-
Medical journals
- Career
Meta-Analysis of BRCA1 Polymorphisms and Breast Cancer Susceptibility
Authors: Ghafouri-Fard Soudeh; Dianatpour Ali; Faramarzi Sepideh
Authors‘ workplace: Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Published in: Klin Onkol 2018; 31(5): 330-338
Category: Review
doi: https://doi.org/10.14735/amko2018330Overview
Background:
BRCA1 codes for a tumor suppressor protein involved in DNA repair. Based on the role of single nucleotide polymorphisms (SNPs) in the modification of gene expression and function and the existence of certain SNPs within 3’-untranslated region of BRCA1 with the ability to change binding sites for mirRNAs, several association studies have been designed to explore the significance of SNPs within BRCA1 gene in conferring breast cancer (BC) risk. This study aims to assess the relationship between BRCA1 SNPs and BC using meta-analysis.
Aim:
To conduct a meta-analysis for retrieving case-control studies on the associations between the rs11655505, rs1799966, rs3737559, rs1799950, rs799917 and rs16941 BRCA1 polymorphisms and BC. The pooled odds ratios and its 95% confidence intervals were measured using fixed and random model to define the association between these polymorphisms and BC risk.
Conclusion:
No significant association was found for any of these polymorphisms and BC risk in the allelic, homozygote, dominant or recessive models. Overall, our study implies that the mentioned polymorphisms are not associated with BC risk. However, our study did not exclude the possible contribution of other SNPs within this gene in BC nor substantial contribution of multiple variants within this gene in conferring BC risk.
Key words:
BRCA1 – breast cancer – meta-analysis
Introduction
Breast cancer (BC) as the most common malignancy among women is associated with high mortality and morbidity [1]. Several susceptibility loci have been detected for this disorder [2–4]. BRCA1, located on chromosome 17q21, has been the first cancer susceptibility gene detected through a linkage study in families with early onset of the disease [5]. BRCA1 gene as a prototype of tumor suppressor genes participates in protection of intact chromosome structure [6]. Highly penetrant variants of this gene explain less than 20% of the genetic risk of BC [7]. The polygenic model for BC emphasizes the existence of numerous low penetrance high risk alleles that totally confer BC risk [8]. Consequently, more common variants in BRCA1 might also been associated with BC risk [8]. Considering the role of single nucleotide polymorphisms (SNPs) in modification of gene expression and function and the existence of certain SNPs within 3’-untranslated region of BRCA1 with the ability to change binding sites for microRNAs (miRNAs), several association studies have been designed to explore the significance of SNPs within the BRCA1 gene in conferring BC risk [9–11]. However, discrepancies have been detected in the results of such studies, which can be attributed to the heterogeneity of patients’ samples, small sample sizes and ethnic origin of patients. Consequently, we conducted a systematic search and meta-analysis to reach a more accurate answer to the question regarding the extent of the contribution of BRCA1 genomic variants in BC susceptibility.
Methods
Search for relevant articles
To find suitable studies for the present meta-analysis, we searched in the PubMed, Google Scholar, EMBASE and Web of Science databases until January 2018 using the following key words – ’breast cancer’ or ‘breast tumor’ with ’BRCA1 gene polymorphism’ or ’BRCA1 gene single nucleotide polymorphism’ or ’BRCA1 gene SNPs’ or ’BRCA1 gene SNP’. Besides, the articles were filtered with the terms ’rs11655505’ or ‘rs1799966’ or ’rs3737559’ or ’rs1799950’ or ’rs799917’ or ’rs16941’. We confined searches to full English articles. The collected studies were entered in the meta-analysis if a) the study provided association between the BRCA1 genetic polymorphisms and the susceptibility to BC, b) the study was designed as a case-control study, c) genotype/allele data of the polymorphism (s) were provided in the study. Case reports, editorials and cell culture experiments were excluded from the meta-analysis. Schema 1 shows the process of selection of studies for inclusion in the meta-analysis.
Schema 1. A systematic flow chart demonstrating the course of selection of articles for this meta-analysis. 
Data extraction
All manuscripts were evaluated by two authors (Dianatpour A. and Faramarzi S.) according to the inclusion and exclusion criteria. The first author’s name, year of publication, ethnicity of study participants, source of DNA (blood or breast tissue) used for SNP genotyping, total number of cases and controls and genotype distribution were collected. The studies were scored based on the Newcastle-Ottawa scale (NOS), [12] and those with NOS scores more than 6 were chosen for Hardy-Weinberg equilibrium (HWE) analysis.
Statistical analysis
The analyses were carried out in SPSS version 20 (IBM Analytics, USA) and RevMan version 5.3. The association between BRCA1 polymorphism (rs11 655505, rs1799966, rs3737559, rs1799 950, rs799917 and rs16941), and susceptibility of BC was assessed from the case-control studies through evaluation of odds ratios and 95% confidence intervals (CI). Moreover, we computed the pooled odds ratios and CIs for each SNP and assessed their significance of association with BC risk using P values in four genetic models including allelic (wild type (W) vs. minor (M)), homo-zygote (WW vs. MM), dominant (WW+WM vs. MM) and recessive (WW vs. WM+MM). Chi-square based Q statistic test and I2 statistics were applied for assessment of the heterogeneity between the selected studies. Based on the value of I2, the random-effects (DerSimonian and Laird‘s method) or fixed effects model were applied. The results were demonstrated as forest and funnel plots, which show the association of BRCA1 genetic polymorphisms with BC and the possible existence of publication bias in the meta-analysis, respectively (Tab. 1).
1. Association between the individual study characteristics and BRCA1 polymorphisms. 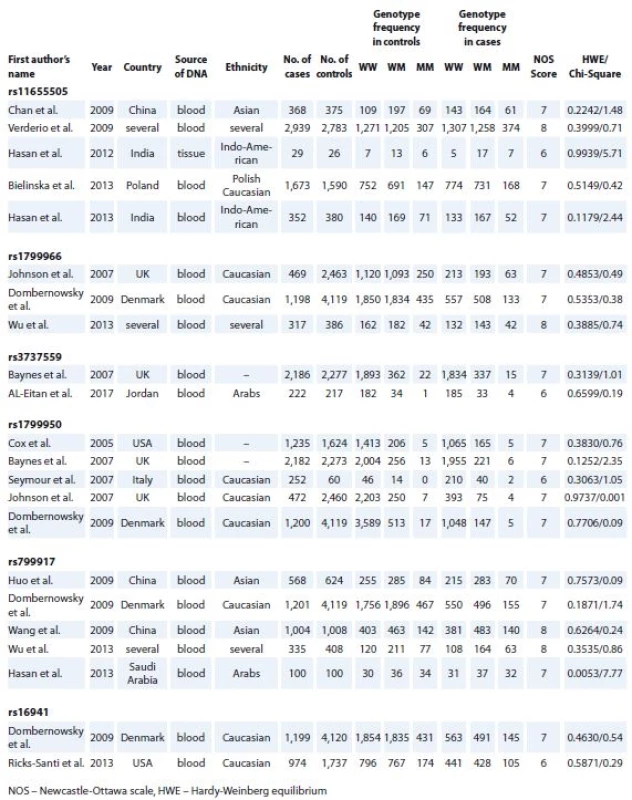
Results
Features of studies included in the meta-analysis
After initial screening of relevant publications, 95 full text original researches were found. After exclusion of papers with insufficient genotype data and matched control groups, 16 studies remained. These studies had NOS scores of 6 to 9 and assessed rs11655505 [9,13–16], rs1799966 [4,8,17], rs3737559 [10,18], rs1799950 [8,10,17,19,20], rs799917 [4,17,21–23] or rs16941 [17,24]. Assessment of genotype distribution in control groups of all included researches showed their compliance with HWE. Tab. 1 shows the detailed information of included studies, such as first author‘s name, year of publication, ethnicity and genotype in the cases and controls in addition to the NOS score, chi-square and HWE P values.
Assessment of association between SNPs and BC risk
None of the assessed SNPs were associated with BC risk in any of allelic, homozygote, dominant or recessive models (Graph 1–4). Besides, the funnel plots were depicted to evaluate the presence of publication bias in the meta-analysis of the mentioned SNPs in 4 genetic models. The overall results of the funnel plot showed fairly symmetrical shapes implying low probability of publication bias.
1. Forest plot of the risk for BRCA1 polymorphisms in allelic model. The error bars indicate 95% CI. Solid squares represent each study in the meta-analysis. Solid diamonds represent pooled OR. CI – confi dence interval, OR – odds ratio, M-H – Mantel-Haenszel method 
2. Forest plot of the risk for BRCA1 polymorphisms in homozygote model. The error bars indicate 95% CI. Solid squares represent each study in the meta-analysis. Solid diamonds represent pooled OR. CI – confi dence interval, OR – odds ratio, M-H – Mantel-Haenszel method 
3. Forest plot of the risk for BRCA1 polymorphisms in dominant model. The error bars indicate 95% CI. Solid squares represent each study in the meta-analysis. Solid diamonds represent pooled OR. CI – confi dence interval, OR – odds ratio, M-H – Mantel-Haenszel method 
4. Forest plot of the risk for BRCA1 polymorphisms in recessive model. The error bars indicate 95% CI. Solid squares represent each study in the meta-analysis. Solid diamonds represent pooled OR. CI – confi dence interval, OR – odds ratio, M-H – Mantel-Haenszel method. 
Discussion
Up to now, several BC predisposition factors including SNPs have been identified with either single-locus or epistatic effects which might be applied for BC risk assessment. Numerous strategies have been suggested to enhance accuracy of risk prediction programs with the hope of inclusion of informative SNPs in population-based risk screening programs [5]. Among putative informative SNPs for such programs are SNPs located in genes with significant role in BC. BRCA1 as the most significant genetic risk factor for BC is a tumor suppressor gene involved in various cellular processes, such as maintenance of X chromosome inactivation as well as the DNA damage response [25]. Other genes implicated in the repair of DNA double strand breaks are also associated with BC risk [4]. Previous studies have assessed the association of BRCA1 SNPs with BC risk in distinct ethnic groups. In the present meta-analysis, we evaluated associations between six BRCA1 SNPs (rs11655505, rs1799966, rs3737559, rs1799950, rs799917 and rs16941) and BC risk using the pooled data of these publications. None of these SNPs resides in a known functional domain of BRCA1. Moreover, online tools for evaluation of the functional importance of missense variants, such as Align-GVGD and SIFT, have predicted them to be benign variants according to the low degree of conservation during evolution and minor biochemical alterations between the reference and variant amino acid [26]. The rs11655505 (c.-2265C>T) is located in the promoter region of BRCA1 gene and enhances promoter activity [15]. The CT genotype of this SNP has been associated with decreased expression of BRCA1 [16]. The rs1799966 (c.4837A>G (p.Ser1613Gly)) is a missense variant classified as a benign variant by expert panel. The rs3737559 (c.4357+117G>A) is an intronic variant located in IVS13. The rs1799950 (c.1067A>G (p.Gln356Arg)) and the rs799917 (c.2612C>G (p.Pro871 Arg)) are other missense variants classified as benign by expert panel. The rs16941 (c.3113A>G (p.Glu1038Gly)) is a missense variant with conflicting interpretations of pathogenicity.
We demonstrated lack of association between 6 SNPs within BRCA1 gene and BC risk. The heterogeneity analysis showed the I2 values very small for rs1799966, rs799917 and rs16941 in all the genetic models, implying lack heterogeneity in these SNPs with BC risk. In addition, I2 values for rs3737559 were zero in allelic and dominant models. However, for the other two SNPs (rs11655505 and rs1799950), I2 values show the inconsistency between studies included in the meta-analysis. In addition to such heterogeneity, there were some other limitations in our study, including lack of ethnicity-based analysis and exclusion of studies written in other languages. In addition, we did not assess BC patients based on the molecular subtypes or environmental factors. However, the stringent quality check of included studies enhanced the reliability of our results.
Our study did not exclude the possible contribution of other SNPs within this gene in BC nor substantial contribution of multiple variants within this gene in conferring BC risk. Evidence supporting the second possibility originated from a genome-wide association study in BC patients, which show-ed that single influence of most of these risk alleles were imperceptibly small. However, the risk conferred by multiple variants was significant suggesting consideration of a risk score through integration of a properly weighted calculation of all possibly functional variants within BRCA1 and other genes [8].
Conclusion
The assessed SNPs within BRCA1 gene are not associated with BC risk. Future studies with larger sample sizes in different ethnic populations and in distinct molecular subtypes of BC are needed to define the association of other SNPs in BRCA1 gene with BC risk.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Soudeh Ghafouri-Fard, MD, PhD
Department of Medical Genetics Shahid Beheshti University of Medical Sciences
Bldg No.2 SBUMS Arabi Ave, Daneshjoo Blvd, Velenjak Tehran, Iran
e-mail: s.ghafourifard@sbmu.ac.ir
Submitted: 21. 5. 2018
Accepted: 27. 6 2018
Sources
1. Seifi-Alan M, Shamsi R, Ghafouri-Fard S et al. Expression analysis of two cancer-testis genes, FBXO39 and TDRD4, in breast cancer tissues and cell lines. Asian Pac J Cancer Prev 2014; 14 (11): 6625–6629.
2. Khorshidi HR, Taheri M, Noroozi R et al. ANRIL genetic variants in Iranian breast cancer patients. Cell J 2017; 19 (Suppl 1): 72–78. doi: 10.22074/cellj.2017.4496.
3. Khorshidi HR, Taheri M, Noroozi R et al. Investigation of the association of HOTAIR single nucleotide polymorphisms and risk of breast cancer in an Iranian population. Iran J Cancer Prev 2017; 10 (5): e7498.
4. Wu HC, Delgado-Cruzata L, Machella N et al. DNA double-strand break repair genotype and phenotype and breast cancer risk within sisters from the New York site of the Breast Cancer Family Registry (BCFR). Cancer Cause Control 2013; 24 (12): 2157–2168. doi: 10.1007/s10552-013-0292-z.
5. Sapkota Y. Germline DNA variations in breast cancer predisposition and prognosis: a systematic review of the literature. Cytogenet Genome Res 2014; 144 (2): 77–91. doi: 10.1159/000369045.
6. Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 2000; 408 (6811): 429–432. doi: 10.1038/35044000.
7. Easton DF. How many more breast cancer predisposition genes are there? Breast Cancer Res 1999; 1 (1): 14–17.
8. Johnson N, Fletcher O, Palles C et al. Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Hum Mol Genet 2007; 16 (9): 1051–1057. doi: 10.1093/hmg/ddm050.
9. Hasan TN, Grace BL, Shafi G et al. rs11655505 (c.-2265 C/T) variant in BRCA1 promoter is not associated with breast cancer risk in south India. Br J Med Med Res 2013; 3 (1): 153–161. doi: 10.9734/BJMMR/2013/2082.
10. Baynes C, Healey CS, Pooley KA et al. Common variants in the ATM, BRCA1, BRCA2, CHEK2 and TP53 cancer susceptibility genes are unlikely to increase breast cancer risk. Breast Cancer Res 2007; 9 (2): R27. doi: 10.1186/bcr1669.
11. Cao JJ, Luo CL, Yan R et al. rs15869 at miRNA binding site in BRCA2 is associated with breast cancer susceptibility. Med Oncol 2016; 33 (12): 135. doi: 10.1007/s12032-016-0849-2.
12. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010; 25 (9): 603–605. doi: 10.1007/s10654-010-9491-z.
13. Bielinska B, Gaj P, Kluska A et al. Association of the BRCA1 promoter polymorphism rs11655505 with the risk of familial breast and/or ovarian cancer. Fam Cancer 2013; 12 (4): 691–698. doi: 10.1007/s10689-013-9647-6.
14. Chan KY, Liu W, Long JR et al. Functional polymorphisms in the BRCA1 promoter influence transcription and are associated with decreased risk for breast cancer in Chinese women. J Med Genet 2009; 46 (1): 32–39. doi: 10.1136/jmg.2007.057174.
15. Verderio P, Pizzamiglio S, Southey MC et al. A BRCA1 promoter variant (rs11655505) and breast cancer risk. J Med Genet 2010; 47 (4): 268–270. doi: 10.1136/jmg.2009.073544.
16. Hasan TN, Grace BL, Shafi G et al. Association of BRCA1 promoter methylation with rs11655505 (c.2265C>T) variants and decreased gene expression in sporadic breast cancer. Clin Transl Oncol 2013; 15 (7): 555–562. doi: 10.1007/s12094-012-0968-y.
17. Dombernowsky SL, Weischer M, Freiberg JJ et al. Missense polymorphisms in BRCA1 and BRCA2 and risk of breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev 2009; 18 (8): 2339–2342. doi: 10.1158/1055-9965.EPI-09-0447.
18. Al-Eitan LN, Jamous RI, Khasawneh RH. Candidate gene analysis of breast cancer in the Jordanian population of Arab descent: a case-control study. Cancer Invest 2017; 35 (4): 256–270. doi: 10.1080/07357907.2017.1289217.
19. Cox DG, Kraft P, Hankinson SE et al. Haplotype analysis of common variants in the BRCA1 gene and risk of sporadic breast cancer. Breast Cancer Res 2005; 7 (2): R171–R175. doi: 10.1186/bcr973.
20. Seymour IJ, Casadei S, Zampiga V et al. Disease family history and modification of breast cancer risk in common BRCA2 variants. Oncol Rep 2008; 19 (3): 783–786.
21. Wang Z, Xu Y, Tang J et al. A polymorphism in Werner syndrome gene is associated with breast cancer susceptibility in Chinese women. Breast Cancer Res Treat 2009; 118 (1): 169–175. doi: 10.1007/s10549-009-0327-z.
22. Huo X, Lu C, Huang X et al. Polymorphisms in BRCA1, BRCA1-interacting genes and susceptibility of breast cancer in Chinese women. J Cancer Res Clin Oncol 2009; 135 (11): 1569–1575. doi: 10.1007/s00432-009-0604-6.
23. Hasan TN, Shafi G, Syed NA et al. Lack of association of BRCA1 and BRCA2 variants with breast cancer in an ethnic population of Saudi Arabia, an emerging high-risk area. Asian Pac J Cancer Prev 2013; 14 (10): 5671–5674.
24. Ricks-Santi LJ, Nie J, Marian C et al. BRCA1 polymorphisms and breast cancer epidemiology in the Western New York exposures and breast cancer (WEB) study. Genet Epidemiol 2013; 37 (5): 504–511. doi: 10.1002/gepi.21730.
25. Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene 2002; 21 (58): 8981–8993. doi: 10.1038/sj.onc.1206176.
26. Cox DG, Simard J, Sinnett D et al. Common variants of the BRCA1 wild-type allele modify the risk of breast cancer in BRCA1 mutation carriers. Hum Mol Genet 2011; 20 (23): 4732–4747. doi: 10.1093/hmg/ddr388.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2018 Issue 5-
All articles in this issue
- Pregnancy and Ovarian Stimulation in the Patients with Breast Cancer
- The Significance of BRAFV600E Mutation in Thyroid Cancer in Terms of Novel Targeted Therapies – Overview of Current Knowledge and Studies
- Psychoneuroimmunology of Cancer – Recent Findings and Perspectives
- Detection of EGFR Mutations in Circulating Tumor DNA (ctDNA) Retrieved from Plasma – Interlaboratory Quality Assessment in the Czech Republic
- Assessment of Incidence and Character of Neurologic/ Neuropsychiatric Complications in a Group of Patients with Malignant Melanoma Treated with Adjuvant High Dose Interferon
- Pituitary Metastasis in a Patient with Pulmonary Adenocarcinoma Presenting with a Disturbance of Consciousness
- Solid Pseudopapillary Tumor of the Pancreas – Rare Neoplastic Disease in 20-Year-Old Woman
- Afatinib in the Treatment of Advanced Non-Small Cell Lung Cancer with Rare EGFR (in exon 18-T179X) Mutation – a Case Report
- Meta-Analysis of BRCA1 Polymorphisms and Breast Cancer Susceptibility
- Tumor-to-Tumor Metastasis – a Unique Case of Clear Cell Renal Cell Carcinoma Harboring Metastasis of Adenocarcinoma of Unknown Origin
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Solid Pseudopapillary Tumor of the Pancreas – Rare Neoplastic Disease in 20-Year-Old Woman
- Psychoneuroimmunology of Cancer – Recent Findings and Perspectives
- The Significance of BRAFV600E Mutation in Thyroid Cancer in Terms of Novel Targeted Therapies – Overview of Current Knowledge and Studies
- Pregnancy and Ovarian Stimulation in the Patients with Breast Cancer
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

