-
Medical journals
- Career
Human rotavirus A detection: Comparison of enzymatic immunoassay and rapid chromatographic test with two quantitative RT-PCR assays
Authors: R. Moutelíková 1; M. Dvořáková Heroldová 2; V. Holá 2; P. Sauer 3; J. Prodělalová 1
Authors‘ workplace: Veterinary Research Institute, Brno, Czech Republic 1; Department of Microbiology, Faculty of Medicine, Masaryk University Brno and St. Anne's Faculty Hospital, Brno, Czech Republic 2; Department of Microbiology, Faculty of Medicine and Dentistry, Palacký University and University Hospital, Olomouc, Czech Republic 3
Published in: Epidemiol. Mikrobiol. Imunol. 67, 2018, č. 3, s. 110-113
Category: Original Papers
Overview
Objective:
The aim of this study was to compare results of two commercially available kits used for routine detection of Rotavirus A in human stool samples with results of commercial quantitative reverse-transcription PCR (RT-qPCR) test and in-house RT-qPCR.
Material and methods:
In total, 749 stool samples were screen - ed with the use of four different methods. The samples were collected from four diagnostic laboratories from March 2016 to June 2017. Diagnose of gastrointestinal disorders was stated in one third of tested patients, the rest of samples was collected from patients with other primary diagnose. The samples were tested with the enzymatic immunoassay (EIA) (RIDASCREEN® Rotavirus) and with rapid diagnostic immunochromatographic test (RDT) (IMMUNOQUICK® No-Rot-Adeno). As a reference method a commercial RT-qPCR test was used (Primerdesign™ Genesig® Kit) and it was compared with in-house RT-qPCR test prepared in our laboratory. The samples which in the reference RT-qPCR gave positive signal of reaction in cycle 28 or higher (Ct ≥ 28) were assessed as negatives in order to include only samples with some clinical relevance into sensitivity determination.
Results:
Diagnostic sensitivity was assessed as 84.2% for EIA and 82.5% for RDT. The specificity of those tests was calculated as 97.8% for EIA and 96.4% for RDT. The performance of both diagnostic tests describing their positive predictive value was determined to be 87.3% for EIA and 80.3% for RDT. Negative predictive value was calculated to be 97.2% for EIA and 96.8% for RDT. Proportion of RVA-positive samples determined with the reference RT-qPCR test with our own cut-off level was 15.2% (n=114). Comparisons of the in-house and reference RT-qPCR tests showed very good agreement of results. The sensitivity of the in-house test was 100% and its specificity 99.7%.
Conclusions:
RT-qPCR is more sensitive for surveillance of rotavirus gastroenteritis than routinely used EIA or RDT methods. The specificity of both evaluated tests was very high. However, EIA was in all performance parameters assessed better than RDT.
keywords:
rotavirus A – enzymatic immunoassay – immunochromato-graphic test – RT-qPCR
INTRODUCTION
Rotavirus A (RVA) is a leading cause of acute gastroenteritis (AGE) in children as well as an important nosocomial pathogen both in hospitalized children and adults. Nearly all children are infected with rotavirus at least once by the age of five years. Estimated number of AGE deaths due to rotavirus infection in children under five years of age in Europe is 970–1750 per year [1]. Rotavirus-associated outpatient visits and hospitalizations in all European countries result in direct medical treatment costs of 25 million dollars each year [2]. Since the WHO recommended universal vaccination of infants against RVA in 2009, the burden of the disease has significantly decreased in countries with RVA vaccination scheme [3].
However, the rate of vaccinated children in the Czech Republic still remains lower than in surrounding countries and as a consequence the incidence rates of RV gastroenteritis-connected hospitalization is higher in the Czech Republic than that reported in other industrialized European countries [4]. RVs are very contagious because a low number of virus particles suffices to productively infect a susceptible individual and also because rotavirus particles are shed in large numbers in faeces (up to 1011 particles/ml) during the acute stage of the infection [5]. Thus, the risk of community-acquired RV infection is significant. Especially endangered facilities are child day-care settings and retirement homes. Immunocompromised persons represent another endangered group of patients as 30% of adults with rotavirus gastroenteritis suffered of some kind of immunosuppression [6]. The proportion of RV gastroenteritis among nosocomial infections is also considerable. With regard to increasing average life expectancy it is probable that RV infections in adults will gain even more importance.
In the last ten years the number of reported gastroenteritis of suspected viral aetiology in the Czech Republic has slightly increased [7] and so the need for sensitive and reliable diagnostic assays is rising. Several methods have been utilized for the detection of rotavirus infection. The diagnosis of rotavirus AGE has initially been done by labour-intensive electron microscopy because these fastidious agents were not readily cultivatable in tissue culture [8]. In vitro cultivation of human rotavirus from stool samples has been achieved, however, it is not practical and not routinely used for detecting rotavirus because of intensive labour requirement and relatively low isolation rates. Of available procedures, ELISA is applied most frequently in the routine diagnostic laboratory due to the ease of use and speed of obtaining a result [9]. However, RT-PCR, which is highly sensitive and specific and also suitable for genotyping [10], has become the ‘gold standard’ of diagnostic discovery. However, rotavirus diarrhoea surveillance according to the WHO recommendations is carried out with the use of enzyme immunoassays (EIA) which are mostly targeted on viral protein VP6 (group-specific antigen).
MATERIAL AND METHODS
Sample collection
In this study we evaluated presence of RVA in frozen stool specimens from all age groups of patients. Samples were collected in two regions of Moravia (South Moravia and Olomouc Region) from patients with gastrointestinal disorders (n = 250) and also from hospitalized patients with other diagnosis (oncological disease, respiratory disorders, etc.). In total, 749 samples were gathered between March 2016 and June 2017 and tested with four different methods (three commercially available tests and one in-house assay).
Laboratory test methods
All samples were tested with RIDASCREEN® Rotavirus (R-Biopharm AG, Germany) and with Immunoquick® NoRotAdeno (Biosynex, France). In order to estimate sensitivity and specificity of those two routinely used commercial tests, they were compared with two real-time reverse transcription PCR (RT-qPCR) which allow the determination of viral load in the sample. One of RT-qPCR assays is commercially available (Primerdesign™ Genesig® Kit, Primerdesign Ltd., UK) and the second was in-house assay previously tested in our laboratory.
RIDASCREEN® Rotavirus is an enzyme immunoassay (EIA) for qualitative identification of rotaviruses in human stool samples. It is a simple and highly sensitive sandwich type EIA which is suited for analyses of large numbers of samples. Monoclonal antibodies used in this method are directed against rotavirus group-specific structural protein VP6. The assay was performed according to manufacturer’s recommendations, results were read using an optical density (OD) spectrophotometer (Sunrise microplate reader, Tecan, Austria) and interpreted according to manufacturer’s instructions.
Immunoquick® NoRotAdeno is a rapid diagnostic test (RDT) which employs lateral flow chromatographic immunoassay for the simultaneous qualitative detection of rotavirus, adenovirus, and norovirus antigens in human stool samples. This test uses specific monoclonal antibodies which are coated on the test membrane. The test was performed according to the manufacturer’s recommendations and the results were read visually within 15 min.
The reference method for the detection of RVA was Primerdesign™ Genesig® Kit (Primerdesign Ltd., UK) which is designed for the in vitro quantification of human rotavirus A genomes with one-step RT-PCR detection protocol. The PCR is targeted on the gene coding non-structural protein 5 (NSP5). According to the manufacturer, under optimal PCR conditions the kit can detect less than 100 copies of target template. However, for the purpose of our study only samples with higher number of target template than 104 copies of viral genome/ml were assessed as positives.
All samples were also tested with in-house RT-qPCR. Briefly, viral RNA was extracted from 10% faecal suspension in phosphate-buffered saline (Biosera, France) using TRI Reagent (Sigma Aldrich, USA) according to manufacturer’s instructions. Viral RNA was reverse transcribed with the use of Transcriptor First Strand cDNA Synthesis Kit (Roche, Germany) and amplified with KAPA Probe Fast Universal qPCR Kit (Kapa Biosystems, USA). The PCR primers and hydrolysis probe were based on previously published qPCR detection system targeted on sequence coding non-structural viral protein NSP3 which is highly conserved in RVAs [11]. For the purpose of our study both primers and the probe were modified (Table 1). The fluorogenic probe was labelled with a HEX reporter on 5´end and a BHQ1 quencher on 3´end. The detection limit of qPCR was assessed with the use of serial 10-fold dilutions (109 to 100 copies/μl) of cDNA prepared from previously quantitated RVA culture (strain OSU) as described by Malenovska [12]. The same serial dilutions were used for the standard curve construction and PCR product quantification. The cut-off level for positive assessment of tested samples was set to 104 copies of viral genome/ml.
1. Sequence of primers and a probe used in the in-house RT-qPCR and their location in RVA NSP3 region (GenBank accession number X81436) 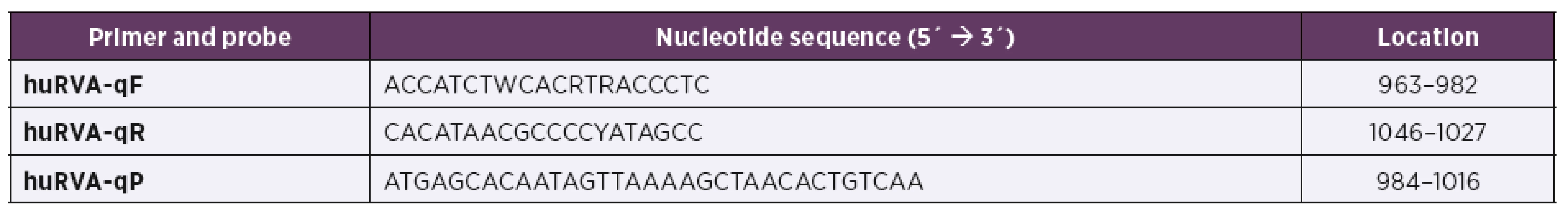
Data analysis
For EIA, RDT and modified in-house RT-qPCR assay, sensitivity and specificity was calculated with RVA Genesig® kit as a reference test. Sensitivity was assessed as a ratio of true positive (TP) results and true positive together with false negative (FN) results; TP/(TP + FN). Specificity of each of the tests was calculated from true negative (TN) and false positive (FP) results; TN/(TN + FP). Moreover, positive and negative predictive value describing the performance and precision of a diagnostic test was calculated with the use of true and false positive (resp. negative) results; TP/(TP + FP), resp. TN/(TN + FN).
RESULTS
Proportion of RVA-positive faecal samples determined with the reference RT-qPCR test with our own cut-off level was 15.2% (n = 114).
The results of the RIDASCREEN® Rotavirus (EIA) and Immunoquick® NoRotAdeno (RDT) compared to the commercial RT-qPCR method (Primerdesign™ Genesig® Kit) as a reference method are shown in Table 2. The true positive rate determined for EIA and RDT showed sensitivity of 84.2% and 82.5%, respectively. Specificity of EIA and RDT tests was calculated as 97.8% and 96.4%, respectively. The performance of both diagnostic tests describing their positive predictive value was determined to be 87.3% for EIA and 80.3% for RDT. The precision of the tests (their negative predictive value) was calculated as 97.2% for EIA and 96.8% for RDT.
2. Results obtained in comparison of the EIA and RDT diagnostic tests with reference RT-qPCR method (Genesig Kit) 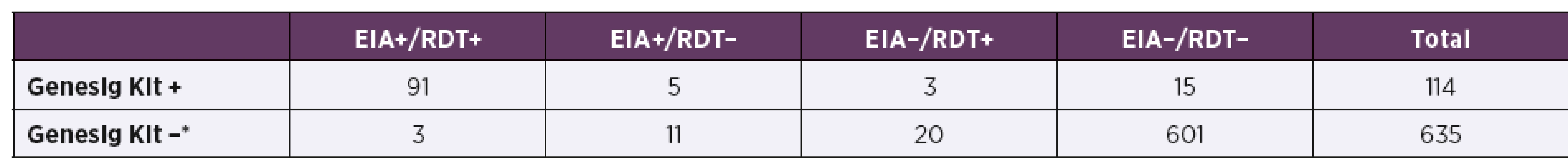
*Samples tested with RT-qPCR method with resulting threshold cycle (Ct) higher than or equal to 28 (Ct ≥ 28) were evaluated as negatives. In-house RT-qPCR assay showed very good agreement with the reference method (Primerdesign™ Genesig® Kit) (Table 3). The sensitivity of the in-house assay was 100% and its specificity was calculated as 99.7%. Similarly good results were obtained in the determination of the positive and negative predictive value of the in-house RT--qPCR. Its performance was 96.6% and the precision was 100%.
3. Comparison of in-house RT-qPCR method with the reference RT-qPCR test (Genesig Kit) 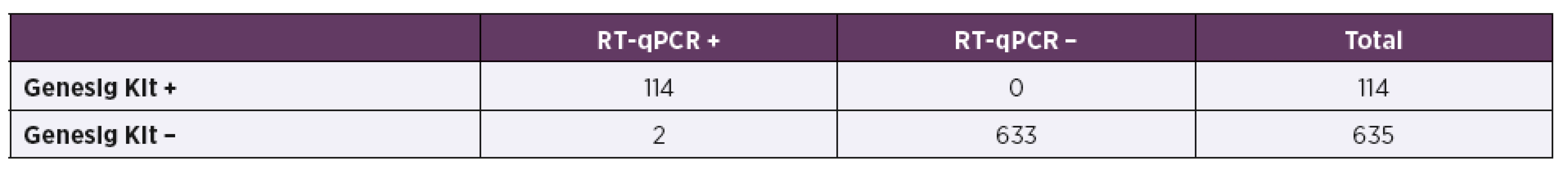
DISCUSSION
The sensitivity and specificity of RIDASCREEN® Rotavirus assay as determined by the producer should be 95.6% and 99.1%, respectively. However, those statistical measures reported in the manufacturer’s insert were determined in comparison to 3 commercial ELISAs. Likewise manufacture’s insert in the Immunoquick® NoRotAdeno test states the sensitivity and specificity as 100% and 98.4% in comparison with another commercially available rapid diagnostic test for rotavirus. This reduced sensitivity calculated in our study compared to producers’ information could be due to the lack of detection of certain genotypes by both EIA and RDT. Similarly, Lagare et al. [13] found lower sensitivity and specificity of evaluated EIA and RDT tests when compared to RT-PCR.
In our study, the 18 false negative specimens by EIA and 20 false negative specimens by RDT amount to 15.8% and 17.4% of missed positive results. It should not be accounted to the high sensitivity of the reference standard method although RT-qPCR is evidently more sensitive than EIA or RDT. As was described earlier by Phillips et al. [14], the cut-off value of highly sensitive RT-qPCR assays for infectious intestinal disease detection should be correlated with the actual clinical symptoms. In our study in order to include only samples with some clinical relevance, the specimens with cycle threshold values higher than 28 (Ct ≥ 28), which amounts to viral load of 104 copies of viral genome/ml or lower, were in reference Genesig® kit assessed as negatives.
Overall, there were 15 results determined as negatives both by EIA and RDT and at the same time positive by both RT-qPCR assays (Genesig Kit and in-house test). The limit of detection stated by the producer is 6.63x103 virus particles/ml for EIA and 1.4x104 PFU/ml for RDT. Out of these discordant results, in 8 samples the detected number of virus particles was very near the limit of detection. However, remaining 7 samples assessed as negatives by EIA as well as RDT contained high viral loads (between 106 and 109 virus particles/ml). These false negative results could be explained either by human factor failure during the testing or by possible presence of less common VP6 genotype (I-type) in the sample which is not correctly recognized by monoclonal antibody used in EIA or RDT [15]. So far, there have been 4 I-types described to be present in RVA strains of human origin most common being I1 and I2, rarely also I3 and I5 [16].
Specificity of both tests, which describes test's ability to correctly reject healthy patients without a condition, was above 95% as well as the precision of EIA and RDT. These results are in agreement with another study comparing ability of immunochromatographic assay for RVA detection in stool samples [17].
The comparison of two RT-qPCR methods (Table 3) confirmed very good performance of the in-house test. The obtained results were nearly identical, the only discrepancy was detection of two RVA-positive samples which were negative when tested with Genesig reference kit. The samples were tested repeatedly with both RT-qPCR assays with the same result. When tested with EIA and RDT, those two questionable samples were assessed as positives. In this case we presume that the commercial RT-qPCR kit failed to detect positive samples which might be caused by certain (even if low) degree of variability in NSP5 genomic segment which was used as a target for the primers and a probe construction [18].
CONCLUSIONS
It was demonstrated that RT-qPCR is more sensitive for surveillance of rotavirus associated gastroenteritis than routinely used EIA or RDT methods. The specificity of both evaluated tests was very high. However, EIA was in all performance parameters assessed better than RDT. In-house RT-qPCR showed very good agreement with commercial real-time RT-PCR. The higher sensitivity of real-time RT--PCR methods may be useful for successful recognition and control of infectious diarrhoea in the intensive care units or retirement homes where rotavirus associated gastroenteritis can have fatal outcome.
Acknowledgments
This study was supported by Ministry of Health of the Czech Republic by the grant No. 16-29937A.
Do redakce došlo dne 10. 2. 2018.
Adresa pro korespondenci:
Mgr. Romana Moutelíková, Ph.D.
Výzkumný ústav veterinárního lékařství
Hudcova 296/70
621 00 Brno
Sources
1. Clark A, Black R, Tate J, et al. Estimating global, regional and national rotavirus deaths in children aged <5 years: Current approaches, new analyses and proposed improvements. PLoS One, 2017;12(9):e0183392.
2. Rheingans RD, Antil L, Dreibelbis R, et al. Economic costs of rotavirus gastroenteritis and cost-effectiveness of vaccination in developing countries. J Infect Dis, 2009;200:S16–S27.
3. Patel MM, Steele D, Gentsch JR, et al. Real-world impact of rotavirus vaccination. Pediatr Infect Dis J, 2011;30:S1–S5.
4. Pazdiora P, Beneš Č. Rotavirus gastroenteritis in the Czech Republic before the start of vaccination. Epidemiol Mikrobiol Imunol, 2013;4 : 131–137.
5. Graham DY, Dufour GR, Estes MK. Minimal infective dose of rotavirus. Arch Virol, 1987;92 : 261–271.
6. Anderson EJ, Shippee DB, Tate JE, et al. Clinical characteristics and genotypes of rotavirus in adults. J Infect, 2015;70(6):683–687.
7. Vybrané infekční nemoci v ČR v letech 2007–2016 – relativně [online]. [cit. 2018-02-12]. Dostupné na http://www.szu.cz/publikace/data/vybrane-infekcni-nemoci-v-cr-v-letech-2007-2016-relativne>
8. Bishop RF, Davidson GP, Holmes IH, Ruck BJ. Detection of a new virus by electron microscopy of faecal extracts from children with acute gastroenteritis. Lancet, 1974;1 : 149–151.
9. Desselberger U. Rotaviruses. Virus Res, 2014;190 : 75–96.
10. Fischer TK, Gentsch JR. Rotavirus typing methods and algorithms. Rev Med Virol, 2004;14 : 71–82.
11. Pang XL, Lee B, Boroumand N, et al. Increased detection of rotavirus using a real time reverse transcription-polymerase chain reaction (RT-PCR) assay in stool specimens from children with diarrhea. J Med Virol, 2004;72 : 496–501.
12. Malenovska, H. Virus quantitation by transmission electron microscopy, TCID50, and the role of timing virus harvesting: A case study of three animal viruses. J Virol Methods, 2013;191 : 136–140.
13. Lagare A, Moumouni A, Kaplon J, et al. Diagnostic accuracy of VIKIA® Rota-Adeno and Premier™ Rotaclone® tests for the detection of rotavirus in Niger. BMC Res Notes, 2017;10 : 505.
14. Phillips G, Lopman B, Tam CC, et al. Diagnosing rotavirus A associated IID: Using ELISA to identify a cut-off for real time RT-PCR. J Clin Virol, 2009;44 : 242–245.
15. Wang H, Liu M, Sugata H, et al. Development of a new enzyme immunoassay for improved detection of rotavirus in fecal specimens of vaccinated infants. J Clin Virol, 2018;99–100 : 44–49.
16. Matthijnssens J, Ciarlet M, Rahman M, et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol, 2008;153 : 1621–1629.
17. Kim J, Kim HS, Kim H-S, et al. Evaluation of an immunochromatographic assay for the rapid and simultaneous detection of rotavirus and adenovirus in stool samples. Ann Lab Med, 2014;34 : 216–222.
18. Jeong S, Than VT, Lim I, Kim W. Whole-genome analysis of a rare human Korean G3P[9] rotavirus strain suggests a complex evolutionary origin potentially involving reassortment events between feline and bovine rotaviruses. PLoS One, 2014; 9(5):e97127.
Labels
Hygiene and epidemiology Medical virology Clinical microbiology
Article was published inEpidemiology, Microbiology, Immunology

2018 Issue 3-
All articles in this issue
- Molecular characterization of Streptococcus pneumoniae isolates recovered from cases of pneumococcal vaccine failure in children under five years of age in the Czech Republic in 2012–2014
- Fecal bacteriotherapy in the treatment of Clostridium difficile infection
- Successful rituximab treatment of granulomatous/lymphocytic interstitial lung disease in common variable immunodeficiency
- Human rotavirus A detection: Comparison of enzymatic immunoassay and rapid chromatographic test with two quantitative RT-PCR assays
- Human Mozdok leptospirosis first diagnosed by serum agglutinin-absorption tests in the Slovak Republic
- Susceptibility of clinical isolates of Bordetella pertussis to chemicals
- Laboratory evaluation of repellency of traditional Czech homemade repellents against Aedes aegypti
- Comparison of the epidemiological patterns of Lyme borreliosis and tick-borne encephalitis in the Czech Republic in 2007–2016
- Epidemiology, Microbiology, Immunology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Comparison of the epidemiological patterns of Lyme borreliosis and tick-borne encephalitis in the Czech Republic in 2007–2016
- Fecal bacteriotherapy in the treatment of Clostridium difficile infection
- Successful rituximab treatment of granulomatous/lymphocytic interstitial lung disease in common variable immunodeficiency
- Susceptibility of clinical isolates of Bordetella pertussis to chemicals
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

