomyces cerevisiae. PLoS Genet 5: e1000535. doi: 10.1371/journal.pgen.1000535 19557187
41. Liti G, Louis EJ (2003) NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol Cell 11 : 1373–1378. 12769859
42. Li B, Lustig AJ (1996) A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev 10 : 1310–1326. 8647430
43. Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119 : 355–368. 15507207
44. Bechter OE, Zou Y, Shay JW, Wright WE (2003) Homologous recombination in human telomerase-positive and ALT cells occurs with the same frequency. EMBO Rep 4 : 1138–1143. 14618159
45. Diede SJ, Gottschling DE (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99 : 723–733. 10619426
46. Signon L, Malkova A, Naylor ML, Klein H, Haber JE (2001) Genetic requirements for RAD51 - and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol 21 : 2048–2056. 11238940
47. Le S, Moore JK, Haber JE, Greider CW (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152 : 143–152. 10224249
48. Malkova A, Ivanov EL, Haber JE (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A 93 : 7131–7136. 8692957
49. Mao DY, Neculai D, Downey M, Orlicky S, Haffani YZ, et al. (2008) Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol Cell 32 : 259–275. doi: 10.1016/j.molcel.2008.10.002 18951093
50. Hecker A, Lopreiato R, Graille M, Collinet B, Forterre P, et al. (2008) Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. EMBO J 27 : 2340–2351. doi: 10.1038/emboj.2008.157 19172740
51. Forstemann K, Hoss M, Lingner J (2000) Telomerase-dependent repeat divergence at the 3' ends of yeast telomeres. Nucleic Acids Res 28 : 2690–2694. 10908324
52. Meng FL, Hu Y, Shen N, Tong XJ, Wang J, et al. (2009) Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J 28 : 1466–1478. doi: 10.1038/emboj.2009.92 19369944
53. Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286 : 117–120. 10506558
54. Kaeberlein M, McVey M, Guarente L (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13 : 2570–2580. 10521401
55. Bertuch AA, Lundblad V (2003) The Ku heterodimer performs separable activities at double-strand breaks and chromosome termini. Mol Cell Biol 23 : 8202–8215. 14585978
56. Petes TD, Botstein D (1977) Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc Natl Acad Sci U S A 74 : 5091–5095. 337310
57. Philippsen P, Thomas M, Kramer RA, Davis RW (1978) Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol 123 : 387–404. 357737
58. Rustchenko EP, Sherman F (1994) Physical constitution of ribosomal genes in common strains of Saccharomyces cerevisiae. Yeast 10 : 1157–1171. 7754705
59. Kobayashi T, Horiuchi T (1996) A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1 : 465–474. 9078378
60. Johzuka K, Horiuchi T (2002) Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7 : 99–113. 11895475
61. Kobayashi T, Heck DJ, Nomura M, Horiuchi T (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12 : 3821–3830. 9869636
62. Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, et al. (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3 : 447–455. 10230397
63. Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, et al. (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev 11 : 255–269. 9009207
64. Smith JS, Boeke JD (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11 : 241–254. 9009206
65. Gottlieb S, Esposito RE (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56 : 771–776. 2647300
66. Anderson RM, Weindruch R (2012) The caloric restriction paradigm: implications for healthy human aging. Am J Hum Biol 24 : 101–106. doi: 10.1002/ajhb.22243 22290875
67. Katewa SD, Kapahi P (2010) Dietary restriction and aging, 2009. Aging Cell 9 : 105–112. doi: 10.1111/j.1474-9726.2010.00552.x 20096035
68. Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292 : 288–290. 11292860
69. Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, et al. (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310 : 1193–1196. 16293764
70. Lin SJ, Defossez PA, Guarente L (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289 : 2126–2128. 11000115
71. Powers RW 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S (2006) Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev 20 : 174–184. 16418483
72. Kaeberlein M, Kirkland KT, Fields S, Kennedy BK (2005) Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev 126 : 491–504. 15722108
73. Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, et al. (2006) Comment on "HST2 mediates SIR2-independent life-span extension by calorie restriction". Science 312 : 1312; author reply 1312. 16741098
74. Gebre S, Connor R, Xia Y, Jawed S, Bush JM, et al. (2012) Osh6 overexpression extends the lifespan of yeast by increasing vacuole fusion. Cell Cycle 11 : 2176–2188. doi: 10.4161/cc.20691 22622083
75. de Lange T (2004) T-loops and the origin of telomeres. Nat Rev Mol Cell Biol 5 : 323–329. 15071557
76. Iyer S, Chadha AD, McEachern MJ (2005) A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol 25 : 8064–8073. 16135798
77. Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 : 19–27. 2659436
Štítky
Genetika Reprodukční medicína
Článek NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY ModuleČlánek HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and MiceČlánek LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male FertilityČlánek The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAsČlánek Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick VectorČlánek Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53Článek The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in NematodesČlánek The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse SpermatocytesČlánek The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 3- Růst a vývoj dětí narozených pomocí IVF
- Délka menstruačního cyklu jako marker ženské plodnosti
- Vztah užívání alkoholu a mužské fertility
- Šanci na úspěšný průběh těhotenství snižují nevhodné hladiny progesteronu vznikající při umělém oplodnění
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
-
Všechny články tohoto čísla
- NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module
- Licensing of Primordial Germ Cells for Gametogenesis Depends on Genital Ridge Signaling
- A Genomic Duplication is Associated with Ectopic Eomesodermin Expression in the Embryonic Chicken Comb and Two Duplex-comb Phenotypes
- Genome-wide Association Study and Meta-Analysis Identify as Genome-wide Significant Susceptibility Gene for Bladder Exstrophy
- Mutations of Human , Encoding the Mitochondrial Asparaginyl-tRNA Synthetase, Cause Nonsyndromic Deafness and Leigh Syndrome
- Exome Sequencing in an Admixed Isolated Population Indicates Variants Confer a Risk for Specific Language Impairment
- Genome-Wide Association Studies in Dogs and Humans Identify as a Risk Variant for Cleft Lip and Palate
- Rapid Evolution of Recombinant for Xylose Fermentation through Formation of Extra-chromosomal Circular DNA
- The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in
- Methyl Farnesoate Plays a Dual Role in Regulating Metamorphosis
- Maternal Co-ordinate Gene Regulation and Axis Polarity in the Scuttle Fly
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Inhibition of Telomere Recombination by Inactivation of KEOPS Subunit Cgi121 Promotes Cell Longevity
- Clonality and Evolutionary History of Rhabdomyosarcoma
- HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice
- Methylation-Sensitive Expression of a DNA Demethylase Gene Serves As an Epigenetic Rheostat
- BREVIPEDICELLUS Interacts with the SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA to Regulate and Expression in Control of Inflorescence Architecture
- Seizures Are Regulated by Ubiquitin-specific Peptidase 9 X-linked (USP9X), a De-Ubiquitinase
- The Fun30 Chromatin Remodeler Fft3 Controls Nuclear Organization and Chromatin Structure of Insulators and Subtelomeres in Fission Yeast
- A Cascade of Iron-Containing Proteins Governs the Genetic Iron Starvation Response to Promote Iron Uptake and Inhibit Iron Storage in Fission Yeast
- Mutation in MRPS34 Compromises Protein Synthesis and Causes Mitochondrial Dysfunction
- LRGUK-1 Is Required for Basal Body and Manchette Function during Spermatogenesis and Male Fertility
- Cis-Regulatory Mechanisms for Robust Olfactory Sensory Neuron Class-restricted Odorant Receptor Gene Expression in
- Effects on Murine Behavior and Lifespan of Selectively Decreasing Expression of Mutant Huntingtin Allele by Supt4h Knockdown
- HDAC4-Myogenin Axis As an Important Marker of HD-Related Skeletal Muscle Atrophy
- A Conserved Domain in the Scc3 Subunit of Cohesin Mediates the Interaction with Both Mcd1 and the Cohesin Loader Complex
- Selective and Genetic Constraints on Pneumococcal Serotype Switching
- Bacterial Infection Drives the Expression Dynamics of microRNAs and Their isomiRs
- The GATA Factor Regulates . Developmental Timing by Promoting Expression of the Family MicroRNAs
- Accumulation of Glucosylceramide in the Absence of the Beta-Glucosidase GBA2 Alters Cytoskeletal Dynamics
- Reproductive Isolation of Hybrid Populations Driven by Genetic Incompatibilities
- The Contribution of Alu Elements to Mutagenic DNA Double-Strand Break Repair
- Systems Biology of Tissue-Specific Response to Reveals Differentiated Apoptosis in the Tick Vector
- Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling
- The Lysine Acetyltransferase Activator Brpf1 Governs Dentate Gyrus Development through Neural Stem Cells and Progenitors
- PHABULOSA Controls the Quiescent Center-Independent Root Meristem Activities in
- DNA Polymerase ζ-Dependent Lesion Bypass in Is Accompanied by Error-Prone Copying of Long Stretches of Adjacent DNA
- Examining the Evolution of the Regulatory Circuit Controlling Secondary Metabolism and Development in the Fungal Genus
- Zinc Finger Independent Genome-Wide Binding of Sp2 Potentiates Recruitment of Histone-Fold Protein Nf-y Distinguishing It from Sp1 and Sp3
- GAGA Factor Maintains Nucleosome-Free Regions and Has a Role in RNA Polymerase II Recruitment to Promoters
- Neurospora Importin α Is Required for Normal Heterochromatic Formation and DNA Methylation
- Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis
- Phenotype Specific Analyses Reveal Distinct Regulatory Mechanism for Chronically Activated p53
- A Systems-Level Interrogation Identifies Regulators of Blood Cell Number and Survival
- Morphological Mutations: Lessons from the Cockscomb
- Genetic Interaction Mapping Reveals a Role for the SWI/SNF Nucleosome Remodeler in Spliceosome Activation in Fission Yeast
- The Role of China in the Global Spread of the Current Cholera Pandemic
- The Nuclear Receptor DAF-12 Regulates Nutrient Metabolism and Reproductive Growth in Nematodes
- A Zinc Finger Motif-Containing Protein Is Essential for Chloroplast RNA Editing
- Resistance to Gray Leaf Spot of Maize: Genetic Architecture and Mechanisms Elucidated through Nested Association Mapping and Near-Isogenic Line Analysis
- Small Regulatory RNA-Induced Growth Rate Heterogeneity of
- Mitochondrial Dysfunction Reveals the Role of mRNA Poly(A) Tail Regulation in Oculopharyngeal Muscular Dystrophy Pathogenesis
- Complex Genomic Rearrangements at the Locus Include Triplication and Quadruplication
- Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10
- Sex Ratio Meiotic Drive as a Plausible Evolutionary Mechanism for Hybrid Male Sterility
- Tertiary siRNAs Mediate Paramutation in .
- RECG Maintains Plastid and Mitochondrial Genome Stability by Suppressing Extensive Recombination between Short Dispersed Repeats
- Escape from X Inactivation Varies in Mouse Tissues
- Opposite Phenotypes of Muscle Strength and Locomotor Function in Mouse Models of Partial Trisomy and Monosomy 21 for the Proximal Region
- Glycosyl Phosphatidylinositol Anchor Biosynthesis Is Essential for Maintaining Epithelial Integrity during Embryogenesis
- Hyperdiverse Gene Cluster in Snail Host Conveys Resistance to Human Schistosome Parasites
- The Class Homeodomain Factors and Cooperate in . Embryonic Progenitor Cells to Regulate Robust Development
- Recombination between Homologous Chromosomes Induced by Unrepaired UV-Generated DNA Damage Requires Mus81p and Is Suppressed by Mms2p
- Synergistic Interactions between Orthologues of Genes Spanned by Human CNVs Support Multiple-Hit Models of Autism
- Gene Networks Underlying Convergent and Pleiotropic Phenotypes in a Large and Systematically-Phenotyped Cohort with Heterogeneous Developmental Disorders
- The ATM Signaling Cascade Promotes Recombination-Dependent Pachytene Arrest in Mouse Spermatocytes
- Combinatorial Control of Light Induced Chromatin Remodeling and Gene Activation in
- Linking Aβ42-Induced Hyperexcitability to Neurodegeneration, Learning and Motor Deficits, and a Shorter Lifespan in an Alzheimer’s Model
- The Complex Contributions of Genetics and Nutrition to Immunity in
- NatB Domain-Containing CRA-1 Antagonizes Hydrolase ACER-1 Linking Acetyl-CoA Metabolism to the Initiation of Recombination during . Meiosis
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
- Osteopetrorickets due to Snx10 Deficiency in Mice Results from Both Failed Osteoclast Activity and Loss of Gastric Acid-Dependent Calcium Absorption
- A Genomic Portrait of Haplotype Diversity and Signatures of Selection in Indigenous Southern African Populations
- Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin
- Inhibits Neuromuscular Junction Growth by Downregulating the BMP Receptor Thickveins
- Replicative DNA Polymerase δ but Not ε Proofreads Errors in and in
- Unsaturation of Very-Long-Chain Ceramides Protects Plant from Hypoxia-Induced Damages by Modulating Ethylene Signaling in
- The Small Protein MntS and Exporter MntP Optimize the Intracellular Concentration of Manganese
- A Meta-analysis of Gene Expression Signatures of Blood Pressure and Hypertension
- Pervasive Variation of Transcription Factor Orthologs Contributes to Regulatory Network Evolution
- Network Analyses Reveal Novel Aspects of ALS Pathogenesis
- A Role for the Budding Yeast Separase, Esp1, in Ty1 Element Retrotransposition
- Nab3 Facilitates the Function of the TRAMP Complex in RNA Processing via Recruitment of Rrp6 Independent of Nrd1
- A RecA Protein Surface Required for Activation of DNA Polymerase V
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Clonality and Evolutionary History of Rhabdomyosarcoma
- Morphological Mutations: Lessons from the Cockscomb
- Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance
- Transcriptomic Profiling of Reveals Reprogramming of the Crp Regulon by Temperature and Uncovers Crp as a Master Regulator of Small RNAs
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



