-
Medical journals
- Career
Nuclear receptors gene polymorphisms and risk of restenosis and clinical events following coronary stenting
Authors: P. Neugebauer 1; M. Goldbergová‑ pávková 2; P. Kala 1; O. Boček 1; P. Jeřábek 1; M. Poloczek 1; M. Vytiska 1; J. Pařenica 1; R. Mikulík 3; Jiří Jarkovský 4; B. Semrád 1; J. Špinar 1; A. Vašků 2
Authors‘ workplace: Interní kardiologická klinika Lékařské fakulty MU a FN Brno, pracoviště Bohunice, přednosta prof. MU Dr. Jindřich Špinar, CSc., FESC 1; Ústav patologické fyziologie Lékařské fakulty MU Brno, přednostka prof. MU Dr. Anna Vašků, CSc. 2; Neurologická klinika Lékařské fakulty MU a FN sv. Anny Brno, přednosta prof. MU Dr. Ivan Rektor, CSc. 3; Institut biostatistiky a analýz Lékařské a Přírodovědecké fakulty MU Brno, přednosta doc. RNDr. Ladislav Dušek, Ph. D. 4
Published in: Vnitř Lék 2009; 55(12): 1135-1140
Category: Original Contributions
Overview
Introduction:
Hereditary factors connected with inflammation and fibroproliferation may play important role in restenotic process after coronary stenting. Peroxisome proliferator‑activated receptors (PPAR) and retinoic X receptors (RXR) regulate the transcription of crucial genes involved in the glucose and lipid metabolism, inflammation and cell differentiation. Methods: In our angiographic and clinical study we assessed the association of gene polymorphisms of L162V for PPAR‑α, C161T for PPAR ‑ γ and A(39526)AA for RXR - α with the risk of restenosis and cardiac events after coronary stenting. Primary endpoint was diameter stenosis ≥ 50% at follow‑up angiography. Secondary endpoints were death, myocardial infarction and/ or target lesion revascularisation at 12 months, and clinical restenosis. The results were adjusted for known predictors of restenosis. The genotypes were analysed by polymerase chains reaction (PCR) and restriction fragment length polymorphism (RFLP) methods. Results: Control angiography was performed in 477 of 565 patients (84.4%) with following restenosis rates in genotype subgroups: CC 29.0% vs GC/ GG 22.6% (p = 0.33) in L162V, CC 29.9% vs TC/ TT 24.6% (p = 0.24) in C161T and A/ A 26.9 % vs A/ AA + AA/ AA 35.0 % (p = 0.14) in A(39526)AA polymorphisms. The T allele of C161T polymorphism was associated with lower frequency of clinical restenosis (p = 0.015). Conclusion: We could not find an association of L162V PPAR‑α, C161T PPAR ‑ γ and A(39526)AA RXR ‑ α gene polymorphisms with angiographic in‑stent restenosis or major cardiac events. However, we found the relationship between C161T PPAR ‑ γ polymorphism and clinical restenosis deserving further study.Key words:
in‑stent restenosis – gene polymorphism – nuclear receptorsIntroduction
Several clinical, angiographic and periprocedural factors have been described to be associated with coronary in stent restenosis (ISR) [1–6]. Inherited factors also explain increased risk for restenosis in some patients [7–8]. Genes connected with fibroproliferation, thrombosis, inflammation or endotelial dysfunction have been under examination with inconsistent results [9–16].
Nuclear peroxisome proliferator activated receptors (PPAR) play a key role in the regulation of glucose and lipid metabolism, homeostasis, cell differentiation and proliferation [17–19]. PPAR heterodimerize with retinoic X receptors (RXR) and modulate the transcription of DNA at specific response elements. Both PPAR α and PPAR γ were found in vascular cells. Clinical trials showed that PPAR γ activators decrease the risk of coronary restenosis and cardiac events [20,21]. L162V (or C162G) and C161T polymorphisms in the genes for PPAR α and PPAR γ, respectivelly, were recognized as risk factors for cardiac events in patients with coronary artery disease [22–25]. A (39526)AA RXR - α polymorphism is predictor of congestive heart failure, its role in the restenosis is still unclear [26].
The goal of our study was to analyze if gene polymorphism of PPAR α, PPAR γ and RXR α is associated with the incidence of ISR.
Methods
Our trial is a prospective, cohort, single center, angiographic and clinical study on the role of PPAR α, PPAR γ and RXR - α gene polymorphisms on the risk of restenosis and cardiac events after coronary stenting for de-novo lesion in native coronary artery.
All consecutive patients, who underwent coronary stenting at the Department of Cardiology and Internal Medicine of the University Hospital Brno, between February 2004 and November 2005, were screened for this study. Patients were excluded for the following reasons: the admission diagnosis of acute myocardial infarction with ST elevation (STEMI), the presence of restenotic lesion at coronary angio, stenosis in arterial or venous bypass graft, treatment of peroral antidiabetic PPAR γ agonists.
Primary endpoint was angiographic restenosis, defined as diameter stenosis ≥ 50% in the stented segment (stent length ± 5mm) at follow up angiography. Secondary endpoints were the presence of death, myocardial infarction and/or target lesion revascularisation (TLR). TLR was defined as repeated percutaneous intervention or coronary artery surgery because of the significant in segment restenosis. Another secondary endpoint was clinical restenosis, defined as symptoms or signs of ischaemia in the presence of angiographic restenosis.
During the procedure, a standardweight-adjusted dose of heparin (80 IU//kg) was given intravenously. Activated clotting time (ACT) value was measured using Hemochron whole blood coagulation system (ITC, Edison, N.J., USA). After coronary stent placement, patients received dual antiplatelet therapy with 100mg aspirin once daily as long as possible and 75mg clopidogrel once daily. Clopidogrel was given for 1 or 6 months depending on the initial diagnosis (acute coronary syndrome or stable angina pectoris) and stent type – bare metal stent (cobalt chromium or stainless steel) or drug eluting stent (Cypher or Taxus). Follow up angiography was performed at 6 months in patients with bare metal stents or 12 months in patients with drug eluting stents. All procedures were performed on the Coroskop T.O.P. and Axiom Artis angiographic systems (Siemens, Erlangen, Germany) in the Department of Invasive and Interventional Cardiology of our institute. Angiographic restenosis was evaluated by offline QCA system Quantcor (Siemens, Erlangen, Germany).
Genomic DNA was isolated from peripheral leukocytes by a standard technique using proteinkinase K. DNA amplification was performed using polymerase chains reactions (PCR) in a 25ml volume containing 50 ng genomic DNA, 20 pmol of each primer, 2.5 mmol/l MgCl2, 50 mmol/l KCl, 25 mmol/l dNTP, 5 mmol/l Tris-HCl (pH 8.8) and 1 Unit Taq polymerase (MBI Ferments, USA). The genotypisation of all three polymorphisms was succesfully performed in 565 patients. In some patients PCR was not valid because they received unfractioned heparin or higher dose of low molecular weight heparin shortly before PCI and these patients were excluded.
The L162V mutation of PPAR α gene is caused by a CG transversion at nucleotide 484 in exon 5. Mismatch PCR method was performed with the primers
5‘-ACTCAAGCTGGTGTATGACAAGT-3‘ and 5‘-CGTTGTGTGACATCCCGACAGAAT-3‘. PCR products were digested with Hinf I (MBI Ferments, USA), electrophoresed through 4% agarose gel, and stained with ethidium bromide. The PCR product of 117 bp was cleaved by Hinf I. This resulted in two fragments 93 bp and 24 bp for G allele and one fragment for C allele. The location of the C161T PPAR γ gene polymorphism is in exon 6. The fragment was amplified using the primer pair 5‘-CAAGACAACCTGCTAGC-3‘ and 5‘-TCCTTGTAGATCTCCTGCAG-3‘. The PCR 200bp products were digested with restriction endonuclease Eco72I (MBI Ferments, USA) at 37 °C overnight and run on 3% agarose gel followed by ethidium bromide staining. This resulted in two fragments (120 and 80 bp) for 161C alleles and one fragment (200 bp) for 161T alleles.
The variants of RXR α gene in introne 5 were detected by RFLP analysis using restriction enzyme EcoO109I (MBI Ferments, USA). Primer pair was 5’-CCTCCTCCTGGCTGTACTT 3’ and 5’-CTCATGACAACTGCCTTGCT-3’. Digested PCR products (183 bp) were loaded into 2% agarose gel. RXR alleles were marked as AA – insertion of A at 39526 (159 + 24 bp), A – without insertion (183 bp).
Categorical data are presented as the percentage or absolute numbers, continuous data are presented as a mean value ± standard deviation (SD) or median (minimum–maximum). Statistical significance for intergroup differences was assessed by χ2 test for categorical variables. For continuous variables, 2-sample Student t test was used to compare the means or the Mann Whitney U test to compare the medians. Adjustment of p value was performed for known predictors of restenosis: treated diabetes, cumulative stent length, stenting ≥ 30mm, number of stents, reference vessel diameter ≤ 2.75mm and the use of drug eluting stent as the restenotic protection. Both unadjusted and adjusted p values are reported. P value ≤ 0.05 was an indicator of statistical significance. The analyses were performed using Statistica for Windows 7.1 (StatSoft Inc., Tulsa, USA), SPSS, version 12.0.1 (SPSS Inc., Chicago, USA), and NCSS 2007 (NCSS, Kaysville, USA).
Patients were enrolled into this trial after giving written informed consent for the genetic blood sample, percutaneous intervention and control angiography. Study protocol was approved by the Local Ethics Committee of University Hospital Brno, and all procedures were performed in accordance with the Declaration of Helsinki.
Results
Altogether 1,953 patients underwent coronary intervention with stenting in the study period. Of these, 1,382 were excluded from the study the following reasons (n = ): no informed consent obtained (194), STEMI (523), acute coronary syndrome treated with unfractioned heparin (141) or low molecular weight heparin (287) given 1–3 hours prior PCI (these patients were excluded because of the initial problems with PCR mentioned above), PCI of venous or arterial bypass grafts (19), PCI for in stent restenosis (38), treatment of diabetes with PPAR γ agonists (2), refused a follow up angiography (123), planned anticoagulation (14), chronical renal failure with blood creatinine ≥ 200 μmol/l (11), bailout PCI with planned cardiosurgery (9), arteriosclerosis of the lower limbs with problematic arterial access (9), known allergic reaction to contrast medium (4), high age, cerebrovascular disease, carcinoma or another severe comorbidities with low probability of performing 6–12 months angiography (8). Total of 571 patients after coronary stenting were finally enrolled into our study and 565 of them had complete genotypisation of all three polymorphisms. Of these, 477 patients underwent control angiography (84.4%) and represent studied patients population. The characteristics of this population are shown in table 1. Patients characteristics according to genotype are shown in table 2. There were interallelic differences of the left ventricle ejection fraction in PPAR α group, treated diabetes in PPAR γ group and cumulative stent length in RXR - α group.
1. Patients characteristics. 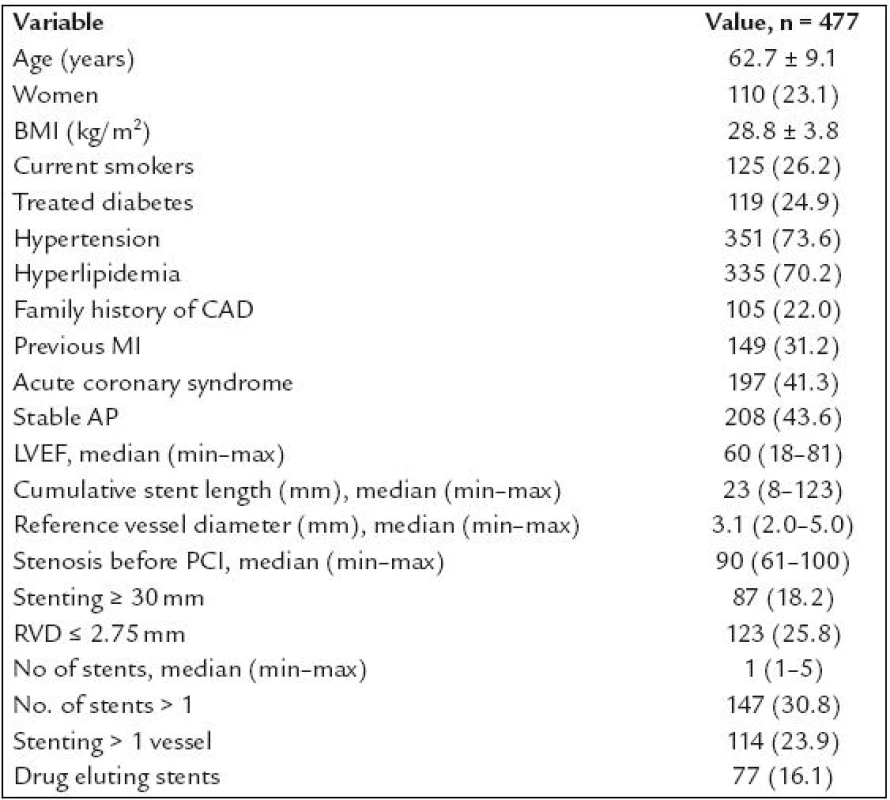
Continuous data are expressed as means ± SD or medians (minimum–maximum), categorical data as numbers (percents). AP – angina pectoris, BMI – body mass index, CAD – coronary artery disease, LVEF – left ventricle ejection fraction, MI – myocardial infarction, PCI – percutaneous coronary intervention, RVD – reference vessel diameter 2. Patients characteristics with respect to genotype. 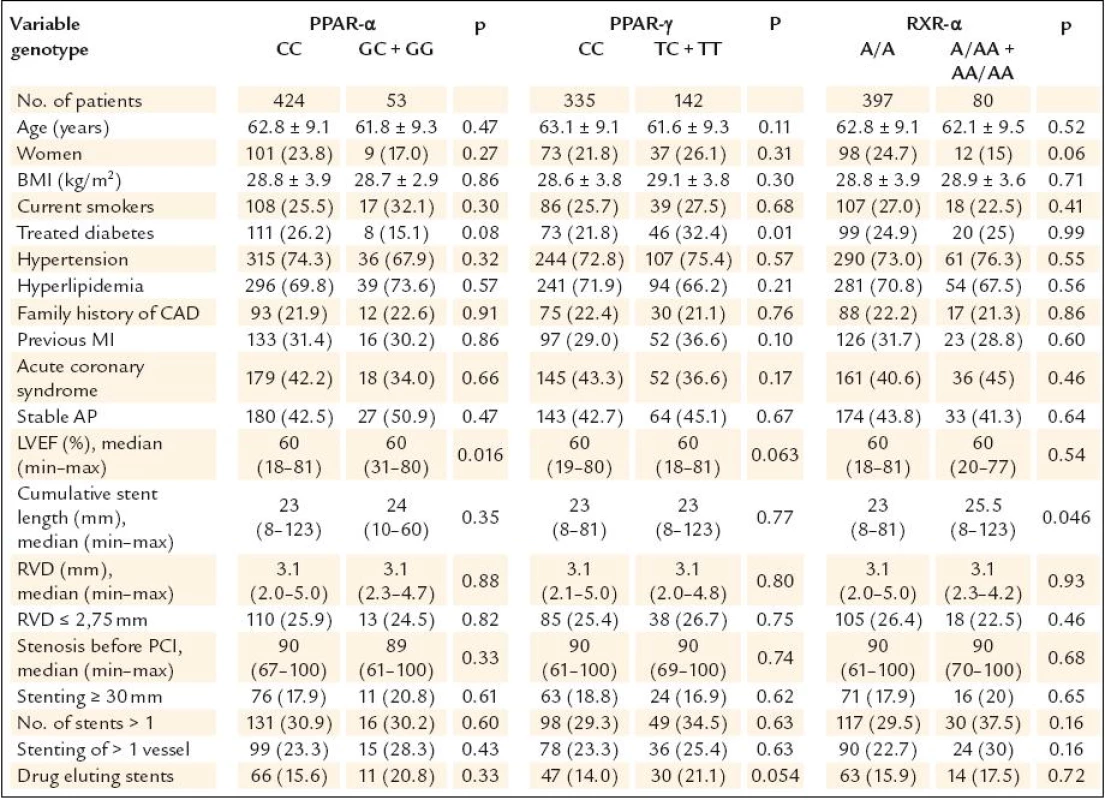
Continuous data are expressed as means ± SD or medians (min–max), categorical data as numbers (percents). AP – angina pectoris, BMI – body mass index, CAD – coronary artery disease, LVEF – left ventricle ejection fraction, MI – myocardial infarction, PCI – percutaneous coronary intervention, RVD – reference vessel diameter The study results are shown in table 3.None of the gene polymorphisms was significantly associated with the primary outcome measurement. The results did not change after adjustment for known ISR predictors. Also the use of drug eluting stents did not have influence on the association between PPAR α, PPAR γ and RXR α gene polymorphisms and ISR (table 4). The analysis of secondary endpoints found the association of C161T PPAR γ polymorphism with clinical restenosis. Incidence of combined major adverse cardiac events (death, MI and/or TLR) was not influenced by any polymorphism.
3. Study endpoints. 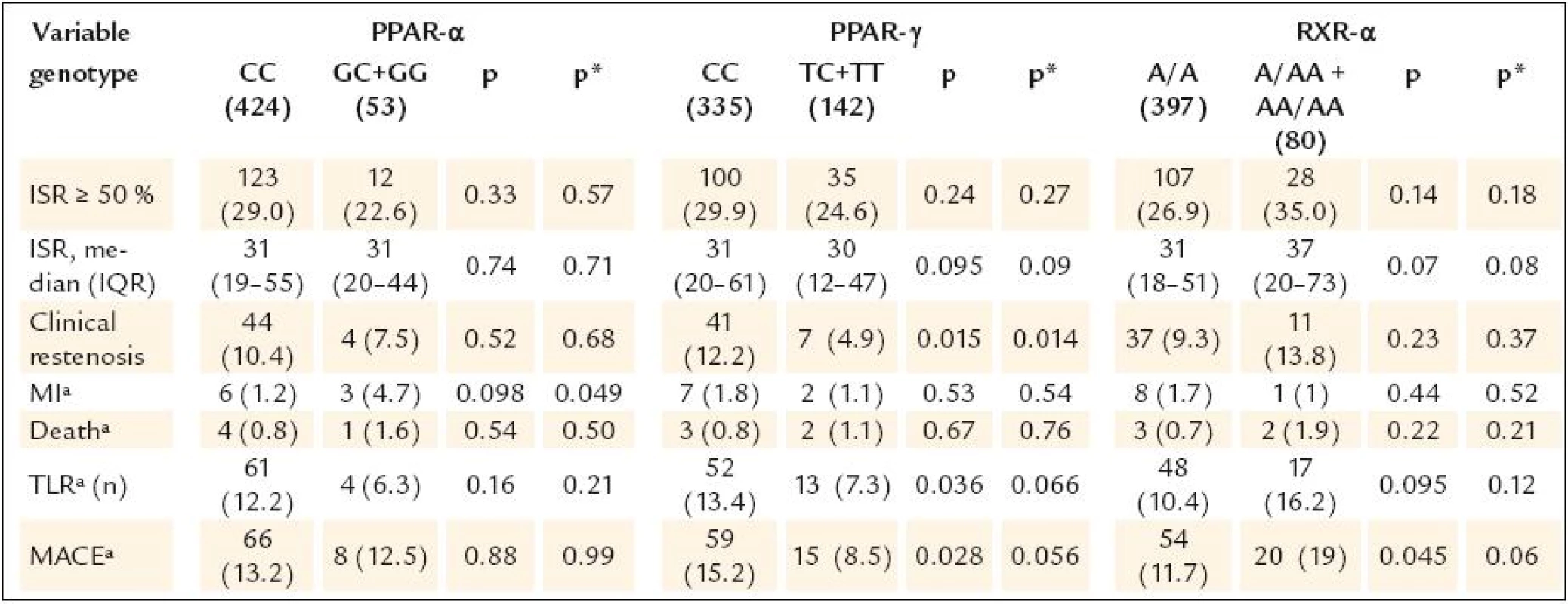
Continuous data are expressed as means ± standard deviation or medians (25.– 75. quartile), categorical data as numbers (percents). p* adjusted for treated diabetes, cumulative stent length, stenting ≥ 30 mm, DES, number of stents, RVD ≤ 2,75 mm; ªanalysed in all 565 patients. IQR – interquartile range, ISR – in-stent restenosis, TLR – target lesion revascularisation, MACE – major adverse cardiac events, MI – myocardial infarction 4. Occurrence of ISR in patients with DES and without DES. 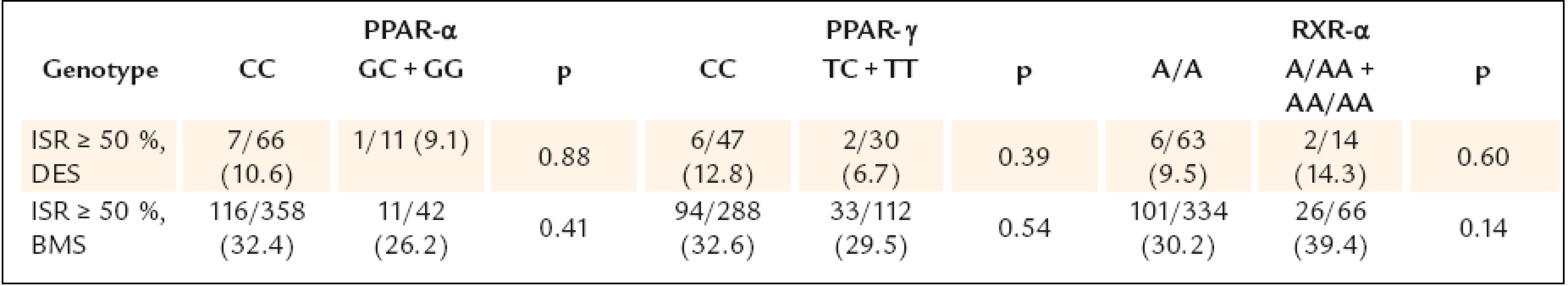
Data are expressed as numbers (percents). BMS – bare metal stents, DES – drug eluting stents, ISR – in-stent restenosis The multivariable logistic regression analysis showed, that the independent predictors of ISR were reference vessel diameter (Odds ratio 0.43, 95% confidence interval 0.28–0.67) and cumulative stent length (OR 1.02, 95% CI 1.01–1.04); the use of drug eluting stent was as expected the protection against restenosis (OR 0.15, 95% CI 0.07–0.34).
Discussion
In our prospective study we found no association of RXR - α, PPAR α, PPAR γ polymorphism with ISR. Although there was 8.1% difference in ISR occurrence in RXR - α genotype, such difference was most likely due to the imbalance in known prognostic factors of ISR. Also, despite the fact that the differences for some secondary endpoints were significant we could not demonstrate consistent findings for any gene polymorphism and significant findings were most likely due to the chance or multiple comparisons.
We analysed both PPAR and RXR gene polymorphisms together, which has not been done before. RXR is alone a specific regulator of cellular proliferation [27–28]. Moreover, PPAR can not work without heterodimerisation with RXR. There are sufficient experimental and clinical data clearly showing the relationship between PPAR γ, muscular proliferation and restenosis, respectively. PPAR γ regulates target genes encoding proteins involved in the regulation of cell-cycle progression, smooth muscle cells proliferation and apoptosis [29–30]. The negative results of our study indicate that the relationship between RXR, PPAR and ISR is more complicated or that studied polymorphisms simply do not belong to the genetic factors related to ISR. Previous large trial analysing different PPAR γ polymorphisms found negative results [31].
One of the reasons for our negative results with restenosis could be the type of studied population. Most of the trials with C161T and L162V PPAR polymorphisms were performed in non European population – either in Australia, Southeastern Asia or in Northern America, as opposed to the population from Central Europe used in our study. Another reason can be different mechanism of in stent restenosis compared to atherosclerotic progression of native coronary artery disease, which has been studied in previous works [22–25]. The third potential explanation for negative results of our study may be the size of the sample group.
From the angiographic perspective, 16.2% of patients, who underwent control angiography, had an implanted drug eluting stent, which could make the whole group not homogeneous. Nevertheless, there was not significant difference in drug eluting stents percentage between the studied alleles. Moreover, the results of primary endpoint remained the same when analysing separately patients with and without drug eluting stent.
It is also necessary to take into account a non linear impact of inherited factors on restenosis. Similarly to the disappointment with results of I/D ACE polymorphism trials, we cannot expect a clear relationship between one gene polymorphism and complex restenotic process with its many levels that could be influenced [32]. Moreover, some crucial genes that are involved in proliferative process can be influenced by others. A good example is a trial of Tiroch et al [33]. The authors found a negative result of an association of heme oxygenase-1 (HO-1) gene promoter polymorphism with restenosis and major cardiac events, despite possible protective role of the HO-1 protein against restenosis. The explanation for this could be another trial, which demonstrates that HO-1 expression is transcriptionally regulated by PPAR α and PPAR γ receptors [34].
In our study, however, we found the association between PPAR γ polymorphism and the risk of clinical restenosis. Although clinical restenosis was a secondary endpoint, we consider the results in this group of patients to be important because we usually treat only those “restenotic” patients who are symptomatic or have positive stress test. Such results generate the hypothesis that PPAR γ C161T polymorphism could be the independent predictor of clinical restenosis.
Limitations
Except for sample size limitation mentioned above, the relative limitation is that incomplete angiographic follow up was conducted in 84.4% of patients. This number is, however, in accordance with other genetic trials with control angiography. Another limitation is the absence of an indepedent CoreLab for angiographic evaluation.
Conclusions
We conclude that none of the three gene polymorphisms for PPAR α, PPAR γ and RXR - α receptors was associated with significant angiographic restenosis after stenting for de-novo lesion in native coronary artery. TC and TT genotypes of C161T gene polymorphism for PPAR γ were associated with a decreased risk of clinical restenosis. Such finding generates the hypothesis that an association between PPAR γ and ISR may exist.
Acknowledgement
We thank the whole team of our institutes, especially Hana Jakubíčková, for technical assistance and data sampling.
This trial was supported by the Research grant from the Czech Society of the Cardiology, Czech Republic.
Tato práce byla podpořena Domácím grantem České kardiologické společnosti (2003–2006).
MUDr. Petr Neugebauer
www.fnbrno.cz
e mail: petr.neug@post.cz, pneugebauer@fnbrno.cz
Sources
1. Antoniucci D, Valenti R, Santoro GM et al. Restenosis after coronary stenting in current clinical practice. Am Heart J 1998; 135 : 510 – 518.
2. Kastrati A, Schömig A, Elezi S et al. Predictive factors of restenosis after coronary stent placement. J Am Coll Cardiol 1997; 30 : 1428 – 1436.
3. Cutlip DE, Chauhan MS, Baim DS et al. Clinical restenosis after coronary stenting: perspectives from multicenter clinical trials. J Am Coll Cardiol 2002; 40 : 2082 – 2089.
4. Mercado N, Boersma E, Wijns W et al. Clinical and quantitative coronary angiographic predictors of coronary restenosis: a comparative analysis from the balloon - to - stent era. J Am Coll Cardiol 2001; 38 : 645 – 652.
5. Kastrati A, Mehilli J, Dirschinger J et al. Restenosis after coronary placement of various stent types. Am J Cardiol 2001; 87 : 34 – 39.
6. Vojáček J. Restenosis after coronary angioplasty: status in 1994. Vnitř Lék 1995; 41 : 325 – 331.
7. Kastrati A, Dirschinger I, Schömig A. Genetic risk factors and restenosis after percutaneous coronary interventions. Herz 2000; 25 : 34 – 46.
8. Agema WR, Jukema JW, Pimstone SN et al. Genetic aspects of restenosis after percutaneous coronary interventions: towards more tailored therapy. Eur Heart J 2001; 22 : 2058 – 2074.
9. Gürlek A, Güleç S, Karabulut H et al. Relation between the insertion/ deletion polymorphism of the angiotensin I converting enzyme gene and restenosis after coronary stenting. J Cardiovasc Risk 2000; 7 : 403 – 407.
10. Koch W, Kastrati A, Mehilli J et al. Insertion/ deletion polymorphism of the angiotensin I‑converting enzyme gene is not associated with restenosis after coronary stent replacement. Circulation 2000; 102 : 197 – 202.
11. von Beckerath N, Koch W, Mehilli J et al. Glycoprotein Ia gene C807T polymorphism and risk for major adverse cardiac events within the first 30 days after coronary artery stenting. Blood 2000; 95 : 3297 – 3301.
12. Rudez G, Pons D, Leebeek F et al. Platelet receptor P2RY12 haplotypes predict restenosis after percutaneous coronary interventions. Hum Mutat 2008; 29 : 375 – 380.
13. Böttiger C, Kastrati A, Koch W et al. Polymorphism of platelet glycoprotein IIb and risk of thrombosis and restenosis after coronary stent placement. Am J Cardiol 1999; 84 : 987 – 991.
14. Monraats PS, Pires NM, Agema WR et al. Genetic inflammatory factors predict restenosis after percutaneous coronary interventions. Circulation 2005; 112 : 2417 – 2425.
15. Gomma AH, Elrayess MA, Knight CJ et al. The endothelial nitric oxide synthase (Glu298Asp and – 786T>C) gene polymorphisms are associated with coronary in‑stent restenosis. Eur Heart J 2002; 23 : 1955 – 1962.
16. Bienertová - Vasků JA, Hlinomaz O, Vasků A. Are common leptin promoter polymorphisms associated with restenosis after coronary stenting? Heart Vessels 2007; 22 : 310 – 315.
17. Bishop - Bailey D. Peroxisome proliferator‑activated receptor in the cardiovascular system. Br J Pharmacol 2000; 129 : 823 – 834.
18. Brown J, Plutzky J. Peroxisome proliferator‑activated receptors as transcriptional nodal points and therapeutic targets. Circulation 2007; 115 : 518 – 533.
19. Marx N, Schönbeck U, Lazar MA et al. Peroxisome proliferator‑activated receptor gamma activators inhibit gene expression and migration in human vascular smooth muscle cells. Circ Res 1998; 83 : 1097 – 1103.
20. Choi D, Kim SK, Choi SH et al. Preventative effects of rosiglitazone on restenosis after coronary stent implantation in patients with type 2 diabetes. Diabetes Care 2004; 27 : 2654 – 2660.
21. Takagi T, Akasaka T, Yamamuro A et al. Troglitazone reduces neointimal tissue proliferation after coronary stent implantation in patients with non‑insulin dependent diabetes mellitus: a serial intravascular ultrasound study. J Am Coll Cardiol 2000; 36 : 1529 – 1535.
22. Wang XL, Oosterhof J, Duarte N. Peroxisome proliferator‑activated receptor gamma C161→T polymorphism and coronary artery disease. Cardiovasc Res 1999; 44 : 588 – 594.
23. Chao TH, Li YH, Chen JH et al. The 161TT genotype in the exon 6 of the peroxisome-proliferator‑activated receptor gamma gene is associated with premature acute myocardial infarction and increased lipid peroxidation in habitual heavy smokers. Clin Sci 2004; 107 : 461 – 466.
24. Tai ES, Collins D, Robins SJ et al. The L162V polymorphism at the peroxisome proliferator activated receptor alpha locus modulates the risk of cardiovascular events associated with insulin resistance and diabetes mellitus: the Veterans Affairs HDL Intervention Trial (VA - HIT). Atherosclerosis 2006; 187 : 153 – 160.
25. Robitaille J, Brouillette C, Houde Aet al. Association between the PPARalpha - L162V polymorphism and components of the metabolic syndrome. J Hum Genet 2004; 49 : 482 – 489.
26. Goldbergová M, Špinarová L, Špinar Jet al. Intron Retinoic X Receptor Alpha Polymorphism in Chronic Heart Failure. Atheroscler 2002; 3 (Suppl 2): 115 – 116.
27. Haraguchi G, Suzuki J, Kosuge H et al. A new RXR agonist, HX630, suppresses intimal hyperplasia in a mouse blood flow cessation model. J Mol Cell Cardiol 2006; 41 : 885 – 892.
28. Ahuja HS, Szanto A, Nagy L et al. The retinoid X receptor and its ligands: versatile regulators of metabolic function, cell differentiation and cell death. J Biol Regul Homeost Agents 2003; 17 : 29 – 45.
29. Meredith D, Panchatcharam M, Miriyala S et al. Dominant ‑ negative loss of PPAR{gamma} function enhances smooth muscle cell proliferation, migration, and vascular remodeling. Arterioscler Thromb Vasc Biol 2009; 29 : 465 – 471.
30. Gizard F, Bruemmer D. Transcriptional control of vascular smooth muscle cell proliferation by peroxisome proliferator‑activated receptor-gamma: therapeutic implications for cardiovascular diseases. PPAR Res 2008; 2008 : 429123.
31. Koch W, Jung V, von Beckerath N et al. Peroxisome proliferator‑activated receptor gamma gene polymorphisms and restenosis in diabetic patients after stenting in coronary arteries. Diabetologia 2004; 47 : 1126 – 1127.
32. Bonnici F, Keavney B, Collins R et al. Angiotensin converting enzyme insertion or deletion polymorphism and coronary restenosis: meta‑analysis of 16 studies. BMJ 2002; 325 : 517 – 520.
33. Tiroch K, Koch W, von Beckerath N et al. Heme oxygenase ‑ 1 gene promoter polymorphism and restenosis following coronary stenting. Eur Heart J 2007; 28 : 968 – 973.
34. Krönke G, Kadl A, Ikonomu E et al. Expression of heme oxygenase ‑ 1 in human vascular cells is regulated by peroxisome proliferator‑activated receptors. Arterioscler Thromb Vasc Biol 2007; 27 : 1276 – 1282.
Labels
Diabetology Endocrinology Internal medicine
Article was published inInternal Medicine

2009 Issue 12-
All articles in this issue
- The programme of managed ambulatory rehabilitation for patients after heart valve defect surgery
- Liver transplantation and peri- operative changes to renal function
- Impact of pandemic H1N1 2009 influenza virus on critical care in Australia: A single centre case series
- The complexity of interactions of the tumour growth process
- The complexity of interactions of the tumour growth process
- Pre‑eclampsia from the perspective of inter- professional collaboration
- Gender differences in pharmacotherapy of chronic heart failure
- Diabetes insipidus followed, after 4 years, with dysarthria and mild right‑ sided hemiparesis – the first clinical signs of Erdheim- Chester disease. Description and depiction of a case with a review of information on the disease
- Symmetrical phlebothrombosis of lower extremities resulting from congenital malformation of vena cava inferior
- Nuclear receptors gene polymorphisms and risk of restenosis and clinical events following coronary stenting
- Internal Medicine
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Pre‑eclampsia from the perspective of inter- professional collaboration
- The programme of managed ambulatory rehabilitation for patients after heart valve defect surgery
- The complexity of interactions of the tumour growth process
- Symmetrical phlebothrombosis of lower extremities resulting from congenital malformation of vena cava inferior
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

