-
Medical journals
- Career
Isocitrate Dehydrogenase Mutations are Better Prognostic Marker than O6-methylguanine-DNA Methyltransferase Promoter Methylation in Glioblastomas – a Retrospective, Single-centre Molecular Genetics Study of Gliomas
Authors: M. Houdova Megova 1; J. Drábek 1; Z. Dwight 2; R. Trojanec 1; V. Koudelakova 1; J. Vrbková 1; O. Kalita 3; S. Mlcochova 1; M. Rabcanova 1; M. Hajdúch 1
Authors‘ workplace: Institute of Molecular and Translational Medicine, Faculty of Medicine and Dentistry Palacky University and University Hospital in Olomouc, Czech republic 1; Department of Pathology, University of Utah, Salt Lake City, Utah, USA 2; Department of Neurooncology, Faculty of Medicine and Dentistry, Palacky University and University Hospital in Olomouc, Czech republic 3
Published in: Klin Onkol 2017; 30(5): 361-371
Category: Original Articles
doi: https://doi.org/10.14735/amko2017361Overview
Background:
Mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) are a promising prognostic biomarker of gliomas. The purpose of our study was to examine the clinical prognostic properties of IDH1/2 mutations in a glioma patient cohort from the Czech Republic using an improved platform for simple and reliable IDH genotyping.Material and Methods:
We retrospectively analyzed a group of 145 glioma patients by testing for the three most frequent IDH mutations, IDH1 R132H, IDH1 R132C, and IDH2 R172K, through the competitive amplification of differentially melting amplicons (CADMA) polymerase chain reaction (PCR). O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation, copy number of EGFR, p53, RB1, MDM2, CDKN2A genes, and deletions in 1p, 19q and 10p chromosomal regions were also analyzed and correlated with clinical characteristics.Results:
Of 145 gliomas, 36 harbored IDH1 R132H mutation and 1 IDH1 R132C mutation. We did not detect any IDH2 R172K mutation. IDH1 mutations were positively associated with MGMT methylation (OR 3.08, 95% CI 1.387–7.282; p = 0.007), 1p/19q co-loss (OR 8.85, 95% CI 2.367–42.786; p = 0.002) and negatively associated with epidermal growth factor receptor amplification (OR 0.12, 95% CI 0.019–0.437; p = 0.006) and 10p loss (OR 0.09, 95% CI 0.005–0.436; p = 0.019). The overall survival of IDH-mutant was 25 months, but only 9 months in IDH-wild type gliomas (p = 0.035); at the same time, survival associated with methylated vs. unmethylated MGMT promoter did not significantly differ (p = 0.166).Conclusion:
Despite IDH1 mutations being closely associated with MGMT methylation in glioma patients, IDH1 mutations in glioblastoma patients are stronger marker of overall survival than MGMT methylation and should be the marker of choice, especially when using genotyping by CADMA PCR.Key words:
isocitrate dehydrogenase – polymerase chain reaction – glioma – glioblastomaIntroduction
Gliomas are the most common brain tumors. They are malignant, highly infiltrating and diffuse [1], and the prognosis of these malignancies remains unfavorable despite aggressive therapy [2]. Mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) were first identified in glioblastomas [3] and lower grade glioma [4]. IDH1/2 mutations probably play significant roles in glioma-genesis [5] and have primarily been detected in heterozygous forms in glioma subtypes of astrocytomas grades 2 and 3, oligodendrogliomas, and secondary glioblastomas [6].
The most frequent IDH1 mutation, IDH1 c.395G>A p. (Arg132His) replaces the arginine with histidine in 90% of IDH1 mutated tumors. IDH2 mutated tumors frequently harbor analogous mutation p. (Arg172Lys). IDH1 and IDH2 mutations are mutually exclusive except for very rare cases [7]. They are associated with 1p/19q co-deletion, mutation in CIC, FUBP1, and TERT genes in oligo-dendroglial tumors [8], and p53 mutation/deletion, ATRX mutation/deletion, and MGMT promoter methylation in astrocytic tumors [9]. In addition, these mutations are inversely associated with EGFR amplification [10]. DNA methylation profiling shows a tight correlation between IDH1/2 mutations and the glioma CpG island methylator phenotype (G-CIMP), characterized by promoter DNA methylation alteration and the epigenetic instability in glioma cells, and favorable prognosis [11].
Mutated IDH1/2 enzymes have de novo aberrant enzymatic activity – the NADPH-dependent reduction of alpha-ketoglutarate to R (-) -2-hydroxyglutarate (2HG) [12], which leads to consumption of alpha-ketoglutarate and NADPH and can inhibit degradation of HIF-1α (hypoxia-inducible factor 1α) and increase of oxidative stress of the tumor cells [13]. Moreover, 2HG is an oncometabolite and may function as a competitive inhibitor of α-ketoglutarate dependent dioxygenases, such as histone demethylases [14]. IDH1/2 mutations are marker of improved prognosis in glioblastoma [15], lower grade glioma [16] and also (together with 1p/19q codeletion) in anaplastic oligodendroglioma and oligoastrocytoma. IDH1/2 mutations are associated with better response to temozolomide treatment [17]. Targeting of mutated IDH1/2 enzymes may be a potential treatment strategy [18]. Each of these findings was reflected and incorporated into new World Health Organization (WHO) 2016 classification of central nervous system tumors [19]. In revised WHO classification, major glioma subtypes are defined by histologic features as well as molecular profiles. All diffusely infiltrating gliomas are grouped together because of sharing the genetic driver mutations in IDH1/2 genes. The appearance of 1p/19q co-deletion together with IDH1/2 mutations is now necessary for diagnosis of oligodendrogliomas. ATRX and p53 mutations assessment helps in diagnosis of astrocytic tumors as well as EGFR amplification in glioblastomas. MGMT promoter methylation also remains an important prognostic marker for glioblastoma patients, treated by concomitant chemoradiotherapy [20]. Mainly, IDH1/2 mutations are screened through immunohistochemistry while Sanger direct sequencing is the most common genotyping method [3,4,16, 21,22]. More sensitive molecular genetics methods like pyrosequencing [23] and/or polymerase chain reaction (PCR) based methods [24–29] have also been tested to overcome limitation of Sanger sequencing that fails to detect mutations that are present in less than 20% of DNA molecules [30].
Here, we analyzed the most common IDH1/2 mutations by using Competitive Amplification of Differentially Melting Amplicons (CADMA PCR). We also investigated the association of IDH mutations with MGMT promoter methylation and other cytogenetic and molecular genetic markers and clinical characteristics of glioma patients of Neurooncology Department of the University Hospital in Olomouc.
Material and methods
Tumor patient samples
A total of 145 solid tumor tissue samples were obtained from patients who had undergone radical resection of glial tumor in the Faculty Hospital in Olomouc between 2005 and 2013. The experimental research presented in this manuscript was performed in compliance with the Helsinki Declaration according to the study ethics proposal approved by the Ethics Board of Palacky University in Olomouc. Written informed consent was obtained from all patients regarding the use of the collected samples in the research projects, including studies for the publication of this report or any accompanied images. The group of gliomas comprised 14 diffuse astrocytoma grade 2, 17 anaplastic astrocytoma grade 3, 3 oligodendroglioma grade 2, 2 anaplastic oligodendroglioma grade 3, 2 oligo-astrocytoma grade 2, 8 anaplastic oligoastrocytoma grade 3, 86 primary glioblastoma grade 4, and 13 secondary glioblastoma grade 4 tumors. All tumors were diagnosed and classified by two independent pathologists according to WHO classification criteria [31] for tumors of the central nervous system immediately after surgery. After reclassification according to new WHO criteria [19] using known IDH and 1p/19q status in 2016, the group consisted of 9 IDH mutant diffused astrocytomas, 5 IDH wild-type diffused astrocytomas, 2 IDH mutant 1p/19q codeleted oligodendrogliomas, 1 not otherwise specified (NOS) oligodendroglioma, 2 NOS oligoastrocytomas, 13 IDH mutant anaplastic astrocytomas, 6 IDH wild-type anaplastic astrocytomas, 4 IDH mutant 1p/19q codeleted anaplastic oligodendrogliomas, 4 NOS anaplastic oligoastrocytomas, 9 IDH mutant glioblastomas, 90 IDH wild-type glioblastomas. Males constituted 62% of the patients (90/145). The median age was 57 years (range 21–85 years), with a standard deviation of 15 years. The time of operation and time of last follow-up or death were recorded as clinical endpoints.
Radiotherapy, concomitant chemoradiotherapy, adjuvant therapy with temozolomide (TMZ) or another chemotherapy were used as oncologic therapy (Tab. 1). Other chemotherapy details are described as follows – in the group of IDH mutant 1p/19q codeleted oligodendrogliomas, one received postoperative radiotherapy (60 Gy, 2 Gy per fraction) and six cycles of PCV chemotherapy (procarbazine, CCNU – lomustine, vincristine). In the group of IDH mutant anaplastic astrocytomas, one received postoperative radiotherapy, six cycles of adjuvant TMZ and other chemotherapy with etoposide, carboplatin and nitrosourea and one received postoperative radiotherapy and adjuvant chemotherapy with nitroso-urea. In the group of IDH mutant, 1p/19q codeleted anaplastic oligodendrogliomas, one received postoperative radiotherapy, six cycles of adjuvant TMZ and another chemotherapy with etoposide and carboplatin. In the group of IDH mutant glioblastomas, two received postoperative radiotherapy, adjuvant TMZ and adjuvant chemotherapy with erlotinib or nitrosourea. In the group of four IDH wild-type glioblastomas who received concomitant chemoradiotherapy, two had another chemotherapy with nitrosourea, one with bevacizumab and one with carboplatin, etoposide and procarbazine.
1. Therapy stratification of glioma patients. 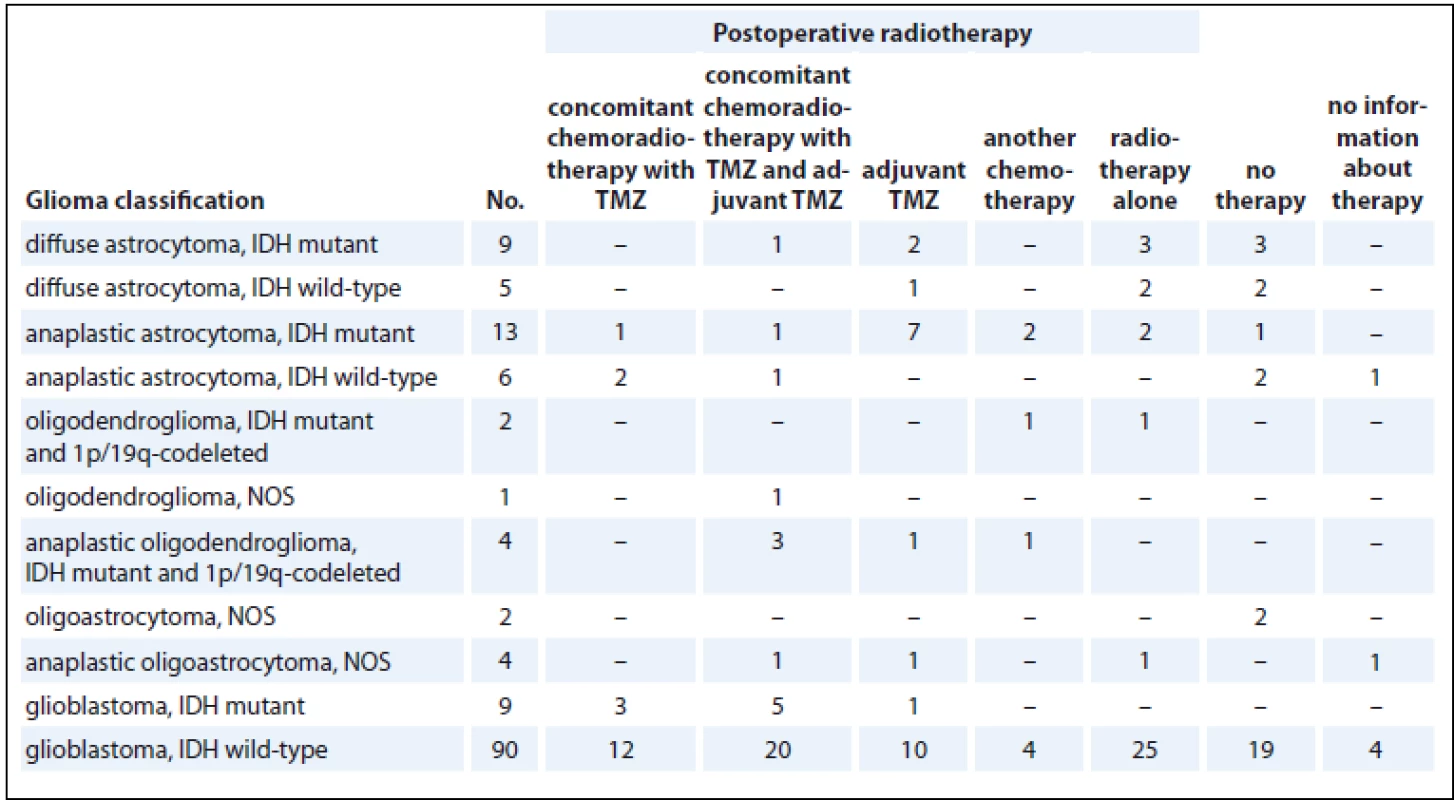
IDH – isocitrate dehydrogenase, TMZ – temozolomide, NOS – not otherwise specified Glial tissue sample in transporting medium (RPMI 1640 medium with L-glutamine, Penicillin/Streptomycin (100 U/mL), 15% fetal bovine serum, insulin (100 IU/mL), transferrin (2 mg/mL), and heparin (25,000 IU/mL)) was transported at room temperature to the laboratory within an hour after surgery. Subsequently, the sample was at once cut into smaller pieces using a scalpel, and the pieces were frozen without any medium at –80 °C until genomic DNA extraction.
The remaining glioma tissue sample was fixed in 10% neutral buffered formalin immediately after surgery and transported to the Department of Clinical and Molecular Pathology, University Hospital in Olomouc, for paraffin embedding. Formalin-fixed paraffin embedded (FFPE) sections of 4–6 µM in thickness were immobilized on positively charged glass slides and transported to Institute of Molecular and Translational Medicine for fluorescent in situ hybridization analysis. The number of FFPE tissue blocks preserved from 2005 to 2016 was inadequate for retrospective analysis.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) analysis was performed to detect the copy numbers of EGFR, p53, RB1, MDM2, and CDKN2A genes and deletions of 1p36.3, 19q.13, and 10p chromosomal regions by using fluorescent probes (IntellMed, Olomouc, Czech Republic) in the corresponding FFPE glioma tissue samples as previously described [32]. The EGFR gene was determined to be amplified if the EGFR/α satellite sequence at the centromere of chromosome 7 (CEP7) ratio was > 2. Other markers were determined as “gain” or “loss” when the copy number average was > 2.3 or < 1.9, resp., or when the gain or loss of the marker was observed in more than 20% of the tumor cells. Because of inconsistent thresholds in the literature [33,34], we previously validated the cut-off ratios for FISH analysis in a group of more than 400 gliomas retrospectively examined in cooperation with the neurooncology department. All FISH analyses were done immediately after acquiring FFPE samples.
DNA extraction
Genomic DNA extraction was performed using a cobas DNA Sample Preparation Kit (Roche Diagnostics Corporation, Basel, Switzerland) according to the manufacturer’s instructions. The concentrations of the DNA samples were spectrophotometrically measured using a NanoDrop ND 1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA) and stored at –80 °C. Archived DNA samples were used for this retrospective study.
Detection of IDH1/2 mutations by CADMA PCR
CADMA detection combines allele-specific PCR with intended mismatch and competition for templates between primers [35]. IDH1 R132H, R132C, and IDH2 R172K primers, which cover 87% of all possible IDH mutations in gliomas, were selectively designed for each mutation to distinguish the mutated and wild-type alleles after high resolution melting (HRM) analysis. The 10µl PCR reaction mixture contained 1× HotStart Taq Plus Master Mix (Qiagen, Hilden, Germany), 1× EVAgreen (Biotinum, Hayward, CA, USA), mutation-specific primers (0.4 nmol), overlapping wild-type primers (0.1 nmol), wild-type primers (0.4 nmol) (Generi-Biotech, Hradec Kralove, Czech Republic) and 4 ng of extracted genomic DNA. PCR protocol included an initial cycle at 95 °C for 15 min, 35 cycles at 95 °C for 10 sec, 60 °C for 20 sec, and 72 °C, followed by 95 °C for 10 sec, 60 °C for 45 sec, and a melting step with an increase to 95 °C at a rate of 0.06 °C/sec. PCR and HRM were performed on a LightCycler 480 System (Roche Diagnostics Corporation, Basel, Switzerland). IDH1 R132H, IDH1 wild-type, IDH2 R172K and IDH2 wild-type reference standards were purchased from Horizon Diagnostics (Horizon Diagnostics, Cambridge, UK), and the IDH1 R132C DNA sample was a gift from the CGB laboratory (CGB laboratory, AGEL company, the Czech Republic). All CADMA PCR analyses were retrospectively performed in 2016 from archived DNA samples.
Real-time MGMT methylation detection by using MethyLight PCR
Real-time methylation specific PCR [36] was performed after the bisulphite conversion of the template DNA according to EZ DNA methylation Gold kit instructions (Zymo Research, Irvine, USA) immediately after DNA extraction from glioma tissue samples. Concurrent detection of the methylation of E-cadherin and Alu-M5 were used as controls for DNA integrity. Commercial methylated and bisulphite-converted DNA (Zymo research, Irvine, CA, USA) was used as controls for bisulfide conversion and the PCR reaction. The 10µL PCR reaction mixture for MGMT promoter methylation detection contained 1× PCR buffer, 3 mM MgCl2, 0.2 mM dNTPs, 0.5 U ThermoTaq, forward primer – CGAATATACTAAAACAACCCGCG (20 µM), reverse primer – GTATTTTTT CGGGAGCGAGGC (20 µM), and probe – FAM-BHQ-CAAATCCTCGCGATACGCACC GTTTACG (4 µM). The 10µL PCR reaction mixture for E-cadherin promoter methylation detection contained 1× PCR buffer, 3 mM MgCl2, 0.2 mM dNTPs, 0.5 U Thermo Taq, forward primer – AATTTTAGGTTAGAGGGTTATCGCGT (20 µM), reverse primer – TCCCCAAAACG AAACTAACGAC (20 µM), and probe – FAM-BHQ-CGCCCACCCGACCTCGCAT (4 µM). The 10µl PCR reaction mixture for Alu-M5 promoter methylation detection contained 1× PCR buffer, 3 mM MgCl2, 0.2 mM dNTPs, 0.5 U ThermoTaq, forward primer – GGTATGATGGCGTATGTTTGT (0.3 µM), reverse primer – GACTCACCA CAACTTCCAC (0.3 µM), and probe – FAM-BHQ-AAACGATTCTCCTACCTCAA CCTCCCGAA (60 nM). The PCR protocol included an initial cycle at 95 °C for 15 min, followed by touchdown PCR for 10 cycles at 95 °C for 15 sec, 65 °C for 50 sec (–1 °C each cycle), and 72 °C for 20 sec, and with 30 PCR cycles at 95 °C for 15 sec, 56 °C for 50 sec, and 72 °C for 20 sec [37–39].
Statistics
Chi-square tests, Fisher tests and logistic regression analyses were used to examine associations among categorical variables referring to the absence or presence of genetic aberrations. The log-rank test was implemented in R statistical software to examine the relation-ship of categorical variables related to the absence or presence of genetic aberrations with overall survival (OS) within distinct tumor groups. OS, defined as the time between surgery and the last check-up or death, was calculated only for patients surviving at least 30 days after surgery.
Results
A total of 145 glioma tissue samples were analyzed for the presence of IDH1 R132H, R132C, and IDH2 R172K by using a new, rapid PCR method based on the CADMA principle. This method was performed within 2 hours, including the results analysis. In total, 36 IDH1 R132H mutations, 1 IDH1 R132C mutation, and 0 IDH2 R172K mutations were detected. The IDH1 gene was mutated (R132H or R132C mutation) in 9 diffuse astrocytomas, 13 anaplastic astrocytomas, 2 oligodendrogliomas, 4 anaplastic oligodendrogliomas and 9 glioblastomas. A single IDH1 R132C mutation was detected in diffuse astrocytoma. The IDH2 R172K mutation was not detected in our glioma samples. The percentage of alleles with the IDH1 R132H mutation was observed for each mutated sample, and the medians of the mutant allele percentage for each glioma subtype are shown in Tab. 2.
2. IDH1 R132H, R132C and IDH2 R172K in glioma samples. 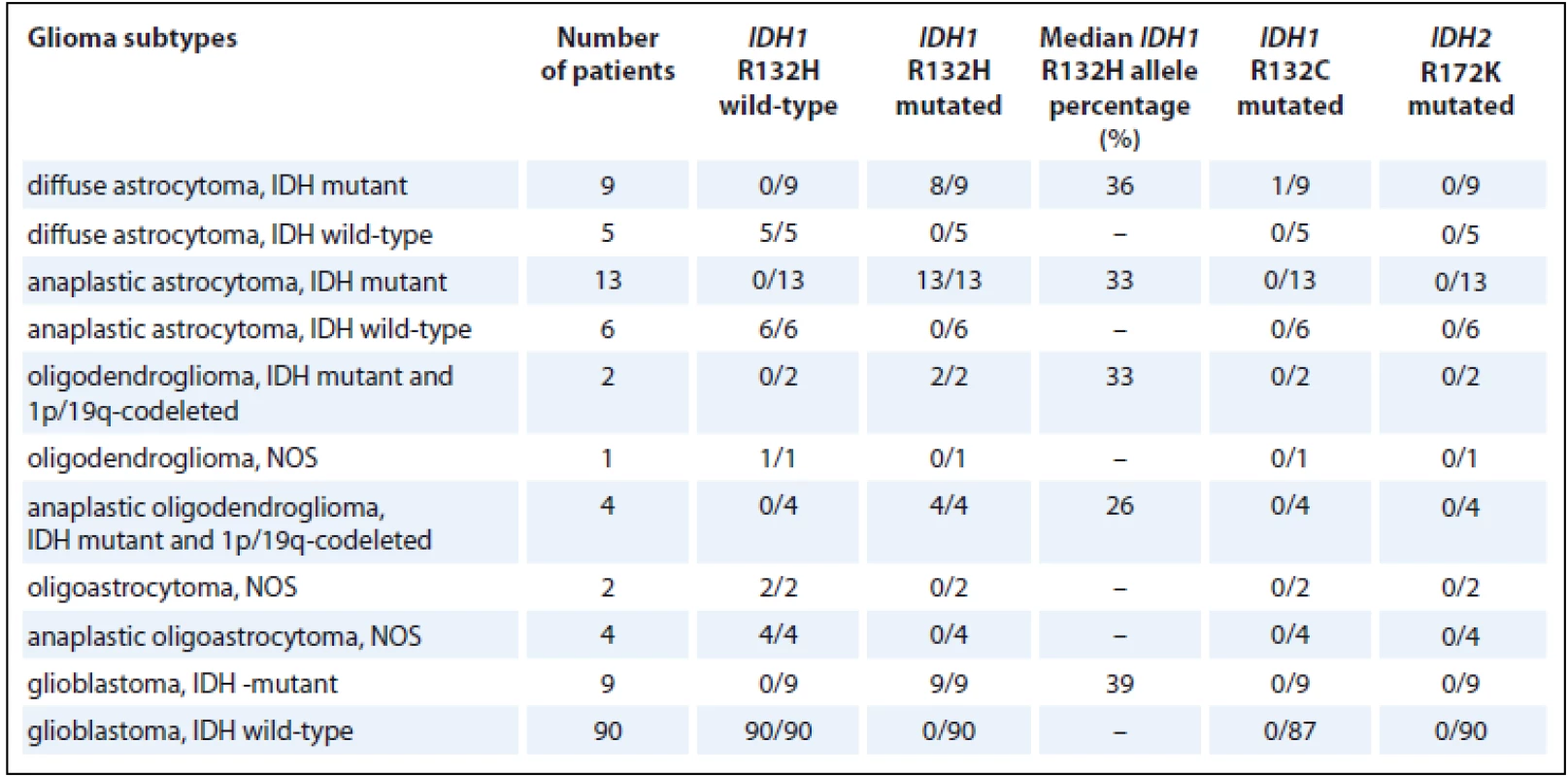
IDH1 – isocitrate dehydrogenase 1, IDH2 – isocitrate dehydrogenase 2, NOS – not otherwise specified In the group of gliomas, MGMT methylation was detected in 52% (75/143) of the samples, whereas EGFR amplification was detected in 27% (32/119) of the samples, CDKN2A loss in 31% (37/119) of the samples, 1p36.3 loss in 29% (35/121) of the samples, RB1 loss in 24% (29/119) of the samples, p53 loss in 15% (18/121) of the samples, 10p loss in 20% (23/115) of the samples, 19q13 loss in 17 % (20/120) of the samples, 1p/19q co-loss in 9% (11/119) of the samples, and MDM2 gain in 25% (29/117) of the samples (Tab. 3).
3. Molecular genetic characteristics of gliomas. 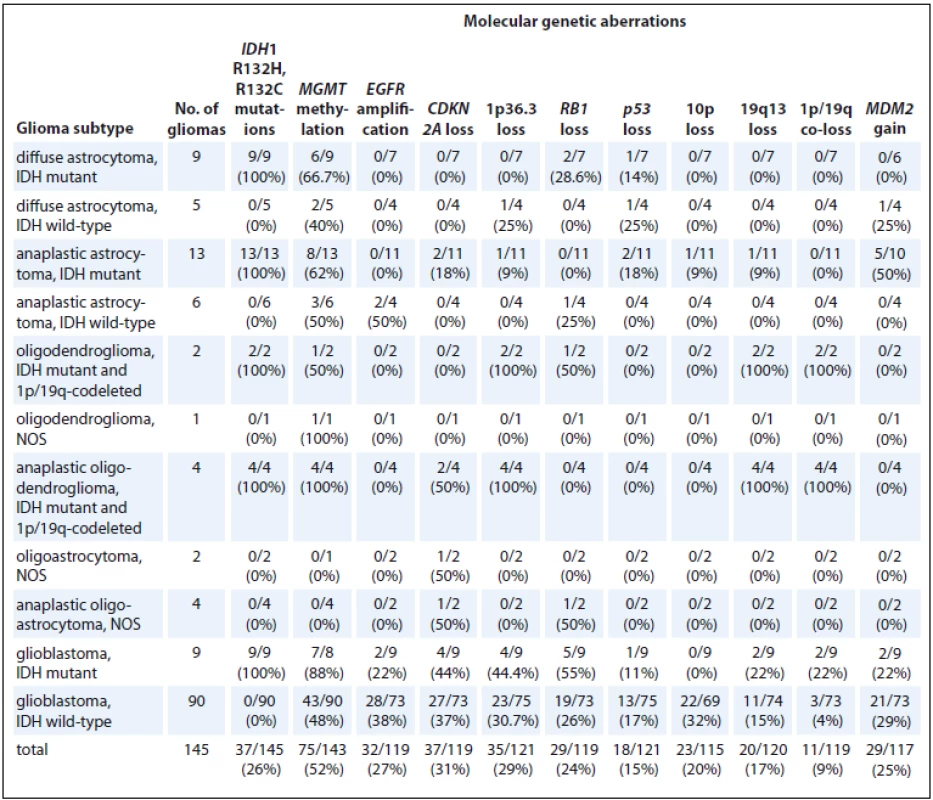
IDH1 – isocitrate dehydrogenase 1, MGMT – O6-methylguanine-DNA methyltransferase, EGFR – epidermal growth factor receptor, CDKN2A – cyclin-dependent kinase inhibitor 2A, MDM2 – mouse double minute 2 homolog, NOS – not otherwise specified IDH1 mutations were associated with MGMT methylation (p = 0.011) and 1p/19q co-loss (p = 0.009) and negatively associated with EGFR amplification (p = 0.003) and 10p loss (p = 0.009) (Tab. 4).
4. Association between IDH1 mutations and other molecular genetic aberrations. 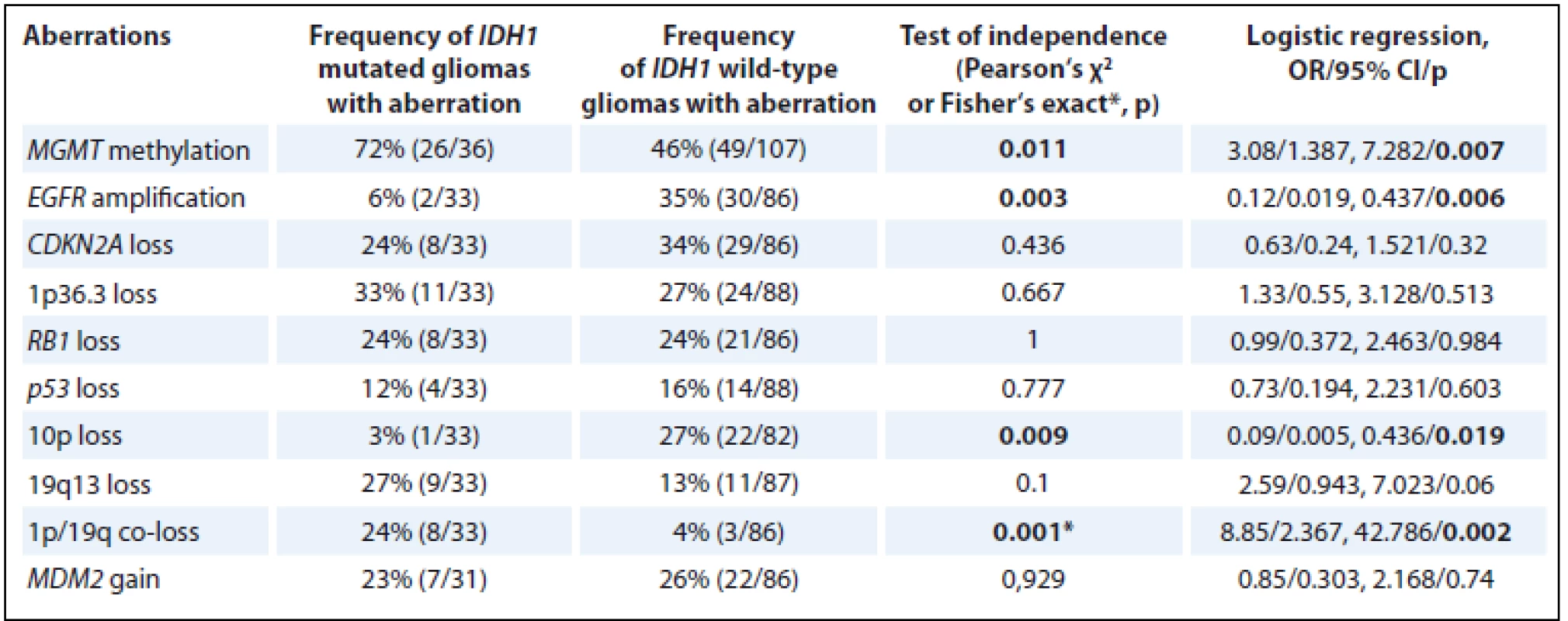
IDH1 – isocitrate dehydrogenase 1, MGMT – O6-methylguanine-DNA methyltransferase, EGFR – epidermal growth factor receptor, CDKN2A – cyclin-dependent kinase inhibitor 2A, MDM2 – mouse double minute 2 homolog Among the group of glioblastomas with OS more than 30 days (89 patients), an association between the OS and their IDH1 mutational status was observed. Glioblastoma patients with IDH1 mutations survived 25 months, while those without IDH1 mutations only survived 9 months (HR 0.4; 95% CI 0.2–0.96; p = 0.035). In contrast, the MGMT methylation status showed no significant association with the OS in this group of glioblastoma patients (HR 0.7; 95% CI 0.46–1.14; p = 0.166) (Graph 1). The characteristics of the patients in the glioblastoma group with respect to the IDH1 mutation status and MGMT methylation status are shown in Tab. 5.
Graph 1. OS of glioblastoma patients. 
The OS of patients with glioblastoma depending on IDH1 mutation status (A) andMGMT methylation status (B). 5. Characteristics of glioblastoma patients with respect to IDH1 mutation and MGMT methylation status. 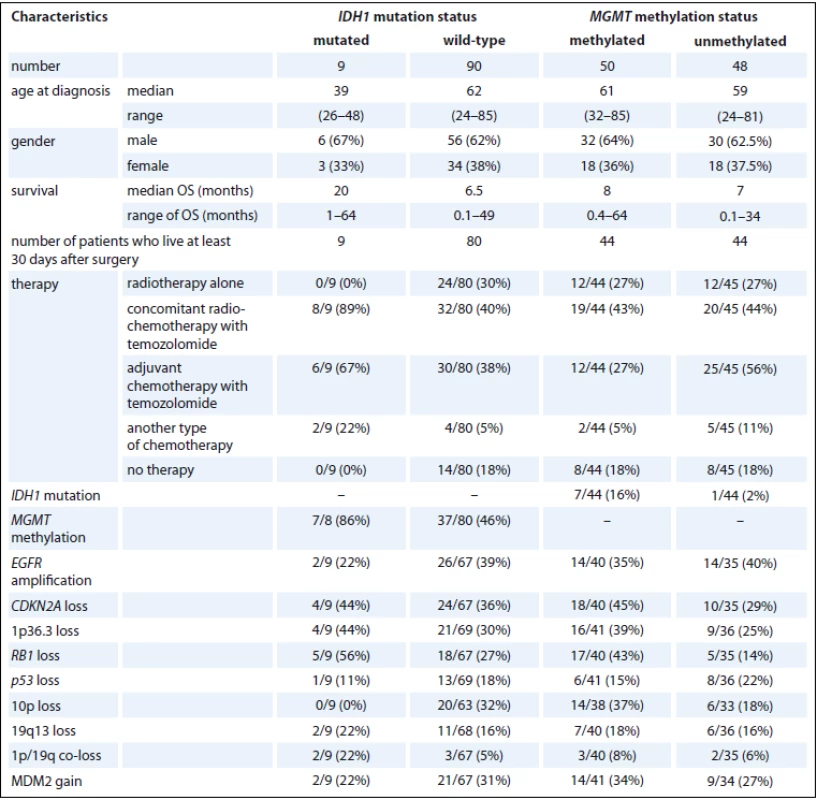
IDH1 – isocitrate dehydrogenase 1, MGMT – O6-methylguanine-DNA methyltransferase, EGFR – epidermal growth factor receptor, CDKN2A – cyclin-dependent kinase inhibitor 2A, MDM2 – mouse double minute 2 homolog, OS – overall survival Discussion
In this single-center study, we investigated the association of IDH mutations with molecular-genetic and clinical characteristics in a group of the Czech glioma patients. We used a CADMA PCR to examine IDH1 (R132H and R132C) and IDH2 (R172K) mutations in 145 glioma samples. We investigated whether IDH testing by CADMA is feasible from both the clinical and laboratory points of view.
One-step CADMA PCR as a 2-hour process including analysis, with sensitivity of 2.5% was more rapid and more sensitive than Sanger sequencing, which is a process lasting at least 5 hours with detection limit about 20% of mutant DNA on the wild-type background [40]. Immunohistochemistry, the most frequently used screening method, requires at least 2 days for analysis and in general practice provides results only for one IDH1 mutation.
Based on these findings, CADMA PCR can be successfully used for the simple and rapid detection of IDH mutations in clinical practice, in which the methodology of IDH detection is not standardized.
We detected 36 (97%) IDH1 R132H mutations and 1 (2.7%) IDH1 R132C mutation from the whole spectrum of detected mutations, concordantly with results from previously studies showing 88–100% and 2.6–7% mutations, resp. [7].
Despite expectations based on previously published data [28,41], we did not detect any IDH2 R172K mutations in the patient group. This finding is consistent with that obtained by the NOA-04 group trial [42], which included randomized WHO grade 3 anaplastic glioma patients. However, IDH mutations are considered primary in growing gliomas, thus suggesting that the differences between the glioma diagnoses in the present and NOA-04 group studies are irrelevant. We did not find any IDH1/2 mutations in a group of three pilocytic astrocytomas and eight ependymomas (data not shown), which were eliminated from analysis for their paediatric and not infiltrating glioma origin.
MGMT methylation was observed in 52% (75/143) of all patients. The frequency of MGMT methylation in individual groups of gliomas corresponded to previously published results [43]. We observed that 72% (26/36) of the IDH1 mutated patients also harbored MGMT methylation in comparison with 46% (49/107) of MGMT methylation in IDH1 wild-type patients. Consequently, among the group of glioma samples examined in the present study, IDH1 mutations were strongly positively associated with the presence of MGMT promoter methylation (OR 3.08, 95% CI 1.387–7.282; p = 0.007). In clinical studies, the association and frequency of MGMT methylation in IDH mutated patients appears to depend on the method of MGMT testing. In previous studies using the same methods as proposed in the present study, methylation-specific PCR and glioma patients with grade 2–4 have shown similar results [40]. In contrast, in studies using pyrosequencing in a group of grade 2–4 gliomas, nearly all IDH mutated patients have also been found to be MGMT methylated [9]. This may reflect higher sensitivity of MGMT methylation testing by pyrosequencing.
EGFR amplification was observed in 27% (32/119) of all glioma patients. EGFR is considered to be the most frequently amplified gene in primary glioblastomas and the EGFR amplification was found in 38% (28/73) of the IDH wild-type glioblastomas in our group, a result that correlated well with the literature findings for primary glioblastomas [45]. Among of the patients examined in the present study, EGFR amplification occurred in 6% (2/33) of IDH1 mutated gliomas in contrast to the 35% (30/86) of patients with IDH wild-type gliomas. Consequently, IDH1 mutations were significantly negatively correlated with EGFR amplification (OR 0.12; 95% CI 0.019–0.437; p = 0.006), as described also in a meta-analysis [46].
CDKN2A loss was observed in 31% (37/119) of all glioma patients, which is in correlation with previously published 37% [47]; 24% (8/33) of the IDH1 mutated gliomas also harbored CDKN2A loss, but there was no significant correlation between these biomarkers.
In the present study, we assessed the status of the chromosomal regions 1p36.3 and 19q13 and co-loss of both regions together, as described in previous studies [48]. 1p36.3 loss was observed in 29% (35/121) and 19q13 loss was observed in 17% (20/120) of patients. Losses in 1p and 19q regions were predominantly observed in oligodendroglial tumors [49,50], and there were only 15 patients with oligodendroglioma and oligoastrocytoma grade 2 and 3 in our group, which may have resulted in the lower observed frequency of the losses of 1p or 19q than reported in the literature [51]. 1p/19q co-loss was observed in 9% (11/119) of all gliomas and was closely positively associated with IDH1 mutations (OR 8.85, 95% CI 2.367–42.786; p = 0.002), similarly to the results from published studies [4,10].
RB1 loss was observed in 24% (29/119) of all gliomas, a value slightly higher than the previously published 2–12% loss [52,53]. Approximately 24% (8/33) of all gliomas harbored both an IDH1 mutation and RB1 loss, but there was no significant association between these aberrations.
P53 loss was observed in 15% (18/121) of all gliomas. The allelic loss of chromosome 17p was observed in approximately one-third of grade 2–4 adult astrocytomas [1] and in mutations of p53 gene with the same frequency [4]. P53 mutations were positively associated with IDH1 mutations in astrocytomas [54], but in the present study, this correlation was not confirmed.
The loss of chromosomal region 10p was observed in 20% (23/115) of all gliomas. Chromosomal region 10p11.1 is located near the centromere of chromosome 10 and was used as a marker of the loss of the entire chromosome 10. The loss of chromosome 10 is primarily observed in high-grade glial tumors. In the present study, frequency of chromosome 10 loss in the IDH wild-type glioblastoma was 32% (22/69), which was less than the 60% frequency reported in other studies [55]. However, there was a strong correlation between chromosome 10 loss and IDH1 mutations. Only 3% (1/33) of the IDH1 mutated gliomas also harbored chromosome 10 loss, whereas 27% (22/82) of the IDH1 wild-type gliomas occurred together with chromosome 10 loss (OR 0.09; 95% CI 0.005–0.436; p = 0.019). Sanson et al. suggested the same association between IDH1 mutations and chromosome 10 loss [44].
MDM2 gain was observed in 25% (29/117) of all gliomas and in 29% (21/73) of glioblastomas, IDH wild-type, in which the MDM2 gain was predominantly observed. The presence of MDM2 high amplification 8% (6/73) in IDH wild-type glioblastomas correlate with 6% in study of Houillier et al. [56]. There was no correlation observed between IDH1 mutations and the MDM2 gain in the gliomas examined in the present study.
We found a statistically significant correlation between the OS of glioma patients and the IDH1 mutational status in a group of glioblastomas with OS more than 30 days – IDH1 mutated glioblastoma patients had almost 3 times longer OS than did IDH1 wild-type patients (HR 0.4; CI 0.2–0.96; p = 0.035) whereas the prognostic relevance of the MGMT promoter methylation was not confirmed (HR 0.7; CI 0.46–1.14; p = 0.166). Polivka et al. showed the same results for the association of IDH mutations with patient outcome in another non-overlapping group of Czech glioblastoma patients [57]. Survival analysis of the selected glioblastomas with all radiotherapy and chemotherapy combinations (75 glioblastomas) showed the same trend that IDH1 mutations are better prognostic marker than MGMT methylation (IDH1 – HR 0.5; CI 0.21–1.06; p = 0.069, MGMT – HR 0.7; CI 0.43–1.18; p = 0.193) as well as in the group with concomitant chemo-radiotherapy (so-called Stupp regimen) (IDH1 – HR 0.6; CI 0.25–1.3; p = 0.183, MGMT – HR 0.8; CI 0.43–1.47; p = 0.457). However, in the multivariate cox regression analysis with age, IDH1 mutation and MGMT methylation as variables, only age was significant. This may be due to the small number of glioblastoma patients. There was no other relevant prognostic association between the screened glioma biomarkers. Neither of these markers showed significant correlations with the IDH mutation status (1p/19q co-loss, EGFR amplification and chromosome 10 loss).
MGMT promoter methylation is considered to be a favorable prognostic factor for survival that is independent of the therapy regime and is also considered to be a predictive factor for alkylating agent chemotherapy in patients with the wild-type IDH1 [42,58]. This suggests that MGMT belongs to the glioma CpG island methylator phenotype (G-CIMP) characterized by promoter DNA methylation alteration) gene set [59] and that there is a tight correlation between the G-CIMP phenotype and absence of IDH1 mutation [60]. On the basis of the G-CIMP study, Wiestler et al. have suggested that IDH1/2 and 1p/19q status is the most relevant for the determination of anaplastic glioma prognosis [61], whereas 1p/19q co-deletion is an indispensable prognostic and predictive marker for oligodendroglial and oligoastrocytic tumors [62]. In addition, Wick et al. found IDH1/2 mutation a convenient proxy for MGMT promoter methylation obviating need for MGMT methylation status testing in IDH mutated gliomas [42,63]. However, a study of 98 primary glioblastomas disagreed and confirmed using 2-log likelihood tests better survival predictions based on both genes than those based on either IDH1 mutations or MGMT methylation alone [64].
We agree with conclusions of Wick et al. also from laboratory point of view. Methylation-specific PCR after bisulphite conversion is the most frequently used method for the analysis of MGMT methylation [63]. If we consider the known complications of bisulphite reaction, i.e. chemical degradation and incomplete bisulphite conversion leading to compromised analytical parameters [60], particularly in degraded DNA from rare patient samples, it is reasonable to examine IDH mutation instead of MGMT promoter methylation in glioma patients. The limitations of our study reflect the design of retrospectively selected patients from only a single institution. The 145 patients were stratified by histopathological criteria into 11 groups with an unequal representation of patients. Only the most numerous group of glioblastomas was sufficient for follow-up clinical data analysis.
Conclusions
In conclusion, we retrospectively analyzed 145 glioma tissues for the presence of IDH1/2 mutations and compared the results with other molecular genetics markers, cytogenetic markers, and clinical characteristics. We confirm positive prognostic effect of IDH1 mutations in contrast with MGMT methylation in a group of 89 glioblastomas. The results suggested that IDH1/2 mutation analysis is a favorable procedure in genetic examination of glioblastoma samples in comparison to MGMT methylation analysis with respect to OS prognosis.
Moreover, we confirmed the known positive association of MGMT and 1p/19q codeletion and negative association of EGFR and chromosome 10 deletion with presence of IDH1 mutations in our glioma dataset. Although the prognostic or predictive role of CDKN2A, P53, RB1, and MDM2 is not clearly established in our set, these cytogenetic markers may advance diagnosis and classification of gliomas and may be appreciated with increasing knowledge in neuro-oncology.
This work was financially supported by grants from the Ministry of Health (NT 13581), Technology Agency (TE02000058) and the Ministry of Education, Youth and Sports (LM2015089) of the Czech Republic. The infrastructural part of this project (Institute of Molecular and Translational Medicine) was supported by a grant from the National Program of Sustainability (NPU LO1304).
Acknowledgements
The authors would like to thank the CGB laboratory for the providing of IDH1 R132C standard DNA and the Nature Publishing Group language editing for English editing services.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
doc. Mgr. Jiri Drabek, Ph.D.
Institute of Molecular and Translational Medicine Faculty of Medicine and Dentistry,
Palacky University and
University Hospital in Olomouc
Hnevotinska 5
779 00 Olomouc
Czech Republic
e-mail: jiri.drabek@upol.cz
Submitted: 3. 5. 2017
Accepted: 23. 7. 2017
On the web site you can find supplement of this article.
Sources
1. Furnari FB, Huang HJ, Cavenee WK. Genetics and malignant progression of human brain tumours. Cancer Surv 1995; 25 : 233–275.
2. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol 2005; 109 (1): 93–108. doi: 10.1007/s00401-005-0991-y.
3. Parsons D, Jones S, Zhang X et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321 (5897): 1807–1812. doi: 10.1126/science.1164382.
4. Yan H, Parsons DW, Jin G et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009; 360 (1533–4406): 765–773. doi: 10.1056/NEJMoa0808710.
5. Horbinski C. What do we know about IDH1/2 mutations so far, and how do we use it? Acta Neuropathol 2013; 125 (5): 621–636. doi: 10.1007/s00401-013-11 06-9.
6. Hartmann C, Meyer J, Balss J et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 2009; 118 (4): 469–474. doi: 10.1007/s00401-009-0561-9.
7. Megova M, Drabek J, Koudelakova V et al. Isocitrate dehydrogenase 1 and 2 mutations in gliomas. J Neurosci Res 2014; 92 (12): 1611–1620. doi: 10.1002/jnr.23456.
8. Bettegowda C, Agrawal N, Jiao Y et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011; 333 (1095–9203): 1453–1455. doi: 10.1126/science.1210557.
9. Mulholland S, Pearson D, Hamoudi R et al. MGMT CpG island is invariably methylated in adult astrocytic and oligodendroglial tumors with IDH1 or IDH2 mutations. Int J Cancer 2012; 131 (5): 1104–1113. doi: 10.1002/ijc.26499.
10. Ichimura K, Pearson D, Kocialkowski S et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol 2009; 11 (4): 341–347. doi: 10.1215/15228517-2009-025.
11. Duncan C, Barwick B, Jin G et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res 2012; 22 (12): 2339–2355. doi: 10.1101/gr.132738.111.
12. Dang L, White D, Gross S et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009; 462 (7274): 739–744. doi: 10.1038/nature08617.
13. Gilbert MR, Liu Y, Neltner J et al. Autophagy and oxidative stress in gliomas with IDH1 mutations. Acta Neuropathol 2014; 127 (2): 221–233. doi: 10.1007/s00401-013-1194-6.
14. Lu C, Ward P, Kapoor G et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012; 483 (7390): 474–478. doi: 10.1038/nature10860.
15. SongTao Q, Lei Y, Si G et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci 2012; 103 (2): 269–273. doi: 10.1111/j.1349-7006.2011.02134.x.
16. Metellus P, Coulibaly B, Colin C et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 2010; 120 (6): 719–729. doi: 10.1007/s00401-010-0777-8.
17. Gorlia T, Delattre JY, Brandes AA et al. New clinical, pathological and molecular prognostic models and calculators in patients with locally diagnosed anaplastic oligodendroglioma or oligoastrocytoma. A prognostic factor analysis of European Organisation for Research and Treatment of Cancer Brain Tumour Group Study 26951. Eur J Cancer 2013; 49 (16): 3477–3485. doi: 10.1016/j.ejca.2013.06.039.
18. Rohle D, Popovici-Muller J, Palaskas N et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013; 340 (6132): 626–630. doi: 10.1126/science.1236062.
19. Louis DN, Ohgaki H, Wiestler OD et al. WHO Classification of Tumours of the Central Nervous System. 4th ed. IARC 2016; 1 (1): 408.
20. Masui K, Mischel PS, Reifenberger G. Molecular classification of gliomas. Handb Clin Neurol 2016; 97–120. doi: 10.1016/B978-0-12-802997-8.00006-2.
21. Balss J, Meyer J, Mueller W et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol 2008; 116 (6): 597–602. doi: 10.1007/s00401-008-0455-2.
22. Horbinski C, Kofler J, Kelly L et al. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues. J Neuropathol Exp Neurol 2009; 68 (12): 1319–1325. doi: 10.1097/NEN.0b013e3181c391be.
23. Arita H, Narita Y, Matsushita Y et al. Development of a robust and sensitive pyrosequencing assay for the detection of IDH1/2 mutations in gliomas. Brain Tumor Pathol 2015; 32 (1): 22–30. doi: 10.1007/s10014-014-0186-0.
24. Watanabe T, Nobusawa S, Kleihues P et al. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 2009; 174 (4): 1149–1153. doi: 10.2353/ajpath.2009.080958.
25. Meyer J, Pusch S, Balss J et al. PCR-and restriction endonuclease-based detection of IDH1 mutations. Brain Pathol 2010; 20 (2): 298–300. doi: 10.1111/j.1750-3639.2009.00327.x.
26. Horbinski C, Kelly L, Nikiforov Y et al. Detection of IDH1 and IDH2 mutations by fluorescence melting curve analysis as a diagnostic tool for brain biopsies. J Mol Diagn 2010; 12 (4): 487–492. doi: 10.2353/jmoldx.2010.090 228.
27. Lv S, Teugels E, Sadones J et al. Correlation between IDH1 gene mutation status and survival of patients treated for recurrent glioma. Anticancer Res 2011; 31 (12): 4457–4463.
28. Perizzolo M, Winkfein B, Hui S et al. IDH mutation detection in formalin-fixed paraffin-embedded gliomas using multiplex PCR and single-base extension. Brain Pathol 2012; 22 (5): 619–624. doi: 10.1111/j.1750-3639.2012.00579.x.
29. Pang B, Durso MB, Hamilton RL et al. A novel COLD-PCR/FMCA assay enhances the detection of low-abundance IDH1 mutations in gliomas. Diagn Mol Pathol 2013; 22 (1): 28–34. doi: 10.1097/PDM.0b013e31826 c7ff8.
30. Jancik S, Drabek J, Berkovcova J et al. A comparison of Direct sequencing, Pyrosequencing, High resolution melting analysis, TheraScreen DxS, and the K-ras StripAssay for detecting KRAS mutations in non small cell lung carcinomas. J Exp Clin Cancer Res 2012; 31 (1756–9966): 79. doi: 10.1186/1756-9966-31-79.
31. Louis DN, Ohgaki H, Wiestler OD et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007; 114 (2): 97–109. doi: 10.1007/s00401-007-0243-4.
32. Bouchalova K, Trojanec R, Kolar Z et al. Analysis of ERBB2 and TOP2A gene status using fluorescence in situ hybridization versus immunohistochemistry in localized breast cancer. Neoplasma 2006; 53 (5): 393–401.
33. Horbinski C, Miller C, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol 2011; 21 (1): 57–73. doi: 10.1111/j.1750-3639.2010.00453.x.
34. Pinkham MB, Telford N, Whitfield GA et al. FISHing Tips: What Every Clinician Should Know About 1p19q Analysis in Gliomas Using Fluorescence in situ Hybridisation. Clin Oncol (R Coll Radiol) 2015; 27 (8): 445–453. doi: 10.1016/j.clon.2015.04.008.
35. Kristensen LS, Andersen GB, Hager H et al. Competitive amplification of differentially melting amplicons (CADMA) enables sensitive and direct detection of all mutation types by high-resolution melting analysis. Hum Mutat 2012; 33 (1098–1004): 264–271. doi: 10.1002/humu. 21598.
36. Parrella P, la Torre A, Copetti M et al. High specificity of quantitative methylation-specific PCR analysis for MGMT promoter hypermethylation detection in gliomas. J Biomed Biotechnol 2009; 531692. doi: 10.1155/ 2009/531692.
37. Baumann S, Keller G, Puhringer F et al. The prognostic impact of O6-Methylguanine-DNA Methyltransferase (MGMT) promotor hypermethylation in esophageal adenocarcinoma. Int J Cancer 2006; 119 (2): 264–268. doi: 10.1002/ijc.21848.
38. Toyooka KO, Toyooka S, Maitra A et al. Establishment and validation of real-time polymerase chain reaction method for CDH1 promoter methylation. Am J Pathol 2002; 161 (2): 629–634. doi: 10.1016/S0002-9440 (10) 64218-6.
39. Weisenberger DJ, Campan M, Long TI et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33 (1362–4962): 6823–6836. doi: 10.1093/nar/gki987.
40. Loussouarn D, Le Loupp AG, Frenel JS et al. Comparison of immunohistochemistry, DNA sequencing and allele-specific PCR for the detection of IDH1 mutations in gliomas. Int J Oncol 2012; 40 (6): 2058–2062. doi: 10.3892/ijo.2012.1404.
41. Das BR, Tangri R, Ahmad F et al. Molecular investigation of isocitrate dehydrogenase gene (IDH) mutations in gliomas: first report of IDH2 mutations in Indian patients. Asian Pac J Cancer Prev 2013; 14 (12): 7261–7264.
42. Wick W, Meisner C, Hentschel B et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology 2013; 81 (17): 1515–1522. doi: 10.1212/WNL.0b013e3182a95 680.
43. Olar A, Aldape K. Biomarkers classification and therapeutic decision-making for malignant gliomas. Curr Treat Options Oncol 2012; 13 (4): 417–436. doi: 10.1007/s11864-012-0210-8.
44. Sanson M, Marie Y, Paris S et al. Isocitrate de-hydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 2009; 27 (25): 4150–4154. doi: 10.1200/JCO.2009.21.9832.
45. Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol 2007; 170 (5): 1445–1453. doi: 10.2353/ajpath.2007.070011.
46. Peng Z, Haitao X, Pin C et al. IDH1/IDH2 Mutations Define the Prognosis and Molecular Profiles of Patients with Gliomas: A Meta-Analysis. PLoS One 2013; 8 (7): e68782.doi: 10.1371/journal.pone.0068782.
47. Reis GF, Pekmezci M, Hansen HM et al. CDKN2A loss is associated with shortened overall survival in lower-grade (World Health Organization Grades II–III) Astrocytomas. J Neuropathol Exp Neurol 2015; 74 (5): 442–452. doi: 10.1097/NEN.0000000000000188.
48. Zhao J, Ma W, Zhao H. Loss of heterozygosity 1p/19q and survival in glioma: a meta-analysis. Neuro Oncol 2014; 16 (1523–5866): 103–112. doi: 10.1093/neuonc/ not145.
49. Smith JS, Perry A, Borell TJ et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol 2000; 18 (0732–183X): 636–645. doi: 10.1200/JCO.2000.18.3.636.
50. Lhotská H, Zemanová Z, Kramář F et al. Molecular cytogenetic analysis of chromosomal aberrations in cells of low grade gliomas and its contribution for tumour classif. Klin Onkol 2014; 27 (3): 183–191. doi: 10.14735/amko2014183.
51. Kuo LT, Kuo KT, Lee MJ et al. Correlation among pathology, genetic and epigenetic profiles, and clinical outcome in oligodendroglial tumors. Int J Cancer 2009; 124 (12): 2872–2879. doi: 10.1002/ijc.24303.
52. Ichimura K, Bolin MB, Goike HM et al. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res 2000; 60 (2): 417–424.
53. Kramar F, Zemanova Z, Michalova K et al. Cytogenetic analyses in 81 patients with brain gliomas: correlation with clinical outcome and morphological data. J Neuro-oncol 2007; 84 (2): 201–211. doi: 10.1007/s11060-007-9358-7.
54. Weller M, Felsberg J, Hartmann C et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol 2009; 27 (34): 5743–5750. doi: 10.1200/ JCO.2009.23.0805.
55. Homma T, Fukushima T, Vaccarella S et al. Correlation among pathology, genotype, and patient outcomes in glioblastoma. J Neuropathol Exp Neurol 2006; 65 (9): 846–854. doi: 10.1097/01.jnen.0000235118.75182.94.
56. Houillier C, Lejeune J, Benouaich-Amiel A et al. Prognostic impact of molecular markers in a series of 220 primary glioblastomas. Cancer 2006; 106 (10): 2218–2223. doi: 10.1002/cncr.21819.
57. Polivka J, Polivka J Jr, Rohan V et al. Isocitrate dehydrogenase-1 mutations as prognostic biomarker in glioblastoma multiforme patients in West Bohemia. Biomed Res Int 2014; 2014 : 735659. doi: 10.1155/2014/735659.
58. Hegi M, Diserens AC, Gorlia T et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005; 352 (10): 997–1003. doi: 10.1056/NEJMoa043331.
59. Bady P, Sciuscio D, Diserens AC et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol 2012; 124 (4): 547–560. doi: 10.1007/s00401-012-1016-2.
60. Noushmehr H, Weisenberger D, Diefes K et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010; 17 (5): 510–522. doi: 10.1016/j.ccr.2010.03.017.
61. Wiestler B, Capper D, Sill M et al. Integrated DNA methylation and copy-number profiling identify three clinically and biologically relevant groups of anaplastic glioma. Acta Neuropathol 2014; 128 (4): 561–571. doi: 10.1007/s00401-014-1315-x.
62. Brandner S, von Deimling A. Diagnostic, prognostic and predictive relevance of molecular markers in gliomas. Neuropathol Appl Neurobiol 2015; 41 (6): 694–720. doi: 10.1111/nan.12246.
63. Wick W, Hartmann C, Engel C et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol 2009; 27 (35): 5874–5880. doi: 10.1200/JCO.2009.23.6497.
64. Molenaar RJ, Verbaan D, Lamba S et al. The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol 2014; 16 (9): 1263–1273. doi: 10.1093/neuonc/nou005.
65. Christians A, Hartmann C, Benner A et al. Prognostic value of three different methods of MGMT promoter methylation analysis in a prospective trial on newly diagnosed glioblastoma. PLoSOne 2012; 7 (3): e33449. doi: 10.1371/journal.pone.0033449
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2017 Issue 5-
All articles in this issue
- Potential of Using Vitamin D as an Adjuvant Treatment of Malignant Melanoma
- Controversy in the Postoperative Treatment of Low-grade Gliomas
- The Role of Chemotherapy in the Treatment of Low-grade Gliomas
- Evaluation of Anti-cancer Therapies with Reimbursement Limited to Comprehensive Cancer Centres Using the European Society for Medical Oncology Magnitude of Clinical Benefit Scale
- Treatment Refusal in Pediatric Oncology
- Isocitrate Dehydrogenase Mutations are Better Prognostic Marker than O6-methylguanine-DNA Methyltransferase Promoter Methylation in Glioblastomas – a Retrospective, Single-centre Molecular Genetics Study of Gliomas
- The Anti-apoptotic Mechanism of Metformin Against Apoptosis Induced by Ionizing Radiation in Human Peripheral Blood Mononuclear Cells
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Controversy in the Postoperative Treatment of Low-grade Gliomas
- The Role of Chemotherapy in the Treatment of Low-grade Gliomas
- Treatment Refusal in Pediatric Oncology
- Isocitrate Dehydrogenase Mutations are Better Prognostic Marker than O6-methylguanine-DNA Methyltransferase Promoter Methylation in Glioblastomas – a Retrospective, Single-centre Molecular Genetics Study of Gliomas
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

