-
Medical journals
- Career
Association of TNF-α -308G>A Polymorphism with Susceptibility to Cervical Cancer and Breast Cancer – a Systematic Review and Meta-analysis
Authors: Meraj Farbod 1; Mojgan Zarchi Karimi 2,3; Naeimeh Heiranizadeh 4; Neda Shalamzari Seifi 5; Javad Bafghi Mohammad Akbarian 6; Hossein Mohammad Jarahzadeh 7; Hossein Neamatzadeh 8,9
Authors‘ workplace: Cancer Institute, Imam Khomeini Hospital, Tehran University of Medical Sciences, Tehran, Iran 1; Abortion Research Center, Yazd Reproductive Sciences Institute, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 2; Department of Obstetrics and Gynecology, Iran University of Medical Sciences, Tehran, Iran 3; Department of Surgery, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 4; Department of Emergency Medicine, Shahrekord University of Medical Sciences, Shahrekord, Iran 5; Department of Healthcare Management, Bam University of Medical Sciences, Bam, Iran 6; Department of Anesthesiology and Critical Care, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 7; Department of Medical Genetics, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 8; Mother and Newborn Health Research Center, Shahid Sadoughi University of Medical Sciences, Yazd, Iran 9
Published in: Klin Onkol 2019; 32(3): 170-180
Category: Review
doi: https://doi.org/10.14735/amko2019170Overview
Background: To date, several studies have been carried out on the association of TNF-α -308G>A with the risk of cervical cancer (CC) and breast cancer (BC). However, their conclusions were not consistent. Thus, we performed a comprehensive meta-analysis to evaluate the association more precisely from all eligible case-control studies.
Methods: We searched the PubMed, Google Scholar and Cochrane Library databases systematically to identify relevant studies up to 1 February 2019. The pooled odds ratio (OR) with 95% confidence interval (CI) was calculated using a fixed-or random-effects model.
Results: A total of 40 case-control studies including 20 studies with 4,780 cases and 4,620 controls on CC and 20 studies with 12,390 cases and 14,910 controls on BC were selected in this meta-analysis. The pooled results showed that the TNF-α -308G>A polymorphism was significantly associated with an increased risk of CC (A vs. G: OR 1.277; 95% CI 1.104–1.477; P = 0.001; AA vs. GG: OR 1.333; 95% CI 1.062–1.674; P = 0.013; AG vs. GG: OR 1.307; 95% CI 1.064–1.605; P = 0.011; and AA + AG vs. GG: OR 1.324; 95% CI 1.104–1.587; P = 0.002) and BC (AA vs. AG + GG: OR 0.094; 95% CI 0.058–0.152; P ≤ 0.001). In the stratified analyses by ethnicity, the TNF-α -308G>A polymorphism was significantly associated with the risk of CC (in Caucasians and Africans) and BC (Caucasians and Asians).
Conclusion: Our findings showed that TNF-α -308G>A polymorphism may be a risk factor for cervical cancer and breast cancer overall and by ethnicity.
Keywords:
breast cancer – cervical cancer – TNF-α gene – polymorphism – meta-analysis
Introduction
There has been a progressive increase in the incidence and mortality of gynaecological cancers and breast cancer (BC). BC and cervical cancer (CC) are the first and second most common malignancies, respectively, in women [1–3]. Moreover, BC and CC rank as the first (15.0%) and fourth (6.6%) leading causes, respectively, of female cancer deaths worldwide [4,5]. In 2017, 255,180 new BC cases and 41,070 BC deaths were estimated to have occurred in the United States [4,6]. In the same year, 12,820 new CC cases were diag-nosed and 4,210 CC deaths were estimated in the US. BC and gynaecological cancers are known to be of multifactorial aetiology. Nulliparity, childbearing age, HPV infection, environment and lifestyle are the most well-established risk factors for BC and gynaecological cancers [7].
Epidemiological and clinical data show that the development of cancer is a multifactorial process [8]. Genetic risks of gynaecological cancers and BC have attracted increasing concern in research on the gene variations involved in immune systems and inflammatory pathways [9]. However, the exact mechanism of BC and/or CC is still largely unexplored. In past decades, apart from genetic variations that have raised major concerns in cancer biology, the role of inflammation factors such as tumour necrosis factor-alpha (TNF-α) has been widely researched in the aetiology of cancer [10,11]. The human TNF-α gene located on chromosome 6p21.3 contains 8 exons and encodes 382 amino acid proteins, in which -308G>A and -238 G>A are the most common polymorphisms [12,13]. TNF-α is a potential pro-inflammatory cytokine, which plays a critical role in a wide range of inflammatory, autoimmune and malignant diseases. To date, more and more studies indicate that TNF-α polymorphisms, especially the -308G>A polymorphism, are associated with gynaecological cancers and BC.
To date, several case-control studies have been performed to investigate the association of TNF-α -308G>A polymorphism with the risk of CC and BC. However, their results are still inconclusive and controversial. These different results may be due to differences in ethnic background, sample size and lifestyle and other factors. Thus, a meta-analysis with a large sample size should be performed to clarify the role of TNF-α -308G>A polymorphism in CC and BC. Therefore, we performed this meta-analysis on all the eligible case-control studies to make a more accurate assessment of the associations.
Materials and methods
Search strategy
We searched the PubMed, Google Scholar, EBSCO, EMBASE, Web of Science, Islamic World Science Citation Center (ISC), Scientific Information Database (SID), Wanfang, Ovid, Weipu and China National Knowledge Infrastructure (CNKI) databases for all articles on the association of the NF-α-308G>A polymorphism in CC and BC risk up to 1 February 2019. We used the following key words and terms for the research: (’Breast cancer’ or ’Breast Carcinoma’) and (’Cervical Cancer’ or ’Cervical Carcinoma’ or ’Cervical tumour’ or ’Uterine Cervix Cancer’) and (’Tumour Necrosis Factor’ or ’TNF-α’ or ’Cachexin’ or ’Cachectin’) and (’TNF -308G>A’ OR ’rs1800629’ or ’c.-488G>A’). In addition, the reference lists of all eligible studies, reviews and previous meta-analyses were also manually screened for additional or missing potential studies. The search was performed without language limitations.
Eligibility criteria
The inclusion criteria of studies in our meta-analysis were as follows: 1) studies with case-control or cohort design; 2) studies that evaluated the association of the TNF-α -308G>A polymorphism with CC and BC risk; and 3) studies with sufficient data to estimate the odds ratio (OR) with 95% confidence interval (CI). Accordingly, the major exclusion criteria were: 1) studies on male BC; 2) case-only studies or no control group was included; 3) family-based and or linkage studies; 4) studies lacking sufficient published data; 5) case reports, reviews, abstracts, letters to editors and posters; 6) duplicates of previous studies.
Data extraction
Two reviewers extracted the data independently and carefully from the eligible studies using a standard protocol in accordance with inclusion criteria. Any disagreement between the included studies and the data was resolved by discussion among the authors and if a conflicting evaluation still existed another author was consulted to resolve the dispute. For each of the eligible case-control studies, we have collected the following data: first authors, year of publication, country, ethnicity (Caucasian, Asian, African and Mixed), source of healthy controls (hospital-based studies and population-based studies), number of cases and controls, the numbers of cases and controls for each genotype, Hardy-Weinberg equilibrium (HWE) in controls and minor allele frequency (MAF).
Statistical analyses
Ethical approval was not required for this study, as it is a systematic review and meta-analysis. The OR and corresponding 95% CI were evaluated to estimate the association of TNF-α -308G>A polymorphism with the risk of CC and BC. The Ztest was used to assess the significance of the pooled OR, in which P < 0.05 was considered as statistically significant. In this meta-analysis, the pooled ORs for TNF-α -308G>A polymorphism were performed under all five genetic models, i.e., allele (A vs. G), homozygote (AA vs. GG), heterozygote (AG vs. GG), dominant (AA + AG vs. GG) and recessive (AA vs. AG + GG), respectively. Between-study heterogeneity was analysed by a chi-squared-based Q-statistic test, in which the P-value < 0.05 was considered significant. In addition, we used the Higgins (I2) test to assess the degree of between-study heterogeneity, in which the I2 values of 25%, 50% and 75% were nominally considered low, moderate and high estimates, respectively. Accordingly, the pooled ORs were calculated using a fixed-effects model (Mantel-Haenszel method) (if P > 0.05 or I2 < 50%); otherwise, a random-effects model (DerSimonian-Laird method) was chosen (if P < 0.05 or I2 > 50%) based on the level of heterogeneity. For each study, the departure of the TNF-α -308G>A polymorphism frequencies in control groups from the HWE was tested using the goodness-of-fit test (i. chi-square test), and deviation was considered when P < 0.05. We performed subgroup analysis according to ethnicity, source of controls (population-based or hospital-based), genotyping methods and HWE. The stability and reliability of the results were evaluated using a sensitivity analysis, in which one study was deleted each time and the analyses were repeated. In addition, a sensitivity analysis was performed by excluding those studies departing from the HWE. Publication bias was tested with the funnel plot and Egger’s linear regression asymmetry test; P < 0.05 suggested statistically significant publication bias. All analyses were performed with the Comprehensive Meta-Analysis (CMA) 2.0 software (Biostat, USA). Two-sided P-values < 0.05 were considered statistically significant.
Results
The selection process of eligible studies is shown in Schema 1. According to our search strategy, 147 articles were screened initially. From these, we excluded 107 articles because these articles did not provide detailed data, were reviews, case reports and previous meta-analyses, and/or had overlapped data. Finally, a total of 40 case-control studies were included in this meta-analysis. The characteristics of the included studies are shown in Tab. 1. Among the 40 studies, there were 20 studies with 4,780 cases and 4,620 controls on CC [14–33] and 20 studies with 12,390 cases and 14,910 controls on BC [34–51]. All of the selected papers were written in English or Chinese. The selected studies included 12 groups of Asians (8 on CC and 4 on BC), 23 groups of Caucasians (9 on CC and 14 on BC) and 4 groups of Africans (3 on CC and one on BC). The TNF-α -308G>A polymorphism frequency in each study, the results of the HWE test in control groups and MAFs are shown in Tab. 1. The distribution of genotypes in all studies was consistent with HWE except for six studies on CC and two studies for BC (Tab. 1).
Schema 1. Flow diagram of selecting eligible studies for the meta-analysis. 
CC – cervical cancer, BC – breast cancer, SNP – single nucleotide polymorphism 1. Characteristics of the case-control studies included in the meta-analyses. 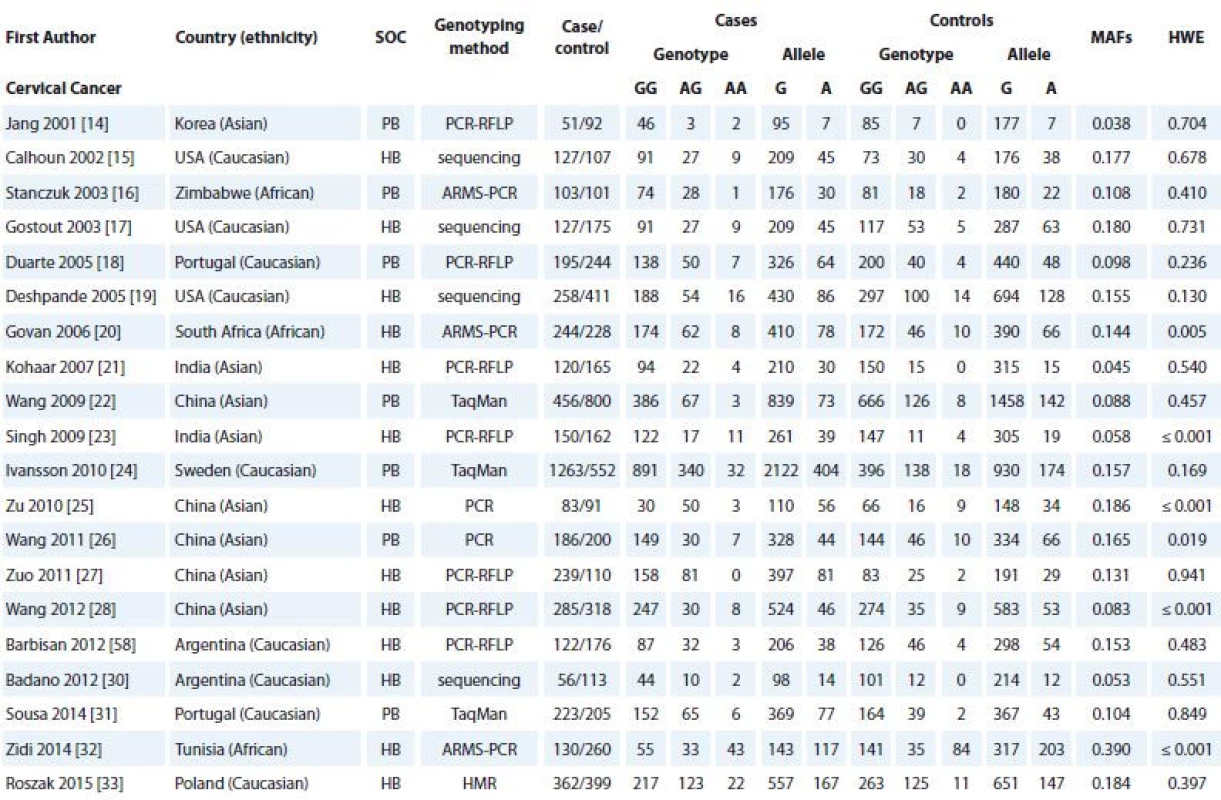
SOC – source of controls, HB – hospital based, PB – population based, PCR – polymerase chain reaction, RFLP – restriction fragment length polymorphism, ARMS – amplifi cation-refractory mutation system, SNP – single nucleotide polymorphism, HMR – high resolution melting, MAF – minor allele frequency, HWE – Hardy-Weinberg equilibrium 
SOC – source of controls, HB – hospital based, PB – population based, PCR – polymerase chain reaction, RFLP – restriction fragment length polymorphism, ARMS – amplifi cation- refractory mutation system, SNP – single nucleotide polymorphism, MAF – minor allele frequency, HWE – Hardy-Weinberg equilibrium Quantitative synthesis
Cervical cancer
Tab. 2 listed the main results of the meta-analysis of TNF-α -308G>A polymorphism and CC risk. The pooled data showed that there was a significant association between TNF-α -308G>A polymorphism and CC risk under four genetic models i.e., allele (A vs. G: OR 1.277; 95% CI 1.104–1.477; P = 0.001, Graph 1A), homozygote (AA vs. GG: OR 1.333; 95% CI 1.062–1.674; P = 0.013), heterozygote (AG vs. GG: OR 1.307; 95% CI 1.064–1.605; P = 0.011) and dominant (AA + AG vs. GG: OR 1.324; 95% CI 1.104–1.587; P = 0.002, Graph 1A). When stratified by ethnicity, there was a significant association between TNF-α -308G>A polymorphism and CC risk in Caucasians (allelic model A vs. G, OR 1.242; 95% CI 1.043–1.478; P = 0.015; homozygote model AA vs. GG, OR 1.586; 95% CI 1.147–2.193; P = 0.005; recessive model: AA vs. AG + GG, OR 1.569; 95% CI 1.137–2.165; P = 0.006) and Africans (heterozygote model AG vs. GG, OR 1.670; 95% CI 1.228–2.270; P = 0.001 and dominant model AA + AG vs. GG, OR 1.453; 95% CI 1.111–1.902; P = 0.006). However, there was no significant association in Asians.
1. Forest plot of TNF-α -308G>A polymorphism with cervical cancer and breast cancer. A. Cervical cancer (recessive model AA + AG vs. GG). B. Breast cancer (dominant model AA vs. AG + GG). CI – confidence interval 
2. Meta-analysis results of association between TNF-α -308G>A polymorphism and cervical cancer risk. 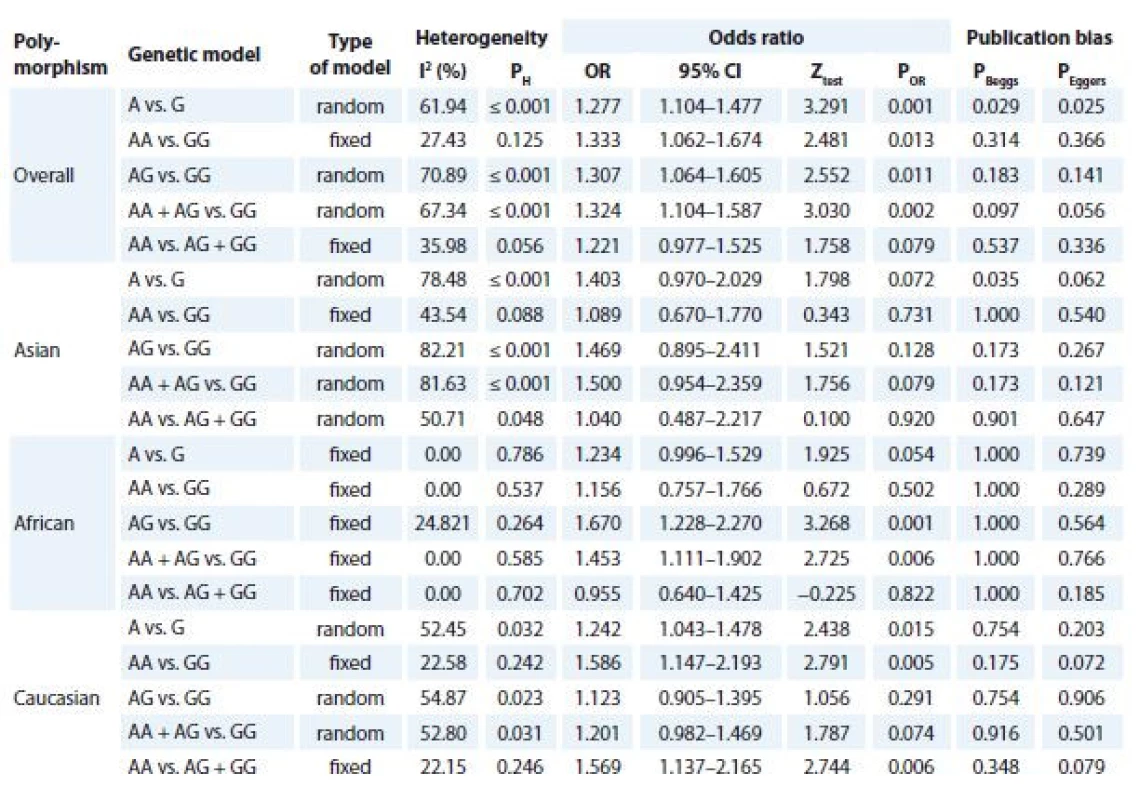
OR – odds ratio, CI – confi dence interval Breast cancer
Tab. 3 summarised the main results of the meta-analysis for TNF-α -308G>A polymorphism and BC. When all eligible studies were pooled together a significant association between TNF-α -308G>A polymorphism and BC risk was found under recessive model (AA vs. AG + GG: OR 0.094; 95% CI 0.058–0.152; P ≤ 0.001, Graph 1B). Similarly, the subgroup analysis results showed a significant association between TNF-α -308G>A polymorphism and increased risk of BC in Asians (AA vs. AG + GG: OR 0.076; 95% CI 0.045–0.128, P ≤ 0.001) and Caucasians (AA vs. AG + GG: OR 0.142; 95% CI 0.024–0.849; P = 0.032) under a recessive model. Furthermore, stratified analyses by source of controls showed that TNF-α -308G>A polymorphism was significantly associated with increased risk of population-based (PB) (AA vs. AG + GG: OR 0.091; 95% CI 0.053–0.155, P ≤ 0.001) and hospital-based (HB) (AA vs. AG + GG: OR 0.103; 95% CI 0.022–0.485; P = 0.004).
3. Meta-analysis results of association between TNF-α -308G>A polymorphism and breast cancer risk. 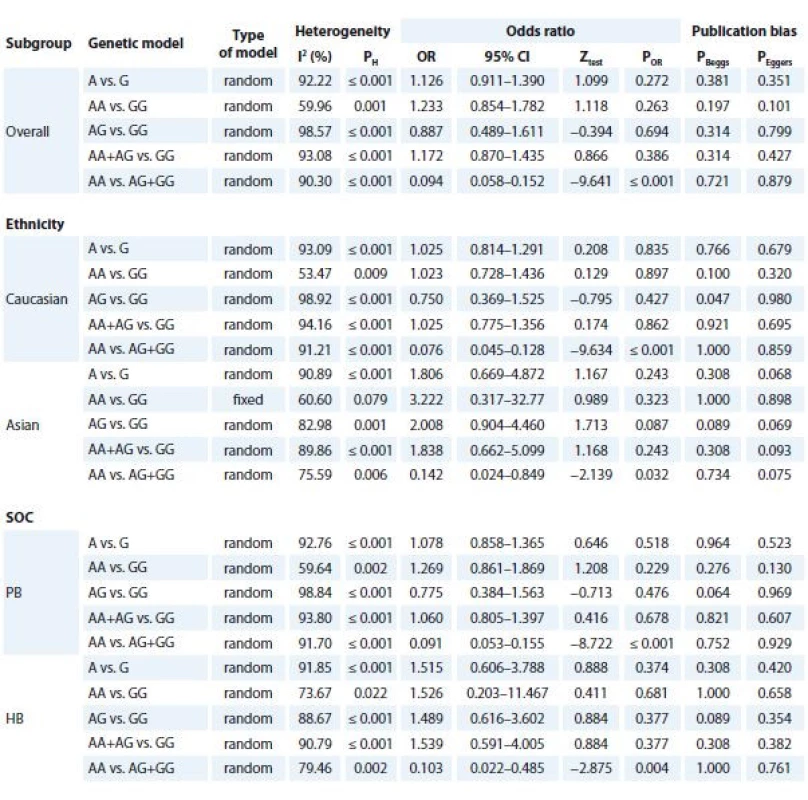
SOC – source of controls, HB – hospital based, PB – population based, OR – odds ratio, CI – confi dence interval 2. Funnel plot for publication bias in the meta-analysis of TNF-α -308G>A polymorphism and cervical cancer risk under the allele model (A vs. G). “Blue” without and “Grey” with trim and fill method. 
Sensitivity analyses and heterogeneity test
There was a significant heterogeneity for CC (under three genetic models) and BC (under five genetic models) in the overall analysis. Thus subgroup analysis was performed to explore the source of the heterogeneity. However, the result indicated that ethnicity, source of controls and publication year were not the main factors responsible for the heterogeneity in this meta-analysis. We performed a sensitivity analysis to assess the influence of the individual study to the pooled ORs by sequentially excluding individual studies. However, the sensitivity analysis showed that the initial results were not considerably adjusted by omitting any individual study (data not shown). In this meta-analysis, we included those HWE-violating studies. However, after those studies were excluded, the TNF-α -308G>A polymorphism association with CC and BC risk was not adjusted.
Publication bias
Begg’s funnel plot and Egger’s test were utilised to evaluate the publication bias of the literature. Neither Begg’s funnel nor Egger’s test showed publication bias for BC under all five genetic models. However, publication bias in the included studies for CC showed evidence of remarkable asymmetry under the allele model and supported by Egger’s test (PBegg’s = 0.029 and PEgger’s = 0.025). Thus, we used the Duval and Tweedie non-parametric ’trim and fill’ method in testing and adjusting the publication bias in meta-analysis. However, the results did not adjust, indicating that the results were statistically robust and reliable (Fig. 3).
Discussion
TNF-α plays a pivotal role in the regulation of immune cells. Genetic variations in the TNF-α gene are thought to modify DNA repair capacity and are suggested to be related to different cancer risks [52]. The human TNF-α gene, encoding an important protein in the regulation of immune cells, plays multiple roles in cell signalling in systemic inflammation, acute phase reaction and disease states [53,54]. In the last decade, epidemiological studies of gynaecological cancers and BC in different ethnicities have showed a significant association between the TNF-α -308G>A polymorphism and the risk of CC and BC [55,56]. However, subsequent replication studies on the association of TNF-α -308G>A polymorphism with CC and BC susceptibility was not consistent. Therefore, to derive a more precise estimation of the associations, we performed a systematic meta-analysis based on 40 case-control studies. The current meta-analysis, which included a total of 20 studies (with 4,780 cases and 4,620 controls) on CC and 20 studies (with 12,390 cases and 14,910 controls) on BC, investigated the association of TNF-α -308G>A polymorphism with susceptibility to CC and BC.
Our pooled data indicated that the AA genotype of TNF-α -308G>A polymorphism may be a risk factor for CC and BC in overall population and by ethnicity. Our results are inconsistent with the most previous meta-analysis on BC. In 2014, Jin et al in meta-analysis reported that the TNF-α -308G>A polymorphism was not associated with BC risk in the overall population. In addition, they have not found a significant association between TNF-α -308G>A polymorphism and BC by ethnicity, control source, genotyping method or HWE status. However, they have found an increased risk of BC in the menopausal status subgroup [56]. Similar to our results, Cai et al in a meta-analysis of 19 studies found that the TNF-α 308G>A polymorphism was significantly associated with CC risk [57]. Furthermore, in another meta-analysis Jin et al have found that -308 G>A and -238 G>A polymorphisms of TNF-α gene may confer susceptibility to CC in an ethnic-specific fashion [55]. However, their meta-analyses did not include all eligible and published studies. Thus, the current study is the most comprehensive meta-analysis on the association of TNF-α -308G>A polymorphism with the risk of CC.
This meta-analysis has several advantages. First, this meta-analysis has pooled the available data from the eligible studies, which has significantly increased the statistical power. In addition, we have concerned the pooled results regarding the source of healthy controls. Second, there was no language or ethnicity limitation in this meta-analysis, thus more original articles that met the criteria were included. However, several limitations of this meta-analysis should be acknowledged. First, we have included only the data of published studies in this meta-analysis. Unpublished studies tend to show more negative results; therefore publication bias may be present at first. Second, the number of studies included in the current meta-analysis for Asians was relatively small and might not have enough statistical power. Third, the numbers of studies as well as sample sizes for other ethnicities such as Africans and Latinos were limited, which might be caused the Type-II error in this meta-analysis. Therefore, data on other ethnicities must be evaluated to determine the potential effects of ethnic variation on CC and BC susceptibility. Fourth, there was high heterogeneity under most genetic models and the ethnicity, genotyping methods and source of controls were not the potential source of the heterogeneity. However, because of limited data, we could not explore other potential sources of heterogeneity such as age, nulliparity, childbearing age, HPV infection, environment, background and lifestyle in the current meta-analysis. Finally, the aetiology of BC and CC is complex and multifactorial; gene-gene or gene-environment interactions contribute to the risk of these malignancies. However, in this meta-analysis we have not addressed these interactions due to the lack of data.
In summary, the present meta-analysis results have indicated that the TNF-α -308G>A polymorphism may be associated with an increased risk of CC and BC in the overall population and in an ethnic-specific fashion. However, taking the limitations into consideration, further well-designed studies with larger sample sizes and more ethnic groups are warranted to verify our findings.
The authors would like to thank Sahel Khajehnoori from the Mother and Newborn Health Research Center for assisting in the revision of this paper.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Prof. Mojgan Karimi-Zarchi, M.D.
Abortion Research Center
Yazd Reproductive Sciences Institute
Shahid Sadoughi University
of Medical Sciences
Yazd, Iran
e-mail: mk.zarchi55@gmail.com
Submitted: 10. 2. 2019
Accepted: 5. 3. 2019
Sources
1. Gupta MK, Singh R, Banerjee M. Cytokine gene polymorphisms and their association with cervical cancer: a North Indian study. Egypt J Med Hum Genet 2016; 17 (2): 155–163. doi: 10.1016/J.EJMHG.2015.10. 005.
2. Karimi Zarchi M, Akhavan A, Gholami H et al. Evaluation of cervical cancer risk-factors in women referred to Yazd-Iran hospitals from 2002 to 2009. Asian Pac J Cancer Prev 2010; 11 (2): 537–538.
3. Behtash N, Karimi Zarchi M, Deldar M. Preoperative prognostic factors and effects of adjuvant therapy on outcomes of early stage cervical cancer in Iran. Asian Pac J Cancer Prev 2009; 10 (4): 613–618.
4. Yan Y, Zhang X. The association between CD28 gene rs3116496 polymorphism and breast cancer risk in Chinese women. Biosci Rep 2017; 37 (6): BSR20170884. doi: 10.1042/BSR20170884.
5. Mao JJ, Wu LX, Wang W et al. Nucleotide variation in ATG4A and susceptibility to cervical cancer in southwestern Chinese women. Oncol Lett 2018; 15 (3): 2992–3000. doi: 10.3892/ol.2017.7663.
6. Karimi Zarchi M, Behtash N, Sekhavat L et al. Effects of tamoxifen on the cervix and uterus in women with breast cancer: experience with Iranian patients and a literature review. Asian Pac J Cancer Prev 2009; 10 (4): 595–598.
7. Doosti M, Bakhshesh M, Zahir ST et al. Lack of evidence for a relationship between high risk human papillomaviruses and breast cancer in Iranian patients. Asian Pac J Cancer Prev 2016; 17 (9): 4357–4361.
8. Karimi-Zarchi M, Forat-Yazdi M, Vafaeenasab MR et al. Evaluation of the effect of GnRH agonist on menstrual reverse in breast cancer cases treated with cyclophosphamide. Eur J Gynaecol Oncol 2014; 35 (1): 59–61.
9. Yazdi MF, Rafieian S, Gholi-Nataj M et al. CYP2D6 genotype and risk of recurrence in tamoxifen treated breast cancer patients. Asian Pac J Cancer Prev 2015; 16 (15): 6783–6787. doi: 10.7314/APJCP.2015.16.15.6783.
10. Sheikhpour E, Noorbakhsh P, Foroughi E et al. A survey on the role of interleukin-10 in breast cancer: a narrative. Rep Biochem Mol Biol 2017; 7 (1): 30–37.
11. Sobhan MR, Mahdinezhad-Yazdi M, Aghili K et al. Association of TNF-α-308 G > A and -238G > A polymorphisms with knee osteoarthritis risk: a case-control study and meta-analysis. J Orthop 2018; 15 (3): 747–753. doi: 10.1016/J.JOR.2018.05.047.
12. Azarpira MR, Ghilian MM, Sobhan MR et al. Association of MTHFR and TNF-α genes polymorphisms with susceptibility to Legg-Calve-Perthes disease in Iranian children: a case-control study. J Orthop 2018; 15 (4): 984–987. doi: 10.1016/j.jor.2018.08.042.
13. Aslebahar F, Neamatzadeh H, Meibodi B et al. Association of tumor necrosis factor-α (TNF-α) -308G>A and -238G>A polymorphisms with recurrent pregnancy loss risk: a meta-analysis. Int J Fertil Steril 2019; 12 (4): 284–292. doi: 10.22074/ijfs.2019.5454.
14. Jang WH, Yang YI, Yea SS et al. The -238 tumor necrosis factor-alpha promoter polymorphism is associated with decreased susceptibility to cancers. Cancer Lett 2001; 166 (1): 41–46.
15. Calhoun ES, McGovern RM, Janney CA et al. Host genetic polymorphism analysis in cervical cancer. Clin Chem 2002; 48 (8): 1218–1224.
16. Stanczuk GA, Sibanda EN, Tswana SA et al. Polymorphism at the -308-promoter position of the tumor necrosis factor-alpha (TNF-αlpha) gene and cervical cancer. Int J Gynecol Cancer 13 (2): 148–153.
17. Gostout BS, Poland GA, Calhoun ES et al. TAP1, TAP2, and HLA-DR2 alleles are predictors of cervical cancer risk. Gynecol Oncol 2003; 88 (3): 326–332.
18. Duarte I, Santos A, Sousa H et al. G-308A TNF-α polymorphism is associated with an increased risk of invasive cervical cancer. Biochem Biophys Res Commun 2005; 334 (2): 588–592. doi: 10.1016/j.bbrc.2005.06.137.
19. Deshpande A, Nolan JP, White PS et al. TNF-α promoter polymorphisms and susceptibility to human papillomavirus 16–associated cervical cancer. J Infect Dis 2005; 191 (6): 969–976. doi: 10.1086/427826.
20. Govan VA, Constant D, Hoffman M et al. The allelic distribution of -308 tumor necrosis factor-alpha gene polymorphism in south African women with cervical cancer and control women. BMC Cancer 2006; 6 (1): 24. doi: 10.1186/1471-2407-6-24.
21. Kohaar I, Thakur N, Salhan S et al. TNFalpha-308G/A polymorphism as a risk factor for HPV associated cervical cancer in Indian population. Cell Oncol 2007; 29 (3): 249–256.
22. Wang SS, Bratti MC, Rodríguez AC et al. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. J Infect Dis 2009; 199 (1): 20–30. doi: 10.1086/595563.
23. Singh H, Jain M, Sachan R et al. Association of TNFA (-308G>A) and IL-10 (-819C>T) promoter polymorphisms with risk of cervical cancer. Int J Gynecol Cancer 2009; 19 (7): 1190–1194. doi: 10.1111/IGC.0b013e3181a3a3af.
24. Ivansson EL, Juko-Pecirep I, Gyllensten UB. Interaction of immunological genes on chromosome 2q33 and IFNG in susceptibility to cervical cancer. Gynecol Oncol 2010; 116 (3): 544–548. doi: 10.1016/j.ygyno.2009.10.084.
25. Zu F, Ai X, La L et al. A preliminary study of TNFa gene 308 single nucleotide polymorphism with cervical cancer risk and HPV subtype infection in southern Xinjiang Uygur patients. Chinese J Obstet Gynecol 2010; 45 (9): 709–711.
26. Wang Q, Zhang C, Walayat S et al. Association between cytokine gene polymorphisms and cervical cancer in a Chinese population. Eur J Obstet Gynecol Reprod Biol 2011; 158 (2): 330–333. doi: 10.1016/j.ejogrb.2011.05.019.
27. Zuo F, Liang W, Ouyang Y et al. Association of TNF-α gene promoter polymorphisms with susceptibility of cervical cancer in southwest China. Lab Med 2011; 42 (5): 287–290. doi: 10.1309/LM532DSPDUXIRJVN.
28. Wang N, Yin D, Zhang S et al. TNF-αlpha rs1800629 polymorphism is not associated with hpv infection or cervical cancer in the Chinese population. PLoS One 2012; 7 (9): e45246. doi: 10.1371/journal.pone.0045246.
29. Barbisan G, Pérez LO, Contreras A et al. TNF-α and IL-10 promoter polymorphisms, HPV infection, and cervical cancer risk. Tumor Biol 2012; 33 (5): 1549–1556. doi: 10.1007/s13277-012-0408-1.
30. Badano I, Stietz SM, Schurr TG et al. Analysis of TNFα promoter SNPs and the risk of cervical cancer in urban populations of Posadas (Misiones, Argentina). J Clin Virol 2012; 53 (1): 54–59. doi: 10.1016/j.jcv.2011.09.030.
31. Sousa H, Oliveira S, Santos AM et al. Tumour necrosis factor alpha 308 G/A is a risk marker for the progression from high-grade lesions to invasive cervical cancer. Tumor Biol 2014; 35 (3): 2561–2564. doi: 10.1007/s13277-013-1337-3.
32. Zidi S, Stayoussef M, Zouidi F et al. Tumor necrosis factor alpha (-238 / -308) and TNFRII-VNTR (-322) polymorphisms as genetic biomarkers of susceptibility to develop cervical cancer among Tunisians. Pathol Oncol Res 2015; 21 (2): 339–345. doi: 10.1007/s12253-014-9826-2.
33. Roszak A, Misztal M, Sowińska A et al. TNF-α -308 G/A as a risk marker of cervical cancer progression in the Polish population. Mol Diagn Ther 2015; 19 (1): 53–57. doi: 10.1007/s40291-015-0130-y.
34. Mestiri S, Bouaouina N, Ahmed SB et al. Genetic variation in the tumor necrosis factor-alpha promoter region and in the stress protein hsp70-2: susceptibility and prognostic implications in breast carcinoma. Cancer 2001; 91 (4): 672–678.
35. Park KS, Mok JW, Ko HE et al. Polymorphisms of tumour necrosis factors A and B in breast cancer. Eur J Immunogenet 2002; 29 (1): 7–10.
36. Giordani L, Bruzzi P, Lasalandra C et al. Association of breast cancer and polymorphisms of interleukin-10 and tumor necrosis factor-alpha genes. Clin Chem 2003; 49 (10): 1664–1667. doi: 10.1373/49.10.1664.
37. Azmy IA, Balasubramanian SP, Wilson AG et al. Role of tumour necrosis factor gene polymorphisms (-308 and -238) in breast cancer susceptibility and severity. Breast Cancer Res 2004; 6 (4): R395–R400. doi: 10.1186/bcr802.
38. Smith KC, Bateman AC, Fussell HM et al. Cytokine gene polymorphisms and breast cancer susceptibility and prognosis. Eur J Immunogenet 2004; 31 (4): 167–173. doi: 10.1111/j.1365-2370.2004.00462.x.
39. Kamali-Sarvestani E, Merat A, Talei AR. Polymorphism in the genes of alpha and beta tumor necrosis factors (TNF-α and TNF-β) and gamma interferon (IFN-γ) among Iranian women with breast cancer. Cancer Lett 2005; 223 (1): 113–119. doi: 10.1016/J.CANLET.2004.09.025.
40. Scola L, Vaglica M, Crivello A et al. Cytokine gene polymorphisms and breast cancer susceptibility. Ann N Y Acad Sci 2006; 1089 (1): 104–109. doi: 10.1196/annals.1386.017.
41. Gallicchio L, McSorley MA, Newschaffer CJ et al. Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Detect Prev 2007; 31 (2): 95–101. doi: 10.1016/j.cdp.2007.02.004.
42. Gaudet MM, Egan KM, Lissowska J et al. Genetic variation in tumor necrosis factor and lymphotoxin-alpha (TNF–LTA) and breast cancer risk. Hum Genet 2007; 121 (3–4): 483–490. doi: 10.1007/s00439-006-0315-x.
43. Sirotkovic-Skerlev M, Cacev T, Krizanac S et al. TNF alpha promoter polymorphisms analysis in benign and malignant breast lesions. Exp Mol Pathol 2007; 83 (1): 54–58. doi: 10.1016/j.yexmp.2006.11.004.
44. Gonullu G, Basturk B, Evrensel T et al. Association of breast cancer and cytokine gene polymorphism in Turkish women. Saudi Med J 2007; 28 (11): 1728–1733.
45. Ostashkin AS, Malivanova TF, Lurchenko VA et al. Tumor necrosis factor gene polymorphisms in breast cancer patients. Genetika 2008; 44 (9): 1275–1280.
46. Kohaar I, Tiwari P, Kumar R et al. Association of single nucleotide polymorphisms (SNPs) in TNF-LTA locus with breast cancer risk in Indian population. Breast Cancer Res Treat 2009; 114 (2): 347–355. doi: 10.1007/s10549-008-0006-5.
47. MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk. Polymorphisms in the BRCA1 and ABCB1 genes modulate menopausal hormone therapy associated breast cancer risk in postmenopausal women. Breast Cancer Res Treat 2010; 120 (3): 727–736. doi: 10.1007/s10549-009-0489-8.
48. Pooja S, Francis A, Bid HK et al. Role of ethnic variations in TNF-α and TNF-β polymorphisms and risk of breast cancer in India. Breast Cancer Res Treat 2011; 126 (3): 739–747. doi: 10.1007/s10549-010-1175-6.
49. Karakus N, Kara N, Ulusoy AN et al. Tumor necrosis factor alpha and beta and interferon gamma gene polymorphisms in Turkish breast cancer patients. DNA Cell Biol 2011; 30 (6): 371–377. doi: 10.1089/dna.2010. 1113.
50. Madeleine MM, Johnson LG, Malkki M et al. Genetic variation in proinflammatory cytokines IL6, IL6R, TNF-region, and TNFRSF1A and risk of breast cancer. Breast Cancer Res Treat 2011; 129 (3): 887–899. doi: 10.1007/s10549-011-1520-4.
51. Gómez Flores-Ramos L, Escoto-De Dios A, Puebla-Pérez AM et al. Association of the tumor necrosis factor-alpha -308G>A polymorphism with breast cancer in Mexican women. Genet Mol Res 2013; 12 (4): 5680–5693. doi: 10.4238/2013.November.18.17.
52. Aslebahar F, Neamatzadeh H, Meibodi B et al. Association of tumor necrosis Factor-α (TNF-α) -308G>A and -238G>A polymorphisms with recurrent pregnancy loss risk: a meta-analysis. Int J Fertil Steril 2019; 12 (4): 284–292. doi: 10.22074/ijfs.2019.5454.
53. Abdel Galil SM, Ezzeldin N, Fawzy F et al. The single-nucleotide polymorphism (SNP) of tumor necrosis factor α−308G/A gene is associated with early-onset primary knee osteoarthritis in an Egyptian female population. Clin Rheumatol 2017; 36 (11): 2525–2530. doi: 10.1007/s10067-017-3727-1.
54. Seifart C, Plagens A, Dempfle A et al. TNF-αlpha, TNF-beta, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers 2005; 21 (3): 157–165.
55. Jin Y. Association of single nucleotide polymorphisms in tumor necrosis factor-alpha with cervical cancer susceptibility. Cell Biochem Biophys 2015; 71 (1): 77–84. doi: 10.1007/s12013-014-0165-4.
56. Jin G, Zhao Y, Sun S et al. Association between the tumor necrosis factor alpha gene -308G> A polymorphism and the risk of breast cancer: a meta-analysis. Tumor Biol 2014; 35 (12): 12091–12098. doi: 10.1007/s13277-014-2510-z.
57. Cai J, Yang MY, Hou N et al. Association of tumor necrosis factor-α 308G/A polymorphism with urogenital cancer risk: a systematic review and meta-analysis. Genet Mol Res 2015; 14 (4): 16102–16112. doi: 10.4238/2015.December.7.22.
58. Barbisan G, Pérez LO, Contreras A et al. TNF-α and IL-10 promoter polymorphisms, HPV infection, and cervical cancer risk. Tumor Biol 2012; 33 (5): 1549–1556. doi: 10.1007/s13277-012-0408-1.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2019 Issue 3-
All articles in this issue
- MicroRNAs in Cerebrospinal Fluid as Biomarkers in Brain Tumor Patients
- Prognosis of HPV-Positive and -Negative Oropharyngeal Cancers Depends on the Treatment Modality
- Nanoparticle-Modified Apoferritin Nanotransfer for Targeted Cytostatic Transport
- Prevalence of Anxiety and Depression and Their Impact on the Quality of Life of Cancer Patients Treated with Palliative Antineoplasic Therapy – Results of the PALINT Trial
- Rhabdomyosarcoma of the Gluteus Maximus – Case Report, Review of Literature and Emerging Therapeutic Targets
- Neoadjuvant Hypertermic Isolated Limb Perfusion in Treatment of Undifferentiated Spindle Cell Sarcoma of Lower Limb with Achieved Complete Pathologic Response
- Primary Intracranial Sarcomas, Myxoid Meningeal Sarcoma – a Case Report and Literature Review
- The Loneliness of Patients in the Pre-Terminal and Terminal Stages of Cancer, the Social Dimension of Dying
- Current FIGO Staging for Carcinoma of the Cervix Uteri and Treatment of Particular Stages
- Association of TNF-α -308G>A Polymorphism with Susceptibility to Cervical Cancer and Breast Cancer – a Systematic Review and Meta-analysis
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Current FIGO Staging for Carcinoma of the Cervix Uteri and Treatment of Particular Stages
- Rhabdomyosarcoma of the Gluteus Maximus – Case Report, Review of Literature and Emerging Therapeutic Targets
- Prevalence of Anxiety and Depression and Their Impact on the Quality of Life of Cancer Patients Treated with Palliative Antineoplasic Therapy – Results of the PALINT Trial
- The Loneliness of Patients in the Pre-Terminal and Terminal Stages of Cancer, the Social Dimension of Dying
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

