-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Assessing Predicted HIV-1 Replicative Capacity in a Clinical Setting
HIV-1 replicative capacity (RC) provides a measure of within-host fitness and is determined in the context of phenotypic drug resistance testing. However it is unclear how these in-vitro measurements relate to in-vivo processes. Here we assess RCs in a clinical setting by combining a previously published machine-learning tool, which predicts RC values from partial pol sequences with genotypic and clinical data from the Swiss HIV Cohort Study. The machine-learning tool is based on a training set consisting of 65000 RC measurements paired with their corresponding partial pol sequences. We find that predicted RC values (pRCs) correlate significantly with the virus load measured in 2073 infected but drug naïve individuals. Furthermore, we find that, for 53 pairs of sequences, each pair sampled in the same infected individual, the pRC was significantly higher for the sequence sampled later in the infection and that the increase in pRC was also significantly correlated with the increase in plasma viral load and with the length of the time-interval between the sampling points. These findings indicate that selection within a patient favors the evolution of higher replicative capacities and that these in-vitro fitness measures are indicative of in-vivo HIV virus load.
Published in the journal: . PLoS Pathog 7(11): e32767. doi:10.1371/journal.ppat.1002321
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002321Summary
HIV-1 replicative capacity (RC) provides a measure of within-host fitness and is determined in the context of phenotypic drug resistance testing. However it is unclear how these in-vitro measurements relate to in-vivo processes. Here we assess RCs in a clinical setting by combining a previously published machine-learning tool, which predicts RC values from partial pol sequences with genotypic and clinical data from the Swiss HIV Cohort Study. The machine-learning tool is based on a training set consisting of 65000 RC measurements paired with their corresponding partial pol sequences. We find that predicted RC values (pRCs) correlate significantly with the virus load measured in 2073 infected but drug naïve individuals. Furthermore, we find that, for 53 pairs of sequences, each pair sampled in the same infected individual, the pRC was significantly higher for the sequence sampled later in the infection and that the increase in pRC was also significantly correlated with the increase in plasma viral load and with the length of the time-interval between the sampling points. These findings indicate that selection within a patient favors the evolution of higher replicative capacities and that these in-vitro fitness measures are indicative of in-vivo HIV virus load.
Introduction
Measuring the fitness of HIV-1 is notoriously difficult. At the between-host level, fitness can be interpreted as the transmission potential which is defined as the expected number of transmissions in the course of an infection [1]. This quantity can however only be measured in cohorts of untreated patients with known infection status that are followed over long time periods [1]. At the within-host level, fitness is determined by the average number of secondary infected cells resulting from a single infected cell in vivo. This hypothetical quantity is difficult to determine [2] but can be approximated by in-vitro measurements of the replicative capacity (RC) (see [3]). However, the in-vivo relevance of such in vitro fitness values is largely unclear.
In a recent publication, some of the authors of this article described a computational method to predict RC values on the basis of viral amino-acid sequences [3]. To this end, a machine-learning algorithm based on a quadratic fitness model was applied to a training data set of 65,000 amino-acid sequences of the pol gene and the associated RC values. The resulting RC-predictor could explain roughly 40% of the deviance of RC values in a test-data set consisting of 5,000 sequences, which had not been used for the inference of this predictor. In the present study, we apply this computational predictor to clinical data from the Swiss HIV Cohort Study (SHCS) (www.shcs.ch) in order to obtain an assessment of the RC-predictor in an independent dataset and to study its correlation with plasma HIV RNA viral load, a known surrogate marker associated with disease progression [3].
Methods
Ethics statement
The Swiss HIV cohort study was approved by individual local institutional review boards of all participating centers (www.shcs.ch). Written informed consent was obtained for each SHCS study participant.
RC-prediction
Fitness is measured as the log replicative capacity of HIV-derived amplicons [representing all of Protease(PR) and most of Reverse Transcriptase (RT)] inserted into a constant backbone of a resistance test vector. The models are then trained to predict this fitness from the amino-acid sequence of the amplicons. Details on the experimental measurement of the RC values and on inferring the predictor have been published in [3]. Here, we briefly reiterate the principles of the models fitted.
In essence, the predictor is based on fitting the data consisting of amino acid sequences s and the corresponding log-RC values (w) with the following model (M1)sij denotes the presence (sij = 1) or absence (sij = 0) of allele j at position i. (or more generally, if an ambiguity in the population sequencing is consistent with several amino acids at a given position, sij denotes the probability of allele j at position i). The model parameters I, mij and εij;kl can be interpreted as intercept, main effects, and epistatic effects. As the number of parameters exceeds the number of data-points, the model M1 has been fitted to the data on the basis of a machine learning approach (generalized kernel ridge regression). With this approach over-fitting is no concern because the sub-dataset on which the predictor is evaluated is independent from the sub-dataset from which the predictor is inferred (see supplementary material of Hinkley et al. [3] for a detailed description of the fitting procedure).
Clinical and sequence data
We assessed the RC-predictor by using two datasets collected from untreated, chronically infected patients. The latter criterion was introduced because HIV RNA levels are usually very high during acute HIV infection, and it was ensured by discarding data points measured within the first 180 days after the first positive HIV test. The patients were enrolled in the Swiss HIV Cohort Study, a longitudinal multicenter observational cohort study (SHCS) (www.shcs.ch) [4]. These datasets consist of clinical data (Table 1) and the corresponding viral amino acid sequences from the SHCS drug resistance database [5]. We focus on patients, for whom amino-acid sequences of the entire protease and the first 303 amino acids of the reverse transcriptase were available. We only consider sequences, which have been obtained from therapy-naïve patients infected with HIV-1 subtype B because the training set originated solely from subtype B strains. The first set consists of nucleotide sequences with the corresponding HIV RNA virus load measurements (plasma viral load set; n = 2073 patients). Selection of viral load measurements is restricted to values obtained within 30 days before or after the genotypic tests, but before initiation of antiretroviral therapy. The second set contains 53 patients for whom genetic sequences are available at two time points, which are at least 6 months apart (median [interquartile] distance between the two measurements: 3.9 [1.9; 7.4] years; longitudinal set) (see [6] for more details on this dataset).
Tab. 1. Multivariable regression model to assess the association of log10 HIV RNA load with the predicted replicative capacity. 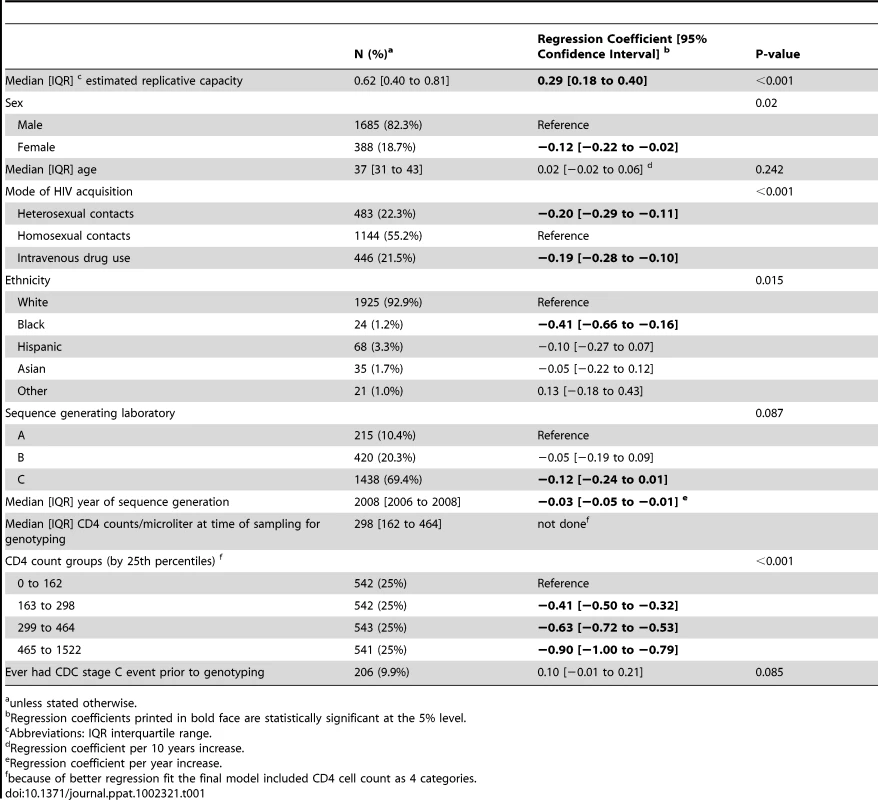
unless stated otherwise. Statistical analyses
Relationships between HIV RNA and pRC were modelled by the use of univariable and multivariable linear regression. Model assumptions were verified by inspecting residual versus fitted plots and by checking for unequal variance across fitted values (heteroskedasticity) and outliers. Because these diagnostics suggested the presence of heteroskedasticity we performed “robust” versions of linear regressions, which estimate a weighted variance based on the Huber−White method.
Statistical calculations were carried out with Stata 11.2 (Stata Corp., College Station, TX, USA). The level of significance was set at 0.05, and all p-values are two sided.
Results
Demographic and clinical characteristics of our study population are displayed in table 1. We assessed the predicted RC (pRC) with respect to two clinically relevant quantities or processes: Firstly, the relation between pRC and virus-load measurements measured around the same time and, secondly, the temporal change of pRC within ART-naive individuals.
In the plasma viral load dataset (2073 patients), values for RC predictions (pRC) were ranging from −1.07 to 1.43 units (median [interquartile range] 0.62 [0.40; 0.81]), and corresponding median [interquartile] HIV RNA levels were 4.7 log10 copies/mL [4.1; 5.2]. Using univariable linear regression analysis, we find a highly significant effect of the pRC value on virus load (F−Test p<0.001; see Figure 1A): a 1 unit increase in pRC is associated with an 0.57 increase [95% confidence interval 0.45; 0.69] in log10 HIV RNA. The fraction of variance in virus load explained through the pRC (R2) is 4.4%. Although somewhat attenuated, this effect of pRC on virus load remains highly significant (p<0.001; 0.29 [0.18; 0.40] log10 copies/mL HIV RNA per 1 unit increase in pRC ;table 1) if we control in a multivariable regression model for age, ethnicity, risk group, sex, CDC C stage and CD4 count at time of viral sequencing, and the laboratory that generated the sequence data. The association between HIV RNA and pRC changes only minimally when the fully adjusted regression model is re-estimated on individuals without any evidence for transmitted drug resistance mutations as defined by the most recent WHO surveillance list [7] (n = 1909; regression coefficient [95% confidence interval] 0.30 [0.18; 0.42] log10 copies HIV RNA per unit change pRC).
Fig. 1. Clinical Relevance of predicted Replicative Capacity (pRC). 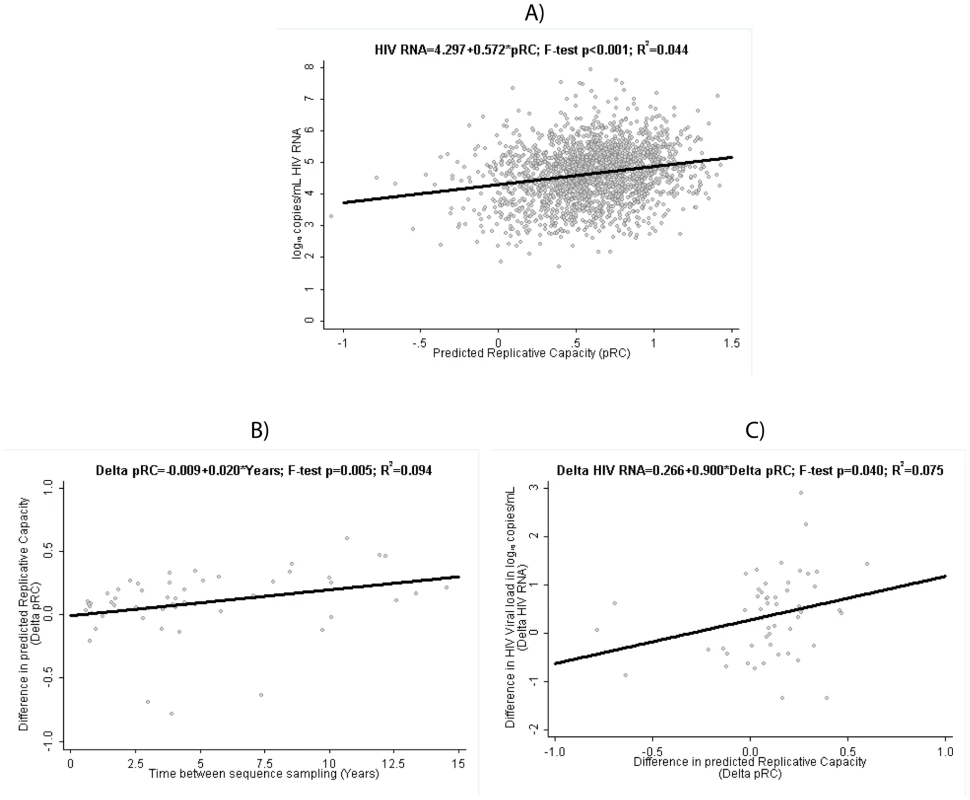
(A) Relation between pRC and virus load (measured as log10(copies of RNA/ml)) in the RNA-load dataset. (B) Temporal increase of pRC in the Longitudinal Dataset: relation between time difference between sequence samples and the change in pRC. (C) Relation between change in pRC and change in RNA-load in the Longitudinal Dataset. For the longitudinal dataset, we find that the pRC value increases in the course of an infection. Among the 53 patients with two viral sequences available taken at least 6 months apart, the median [interquartile] difference in pRC is 0.10 units [0.04; 0.25] and is statistically significantly different from 0 (p sign rank<0.001). Unadjusted linear regression estimates this increase in pRC at 0.020 units per year [95% confidence interval 0.006; 0.035] (figure 1B). At the same time, HIV RNA also tended to be higher at the second, later time point, with a median of 0.42 log10 copies/mL [−0.28; 0.88] (sign rank p = 0.005). Consequently, we find a statistically significant association between the change in pRC correlates and the change in HIV RNA over time in these 53 patients when applying a linear regression model to the data, which predicts a rise of 0.90 [0.01; 1.79] log10 copies/mL in HIV RNA per 1 unit increase in pRC over time (figure 1C). This finding suggests that within-host evolution seems to be characterized by a trend towards higher replication rates, and consequently higher plasma HIV RNA viral loads.
The above analyses were based on untreated patients sampled after the acute phase of the infection. We find similar results if we exclude patients, which have been sampled in the AIDS phase (defined as patients with at least one CDC stage C event, n = 206). In particular, we still find a highly significant (p<0.001) correlation between pRC and RNA load (slope: 1 unit increase in pRC is associated with an 0.54 increase [95% confidence interval 0.41; 0.66] in log10 HIV RNA) and a significant (p = 0.0058) increase of RC over time (increase in pRC at 0.020 units per year [95% confidence interval 0.006; 0.035]). Only the significance-level of the correlation between the temporal change of pRC and the temporal change of RNA load changes from ‘significant’ (p = 0.04) to ‘trend’ (p = 0.058); however even in this case the point estimates for the regression coefficient are very similar in both cases (0.9[0.01; 1.79] vs. 0.84[−0.03; 1.70]).
Discussion
How do the pRCs analyzed here relate to previous findings? For example, the 6 sequences (in our data-set) carrying the lamivudine mutation M184V, which has a large negative fitness effect on the virus [8] and has been associated with an 0.3 log10 copies lower HIV RNA relative to wild type [9], had a median [interquartile range] pRC of 0.1 [−1.3; 0.6], compared to 0.6 [0.4; 0.8] in the 1909 sequences without any transmitted resistance mutations (Wilcoxon rank sum p<0.001). Overall, the pRC varied over a range of 2.5 units from minimum to maximum. Our unadjusted and adjusted regression models would therefore predict a difference in HIV RNA of approximately 1.4 and 0.73 log10 copies/mL between the lowest and the highest pRC value. Yet HIV RNA viral loads varied over 6 logs from 1.9 to 7.9 log10 copies/mL in our dataset. This discrepancy is not very surprising given that our predictor for RC only takes the variation of 400 amino acid positions (roughly 10% of the genome of HIV) into account. However, the finding of a correlation of pRC and HIV RNA is robust, as confirmed by several sensitivity analyses, and it is consistent with a number of previous studies, which have also shown a correlation between in vitro measurements of RC and virus load [10], [11], [12], [13], [14].
Our findings thus support the notion that virus load is to a large extent controlled by virus genetics [15], [16], [17]. The fraction of variance explained by pRC (4.4%) is much lower than the fraction of variance in virus load explained by virus genetics in previous studies [15], [16], [17], but it should be borne in mind that the estimates of studies [15], [16], [17] are based on the variation in the entire genome (Note that this is the case even for Alizon et al.[15], because, even though the phylogenies used in that study were inferred from the pol-gene, they reflect the relatedness of the entire genome provided that recombination is not too common on an epidemiological level). It should also be noted that our results argue that at least a part of the virus' genetic control of the virus load established in patients appears to be mediated by the replicative capacity of the virus. This finding that virus load is controlled by RC contrasts the interpretation that virus load is mainly determined by the activation-rate of CD4 cells[18]. However, the relative importance of these different factors remains an open question. The increase of pRCs over time is also consistent with previous observations [19], and supports the view that, within a single host, HIV is selected for higher replicative capacities over time.
Overall our results show on the basis of a computational predictor, firstly that in vitro replicative capacity increases in the course of infection, which is consistent with the interpretation that RC is a determinant of fitness at the within-host level, and secondly that RC is linked to virus load, which has been shown to be a in vivo determinant of viral fitness at an epidemiological level [1]. In our view, it is remarkable that predicted RC based on partial pol sequences representing only 10% of HIVs genome correlates with virus load. Accordingly, taking into account the variation in the entire HIV genome (as will become possible in the future) may help to develop much more accurate predictors of virus fitness and virus load.
Zdroje
1. FraserCHollingsworthTDChapmanRde WolfFHanageWP 2007 Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci U S A 104 17441 17446
2. RibeiroRMQinLChavezLLLiDSelfSG 2010 Estimation of the initial viral growth rate and basic reproductive number during acute HIV-1 infection. J Virol 84 6096 6102
3. HinkleyTMartinsJChappeyCHaddadMStawiskiE 2011 A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nat Genet 43 487 489
4. Schoeni-AffolterFLedergerberBRickenbachMRudinCGunthardHF Cohort profile: the Swiss HIV Cohort study. Int J Epidemiol 39 1176 1178
5. von WylVYerlySBoniJBurgisserPKlimkaitT 2007 Emergence of HIV-1 drug resistance in previously untreated patients initiating combination antiretroviral treatment: a comparison of different regimen types. Arch Intern Med 167 1782 1790
6. KouyosRDvon WylVYerlySBoniJRiederP 2011 Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis 52 532 539
7. BennettDECamachoRJOteleaDKuritzkesDRFleuryH 2009 Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 4 e4724
8. Martinez-PicadoJMartinezMA 2008 HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res 134 104 123
9. HarrisonLCastroHCanePPillayDBoothC 2010 The effect of transmitted HIV-1 drug resistance on pre-therapy viral load. AIDS 24 1917 1922
10. Quinones-MateuMEBallSCMarozsanAJTorreVSAlbrightJL 2000 A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol 74 9222 9233
11. TrkolaAKusterHLeemannCRuprechtCJoosB 2003 Human immunodeficiency virus type 1 fitness is a determining factor in viral rebound and set point in chronic infection. J Virol 77 13146 13155
12. JoosBRiederPFischerMKusterHRusertP 2010 Association between specific HIV-1 Env traits and virologic control in vivo. Infect Genet Evol 10 365 372
13. JoosBTrkolaAFischerMKusterHRusertP 2005 Low human immunodeficiency virus envelope diversity correlates with low in vitro replication capacity and predicts spontaneous control of plasma viremia after treatment interruptions. J Virol 79 9026 9037
14. DaarESKeslerKLWrinTPetropouloCJBatesM 2005 HIV-1 pol replication capacity predicts disease progression. AIDS 19 871 877
15. AlizonSvon WylVStadlerTKouyosRDYerlyS 2010 Phylogenetic approach reveals that virus genotype largely determines HIV set-point viral load. PLoS Pathog 6 e1001123
16. HollingsworthTDLaeyendeckerOShirreffGDonnellyCASerwaddaD 2010 HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog 6 e1000876
17. HechtFMHartogensisWBraggLBacchettiPAtchisonR 2010 HIV RNA level in early infection is predicted by viral load in the transmission source. AIDS 24 941 945
18. BonhoefferSFunkGAGunthardHFFischerMMullerV 2003 Glancing behind virus load variation in HIV-1 infection. Trends Microbiol 11 499 504
19. TroyerRMCollinsKRAbrahaAFraundorfEMooreDM 2005 Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol 79 9006 9018
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Microbial Spy Games and Host Response: Roles of a Small Molecule in Communication with Other SpeciesČlánek Sequence-Based Analysis Uncovers an Abundance of Non-Coding RNA in the Total Transcriptome ofČlánek The Splicing Factor Proline-Glutamine Rich (SFPQ/PSF) Is Involved in Influenza Virus Transcription
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 11- Stillova choroba: vzácné a závažné systémové onemocnění
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Choroby jater v ordinaci praktického lékaře – význam jaterních testů
- Diagnostický algoritmus při podezření na syndrom periodické horečky
-
Všechny články tohoto čísla
- Microbial Spy Games and Host Response: Roles of a Small Molecule in Communication with Other Species
- Simple Rapid Near-Patient Diagnostics for Tuberculosis Remain Elusive—Is a “Treat-to-Test” Strategy More Realistic?
- Ultra-Efficient PrP Amplification Highlights Potentialities and Pitfalls of PMCA Technology
- Assessing Predicted HIV-1 Replicative Capacity in a Clinical Setting
- Inhibition of IL-10 Production by Maternal Antibodies against Group B Streptococcus GAPDH Confers Immunity to Offspring by Favoring Neutrophil Recruitment
- Anti-filarial Activity of Antibiotic Therapy Is Due to Extensive Apoptosis after Depletion from Filarial Nematodes
- West Nile Virus Experimental Evolution and the Trade-off Hypothesis
- Shedding Light on the Elusive Role of Endothelial Cells in Cytomegalovirus Dissemination
- Galactosaminogalactan, a New Immunosuppressive Polysaccharide of
- Fatal Prion Disease in a Mouse Model of Genetic E200K Creutzfeldt-Jakob Disease
- BST2/Tetherin Enhances Entry of Human Cytomegalovirus
- Metagenomic Analysis of Fever, Thrombocytopenia and Leukopenia Syndrome (FTLS) in Henan Province, China: Discovery of a New Bunyavirus
- Neurons are MHC Class I-Dependent Targets for CD8 T Cells upon Neurotropic Viral Infection
- Sap Transporter Mediated Import and Subsequent Degradation of Antimicrobial Peptides in
- A Molecular Mechanism for Bacterial Susceptibility to Zinc
- Genomic Transition to Pathogenicity in Chytrid Fungi
- Evolution of Multidrug Resistance during Infection Involves Mutation of the Essential Two Component Regulator WalKR
- ChemR23 Dampens Lung Inflammation and Enhances Anti-viral Immunity in a Mouse Model of Acute Viral Pneumonia
- SH3 Domain-Mediated Recruitment of Host Cell Amphiphysins by Alphavirus nsP3 Promotes Viral RNA Replication
- A Gammaherpesvirus Cooperates with Interferon-alpha/beta-Induced IRF2 to Halt Viral Replication, Control Reactivation, and Minimize Host Lethality
- Early Secreted Antigen ESAT-6 of Promotes Protective T Helper 17 Cell Responses in a Toll-Like Receptor-2-dependent Manner
- CD4 T Cell Immunity Is Critical for the Control of Simian Varicella Virus Infection in a Nonhuman Primate Model of VZV Infection
- The Role of the P2X Receptor in Infectious Diseases
- Down-Regulation of Shadoo in Prion Infections Traces a Pre-Clinical Event Inversely Related to PrP Accumulation
- Cross-Reactive T Cells Are Involved in Rapid Clearance of 2009 Pandemic H1N1 Influenza Virus in Nonhuman Primates
- Single Molecule Analysis of Replicated DNA Reveals the Usage of Multiple KSHV Genome Regions for Latent Replication
- The Critical Role of Notch Ligand Delta-like 1 in the Pathogenesis of Influenza A Virus (H1N1) Infection
- Sequence-Based Analysis Uncovers an Abundance of Non-Coding RNA in the Total Transcriptome of
- Murine Gamma Herpesvirus 68 Hijacks MAVS and IKKβ to Abrogate NFκB Activation and Antiviral Cytokine Production
- EBV Tegument Protein BNRF1 Disrupts DAXX-ATRX to Activate Viral Early Gene Transcription
- SAG101 Forms a Ternary Complex with EDS1 and PAD4 and Is Required for Resistance Signaling against Turnip Crinkle Virus
- Multiple Candidate Effectors from the Oomycete Pathogen Suppress Host Plant Immunity
- Rab7A Is Required for Efficient Production of Infectious HIV-1
- A TNF-Regulated Recombinatorial Macrophage Immune Receptor Implicated in Granuloma Formation in Tuberculosis
- Novel Anti-bacterial Activities of β-defensin 1 in Human Platelets: Suppression of Pathogen Growth and Signaling of Neutrophil Extracellular Trap Formation
- CD11b, Ly6G Cells Produce Type I Interferon and Exhibit Tissue Protective Properties Following Peripheral Virus Infection
- The Splicing Factor Proline-Glutamine Rich (SFPQ/PSF) Is Involved in Influenza Virus Transcription
- A Kinase Chaperones Hepatitis B Virus Capsid Assembly and Captures Capsid Dynamics
- Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Pathovar
- Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler
- The Pore-Forming Toxin Listeriolysin O Mediates a Novel Entry Pathway of into Human Hepatocytes
- The Human Herpesvirus-7 (HHV-7) U21 Immunoevasin Subverts NK-Mediated Cytoxicity through Modulation of MICA and MICB
- Avirulence Effector Avr3b is a Secreted NADH and ADP-ribose Pyrophosphorylase that Modulates Plant Immunity
- Murid Herpesvirus-4 Exploits Dendritic Cells to Infect B Cells
- Unique Type I Interferon Responses Determine the Functional Fate of Migratory Lung Dendritic Cells during Influenza Virus Infection
- Evolution of a Species-Specific Determinant within Human CRM1 that Regulates the Post-transcriptional Phases of HIV-1 Replication
- Transcriptome Analysis of Transgenic Mosquitoes with Altered Immunity
- Antibody Evasion by a Gammaherpesvirus O-Glycan Shield
- UDP-glucose 4, 6-dehydratase Activity Plays an Important Role in Maintaining Cell Wall Integrity and Virulence of
- Protease-Resistant Prions Selectively Decrease Shadoo Protein
- A LysM and SH3-Domain Containing Region of the p60 Protein Stimulates Accessory Cells to Promote Activation of Host NK Cells
- Deletion of AIF Ortholog Promotes Chromosome Aneuploidy and Fluconazole-Resistance in a Metacaspase-Independent Manner
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Multiple Candidate Effectors from the Oomycete Pathogen Suppress Host Plant Immunity
- The Splicing Factor Proline-Glutamine Rich (SFPQ/PSF) Is Involved in Influenza Virus Transcription
- A TNF-Regulated Recombinatorial Macrophage Immune Receptor Implicated in Granuloma Formation in Tuberculosis
- SH3 Domain-Mediated Recruitment of Host Cell Amphiphysins by Alphavirus nsP3 Promotes Viral RNA Replication
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



