-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Conflict within and the Escalating War between the Sex Chromosomes
article has not abstract
Published in the journal: . PLoS Genet 8(9): e32767. doi:10.1371/journal.pgen.1002955
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1002955Summary
article has not abstract
Selfish genetic elements that distort Mendelian segregation to favor their own transmission are common in eukaryotic genomes [1], [2]. Segregation distortion can reduce whole organism fitness, resulting in strong counter selection for genes that suppress distorters. Such intragenomic conflicts have the potential to drive recurrent bouts of antagonistic co-evolution [3]. Theory predicts that genetic conflicts should be particularly intense between the sex chromosomes [4], [5]. The expectation that sex-linked conflict should be rampant has led to a renewed emphasis on the importance of antagonistic co-evolution for driving genome evolution [6]. However, while numerous examples of genes involved in intragenomic conflict now exist [1], evidence for antagonistic co-evolution between the mammalian X and Y chromosomes has remained elusive.
In this issue of PLOS Genetics, Cocquet et al. have demonstrated a genetic basis for X–Y conflict acting during a crucial stage of mouse spermatogenesis [7]. The sex chromosomes are silenced via chromatin remodeling during the initiation of meiosis (meiotic sex chromosome inactivation or MSCI) [8]. Gene silencing persists through the remainder of spermatogenesis (postmeiotic sex chromatin or PMSC), save for a subset of genes that escape inactivation [9].
Considerable progress has been made recently on the epigenetic regulation of MSCI and PMSC, including the identification of a multicopy Y-linked gene, Sly, involved in the maintenance of PMSC [10]. Male mice with Sly deficiency show up-regulation of several sex-linked genes during PMSC, are sub-fertile, and produce female-biased litters. Thus, Sly is a strong candidate for being involved in X–Y conflict due to its repressive interaction with other genes and its potential to bias sex chromosome transmission. Intriguingly, there are two X-linked genes (Slx and Slxl1; hereafter Slx/Slxl1) that are closely related to Sly. Both are regulated by Sly, occur in large multicopy clusters on the X, and are crucial for spermatogenesis [11].
To test for genetic conflict between these genes, Cocquet et al. generated transgenic mice expressing short hairpin RNA (shRNA) that knockdown Sly or Slx/Slxl1 transcript levels without completely knocking out gene function [7]. Both Sly - and Slx/Slxl1-deficient mice showed impaired spermatogenesis, but Slx/Slxl1 deficiency led to a slight reduction in sex-linked gene expression in postmeiotic cells and male-biased litters (Figure 1A). Strikingly, mice deficient for both Sly and Slx/Slxl1I showed a complete rescue of XY expression, male fertility, and sex ratio phenotypes. That is, the genes have antagonistic roles during spermatogenesis: Sly represses XY expression during PMSC and promotes the transmission of the Y, while Slx/Slxl1 activates XY expression and promotes the transmission of the X. The surprising conclusion is that antagonism depends on the relative expression of these genes and not their total abundance.
Fig. 1. The interaction and evolution of Sly and Slx/Slxl1. 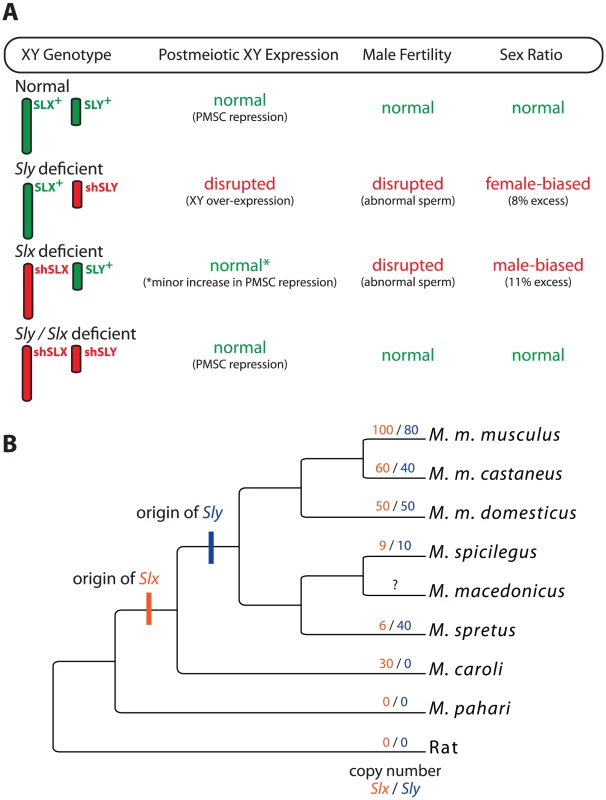
(A) A summary of the results from the various deficiency models generated by Cocquet et al. [7]. X and Y chromosome genotypes are given along the margin with wild-type genotypes in green and deficiency genotypes in red (shSLX and shSLY). Two transgenic constructs were made to target Slx/Slxl1 but are presented together here for clarity. For each genotype, the general status of XY expression, male fertility, and sex ratio are given. Phenotypes falling severely outside the wild-type range are in red. (B) Evolutionary relationships among some of the mouse species in the genus Mus, following [19]. The branches are not to scale and not all species of Mus are shown. Most standard laboratory strains, including those used by Cocquet et al. [7], are derived from M. m. domesticus. Inferred copy numbers for Slx (orange) and Sly (blue) [13] are given for each lineage. Slxl1 is not shown. Several questions remain regarding the mechanistic and genetic bases of distortion. For example, segregation distortion in the Sly-Slx/Slxl1 system appears to be caused by the differential fertilization ability of X - and Y-bearing sperm. Distorter genes often skew transmission through epistatic interactions with one or more responder genes [12]. In this context both Sly and Slx/Slxl1 appear to be distorters acting on one or more responder genes to impair the function of the X - or Y-bearing sperm, respectively [7]. Which raises the question, what are the responders?
Even more interesting are the evolutionary consequences of recurrent sex-linked conflict. If Sly and Slx/Slxl1I were locked in an antagonistic conflict, then we would predict that each would be rapidly evolving on some level. The relevant metric here seems to be gene copy number. Sly and Slx/Slxl1I are recent additions to the mouse genome, appearing within the past 3 million years (Figure 1B). Since that time they have rapidly expanded in some, but not all, lineages [13]. Why? Is genetic conflict more intense in some species? Or is the antagonistic interaction a consequence of novel functions that have evolved more recently? The Mus musculus X is enriched for dozens of other multicopy gene families expressed primarily in postmeiotic cells, which is thought to be a mechanism for escaping PMSC [14]. This interpretation now appears to be correct, with the added caveat that the entire process may be a side effect of genetic conflict between Sly and Slx/Slxl1I. Most of these X-linked amplicons are repressed by Sly during PMSC. Thus, the rapid expansion of Sly—driven by conflict with Slx/Slxl1I—may in turn drive compensatory expansion of other sex-linked genes in order to maintain proper expression levels [13].
One important consequence of recurrent sex-linked conflict is its potential to drive speciation [6]. Several of the mice presented in Figure 1B can hybridize, often resulting in hybrid male sterility (HMS). In particular, some reciprocal crosses between M. m. musculus and M. m. domesticus yield asymmetric HMS; males are only sterile when a M. m. musculus female is crossed with an M. m. domesticus male. Moreover, sterile males show widespread over-expression of the X, presumably due to a failure of MSCI and/or PMSC [15]. Cocquet et al. [7] propose that interactions between Sly and Slx/Slxl1I may be the cause of this HMS because copy number differences between the subspecies will yield hybrid males that are Sly deficient [7]. While this model is intriguing, it must be considered in light of recent work showing that HMS between M. m. musculus and M. m. domesticus is genetically complex and not strongly dependent on the origin of Y [16], and that other genetic interactions causing HMS also disrupt XY gene expression [17]. Nonetheless, these data do not exclude an important contribution of Sly/Slx mismatch to HMS in this or any other mouse hybrid crosses. If true, this would provide the first direct evidence that sex-linked genetic conflict can drive mammalian speciation.
Finally, the finding that a few novel genes control epigenetic regulation of a key step in spermatogenesis is quite remarkable. The basic epigenetic processes underlying PMSC appear to be conserved within mammals, yet its genetic regulation has only been elucidated in mice [10]. These insights are exciting, but are tempered by the fact that the key genes regulating PMSC in mice do not exist in the vast majority of mammals. The human X and Y show similar patterns of PMSC repression, including escape from silencing of several single and multicopy genes [18]. However, fewer than 20% of these genes are shared with mouse. Collectively, these findings illustrate the power of evolution to generate novelty in the face of developmental constraint and call into question the notion that research on a few model systems will be sufficient to elucidate the general molecular underpinnings of reproduction. When it comes to the evolution of reproduction and the sex chromosomes, exceptions may prove to be the rule.
Zdroje
1. BurtA, TriversR (2006) Genes in conflict: the biology of selfish genetic elements. Belknap Press of Harvard University Press
2. WerrenJH (2011) Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci U S A 108 : 10863–10870.
3. RiceWR, HollandB (1997) The enemies within: intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific Red Queen. Behav Ecol Sociobiol 41 : 1–10.
4. FrankSA (1991) Divergence of meiotic drive suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45 : 262–267.
5. HurstLD, PomiankowskiA (1991) Causes of sex-ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's Rule and related phenomena. Genetics 128 : 841–858.
6. MeiklejohnCD, TaoY (2010) Genetic conflict and sex chromosome evolution. Trends Ecol Evol 25 : 215–223.
7. CocquetJ, EllisP, MahadevaiahS, AffaraN, VaimanD, et al. (2012) A genetic basis for a postmeiotic X vs. Y chromosome intragenomic conflict in the mouse. PLoS Genet 8: e1002900 doi:10.1371/journal.pgen.1002900.
8. TurnerJMA (2007) Meiotic sex chromosome inactivation. Development 134 : 1823–1831.
9. NamekawaSH, ParkPJ, ZhangL-F, ShimaJE, McCarreyJR, et al. (2006) Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16 : 660–667.
10. CocquetJ, EllisPJI, YamauchiY, MahadevaiahSK, AffaraNA, et al. (2009) The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 7: e1000244 doi:10.1371/journal.pbio.1000244.
11. CocquetJ, EllisPJI, YamauchiY, RielJM, KaracsTPS, et al. (2010) Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol Biol Cell 21 : 3497–3505.
12. LyonMF (1984) Transmission ratio distortion in mouse t-haplotypes is due to multiple distorter genes acting on a responder locus. Cell 37 : 621–628.
13. EllisPJI, BaconJ, AffaraNA (2011) Association of Sly with sex-linked gene amplification during mouse evolution: a side effect of genomic conflict in spermatids? Hum Mol Genet 20 : 3010–3021.
14. MuellerJL, MahadevaiahSK, ParkPJ, WarburtonPE, PageDC, et al. (2008) The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 40 : 794–799.
15. GoodJM, GigerT, DeanMD, NachmanMW (2010) Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet 6: e1001148 doi:10.1371/journal.pgen.1001148.
16. CampbellP, GoodJM, DeanMD, TuckerPK, NachmanMW (2012) The contribution of the Y chromosome to hybrid male sterility in house mice. Genetics 191 : 1271–1281.
17. MiholaO, TrachtulecZ, VlcekC, SchimentiJC, ForejtJ (2009) A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323 : 373–375.
18. SinH-S, IchijimaY, KohE, NamikiM, NamekawaSH (2012) Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res 22 : 827–836.
19. LundriganBL, JansaSA, TuckerPK (2002) Phylogenetic relationships in the genus Mus, based on paternally, maternally, and biparentally inherited characters. Syst Biol 51 : 410–431.
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese SubjectsČlánek Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in RiceČlánek Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System inČlánek An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural CrestČlánek A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of BacteriaČlánek A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 9- Akutní intermitentní porfyrie
- Růst a vývoj dětí narozených pomocí IVF
- Vliv melatoninu a cirkadiálního rytmu na ženskou reprodukci
- Intrauterinní inseminace a její úspěšnost
- Délka menstruačního cyklu jako marker ženské plodnosti
-
Všechny články tohoto čísla
- Heterozygous Mutations in DNA Repair Genes and Hereditary Breast Cancer: A Question of Power
- GWAS of Diabetic Nephropathy: Is the GENIE out of the Bottle?
- The Conflict within and the Escalating War between the Sex Chromosomes
- Proteome-Wide Analysis of Disease-Associated SNPs That Show Allele-Specific Transcription Factor Binding
- Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes and as Potential Breast Cancer Susceptibility Alleles
- A Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in
- Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese Subjects
- Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in Rice
- Co-Evolution of Mitochondrial tRNA Import and Codon Usage Determines Translational Efficiency in the Green Alga
- SIRT6/7 Homolog SIR-2.4 Promotes DAF-16 Relocalization and Function during Stress
- CNV Formation in Mouse Embryonic Stem Cells Occurs in the Absence of Xrcc4-Dependent Nonhomologous End Joining
- Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System in
- Citrullination of Histone H3 Interferes with HP1-Mediated Transcriptional Repression
- Variation in Genes Related to Cochlear Biology Is Strongly Associated with Adult-Onset Deafness in Border Collies
- The Long Non-Coding RNA Affects Chromatin Conformation and Expression of , but Does Not Regulate Its Imprinting in the Developing Heart
- Rif2 Promotes a Telomere Fold-Back Structure through Rpd3L Recruitment in Budding Yeast
- Is a Metastasis Susceptibility Gene That Suppresses Metastasis by Modifying Tumor Interaction with the Cell-Mediated Immunity
- The p38/MK2-Driven Exchange between Tristetraprolin and HuR Regulates AU–Rich Element–Dependent Translation
- Rare Copy Number Variants Contribute to Congenital Left-Sided Heart Disease
- A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse
- An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural Crest
- Characterization of Inducible Models of Tay-Sachs and Related Disease
- Hominoid-Specific Protein-Coding Genes Originating from Long Non-Coding RNAs
- Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24
- Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Virulence
- Convergence of the Transcriptional Responses to Heat Shock and Singlet Oxygen Stresses
- Genomics of Adaptation during Experimental Evolution of the Opportunistic Pathogen
- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Vsx2 Controls Eye Organogenesis and Retinal Progenitor Identity Via Homeodomain and Non-Homeodomain Residues Required for High Affinity DNA Binding
- The Long Path from QTL to Gene
- TCF7L2 Modulates Glucose Homeostasis by Regulating CREB- and FoxO1-Dependent Transcriptional Pathway in the Liver
- The Non-Flagellar Type III Secretion System Evolved from the Bacterial Flagellum and Diversified into Host-Cell Adapted Systems
- Complex Chromosomal Rearrangements Mediated by Break-Induced Replication Involve Structure-Selective Endonucleases
- Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-A in Fission Yeast
- A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of Bacteria
- Determinants of Human Adipose Tissue Gene Expression: Impact of Diet, Sex, Metabolic Status, and Genetic Regulation
- Genome-Wide Association Studies Identify Heavy Metal ATPase3 as the Primary Determinant of Natural Variation in Leaf Cadmium in
- Tethering of the Conserved piggyBac Transposase Fusion Protein CSB-PGBD3 to Chromosomal AP-1 Proteins Regulates Expression of Nearby Genes in Humans
- A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- H4K20me1 Contributes to Downregulation of X-Linked Genes for Dosage Compensation
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Coevolution within and between Regulatory Loci Can Preserve Promoter Function Despite Evolutionary Rate Acceleration
- New Susceptibility Loci Associated with Kidney Disease in Type 1 Diabetes
- SWI/SNF-Like Chromatin Remodeling Factor Fun30 Supports Point Centromere Function in
- A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
- Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription
- Genetic Modifiers of Chromatin Acetylation Antagonize the Reprogramming of Epi-Polymorphisms
- UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development
- A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



