-
Medical journals
- Career
Droplet digital PCR as a novel diagnostic tool
Authors: B. Váňová 1,2; B. Malicherova 2; T. Burjanivová 3; A. Liskova 4; K. Janikova 2,5; K. Jasek 2; Z. Lasabová 3; M. Tatár 5; L. Plank 6
Authors‘ workplace: Martin´s Center of Immunology, Ltd., Martin, Slovakia 1; Biomedical Center Martin JFM CU, Martin, Slovakia 2; Department of Molecular Biology JFM CU, Martin, Slovakia 3; Clinic of Obstetrics and Gynecology JFM CU and University Hospital in Martin, Slovakia 4; Department of Pathological Physiology JFM CU, Martin, Slovakia 5; Department of Pathological Anatomy, JFM CU and University Hospital in Martin, Slovakia 6
Published in: Klin Onkol 2021; 34(1): 33-39
Category: Review
doi: https://doi.org/10.48095/ccko202133Overview
Background: Nowadays, modern treatment methods for cancer patients are based on targeting specific molecules involved in cellular signaling system associated with tumor initiation and progression. The success of such approach depends on a correctly chosen diagnostic test with high sensitivity that identifies the occurrence and level of biomarkers in patients to select those who will respond and benefit from the treatment. The development of new technologies and the upgrades of the known ones contribute to the innovations in molecular characterization of cancer, which allows the detection of patient’s mutational status with high sensitivity and specificity. Purpose: Here, we discuss the utilization of the third-generation type of polymerase chain reaction (PCR), droplet digital PCR (ddPCR), in the molecular diagnostics of oncology diseases. According to the studies reported in our review, ddPCR represents a promising tool in genetic profiling of cancer patients. Therefore, the optimization and precise validation may enable gradual implementation of ddPCR into clinical practice in the field of oncology.
Keywords:
cancer – tumor biomarkers – molecular diagnostics – ddPCR
Introduction
Cancer diseases represent a serious global problem. Cancer is currently one of the leading causes of death in developed as well as developing countries [1]. Environmental and modern lifestyle factors, together with genetic/epigenetic alterations, contribute to the genesis of genetically unstable cells that, in cooperation with local microenvironment, trigger the process of tumorigenesis in the organism. It is characterized by uncontrolled cell division, which may lead to the formation of an abnormal mass of tissue called tumor. Malignant tumors grow infiltratively through the tissue of a primary organ. When cancer cells infiltrate blood or lymph vessels, they can spread to lymph nodes or distant organs and establish secondary tumors (metastasis) [2,3].
Modern treatment methods for cancer patients are based on targeting specific molecules involved in cellular signaling system. The most common targeted molecules are growth factor receptors or proteins involved in signaling pathways, which affect cell processes such as proliferation, angiogenesis, etc. [4]. Due to the fact that patients with different molecular conditions do not respond to particular treatment, the precise selection of patients for specific therapeutic approach is strongly recommended [5–7]. The success of such approach depends on correctly chosen diagnostic test with high sensitivity that identifies the occurrence and level of biomarkers in patients to select those who will respond and benefit from the treatment [8,9]. The development of new technologies and upgrades of the known ones (genomic sequencing, polymerase chain reaction – PCR, etc.) lead to the innovations in molecular characterization of cancer and allow the detection of mutational status of tumors with higher sensitivity and specificity in comparison to golden standards in molecular diagnostics such as Sanger sequencing, real-time PCR, etc. [10].
Digital PCR
The end of the 20th century is considered as an important milestone in the development of molecular biology. At that time, Dr. Kary Mullis introduced the polymerase chain reaction (PCR) as we know it today, which was the first generation of PCR. Gradually, the second generation of this technique, quantitative PCR (qPCR), was introduced several years later [11,12]. The next milestone leading to the development of the third generation of PCR represents the description of the possibility of digitizing the reaction. In principle, the methodology is based on classical PCR. However, the division of individual DNA templates into ‘microparticles’ with amplification reaction occurring for each DNA molecule separately and independently represents a significant difference. As a result, the absolute quantification of the amount of mutations in the sample can be performed without the need for prior calibration with a dilution series (Tab. 1) [13].
1. Comparison of PCR, qPCR and ddPCR methods. 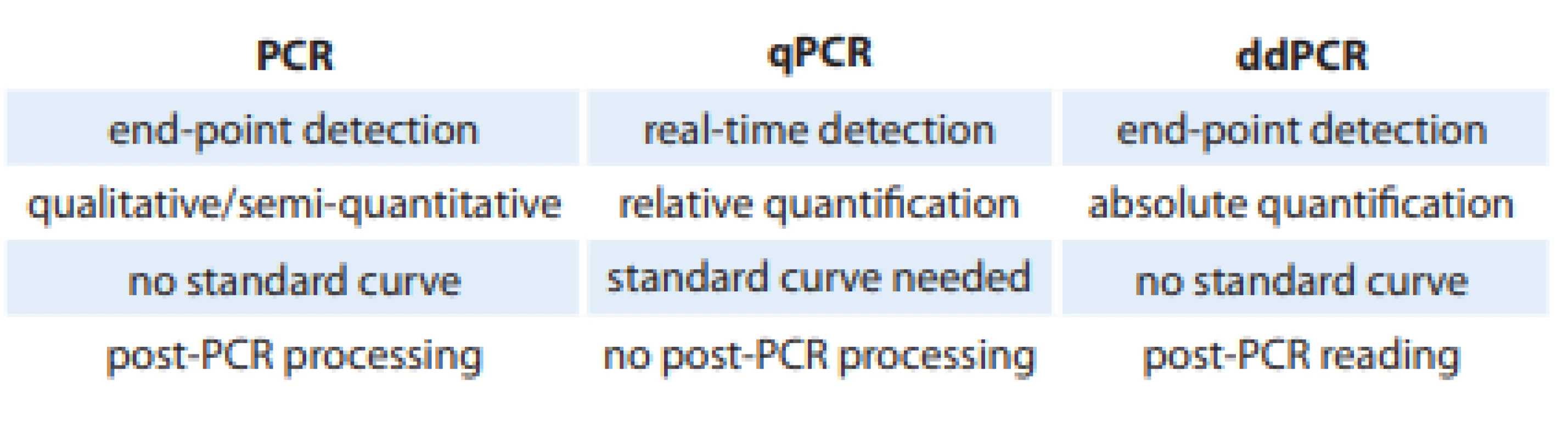
PCR – polymerase chain reaction, qPCR – quantitative PCR, ddPCR – droplet digital PCR Today, three basic platforms of the digital PCR (dPCR) are recognized: emulsion or droplet dPCR (ddPCR), dPCR on a microtiter plate, and microfluidic dPCR (cdPCR). Each of them is characterized by its own instrumentation, specific method of sample separation and the format for PCR amplification itself (microplate, chip, oil emulsion, etc.) [14].
The ThermoFisher Scientific offers digital PCR methodology using a microtiter plate composed of 96 or 384 wells into which a mixture of template, PCR mix, and TaqMan reagents are pipetted sequentially. After amplification, the sample is evaluated using a digital reader based on TaqMan probes that detects the emitted signal in each well. However, such a format is very disadvantageous in terms of the cost, difficult workflow, and limited capacity in the number of analyzed particles/wells [15,16].
The microfluidic dPCR format uses a specially designed chip that is divided into hundreds to thousands of small wells. The reagents, already prepared in these wells, are mixed with the sample by hydraulic or pneumatic pressure and divided into thousands of PCR chambers through channels on the chip in which the analyzed templates are amplified separately. Today, there are devices automated for such pipetting that contain a thermocycler to amplify the template DNA. The evaluation is also based on the detection of the fluorescence signal of the individual chambers on the chip. However, the disadvantage is, once again, high financial cost compared to the ddPCR format [15,17].
Droplet digital PCR
The emulsion or droplet dPCR (ddPCR) is performed in microreactors in the form of nano - to picoliter droplets [18]. ddPCR was first introduced by QuantaLife, later Bio-Rad Laboratories. The newer platform came from RainDanceTM Technologies, which was also purchased by Bio-Rad Laboratories, Inc.
ddPCR has been commercialized relatively recently and offers a very simple ’workflow’ for the consistent distribution of DNA molecules into micro-droplets (Fig.1). When using a Bio-Rad platform (QX100TM or QX200TM), 20 µL of the sample mix is fractionated into 20,000 droplets. The high-sensitivity RainDance platform (RainDropTM), in turn, allows the generation of 5–10 million droplets from 25–50 µL of the reaction mix - ture [19,20].
1. Workflow for the consistent distribution of DNA molecules into micro-droplets. 
FAM/HEX /TET/VIC – fl uorescent dyes, mtDNA – mutated DNA, PCR – polymerase chain reaction, WT DNA– wild type DNA Eight samples and droplet generation oil, which contains emulsion stabilizer and biocompatible surfactant, are loaded to specially designed cassettes. The prepared cartridge is then inserted into a droplet generator in which the oil and water phases are mixed under vacuum conditions and monodisperse droplets are formed. Ideally, the generated droplets are uniform in size, thermally stable, and do not overlap or crack during the preparation process. After the transfer to a thermocycler and end-point PCR amplification, a special reading device, based on flow cytometric and using TaqMan probes, senses the fluorescence of the individual particles in two channels, which correspond to the fluorescent dyes – FAM for mutated DNA and HEX/VIC for wild type DNA (WT DNA) for Bio-Rad platforms and FAM and VIC/TET with RainDropTM technology [20,21].
BEAMing PCR
A special variant of ddPCR, so-called BEAMing PCR, also uses, in addition to emulsification of the sample by oil, magnetic beads in which the template is amplified [22]. When the emulsion of droplets is established, each particle contains, ideally, one fragment of DNA template, a magnetic bead, and reagents for PCR reaction. The individual DNA molecule is then covalently attached to the bead through primers and amplified. After PCR amplification, the magnetic beads coated by DNA products are magnetically purified, and fluorescent probes, specifically designed for the mutated DNA or WT DNA, are hybridized. The fluorescence signal of individual particles is then detected on the basis of flow cytometry with Sysmex Partec CyFlow® Cube 6i Flow Cytometer (Sysmex Partec, Görlitz, Germany) and evaluated by FCS Express V5.0 software (Sysmex Partec, Görlitz, Germany) [23].
The evaluation of ddPCR data
Depending on the capacity of the chip/plate or the amount of generated droplets, we obtain partial qualitative results based on fluorescent signals for the mutated and wild-type versions of the template DNA [14]. However, there is a possibility that after the distribution of DNA molecules, the individual micro-particles/wells contain either none or several fragments of the analyzed template. Thus, after the amplification into the terminal plateau phase of the PCR reaction, a part of the micro-reactors emits a signal and a part is negative without a signal. Therefore, the evaluation is based on the so-called Poisson distribution method. Using equation (I), where p represents the number (fraction) of positive particles, we can determine the average number of targeted DNA molecules per replication reaction or λ. Together with the reaction volume and the total number of analyzed particles/droplets, the λ value is used for the calculation of the absolute concentration of targeted DNA.
(I) λ = –ln (1 – p)
In general, the more micro-reactors are analyzed, the more accurate is the detection and the identification of the mutated vs. the wild-type DNA sequences in the sample. This suggests that the format on the micro-titer plate significantly reduces the efficiency of the assay compared to the ddPCR but it is still used today [18].
However, there are two other methods for data interpretation: the Tzonev’s approach [24] and the calibration line with a control of type I and type II errors [25]. Whereas the Tzonev’s method determines the limit of detection (LoD) in terms of a number of events, another method determines LoD in terms of a number of copies per microliter (copies/μL).
ddPCR in diagnostics sphere
The commercial usage of ddPCR is dated to 2011 when Hindson et al described the application of ddPCR for absolute quantification of circulating DNA and for the detection of several genetic changes [18]. Through the years, the method has become popular. Nowadays, we use it as a tool for the detection of viruses and other pathogens [26–28], fetal screening [29,30], and analysis of genetic alterations in patients with various disorders [31–33].
The diagnostics of pathogens, based on their detection in blood/plasma, may be difficult in some cases. The hepatitis B virus, human-immunodeficiency virus, herpes-simplex virus or pathogens like Mycobacterium tuberculosis and Plasmodium malariae can induce a latent type of infection. Therefore, its concentration in a sample is too low and traditional methods (e. g. blood smear, ELISA, qPCR, etc.) are not capable to detect the presence of contagion [34]. On the other hand, using ddPCR for the detection of infectious agents shows higher sensitivity and accuracy than conventional methods [26,35–37]. The same problem is currently discussed in relation to SARS-CoV-2 confirmation. The results presented by Suo et al show high differences between qPCR vs. ddPCR outcomes in which 26 patients with negative reports assessed by qPCR were SARS-CoV-2 positive in ddPCR testing. The reason for such results is probably associated with relatively low viral load in the throat swabs of patients and also with limitations of qPCR, which could not capture the viral RNA. According to that, ddPCR should be used as a complement method for standard technique to reduce the false negatives, which may decrease the risk of viral spreading [28].
Today, the prenatal testing is still mostly performed by invasive tests such as amniocentesis and chorionic villus sampling, even with a 1% risk of the miscarriage [38]. However, cell-free fetal DNA isolated from maternal plasma can be used for prenatal diagnosis of fetal genetic profile in a non-invasive manner. The analysis of maternal peripheral blood by ddPCR for fetal sex and RhD determination, detection of sickle cell anemia, identification of fetal aneuploidies (e. g. trisomy 21), and assessment of fetal genotypes for hemophilia and β-thalassemia mutations was described in the last decade. In all studies, the results were correctly determined by ddPCR when compared to clinically validated methods [39–44]. These facts indicate that ddPCR is an effective and reliable method suitable also for prenatal diagnostics.
Nevertheless, the most common usage of ddPCR is related to the detection of genetic alteration, especially in cancer-associated genes.
ddPCR and cancer
Various molecular variations at the genetic and epigenetic level may be detected by ddPCR. Single nucleotide variations (SNVs), copy number variations (CNVs), and hyper - or hypo - -methylation of genes represent the most frequent changes in patients with different kinds of cancer which could be identified by ddPCR with high sensitivity and specificity [45,46]. In addition, the level of expression in some genes can be also diagnosed by this method [47].
The alterations in gene copy numbers are a phenomenon that may drive the process of tumorigenesis. For example, the detection of some CNVs allows to distinguish benign noncancerous tissue from precancerous/cancerous tissue in patients with oral lesions [48]. In some cancers, the presence of CNVs affects the response to therapy and can correlate with patient’s prognosis [49,50]. It is typical for MET gene copy number detection when MET-inhibitor therapy is set up for patients with positive CNV status. When evaluating MET genetic alteration, Zhang et al compared fluorescence in situ hybridization (FISH) vs. SNP 6.0 vs. ddPCR. The results showed the potential of ddPCR technique to quantify the CNV of MET gene with high precision and accuracy in comparison to FISH and SNP 6.0 in cancer cell lines and formalin-fixed paraffin-embedded (FFPE) DNA, without the need for standard curves or endogenous control [51]. The standard methods like qPCR are also limited in the measurement of CNVs due to the systematic errors, which can occur during normalization of DNA concentrations with standard samples and by using cycle threshold (CT). On the other hand, ddPCR does not need standard samples and CT values for the detection and quantification of CNVs leading to less ambiguous and more accurate results [45].
Modifications based on DNA methylation, especially hyper-methylation of promoter regions, arise in many types of cancer. These changes have a potential to become dynamic diagnostic, prognostic, and predictive biomarkers. Today, 14 of these biomarkers are used in clinical diagnostics [52,53]. The utilization of ddPCR for the detection of DNA methylation status has appeared only recently. The analysis of fresh surgical margins of head and neck squamous cell carcinoma using quantitative methylation-specific ddPCR (ddQMSP) for PAX5 gene status confirmed high sensitivity of the method [54]. However, the samples are mostly derived from FFPE tissue, therefore, the epigenomic analyses are limited by a fixation process and long storage, which cause DNA fragmentation. Van Wesenbeeck et al demonstrated high accuracy of ddPCR for methylation testing in FFPE samples and, in comparison to real-time PCR, also with a wide range of DNA input [46]. Moreover, a new type of ddPCR, so-called optimized bias-based pre-amplification-digital droplet PCR (OBBPA-ddPCR), was established by Menschikowski. The serum samples from patients with prostate carcinoma were analyzed and methylation status of PLA2R1 gene was observed by this technique. The outcomes indicated that the analytical sensitivity of OBBPA-ddPCR is significantly higher in comparison with routine methods such as methylation-sensitive high-resolution melting analysis or classic ddPCR. However, further studies are needed to confirm the use of this new two-step ddPCR technique as an analytical tool [55].
The assessment of expression profile in cancer cells, focusing on selected genes, is extensively used and allows to define the subtypes of tumors and also to stratify patients to particular subgroups. Some of these changes are already validated in the sphere of diagnosis, prognosis, and prediction of the response to therapy [56,57]. The best-known biomarker, expression level of which is observed in cancer cells, is the human epidermal growth factor receptor 2 (HER2). Generally, the over-expression of HER2 is detected mostly by immunohistochemistry (IHC) or FISH using FFPE tissue [58]. However, these methods are affected by the subjective view of the evaluator, which may lead to false results. On the other hand, ddPCR is an objective tool with possibility to detect gene expression in FFPE samples and also in cell-free tumor DNA (cftDNA) through the analysis of gene amplification [31].
Regarding SNVs together with small INDEL mutations, a lot of them have confirmed their clinical importance and ability to affect the process of anti-cancer therapy. The critical parameter for precise identification of mutations, especially those of somatic origin, is represented by their frequency in a sample. The variants with low allele frequencies are difficult to distinguish and detect [59]. The potential of ddPCR to capture mutation, even if its concentration is under LoD of certain standard methods, provides new opportunities in cancer research and diagnostics [60,61]. The monitoring of mutations in clinical practice includes e. g. V600E in BRAF, G12/G13 mutations in exon 2 of KRAS, exon 19 mutations in EGFR, etc. Many authors dealt with the optimization of ddPCR for the testing of mentioned genetic changes using different kind of examined material, such as FFPE, circulating tumor cells, or plasma. LoD was assessed for each assay and allowed to detect these variations even if the number of mutated copies in samples was low [61–63].
Detection of minimal residual disease
The minimal residual disease (MRD) is characterized as an existence of occult cancer cells, micro-metastasis in bone marrow, or tumor biomarkers (e. g. cftDNA) in the organism after undergoing the treatment. Its detection after primary therapy may identify high-risk patients who need to be treated by adjuvant therapy. However, many MRD subtypes are known and therefore it is necessary to understand their relevance for patient’s further curative process [64].
The typical diseases, in which the detection of MRD represents valuable prognostic information, are lymphoid malignancies. The mostly used methods for MRD diagnosis are flow cytometry and qPCR in which immunoglobulin gene (IGH) rearrangements are observed. Drandi et al proved that even if the qPCR is still considered as a standard tool for MRD detection [66], ddPCR may most accurately assess the presence of occult tumor biomarkers in mature B-cell lymphoid malignancies [65].
In solid tumors, the most potential and promising marker of MRD is cftDNA, which is released into the blood and other bodily fluids from cancer cells. Due to the low concentration of cftDNA in blood samples, methods for its detection must consistently determine mutations in plasma with mutant allele frequency of < 0.1% [67]. Therefore, several authors published papers dealing with the analysis of cftDNA by ddPCR in patients with different types of cancer (breast, pancreatic, colorectal, bladder, or non-small cell lung carcinoma, etc.) with a high sensitivity detection threshold of ~ 0.01% [68–72]. Despite the fact that ddPCR approach of MRD detection is highly sensitive, it still requires assay customization specifically to examined patient and knowledge of previously detected tumor-specific somatic mutations [70].
Conclusion
The detection of particular biomarkers (SNV, CNV, methylation, cftDNA, etc.) is critical for its clinical application in cancer management. High specificity and sensitivity of the described technology, which have been established in every type of experiment focused on these markers, advance ddPCR to the next level in the field of oncology. As discussed in our review, ddPCR represents a new practical and relatively easy method with high potential in the diagnostic area and may help to improve the genetic profiling of patients.
In summary, the detection of the molecular background of patients with cancer is highly necessary, especially for the therapy design. Since older methods, still used today, are not sensitive enough, the role of ddPCR as a diagnostic tool may become a useful replacement.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do bi omedicínských časopisů.
Mgr. Barbora Vanova, PhD
Martin’s Center of Immunology, Ltd.
Pavla Mudroňa 12
036 01 Martin
Slovakia
Submitted/Obdŕžané: 18. 8. 2020
Accepted/Prijaté: 22. 10. 2020
Sources
1. World Health Organization. Cancer. [online]. Available from: https: //www.who.int/health-topics/cancer#tab=tab_1.
2. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000; 100 (1): 57–70. doi: 10.1016/s0092-8674 (00) 81 683-9.
3. American Cancer Society. Cancer facts & figures 2018. [online]. Available from: https: //www.cancer.org/cancer/colon-rectal-cancer/about/what-is-colorectal-cancer.html.
4. Marks EI, Yee NS. Molecular genetics and targeted therapeutics in biliary tract carcinoma. World J Gastroenterol 2016; 22 (4): 1335–1347. doi: 10.3748/wjg.v22.i4.1335.
5. Verma M. Personalized medicine and cancer. J Pers Med 2012; 2 (1): 1–14. doi: 10.3390/jpm2010001.
6. Gil J, Laczmanska I, Pesz KA et al. Personalized medicine in oncology. New perspectives in management of gliomas. Contemp Oncol 2018; 22 (1A): 1–2. doi: 10.5114/wo.2018.73872.
7. National Comprehensive Cancer Network. Targeted therapy. [online]. Available from: https: //www.nccn.org/patients/resources/life_with_cancer/treatment/targeted_therapy.aspx.
8. Taube SE. Biomarkers in oncology: trials and tribulations. Ann N Y Acad Sci 2009; 1180 : 111–118. doi: 10.1111/j.1749-6632.2009.05019.x.
9. Simon R. Clinical trial designs for evaluating the medical utility of prognostic and predictive biomarkers in oncology. Pers Med 2010; 7 (1): 33–47. doi: 10.2217/pme. 09.49.
10. Deschoolmeester V, Baay M, Specenier P et al. A review of the most promising biomarkers in colorectal cancer: one step closer to targeted therapy. The Oncologist 2010; 15 (7): 699–731. doi: 10.1634/theoncologist.2010-0025.
11. Mullis K, Faloona F, Scharf S et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 1986; 51 (Pt 1): 263–273. doi: 10.1101/sqb.1986.051.01.032.
12. Schaad NW, Frederick RD. Real-time PCR and its application for rapid plant disease diagnostics. Can J Plant Pathol 2002; 24 (3): 250–258.
13. Baker M. Digital PCR hits its stride. Nat Methods 2012; 9 (6): 541–544.
14. Hrstka R, Kolářová T, Michalová E et al. Vývoj metod založených na PCR a jejich aplikace v onkologickém výzkumu a praxi. Klin Onkol 2014; 27 (Suppl 1): S69–S74. doi: 10.14735/amko20141s69.
15. Liao P, Huang Y. Digital PCR: endless frontier of ‘divide and conquer.’ Micromachines (Basel) 2017; 8 (8): 231. doi: 10.3390/mi8080231.
16. Digital PCR – SK. [online]. Available from: //www.thermofisher.com/uk/en/home/life-science/pcr/digital-pcr.html.
17. Zhang C, Xing D. Miniaturized PCR chips for nucleic acid amplification and analysis: latest advances and future trends. Nucleic Acids Res 2007; 35 (13): 4223–4237. doi: 10.1093/nar/gkm389.
18. Hindson BJ, Ness KD, Masquelier DA et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011; 83 (22): 8604–8610. doi: 10.1021/ac202028g.
19. Pinheiro LB, Coleman VA, Hindson CM et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 2012; 84 (2): 1003–1011. doi: 10.1021/ac202578x.
20. RainDance Digital PCR Reagents and Consumables. Life Science Research | Bio-Rad. [online]. Available from: https: //www.bio-rad.com/en-dk/product/raindance-digital-pcr-reagents-consumables?ID=Q8L21TE08O1Y.
21. Droplet DigitalTM PCR (ddPCRTM) Technology | LSR | Bio-Rad. [online]. Available from: https: //www.bio-rad.com/en-dk/applications-technologies/droplet-digital-pcr-ddpcr-technology?ID=MDV31M4VY.
22. Diehl F, Li M, He Y et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods 2006; 3 (7): 551–559. doi: 10.1038/nmeth898.
23. BEAMing Technology Overview | OncoBEAMTM. [online]. Available from: https: //www.oncobeam.com/oncobeam-technology/technology-overview.
24. Sensitivity, specificity and limit of detection in dPCR BIO-RAD2016. [online]. Available from: https: //www.bio-rad.com/en-dk/category/genomics?ID=2d11dcf8-2dbe-47a5-a1de-8315abd3c17e.
25. Slutsky B. Handbook of chemometrics and qualimetrics: part A. In: Massart DL, Vandeginste BG, Buydens LM et al. Data handling in science and technology, vol. 20A. Amsterdam: Elsevier 1997: Xvii + 867.
26. Strain MC, Lada SM, Luong T et al. Highly precise measurement of HIV DNA by droplet digital PCR. PloS One 2013; 8 (4): e55943. doi: 10.1371/journal.pone.0055943.
27. Maheshwari Y, Selvaraj V, Hajeri S et al. Application of droplet digital PCR for quantitative detection of Spiroplasma citri in comparison with real time PCR. PLOS One 2017; 12 (9): e0184751. doi: 10.1371/journal.pone.0184751.
28. Suo T, Liu X, Feng J et al. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg Microbes Infect 2020; 9 (1): 1259–1268. doi: 10.1080/22221751.2020.1772678.
29. Hudecova I, Jiang P, Davies J et al. Noninvasive detection of F8 int22h-related inversions and sequence variants in maternal plasma of hemophilia carriers. Blood 2017; 130 (3): 340–347. doi: 10.1182/blood-2016-12-755017.
30. Camunas-Soler J, Lee H, Hudgins L et al. Noninvasive prenatal diagnosis of single-gene disorders by use of droplet digital PCR. Clin Chem 2018; 64 (2): 336–345. doi: 10.1373/clinchem.2017.278101.
31. Kinugasa H, Nouso K, Tanaka T et al. Droplet digital PCR measurement of HER2 in patients with gastric cancer. Br J Cancer 2015; 112 (10): 1652–1625. doi: 10.1038/bjc.2015.129.
32. Malicherova B, Burjanivova T, Grendar M et al. Droplet digital PCR for detection of BRAF V600E mutation in formalin-fixed, paraffin-embedded melanoma tissues: a comparison with Cobas® 4800, Sanger sequencing, and allele-specific PCR. Am J Transl Res 2018; 10 (11): 3773–3781.
33. Vanova B, Kalman M, Jasek K et al. Droplet digital PCR revealed high concordance between primary tumors and lymph node metastases in multiplex screening of KRAS mutations in colorectal cancer. Clin Exp Med 2019; 19 (2): 219–224. doi: 10.1007/s10238-019-00545-y.
34. Li H, Bai R, Zhao Z et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci Rep 2018; 38 (6): BSR20181170. doi: 10.1042/BSR20181170.
35. Mu D, Yan L, Tang H et al. A sensitive and accurate quantification method for the detection of hepatitis B virus covalently closed circular DNA by the application of a droplet digital polymerase chain reaction amplification system. Biotechnol Lett 2015; 37 (10): 2063–2073. doi: 10.1007/s10529-015-1890-5.
36. Yang J, Han X, Liu A et al. Use of digital droplet PCR to detect mycobacterium tuberculosis DNA in whole blood-derived DNA samples from patients with pulmonary and extrapulmonary tuberculosis. Front Cell Infect Microbiol 2017; 7 : 369. doi: 10.3389/fcimb.2017.00369.
37. Koepfli C, Nguitragool W, Hofmann NE et al. Sensitive and accurate quantification of human malaria parasites using droplet digital PCR (ddPCR). Sci Rep 2016; 16 (6): 39183.
38. Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol 2007; 110 (3): 687–694. doi: 10.1097/01.AOG.0000278820.54029.e3.
39. Lun FMF, Tsui NBY, Chan KCA et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci USA 2008; 105 (50): 19920–19925. doi: 10.1073/pnas.0810373105.
40. Tsui NBY, Kadir RA, Chan KCA et al. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood 2011; 117 (13): 3684–3691. doi: 10.1182/blood-2010-10-310789.
41. Barrett AN, McDonnell TCR, Chan KCA et al. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin Chem 2012; 58 (6): 1026–1032.
42. El Khattabi LA, Rouillac-Le Sciellour C, Le Tessier D et al. Could digital PCR be an alternative as a non-invasive prenatal test for trisomy 21: a proof of concept study. PLoS One 2016; 11 (5): e0155009. doi: 10.1371/journal.pone.0155009.
43. D’Aversa E, Breveglieri G, Pellegatti P et al. Non-invasive fetal sex diagnosis in plasma of early weeks pregnants using droplet digital PCR. Mol Med 2018; 24 (1): 14.
44. O’Brien H, Hyland C, Schoeman E et al. Non-invasive prenatal testing (NIPT) for fetal Kell, Duffy and Rh blood group antigen prediction in alloimmunised pregnant women: power of droplet digital PCR. Br J Haematol 2020; 189 (3): e90–e94. doi: 10.1111/bjh.16500.
45. Mazaika E, Homsy J. Digital droplet PCR: CNV analysis and other applications. Curr Protoc Hum Genet 2014; 82 : 7.24.1–13. doi: 10.1002/0471142905.hg0724s82.
46. Van Wesenbeeck L, Janssens L, Meeuws H et al. Droplet digital PCR is an accurate method to assess methylation status on FFPE samples. Epigenetics 2018; 13 (3): 207–213. doi: 10.1080/15592294.2018.1448679.
47. Kinugasa H, Nouso K, Miyahara K et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015; 121 (13): 2271–2280. doi: 10.1002/cncr.29364.
48. Hughesman CB, Lu XJD, Liu KYP et al. Detection of clinically relevant copy number alterations in oral cancer progression using multiplexed droplet digital PCR. Sci Rep 2017; 7 (1): 11855. doi: 10.1038/s41598-017-11201-4.
49. Shlien A, Malkin D. Copy number variations and cancer. Genome Med 2009; 1 (6): 62.
50. Liu YJ, Shen D, Yin X et al. HER2, MET and FGFR2 oncogenic driver alterations define distinct molecular segments for targeted therapies in gastric carcinoma. Br J Cancer 2014; 110 (5): 1169–1178. doi: 10.1038/bjc.2014.61.
51. Zhang Y, Tang E-T, Du Z. Detection of MET gene copy number in cancer samples using the droplet digital PCR method. PloS One 2016; 11 (1): e0146784. doi: 10.1371/journal.pone.0146784.
52. Lewandowska J, Bartoszek A. DNA methylation in cancer development, diagnosis and therapy--multiple opportunities for genotoxic agents to act as methylome disruptors or remediators. Mutagenesis 2011; 26 (4): 475–487. doi: 10.1093/mutage/ger019.
53. Koch A, Joosten SC, Feng Z et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol 2018; 15 (7): 459–466. doi: 10.1038/s41571-018-0004-4.
54. Hayashi M, Guerrero-Preston R, Sidransky D et al. PAX5 methylation detection by droplet digital PCR for ultra-sensitive deep surgical margins analysis of head and neck squamous cell carcinoma. Cancer Prev Res (Phila) 2015; 8 (11): 1017–1026. doi: 10.1158/1940-6207.CAPR-15-0180.
55. Menschikowski M, Jandeck C, Friedemann M et al. Identification of rare levels of methylated tumor DNA fragments using an optimized bias based pre-amplification-digital droplet PCR (OBBPA-ddPCR). Oncotarget 2018; 9 (90): 36137–36150. doi: 10.18632/oncotarget.26315.
56. Chibon F. Cancer gene expression signatures – the rise and fall? Eur J Cancer Oxf Engl (1990) 2013; 49 (8): 2000–2009. doi: 10.1016/j.ejca.2013.02.021.
57. Rapin N, Bagger FO, Jendholm J et al. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood 2014; 123 (6): 894–904. doi: 10.1182/blood-2013-02-485771.
58. Yan M, Schwaederle M, Arguello D et al. HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev 2015; 34 (1): 157–164. doi: 10.1007/s10555-015-9552-6.
59. Wang Q, Jia P, Li F et al. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med 2013; 5 (10): 91. doi: 10.1186/gm495.
60. Milbury CA, Zhong Q, Lin J et al. Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif 2014; 1 (1): 8–22. doi: 10.1016/j.bdq.2014.08.001.
61. Decraene C, Silveira AB, Bidard F-C et al. Multiple hotspot mutations scanning by single droplet digital PCR. Clin Chem 2018; 64 (2): 317–328. doi: 10.1373/clinchem.2017.272518.
62. Denis JA, Patroni A, Guillerm E et al. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol Oncol 2016; 10 (8): 1221–1231. doi: 10.1016/j.molonc.2016.05.009.
63. Burjanivova T, Malicherova B, Grendar M et al. Detection of BRAFV600E mutation in melanoma patients by digital PCR of circulating DNA. Genet Test Mol Biomark 2019; 23 (4): 241–245. doi: 10.1089/gtmb.2018. 0193.
64. Murray NP. Biomarkers detecting minimal residual disease in solid tumors: what do they mean in the clinical management of patients? Biomark Med 2019; 13 (18): 1535–1538. doi: 10.2217/bmm-2019-0401.
65. Drandi D, Kubiczkova-Besse L, Ferrero S et al. Minimal residual disease detection by droplet digital PCR in multiple myeloma, mantle cell lymphoma, and follicular lymphoma: a comparison with real-time PCR. J Mol Diagn JMD 2015; 17 (6): 652–660. doi: 10.1016/j.jmoldx.2015.05.007.
66. Dudová S, Hájek R. Využití metody real-time PCR (kvantitativní PCR, PCR v reálném čase) v hematologii a studiu mnohočetného myelomu. Klin Onkol 2018; 21 (Suppl 1): 220–222.
67. Chin R-I, Chen K, Usmani A et al. Detection of solid tumor molecular residual disease (MRD) using circulating tumor DNA (ctDNA). Mol Diagn Ther 2019; 23 (3): 311–331. doi: 10.1007/s40291-019-00390-5.
68. Taniguchi K, Uchida J, Nishino K et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res Off J Am Assoc Cancer Res 2011; 17 (24): 7808–7815. doi: 10.1158/1078-0432.CCR-11-1712.
69. Taly V, Pekin D, Benhaim L et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem 2013; 59 (12): 1722–1731. doi: 10.1373/clinchem.2013.206359.
70. Garcia-Murillas I, Schiavon G, Weigelt B et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015; 7 (302): 302ra133. doi: 10.1126/scitranslmed.aab0021.
71. Sausen M, Phallen J, Adleff V et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 2015; 6 : 7686. doi: 10.1038/ncomms8686.
72. Birkenkamp-Demtröder K, Nordentoft I, Christensen E et al. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur Urol 2016; 70 (1): 75–82. doi: 10.1016/j.eururo.2016.01.007.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2021 Issue 1-
All articles in this issue
- Patnáct let Národního onkologického programu České republiky a jeho vliv na výsledky u solidních nádorů dospělých v přehledu a číslech
- Breast cancer rehabilitation
- The development and significance of microRNA sequence variants in carcinogenesis
- Germline mutations in RAD51C and RAD51D and hereditary predisposition to ovarian cancer
- Patient delay in presenting symptoms of breast cancer in women in the Czech Republic
- Pembrolizumab as a first-line therapy in spinocellular esophageal cancer
- Squamous cell rectal cancer in a young woman
- Failure of adjuvant treatment for malignant melanoma – what next?
- Aktuality z odborného tisku
- Král Z, Adam Z et al. Histiocytární neoplazie a další vybrané velmi vzácné krevní nemoci.
- Zemřela Mgr. Jana Drexlerová
- Droplet digital PCR as a novel diagnostic tool
- Pathological response and clinical outcomes in operable triple-negative breast cancer with cisplatin added to standard neoadjuvant chemotherapy
- Post-treatment persistent alpha-fetoprotein elevation in a patient with testicular cancer
- Four years of low dose enzalutamide for metastatic castration-resistant prostate cancer
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Breast cancer rehabilitation
- Germline mutations in RAD51C and RAD51D and hereditary predisposition to ovarian cancer
- Droplet digital PCR as a novel diagnostic tool
- Squamous cell rectal cancer in a young woman
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

