-
Medical journals
- Career
Twenty Years of BRCA1 and BRCA2 Molecular Analysis at MMCI – Current Developments for the Classification of Variants
Authors: Eva Machackova 1; Kathleen Claes 2; Miroslava Mikova 1; Jana Házová 1; Eva Hrabincová Sťahlová 1; Petra Vasickova 1; Martin Trbusek 3; Marie Navrátilová 1; Marek Svoboda 1; Lenka Foretová 1
Authors‘ workplace: Department of Cancer Epidemiology and Genetics, Masaryk Memorial Cancer Institute, Brno 1; Center for Medical Genetics, Ghent University Hospital, Ghent, Belgium 2; Department of Internal Medicine Haematology and Oncology, University Hospital Brno 3
Published in: Klin Onkol 2019; 32(Supplementum2): 51-71
Category: Original Articles
doi: https://doi.org/10.14735/amko2019S51Overview
Background: Deleterious mutations in the BRCA1 and BRCA2 genes account for a considerable proportion of dominantly inherited breast and ovarian cancer susceptibility. The laboratory interpretation has always been dependent on the information available at the time of the report conclusion. The aim of this study has been to review the results from the BRCA testing at Masaryk Memorial Cancer Institute (MMCI).
Patients and methods: Patients with suspected hereditary predisposition to breast/ovarian cancer, belonging to 7,400 families, were referred by genetic counsellors for BRCA1 and BRCA2 mutation testing at the MMCI from 1999 to the beginning of 2018. Various methods have been used over 20 years of laboratory practice – starting with the Protein Truncation Test and Heteroduplex Analysis via the High Resolution Melting analysis and Sanger sequencing up to Next Generation Sequencing.
Results: BRCA1 and BRCA2 mutation screening resulted in the identification of 1,021 families with a germline high-risk BRCA1 mutation and 497 families carrying a high-risk BRCA2 mutation, representing a mutation detection rate of 20.5%. A broad spectrum of unique mutations classified as pathogenic or likely pathogenic has been detected in both genes – 124 in the BRCA1 and 123 in the BRCA2 gene. Other sequence variants (96 unique variants in the BRCA1 and 126 in the BRCA2 gene) have been revised and classified as benign or likely benign. The other 82 unique variants remain classified as of uncertain significance mainly due to a lack of information for inclusion in other groups. All the results are summarised in the tables, including the reasons for their classification.
Conclusion: The clinical classification of rare sequence variants identified in the high-risk breast cancer susceptibility genes BRCA1 and BRCA2 is essential for appropriate genetic counselling. Here we present an overview of BRCA mutation frequencies in our region and the retrospective evaluation and eventually reclassification of previously reported rare variants in light of recent findings.
Keywords:
breast cancer – ovarian cancer – BRCA1 gene – BRCA2 gene – germline mutation
Introduction
Several breast and ovarian cancer susceptibility genes have been identified to date. The most important genes in the context of genetic counselling remain the BRCA1 and BRCA2. The germline BRCA1/2 heterozygote frequency in individuals of European non-Finnish descent – BRCA1 mutation frequency 0.21% (1 : 480) and BRCA2 mutation frequency 0.31% (1 : 327); both combined 0.51% (1 : 195) [1] was calculated by examining publicly available data from the Exome Variant Server and the Exome Aggregation Consortium database. However, these calculations do not incorporate large genomic rearrangements or uncharacterised, but potentially pathogenic, missense mutations and, therefore, could be underestimates of true population frequencies of BRCA1/2 heterozygotes [1]. These figures are in agreement with those obtained in an unselected population cohort of 50,726 adults who underwent exome sequencing: 0.52% (n = 267) were found to be BRCA1/2 mutation carriers [2].
Mutations in the BRCA1 (MIM#113705) and BRCA2 (MIM#600185) account for an autosomal dominant transmission of susceptibility to breast and ovarian cancers. BRCA1/2 genes have been studied very well since their discovery in 1994 and 1995. The cumulative breast cancer risk up to the age of 80 was determined at 72% for BRCA1 and 69% for BRCA2 mutation carriers [3]. Breast cancer incidences increased rapidly in early adulthood until the age of 30 to 40 for BRCA1 and until the age of 40 to 50 for BRCA2 mutation carriers, then remained relatively constant throughout the remainder of the patient’s lifetime. The cumulative ovarian cancer risk up to the age of 80 was determined at 44% for BRCA1 and 17% for BRCA2 mutation carriers [3].
Genetic counselling has become an integral part of BRCA1/2 testing and helps patients in making informed decisions about undergoing testing. The results are used to plan optimal women’s treatment or clinical management options, which involve a combination of early cancer screening, prophylactic surgery and other risk reduction strategies.
Genetic testing may detect changes that are clearly pathogenic, clearly neutral or variants of uncertain clinical significance (VUS). Such variants present a considerable challenge to the diagnostic laboratory and the receiving clinician in terms of interpretation. Adequate classification of rare sequence changes identified in the high-risk breast cancer susceptibility genes BRCA1 and BRCA2 is essential for appropriate genetic counselling of individuals carrying these variants.
The aim of this study has been to review the results from the BRCA testing at Masaryk Memorial Cancer Institute (MMCI) during the last 20 years of diagnostic practice. This will give a necessary overview of mutation frequencies in our region and the retrospective evaluation and eventually reclassification of previously reported rare variants in light of recent findings. The reporting of novel sequence variants included a clinical interpretation based on the best data available at the time of testing. Often, as subsequent studies were done, either within the same family in our region or others reported in literature, this clinical interpretation may need to be modified or changed. When the causal status of a sequence variant is indeterminate, follow-up activities may be useful to clarify this relationship and assist risk assessment.
A system of five classes of variants based on the degree of likelihood of pathogenicity is used as recommended [4] in agreement with the ACMG (American College of Medical Genetics) [5] / ENIGMA (Evidence-based Network for the Interpretation of Germline Mutant Alleles) criteria [6]. Each class is associated with specific recommendations for clinical management of at-risk relatives.
Materials and methods
Patients, controls, and criteria for testing
The patients were referred to the MMCI in Brno for genetic counselling by physicians from various specialisations or were sent for testing by other medical geneticists from various parts of the Czech Republic between 1999 and 2018. All the tested individuals provided a signed informed consent following appropriate genetic counselling. Genetic testing was offered to high-risk individuals meeting the recommended criteria for BRCA testing. In this study, “family” was defined by the practice of giving an index patient (proband) a separate family number if he/she did not already have family members registered in our laboratory. If another relative came, this person was included in the already registered family. The results of the testing from 1999–2018 include 7,400 high-risk families referred for genetic testing in the context of a presumed genetic predisposition for breast and/or ovarian cancer. The criteria for genetic testing have been revised and edited over 20 years of genetic practice and have been published elsewhere in the framework of the guidelines for the Czech Republic [7,8].
The control “cancer-free” group was composed of healthy individuals (150) above 60 years of age without the occurrence of cancer in their personal history and without the occurrence of tumours of the breast, ovaries, prostate or colon cancer in their first-or second-degree relatives. All the control individuals provided a signed informed consent with participation for the purposes of research.
Mutation screening
Genomic DNA was isolated from blood samples with a QIAamp DNA blood purification kit (Qiagen). Initially, individuals from approximately the first 1,000 families were analysed during 1999–2006 using the Protein Truncation Test and Heteroduplex Analysis followed by Sanger sequencing on the ALF express™ DNA sequencer (Pharmacia) described elsewhere [8].
High-Resolution Melting (HRM) curve analysis was used to analyse individuals from 5,900 families during 2007–2017. To cover the complete coding region and splice sites of BRCA1 and BRCA2, 89 polymerase chain reaction (PCR) amplicons (200–540 bp) were amplified. LCGreen® Plus was used as the intercalating dye for HRM analysis performed on a 96-well LightScanner™ (Idaho Technology Inc.). Later (starting in 2012), the analysis of several highly polymorphic amplicons was transferred to dHPLC (Denaturing High-Performance Liquid Chromatography; Transgenomic Wave system 4500) for better resolution of polymorphic regions. Sanger sequencing was performed on 3130 Genetic Analyser (Applied Biosystems) from 2007 with a BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems).
In addition, we evaluated the frequency of large genomic rearrangements in the BRCA1 gene with Multiplex Ligation-dependent Probe Amplification (MLPA) (MRC-Holland). More details about our pilot study have been reported elsewhere [9]. MLPA for the BRCA1 gene (SALSA MLPA Probemix P002; and confirmation Probemix P087; MRC Holland) has been the standard test since 2005. Focused on BRCA2, after testing 1,000 high-risk individuals without the detection of any exon-spanning deletion/duplication, MLPA for the BRCA2 gene has been removed from the standard protocol. MLPA for BRCA2 (SALSA MLPA Probemix P045; MRC Holland) was re-introduced as a standard test in 2015. Fragment analysis was performed on a 3130 Genetic Analyser (Applied Biosystems); free MLPA analysis software, Coffalyser.net was used for data analysis (MRC-Holland) together with a visual inspection of the fragment analysis profile.
Next generation sequencing methods were introduced in our laboratory in 2014 and several hundred individuals from 500 unrelated families referred for hereditary breast and/or ovarian cancer predisposition were tested up to mid-2018. Next generation sequencing was performed on a MiSeq system (Illumina).
We started with a commercially available targeted enrichment TruSight Cancer panel (Illumina) including 94 cancer predisposition genes along with the BRCA1/2 genes. All the procedures were performed according to the manufacturers’ instructions – Trusight-rapid-capture-sample-prep protocol (Illumina). The first experiences with the TruSight Cancer panel have been published elsewhere [10]. Two hundred families were analysed with the TruSight cancer panel.
From 2016 onwards we have used the NimbleGen SeqCap EZ Choice (Roche) to create a sequencing library with a multi-gene panel called CZECANCA (CZEch CAncer paNel for Clinical Application) according to NimbleGen SeqCap EZ Library SR User’s Guide [11]. Three hundred families were analysed with the CZECANCA cancer panel.
A FinalistDX integrated bioinformatics computing system (Institute of Applied Biotechnologies) with a Linux operating system (Ubuntu) was used for data processing and analysis of MiSeq FASTQ files. FinalistDX software allows fast and comprehensive bioinformatics analysis from raw FASTQ files to quality control, alignment to the reference genome (bam, bai), coverage analysis, variants calling (VCF files) and recently also copy number variations (CNV) analysis with detailed reports in a variety of formats (tsv, xls, pdf). The alternative bioinformatics analysis, which was used in parallel, has been described elsewhere [11].
Nomenclature and variant classification
All sequence variants have been named and are referred to in the text according to the nomenclature used by the Human Genome Variation Society recommendation guidelines [12], using the A of the ATG-translation initiation codon as nucleotide +1 [13]. Detected sequence alterations are described at the coding DNA reference sequence (cDNA) level according to the BRCA1 most common human transcript (NM_007294.3) with the traditional numbering of exons 1–24 without the presence of exon 4, and according to the BRCA2 (NM_000059.3) reference sequences.
The variants were assessed in Alamut® Visual software (Interactive Biosoftware) and other public databases (BIC, LOVD, UMD, ClinVar) to determine whether they were known in other populations. All variants were evaluated regarding pathogenicity following the recommended terminology for classification [4–6]:
- Class 5: pathogenic (probability of being pathogenic > 0.99);
- Class 4: likely pathogenic (probability of being pathogenic 0.95–0.99);
- Class 3: uncertain significance (probability of being pathogenic 0.05–0.949);
- Class 2: likely benign (probability of being pathogenic 0.001–0.049);
- Class 1: benign (probability of being pathogenic < 0.001).
Prediction of putative splice site variants and mRNA (cDNA) analysis
All putative splice site variants were tested using the Splice Site Prediction Programs for their potential to alter splicing. Several predictive programs were used as the NNSplice [14] or NetGene2 [15]. Later, Alamut® Visual (Interactive Biosoftware) predictions on ribonucleic acid (RNA) splicing, allowing the assessment of their potential impact on splice junctions and visualization of cryptic or de novo splice sites, was used. Splicing prediction tools included in Alamut are represented by NNSplice [14]; GeneSplicer [16]; MaxEntScan [17]; SpliceSiteFinder-like [18]. Subsequently, messenger RNA (mRNA) / cDNA analysis was performed to verify ’in silico’ predictions in cases of previously not characterised putative splice variants as described elsewhere [8,19,20].
In silico analyses for missense and in-frame indel variants
Several prediction software were used including Grantham Variation (GV) and Grantham Deviation (GD) scores, later Align-GVGD [21], which combines the biophysical characteristics (side-chain composition, polarity and volume) of amino acids and protein multiple sequence alignments. We also applied PRIORS V2.0. The BRCA1 and BRCA2 Prior Probabilities database combines Prior Probabilities of pathogenicity from missense substitution severity and spiceogenity [22].
Alamut® Visual software (Interactive Biosoftware) has been used since 2015, which integrates several missense variant pathogenicity prediction tools and algorithms such as SIFT, PolyPhen2, Align-GVGD or MutationTaster; as well as the splicing prediction tools mentioned above.
For in-frame deletions/insertions we used PROVEAN (Protein Variation Effect Analyser) software [23], which predicts whether an amino acid substitution or indel will have an impact on the biological function of a protein [24].
More recently, the free interactive database VarSome has been used as well, created by Saphetor SA for the human genomics annotation tool [25].
Definition of deleterious mutations
Sequence variants were categorised on the basis of their predicted effect on the mRNA and amino acid level and defined as deleterious mutations according to the ACMG/ENIGMA criteria [5,6]:
- Frameshift and nonsense variants in both genes, with the exception of BRCA2 variants leading to a stop codon 3’of codon 3326 as BRCA2 c.9976A>T; p.Lys3326* (rs11571833) has been found to be of clinically low significance, associated with only very slightly increased risk of breast cancer (ORw 1.28; 95% CI 1.17–1.40, P = 5.9 × 10−6) [26]. Therefore, other variants leading to a stop codon 3’ of codon 3326 are considered as class 2 variants.
- Variants occurring in the consensus splice acceptor or donor sequence sites, either within 2 bp of exon-intron junctions, when they are experimentally demonstrated to result in abnormal mRNA transcript and found to produce only transcript (s) carrying a premature termination codon, or an in-frame deletion disrupting the expression of one or more known clinically important residues.
- Missense variants that have been conclusively demonstrated, on the basis of data from linkage analysis of high-risk families, functional assays or biochemical evidence, to have a deleterious effect on known functional domains.
- Copy number deletion/duplication variant that removes/duplicates one or more exons spanning a known clinically important functional or is proven by laboratory studies to result in a frameshift alteration predicted to disrupt the expression of one or more known clinically important functional residues.
Results
BRCA1 findings
In total, there were 1,021 families confirmed to carry a high-risk BRCA1 mutation (13.8% of the 7,400 families analysed). The majority of clinically deleterious mutations detected in the BRCA1 gene were protein-truncating mutations (731 families with frameshift or nonsense mutations), followed by missense mutations located in the RING domain and less frequently by BRCT (BRCA1 C-terminal) domains (118 families), large intragenic rearrangements (102 families) and splice site mutations (70 families).
An overview of the BRCA1 frameshift and nonsense mutations detected in Czech high-risk families is provided in Tab. 1. We identified 731 families with frameshift or nonsense mutations in the BRCA1 gene, which accounted for 79 different unique deleterious mutations. The most common mutations are c.5266dup (p.Gln1756Profs*74) detected in 329 families; c.3700_3704del (p.Val1234Glnfs*8), detected in 114 families; c.1687C>T (p.Gln563*), detected in 36 families; c.68_69del (p.Glu23Valfs*17), detected in 24 families.
1. BRCA1 nonsense and frame-shift mutations detected in Czech patients. 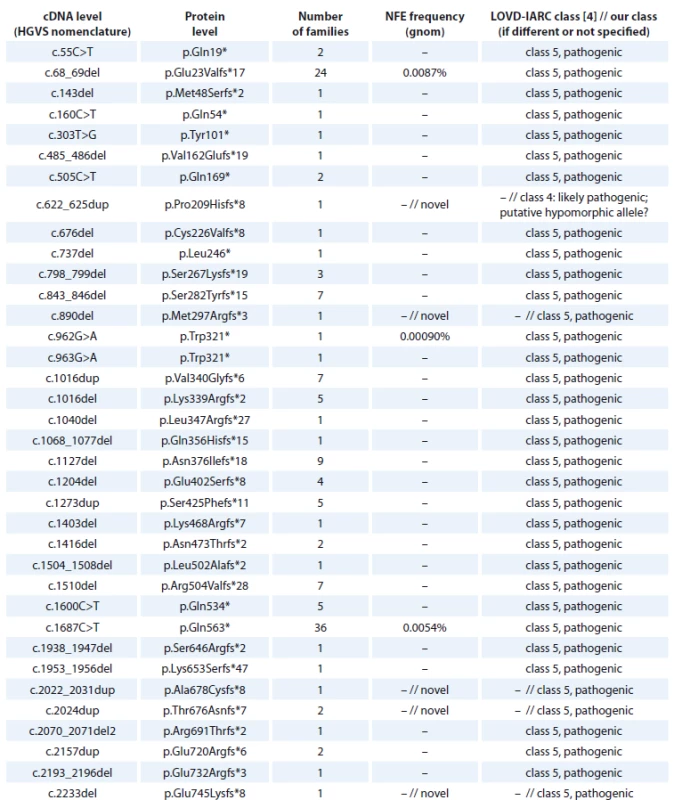
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, NFE – non-Finnish European, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, NFE – non-Finnish European, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, NFE – non-Finnish European, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer The BRCA1 splice site mutations detected in high-risk Czech families are summarised in Tab. 2. They concern 71 families, representing 16 different deleterious splice site mutations classified as pathogenic (class 5) or likely pathogenic (class 4). One splice site alteration c.4096+3A>T leading to 2 in-frame transcripts at the cDNA level (exon 11 skipping and Δ3309nt 3’end of exon 11) and previously classified as pathogenic by [27] was reclassified as a variant of VUS, based on segregation data and the finding of a healthy 58-year-old homozygous woman for c.4096+3A>G variant in a consanguineous Danish family with several cases of breast/ovarian cancer [28]. However, in the Danish population BRCA1 c.4096+3A>G is now considered as likely benign [28]. The most common splice site mutation was c.213-12A>G (detected in 33 families), activating a cryptic splice site and causing a frameshift [29].
2. BRCA1 splice site alterations detected in Czech patients. 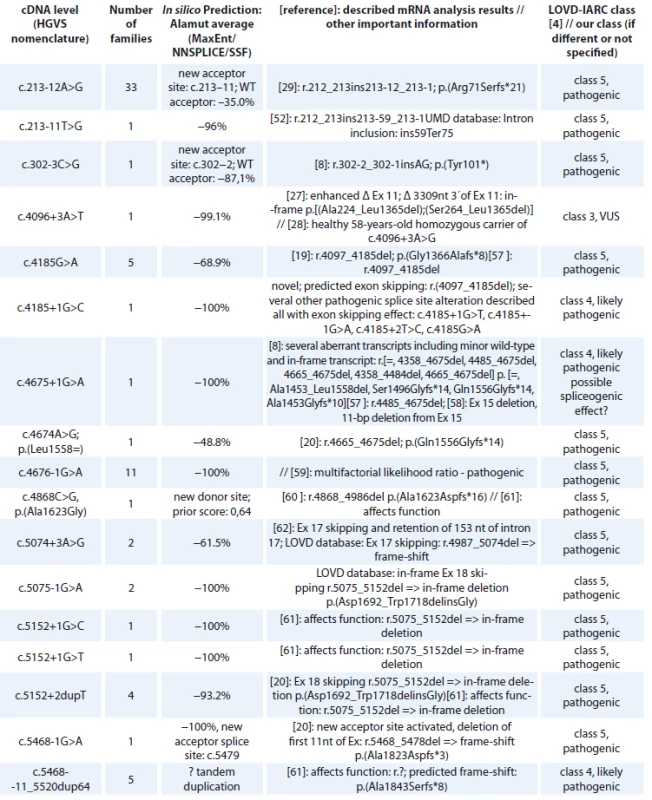
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, mRNA – messenger RNA, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, WT – wild type, VUS – variants of uncertain clinical signifi cance All BRCA1 pathogenic missense mutations are shown in Tab. 3. In 118 families, 13 different unique missense alterations, classified as pathogenic (class 5) or likely pathogenic (class 4) were detected. The most common missense mutations were in the RING domain affecting the Zn2+ ligand residues of C3HC4 finger domain – p.Cys61Gly (detected in 69 families) and p.Cys39Arg (detected in 21 families).
3. BRCA1 missense variants detected in Czech patients classifi ed as pathogenic (class-5) and likely pathogenic (class-4) 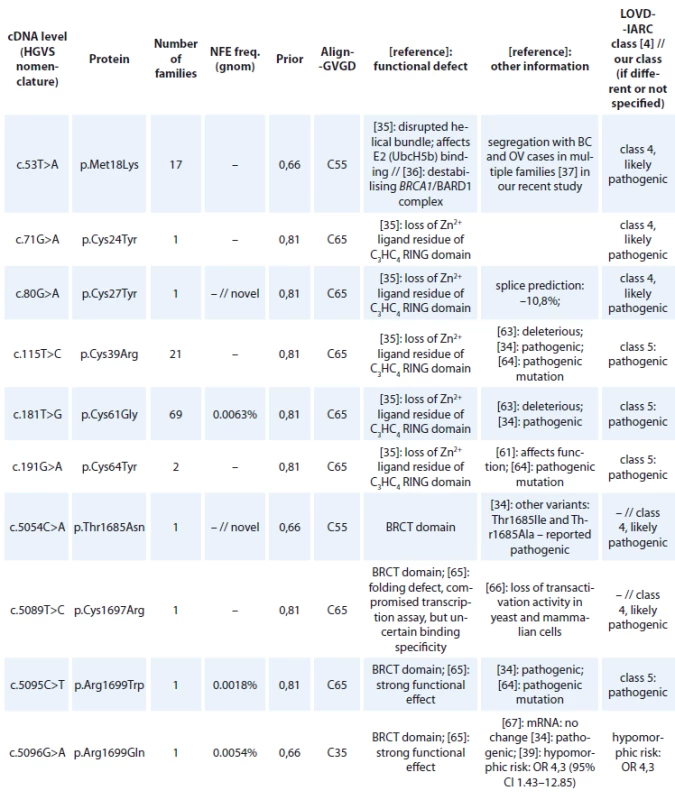
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, GVGD – Grantham Variation and Grantham Deviation, NFE – non-Finnish European, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, BRCT – BRCA1 C-terminal, OR – odds ratio, VUS – variants of uncertain clinical signifi cance 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, GVGD – Grantham Variation and Grantham Deviation, NFE – non-Finnish European, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, BRCT – BRCA1 C-terminal, OR – odds ratio, VUS – variants of uncertain clinical signifi cance An overview of all large intragenic rearrangements detected in the BRCA1 gene have been provided in Tab. 4. There were 102 families with large intragenic rearrangements, representing 16 different unique mutations classified as pathogenic (class 5) or likely pathogenic (class 4). The most common large intragenic rearrangements were deletions spanning exons 5_14 (c.135-485_4485-913del), detected in 36 families and spanning exons 21_22 (c.5278-492_5406+1290delins236), detected in 21 families.
4. BRCA1 large intragenic rearrangements detected in Czech patients. 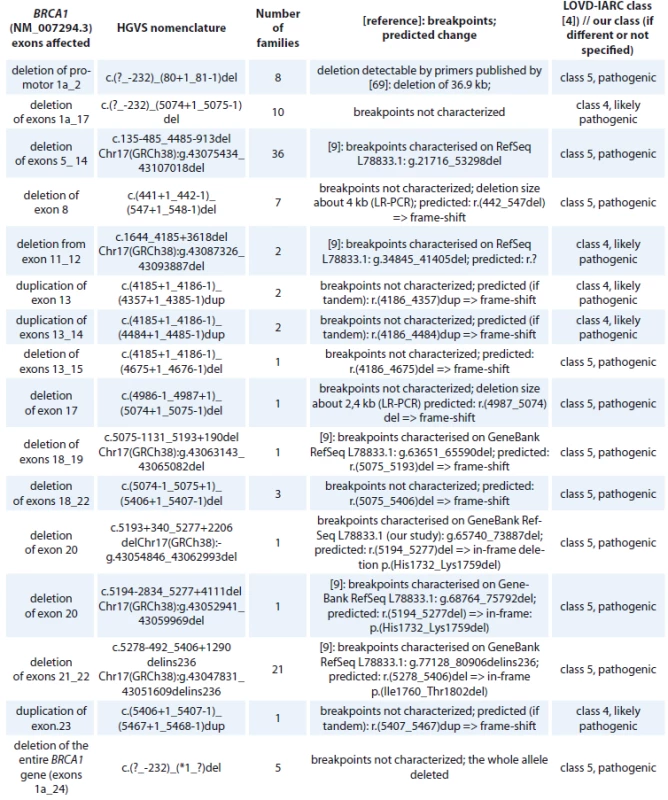
HGVS – Human Genome Variation Society, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, LR-PCR – long-range polymerase chain reaction A summary of BRCA1 in-frame deletions/insertions detected in Czech high-risk families is provided in the supplementary Tab. 5. All of them are considered as VUS (class 3).
The remaining BRCA1 missense alterations classified as VUS (class 3), likely benign (class 2) and benign (class 1) are summarised in the supplementary Tab. 6, including the reasons for their classification. Silent variants were not included if, on the basis of the prediction the chance of affecting splicing was very low. A summary of BRCA1 intronic variants targeted mainly up to position +/−50 to exon is provided in the supplementary Tab. 7.
BRCA2 findings
In total, there were 497 families confirmed to carry high-risk BRCA2 mutation (6.7% of the 7,400 families analysed). Most clinically deleterious mutations detected in the BRCA2 gene were protein-truncating mutations (404 families with deleterious frameshift or nonsense mutation), followed by splice site mutations (45 families), missense mutations with the exception of the mutation in the initiation codon located in the DBD (DNA/DSS1 binding) domain (44 families) and large intragenic rearrangements turned out to be very rare (4 families).
A summary of the frameshift and nonsense mutations detected in Czech high-risk families in BRCA2 is provided in Tab. 8. In total, 404 families were identified with a truncating mutation, representing 95 different deleterious mutations. The most common mutations were: c.8537_8538del (p.Glu2846Glyfs*22), detected in 61 families; c.7913_7917del (p.Phe2638*), detected in 37 families; c.2808_2811del (p.Ala938Profs*21), detected in 26 families. Two variants leading to a stop codon 3’ of codon 3326 were very frequent – p.Lys3326* and c.10095delC insGAATTATATCT (p.Ser3366Asnfs*4).
Tab. 8. BRCA2 nonsense and frame-shift mutations detected in Czech patients 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, NC – not counted, NFE – non-Finnish European, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, mRNA – messenger RNA 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, NC – not counted, NFE – non-Finnish European, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, mRNA – messenger RNA An overview of the BRCA2 splice site mutations detected in high-risk Czech families is shown in Tab. 9. There were 46 families carrying a splice site alteration, representing 9 different deleterious splice site mutations classified as pathogenic (class 5) or likely pathogenic (class 4). One splice site alteration, c.9501+3A>T, previously described as pathogenic and causing partial exon skipping (the aberrant transcript represented only 13% of the wild type transcript [30]) was reclassified as a variant of VUS. The most common splice site mutation was c.475G>A, a substitution affecting the last nucleotide of exon 5 for which RNA analysis has demonstrated that it causes abnormal splicing and results in a frameshift and a truncated protein [8], was detected in 10 families.
Tab. 9. BRCA2 splice site alterations detected in Czech patients. 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, mRNA – messenger RNA, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, VUS – variants of uncertain clinical signifi cance The deleterious BRCA2 missense mutations detected in high-risk Czech families can be found in Tab. 10. There were 44 families carrying a deleterious missense mutation, representing 15 different mutations classified as pathogenic (class 5) or likely pathogenic (class 4). With the exception of one mutation in the initiation codon, all deleterious missense mutations were located in the DBD domain. The most common missense mutation, classified as likely pathogenic was c.9371A>T (p.Asn3124Ile), detected in 16 families.
Tab. 10. BRCA2 missense variants detected in Czech patients classifi ed as pathogenic (class 5) and likely pathogenic (class 4). 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, NFE – non-Finnish European, mRNA – messenger RNA, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, HDR – homology directed repair, mouse ES – embryonic stem cells 
cDNA – coding DNA reference sequence, HGVS – Human Genome Variation Society, NFE – non-Finnish European, mRNA – messenger RNA, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, HDR – homology directed repair, mouse ES – embryonic stem cells Of the total study cohort, only about 3,000 families were analysed for large intragenic rearrangements by MLPA (or later CNV analysis). Only 4 families were found to carry a large intragenic rearrangement in BRCA2 (Tab. 11). In one of the complex BRCA2 rearrangements we have not been able to reveal the exact character of the complex changes (Fig. 1). Because of the low frequency of BRCA2 large intragenic rearrangements, we did not perform a retrospective analysis of all previously unsolved cases.
Tab. 11. BRCA2 large intragenic rearrangements detected in Czech patients. 
HGVS – Human Genome Variation Society, LOVD – Leiden Open Variation Database, IARC – International Agency for Research on Cancer, CNV – copy number variations, MLPA – Multiplex Ligation-dependent Probe Amplifi cation 1. Results of Multiplex Ligation-dependent Probe Amplifi cation analysis using the Coff alyser software – family-96 with complex BRCA2 rearrangements (identical profi le detected in mother and in her son). Duplication of exon 21 and 3´end of exon 24 to exon 27 combined with deletion aff ecting the coding sequence of exons 22 and 5´end of exon 24: c.8886_9195del310 – confi rmed by polymerase chain reaction analysis (amplifi ed with primers for exon 21.Forward and exon 24.Reverse) and Sanger sequencing. BRCA2 Probemix P045 (MRC Holland): probe 08267-L23772 of exon 24 (9455-9454 reverse GAAACGACAAAT-CCTATTAGGTCC ) corresponds to the systematic position c.9227-9228. 
The remaining BRCA2 missense alterations, classified as VUS (class 3), likely benign (class 2) and benign (class 1) are summarised in the supplementary Tab. 12, including the reasons for their classification. Silent variants were not included if, on the basis of the prediction the chance of affecting splicing was very low. A summary of the BRCA2 intronic variants is provided in the supplementary Tab. 13.
During these 20 years the majority of variants, which were originally reported as of VUS, could be reclassified as likely benign or benign. As soon as information confirming the neutrality of any missense or intronic variant was available, we ceased to register their frequency and to report them. The frequency of neutral variants was not monitored because homozygotes of frequent variants were not detectable by screening with Heteroduplex nor HRM analysis and therefore, their frequency would be underestimated.
Discussion
Deleterious mutations in the BRCA1 and BRCA2 genes account for a considerable proportion of dominantly inherited breast and ovarian cancer susceptibility and have received wide acceptance in diagnostic testing and prevention. The classification of sequence variants into high-risk or low-risk categories is both challenging and critical for clarification of the causative status. Only class 5 (pathogenic) and class 4 (likely pathogenic) can be used for guidance of clinical management. However, this classification system does not allow distinguishing between highly penetrant and hypomorphic alleles (like BRCA1 c.5096G>A; p.Arg1699Gln) [31].
BRCA1 and BRCA2 mutation screening in our laboratory resulted in the identification of 1021 families with deleterious BRCA1 mutations and 497 families with a BRCA2 mutation, resulting in a mutation detection ratio of 20.5%. A broad spectrum of different deleterious mutations was found in both genes – 124 unique mutations in BRCA1 and 123 unique mutations in BRCA2. The most frequent were small frameshift and nonsense mutations – 174 unique mutations (79 in BRCA1 and 95 in BRCA2) scattered in all regions of the coding sequences.
A missense mutation that leads to a non-conservative substitution of an evolutionarily conserved amino acid is more likely to be causative than a missense mutation that leads to a conservative substitution or alters an amino acid that is not evolutionarily conserved [5]. The PRIORS probability tool seems to have the strongest predictive value in agreement with already known deleterious missense mutations. The extent to which a sequence variation is considered causative of disease may be influenced by multiple parameters such as family history, segregation of the variant with affected relatives in a family, nature, and position of the amino acid substitution, evolutionary conservation of the affected residue, co-occurrence with a deleterious mutation, epidemiological and case/control studies, functional in vitro studies or knock-out animal models [5]. A novel variant of VUS with neutral predictions in less conserved regions that are unlikely to affect splicing were always mentioned in the report but interpreted as a variant suspected to be of low clinical significance.
For BRCA1 only missense variants located in the highly conserved RING finger domain at the N-terminal region (amino acids 1–109) and in the transcriptional activation domain with two BRCT repeats (amino acids 1640–1729 and 1760–1821) at the C-terminal region are considered high-risk deleterious mutations [32], (LOVD database). In the BRCA2 gene only some missense variants located in the most conserved C-terminal DNA binding DBD domain (amino acids 2460–3170) have been confirmed to be high-risk deleterious mutations [33,34], (LOVD database).
Besides the five deleterious missense mutations at the strongly conserved C3HC4 Zn2+ ligand cysteine residues of the RING domain (Cys24, Cys27, Cys39, Cys61, Cys64), only p.Met18Lys, disrupting the helical bundle of the RING domain, affecting E2 (UbcH5b) binding and destabilising the heterodimerisation of BRCA1/BARD1 complex, was considered as a likely pathogenic variant [35–37]. BRCA1 p.Met18Lys is a Czech founder mutation detected in 17 unrelated Czech families, with multiple breast and/or ovarian cancer patients, but is rare in other populations (ClinVar, LOVD databases). More frequently p.Met18Thr has been reported, which is also classified as likely pathogenic [38] even with milder Align-GVGD predictions (C45 vs. C55 for p.Met18Lys).
Several previously classified deleterious missense mutations have been found in the BRCT domains of the BRCA1 gene: p.Arg1699Trp, p.Ser1715Arg, classified as definitively pathogenic (class 5); p.Cys1697Arg, p.Ala1708Val classified as likely pathogenic (class 4) on the basis of reported functional tests and strong ’in silico’ predictions (Tab. 3). Two novel variants, p.Thr1685Asn, p.Cys1787Tyr were classified as likely pathogenic (class 4), because they are altering highly conserved residues, p.Thr1685 and p.Cys1787 with the same strong ’in silico’ predictions as for both previously reported definitively pathogenic variants (p.Thr1685Ile, p.Thr1685Ala and p.Cys1787Ser, respectively) [34,38]. However, the penetrance of some missense variants may be lower, as a hypomorphic effect was shown for BRCA1 p.Arg1699Gln, demonstrating ambiguous functional deficiency across multiple assays and calculated to be associated with reduced penetrance with estimated cumulative risks to age 70 of breast or ovarian cancer of 24% [39,31].
Besides BRCA2 c.3G>A (p.Met1?) disrupting the translation initiation codon and classified as likely pathogenic (class 4) using a multifactorial analysis approach [40], several known deleterious missense mutations were detected in the conserved DBD domain of the BRCA2 gene (Tab. 10): p.Trp2626Cys, p.Ile2627Phe, p.Asp2723His, p.Asp2723Gly, p.Arg3052Trp were classified as definitively pathogenic (class 5; LOVD, Enigma Rules); p.Gly2596Glu, p.His2623Arg, p.Lys2630Gln, p.Ser2670 Leu, p.Arg2784Trp, p.Glu3002Lys, p.Gly 3076Arg, p.Asn3124Ile were classified as likely pathogenic (class 4) on the basis of reported functional tests and strong ’in silico’ predictions (Tab. 10). The novel BRCA2 p.Glu2663Gly was classified as likely pathogenic (class 4) because it alters a highly conserved residue, p.Glu2663, with the same strong ’in silico’ predictions as for previously reported definitively pathogenic variants altering p.Glu2663Val [34]. As for BRCA1, some of the BRCA2 missense alterations might exert a hypomorphic effect. BRCA2 p.Tyr3035Ser is associated with only a moderate risk of breast cancer – (OR 2.52; P = 0.04), similar to CHEK2 inactivating mutations [39]. In our family-1338 the p.Tyr3035Ser variant was detected in a woman also carrying the BRCA2 nonsense mutation p.Ser1882* ’in trans’. This patient was diagnosed with bilateral breast cancer at the ages of 36 and 42 without symptoms of Fanconi anaemia.
Some variants were found in the nuclear localisation signals of BRCA1 (amino acids 503–508; 606–615) [41]: p.Arg 504His; p.Lys608del; or in the nuclear export sequences (amino acids 22–39 and amino acids 81–99) [32]: p.Pro25Leu; p.Lys38Arg – all of them classified as VUS (class 3). No variant was found in the nuclear localisation signals of BRCA2 (NLS1 spanning amino acids 3263–3269; NLS2 spanning amino acids 3311–3317 and putative NLS3 spanning residues 3381-3385, which was found irrelevant) [42]. Thus, the most carboxy-terminal deleterious mutation in BRCA2 was c.9463_9467delinsGAATGATC p. (Phe3155Glufs*2) removing both essential NLSs.
Some variants were found which affected the numerous phosphorylation sites in the BRCA1 gene: p.Ser616del, Ser1542Cys and in BRCA2 p.Pro3292Leu classified as VUS (class 3). BRCA1 p.Ser1542Cys has been shown to eliminate ATM kinase binding, which is necessary for the phosphorylation of BRCA1 in response to double-stranded breaks induced by γ irradiation [43]. BRCA2 p.Pro3292Leu altered the sequence for CDK2 binding for Ser3291, and completely abolished kinase binding. It was shown previously that phosphorylation of Ser3291 by CDKs blocks the interaction between BRCA2 and RAD51, serving as a molecular switch for the regulation of recombination activity, which suggests potential significance by negatively affecting the interaction between BRCA2 and RAD51 [43].
All VUS variants summarised in our study may not have been detected in patients analysed with Heteroduplex Analysis and Protein Truncation Test at the beginning and later with High Resolution Melting analysis when the analysis was terminated after the detection of pathogenic mutation. Therefore, the presence of some VUS in our analysed group may be underestimated. Only patients without detected pathogenic mutations or with a double-sided risk were completely analysed even if the high-risk mutation (class 5, definitively pathogenic) was detected. In two families with double-sided risk of breast and ovarian cancers a case was found with double BRCA1 and BRCA2 risk mutation – (family-340: BRCA1 p.Met18Lys together with BRCA2 p.Arg3052Trp – in a woman diagnosed with breast cancer at the age of 41 and with Fallopian tube cancer at age of 52) and (family-5597: BRCA1 p.Met18Lys together with splice BRCA2 c.7007G>A – in woman diagnosed with triple negative breast cancer at the age of 40).
The case report about one of our families with a double-sided risk of breast cancer and three affected children diag-nosed with Fanconi anaemia carrying bi-allelic FANCD1/BRCA2 mutations: c.[ (658_659del); (9366_9367del) ] has been described elsewhere [44]. The breast cancer genes, BRCA1 and BRCA2, are both essential for viability, therefore at least one of these bi-allelic mutations in BRCA2 in patients with Fanconi anaemia-D1 subtype should have a hypomorphic nature. BRCA2 c.658_659del is described repeatedly in association with eight cases of Fanconi anaemia-D1 patients (LOVD database: Variants associated with Fanconi anaemia) whereas c.9366_9367del is submitted only once there for association with FA (our case; [44]). Therefore, the strongest candidate for BRCA2 hypomorphic mutation is BRCA2 c.658_659del (previously 886delGT by BIC nomenclature) in exon 8 described in association with Fanconi anaemia and medulloblastoma in several cases [44–46]. Several naturally occurring alternative transcripts in BRCA2 were described [47] which might bypass the lethality of deleterious mutation by residual alternative splicing in some regions [48], however no natural alternative transcript interfering with exon 8 of BRCA2 was described.
The first hypomorphic missense mutations with reduced penetrance were recently described with moderately increased risks of breast cancer among Europeans [39]. A similar effect is obvious for the spliceogenic variant that retains the ability to produce in-frame isoforms or residual full-length transcript [49–51], which has been repeatedly found as Fanconi anaemia-D1 associated BRCA2 mutations. A possible reduced risk of spliceogenic and hypomorphic variants should be taken into consideration for clinical follow-up. Possible spliceogenic splice site alterations in the BRCA2 gene detected in our study are marked in Tab. 9. Residual full-length transcript was detected in BRCA2 c.476-2A>G and c.8755-1G>A classified as pathogenic [52], whereas in c.9501+3A>T the full-length transcript was predominant and therefore the role in breast cancer risk is questionable and variant c.9501+3A>T was reclassified as VUS [49]. Several BRCA2 splice variants retain the ability to produce partially in-frame transcripts, which might have a residual function: c.7007G>A and c.8486A>G.
Spliceogenic variants are more frequent in BRCA2, but naturally occurring alternative splicing at the BRCA1 might influence the significance of mutations in particular regions as well. According to several reports, the relative expression levels of the most abundant alternative transcripts Δ (9,10), Δ11q and Δ (9,10,11q) are tissue-specific, cell-cycle regulated and markedly altered in tumour samples [53]. A few years ago a case was described of a woman identified with a bi-allelic mutation in BRCA1 c.[594-2A>C; 2681_2682delAA] with-out symptoms of Fanconi anaemia, which led to the later revelation that c.594-2A>C was not a high-risk mutation [54]. The spliceogenic effect of c.594-2A>C was confirmed to upregulate viable Δ (9,10) in-frame isoform and a previously described pathogenic mutation specified as causing exon 10 skipping (a truncating alteration) was reclassified as a variant of VUS [54,55]. A naturally occurring isoform Δ (9,10) might be the cause of survival in a Fanconi anaemia-like case with bi-allelic BRCA1 mutations c.594_597del (localised in exon 10) and p.Arg1699Trp [56]. Therefore we classified our novel frameshift mutation located in exon 10 of the BRCA1 gene: c.622_625dup as likely pathogenic (class 4), because of the possibility to bypass this truncating mutation by a naturally occurring isoform Δ (9,10) and its putative hypomorphic effect.
The comprehensive description of BRCA1 alternative splicing is highly relevant for diagnosis, in particular when assessing the impact of BRCA1 germline variants on splicing. Recently in a consanguineous Danish family with several cases of breast/ovarian cancer a 58-year-old healthy homozygous carrier of the BRCA1 c.4096+3A>G was identified which led to the reclassification of this splice site mutation to a ’variant of uncertain significance’ or even to a ’likely benign variant’ [28]. BRCA1 c.4096+3A>G has been shown to enhance the abundance of the naturally occurring isoforms: skipping of exon 11 and lacking 3309 nucleotides from exon 11 (Δ11q): c.[671_4096del,787_4096del] p.[Ala224_Leu1365del,Ser264_Leu1365del], which was previously assumed to affect function [27]. Currently, there is evidence that in-frame (naturally occurring) alternative transcripts may rescue gene functionality. If the cells are viable to overcome the loss of large segments of the coding sequences in the central part of the BRCA1 gene in naturally occurring isoforms Δ (9,10), Δ11q and Δ (9,10,11q), it is unlikely that there could be any clinically significant missense mutation in this region. A potentially spliceogenic variant in the BRCA1 gene c.4675+1G>A was confirmed to have residual full-length transcript [8] (Tab. 2). With the exception of one novel mutation all BRCA splice site mutations were characterised at the mRNA level (Tab. 2, 9). Novel BRCA1 c.4185+1G>C was classified as likely pathogenic (class 4) on the basis of the prediction to cause exon skipping and confirmed exon skipping for several other variants detected in this splice site: c.4185+1G>T, c.4185+1G>A, c.4185+2T>C, c.4185G>A (Tab. 2).
The use of MLPA to detect large-scale rearrangements is now a standard component of BRCA1 and BRCA2 gene testing in the clinical setting even if it is currently also widely used CNV analysis with next generation sequencing (NGS) data available. Genomic rearrangements accounted for 10.4% of all BRCA mutations detected in our study – 102 in BRCA1, 16 unique (Tab. 4), and only 4 in BRCA2, 4 unique (Tab. 11). Several breakpoints in BRCA1 rearrangements were characterised [8] and a range of several deletions was confirmed by long-range PCR (Tab. 4). Genomic rearrangements are probably frequent in the BRCA1 gene because of the extremely high density of intronic Alu repeats and the presence of a duplicated promoter region containing a BRCA1 pseudogene that could provide hotspots for unequal homologous recombination [9]. However, genomic rearrangements in the BRCA2 gene are very rare in our region. BRCA2 deletion of exons 22 to 24 was detected by MLPA. The deletion of BRCA2 exon 18 was originally detected by CNV analysis from NGS data and confirmed on two DNA samples by MLPA analysis. Suspected duplication of BRCA2 exons 22–27 come originally from the Children’s Clinic after comparative genomic hybridisation analysis beyond more 3’ distant region and was confirmed by MLPA. In one of the complex BRCA2 rearrangements we have not been able to reveal the exact character of the changes (Fig. 1). MLPA analysis revealed a duplication of exon 21; deletion of exons 22 and 23 followed by duplication of exons 24 to exon 27. The deleted sequence was confirmed by PCR amplification and Sanger sequencing as c.8886_9195del310. The same MLPA profile (and CNV profile) was detected in two individuals – in a mother and in her son. Probably all detected BRCA2 rearrangements are novel, not reported in public databases from other populations.
We may discuss whether MLPA will remain the standard technology to detect CNVs. Has it ever evaluated whether the data generated by the NGS panel are robust enough to evaluate CNVs?
During the 20 years of BRCA analysis a number of variants, which were originally reported as of VUS, was gradually reclassified as likely benign or benign (supplementary Tab. 6, 7, 12, 13). As soon as information confirming the neutrality of any missense or intronic variant was available, we ceased to register their frequency and stopped reporting them. Reports indicate that common polymorphisms have not been included in the report and clinicians may not be aware of what common polymorphisms are in our population. Our clinicians were always informed when a variant was reclassified as pathogenic, but they were not always aware of the fact of neutrality. The laboratory’s interpretation has always been dependent on the information available at the time of the conclusion of the report. Thus, we decided to publish all detected variants, even those already confirmed as neutral. This can serve clinicians in our region to review data in the patient records.
Some variants of VUS could not be classified unequivocally recently. Several new potentially significant missense variants have been detected for further analysis. The limited number of individual variants and lack of experimental validation lead to inconclusive interpretations but data sharing can help to speed up clarification of significance for some of them. Functional studies of potentially significant variants and population-level data with accurate phenotyping will improve variant classification and reduce uncertainties in future. By understanding more about the VUS interpretation, clinicians can help navigate medical decision-making using the best available information and become comfortable with accepting the fact that many DNA results cannot be interpreted with the tools and data available today.
Thanks to the technical laboratory staff at the MMCI involved in laboratory analysis: Hana Pavlu, Jitka Kuklova, Veronika Kosinova, Zuzana Juraskova and Marcela Macku.
Thanks to other genetic counselling centres in the Czech Republic participating in the recruitment of patients examined in the MMCI: Institute of Biology and Medical Genetics of the 2nd Faculty of Medicine and Faculty Hospital Motol in Prague; Department of Medical Genetics, Thomayer Hospital in Prague; Department of Medical Genetics, Faculty Hospital Hradec Kralove; Department of Medical Genetics, Faculty Hospital Olomouc; Department of Medical Genetics, Faculty Hospital Ostrava; Department of Medical Genetics, Faculty Hospital Brno; Department of Clinical Genetics, Hospital Ceske Budejovice, Department of Clinical Genetics, Masaryk Hospital Usti nad Labem; and other genetic counsellors from different regions of the Czech Republic.
This work was supported by the Czech Ministry of Health MH CZ – DRO (MMCI, 00209805) and by grants NV15-28830A, NV15-27695A.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
RNDr. Eva Macháčková, Ph.D.
Department of Cancer
Epidemiology and Genetics
Masaryk Memorial Cancer Institute
Zluty kopec 7
656 53 Brno
e-mail: emachack@mou.cz
Submitted: 27. 2. 2019
Accepted: 18. 4. 2019
Sources
1. Maxwell KN, Domchek SM, Nathanson KL et al. Population frequency of germline BRCA1/2 mutations. J Clin Oncol 2016; 34 (34): 4183–4185. doi: 10.1200/JCO.2016.67.0554.
2. Manickam K, Buchanan AH, Schwartz MLB et al. Exome sequencing–based screening for BRCA1/2 expected pathogenic variants among adult biobank participants. JAMA Netw Open 2018; 1 (5): e182140. doi: 10.1001/jamanetworkopen.2018.2140.
3. Kuchenbaecker KB, Hopper JL, Barnes DR et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017; 317 (23): 2402–2416. doi: 10.1001/jama.2017.7112.
4. Plon SE, Eccles DM, Easton D et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008; 29 (11): 1282–1291. doi: 10.1002/humu.20880.
5. Richards CS, Bale S, Bellissimo DB et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med 2008; 10 (4): 294–300. doi: 10.1097/GIM.0b013e31816b5cae.
6. Enigma consortium.org. Evidence-based Network for the Interpretation of Germline Mutant Alleles: ENIGMA criteria. [online]. Available from: https: //enigmaconsortium.org/library/general-documents/enigma-classification-criteria.
7. Foretova L, Machackova E, Palacova M et al. Recommended extension of indication criteria for genetic testing of BRCA1 and BRCA2 mutations in hereditary breast and ovarian cancer syndrome. Klin Onkol 2016; 29 (Suppl 1): 9–13. doi: 10.14735/amko2016S9.
8. Machackova E, Foretova L, Lukesova M et al. Spectrum and characterisation of BRCA1 and BRCA2 deleterious mutations in high-risk Czech patients with breast and/or ovarian cancer. BMC Cancer 2008; 8 : 140. doi: 10.1186/1471-2407-8-140.
9. Vasickova P, Machackova E, Lukesova M et al. High occurrence of BRCA1 intragenic rearrangements in hereditary breast and ovarian cancer syndrome in the Czech Republic. BMC Med Genet 2007; 8 : 32. doi: 10.1186/1471-2350-8-32.
10. Machackova E, Hazova J, Stahlova Hrabincova E at el. Retrospective NGS study in high-risk hereditary cancer patients at Masaryk Memorial Cancer Institute. Klin Onkol 2016; 29 (Suppl 1): 35–45. doi: 10.14735/amko2016S35.
11. Soukupova J, Zemankova P, Lhotova K et al. Validation of CZECANCA (CZEch CAncer paNel for Clinical Application) for targeted NGS-based analysis of hereditary cancer syndromes. PLoS One 2018; 13 (4): e0195761. doi: 10.1371/journal.pone.0195761.
12. HGVS.org. Human Genome Variation Society. [online]. Available from: http: //varnomen.hgvs.org/.
13. den Dunnen JT, Dalgleish R, Maglott DR et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 2016; 37 (6): 564–569. doi: 10.1002/humu.22981.
14. NNSplice. BDGP Splice Site Prediction by Neural Network, National Human Genome Research Institute. [online]. Available from: http: //www.fruitfly.org/seq_tools/splice.html.
15. NetGene2. Technical University of Denmark, DTU Bioinformatics. [online]. Available from: http: //www.cbs.dtu.dk/services/NetGene2/.
16. GeneSplicer. University of Maryland, CBCB: Centre for Bioinformatics and Computational Biology. [online]. Available from: http: //www.cbcb.umd.edu/software/GeneSplicer/gene_spl.shtml.
17. MaxEntScan. Massachusetts Institute of Technology, Burge Laboratory web server. [online]. Available from: http: //genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html.
18. SpliceSiteFinder-like. Aix-Marseille University, Bioinformatics and Genetics Team, Human Spicing Finder. [online]. Available from: http: //www.umd.be/HSF3/HSF.shtml.
19. Claes K, Poppe B, Machackova E et al. Differentiating pathogenic mutations from polymorphic alterations in the splice sites of BRCA1 and BRCA2. Genes Chromosomes Cancer 2003; 37 (3): 314–320. doi: 10.1002/gcc.10221.
20. Baert A, Machackova E, Coene I et al. Thorough in silico and in vitro cDNA analysis of 21 putative BRCA1 and BRCA2 splice variants and a complex tandem duplication in BRCA2 allowing the identification of activated cryptic splice donor sites in BRCA2 exon 11. Hum Mutat 2018; 39 (4): 515–526. doi: 10.1002/humu.23390.
21. Align-GVGD. Huntsman Cancer Institute University of Utah. [online]. Available from: http: //agvgd.hci.utah.edu.
22. Vallée MP, Di Sera TL, Nix DA et al. Adding in silico assessment of potential splice aberration to the integrated evaluation of BRCA gene unclassified variants. Hum Mutat 2016; 37 (7): 627–639. doi: 10.1002/humu.22973.
23. PROVEAN. Protein Variation Effect Analyser, J. Craig Venter Institute. [online]. Available from: http: //provean.jcvi.org/seq_submit.php.
24. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015; 31 (16): 2745–2747. doi: 10.1093/bioinformatics/btv195.
25. Kopanos C, Tsiolkas V, Kouris A et al. VarSome: the human genomic variant search engine. Bioinformatics 2019; 35 (11): 1978–1980. doi: 10.1093/bioinformatics/bty897.
26. Meeks HD, Song H, Michailidou K et al. BRCA2 polymorphic stop codon K3326X and the risk of breast, prostate, and ovarian cancers. J Nat Cancer Inst 2016; 108 (2): djv315. doi: 10.1093/jnci/djv315.
27. Wappenschmidt B, Becker AA, Hauke J et al. Analysis of 30 putative BRCA1 splicing mutations in hereditary breast and ovarian cancer families identifies exonic splice site mutations that escape in silico prediction. PLoS One 2012; 7 (12): e50800. doi: 10.1371/journal.pone.0050800.
28. Byrjalsen A, Steffensen AY, Hansen TO et al. Classification of the spliceogenic BRCA1 c.4096+3A>G variant as likely benign based on cosegregation data and identification of a healthy homozygous carrier. Clin Case Rep 2017; 5 (6): 876–879. doi: 10.1002/ccr3.944.
29. Hoffman JD, Hallam SE, Venne VL, Lyon E et al. Implications of a novel cryptic splice site in the BRCA1 gene. Am J Med Genet 1998; 2; 80 (2): 140–144.
30. Sanz DJ, Acedo A, Infante M et al. A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin Cancer Res 2010; 16 (6): 1957–1967. doi: 10.1158/1078-0432.CCR-09-2564.
31. Spurdle AB, Whiley PJ, Thompson B et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet 2012; 49 (8): 525–532. doi: 10.1136/jmedgenet-2012-101037.
32. Thompson ME. BRCA1 16 years later: nuclear import and export processes. FEBS J. 2010; 277 (15): 3072–3078. doi: 10.1111/j.1742-4658.2010.07733.x.
33. Guidugli L, Shimelis H, Masica DL et al. Assessment of the clinical relevance of BRCA2 missense variants by functional and computational approaches. Am J Hum Genet 2018; 102 (2): 233–248. doi: 10.1016/j.ajhg.2017.12.013.
34. Lindor MN, Guidugli L, Wang X et al. A review of a multifactorial probability based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS) Hum Mutat 2012 Jan; 33 (1): 8–21. doi: 10.1002/humu.21627.
35. Morris JR, Pangon L, Boutell C, et at. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum Mol Genet 2006; 15 (4): 599–606. doi: 10.1093/hmg/ddi476.
36. Sarkar M, Magliery TJ. Re-engineering a split-GFP reassembly screen to examine RING-domain interactions between BARD1 and BRCA1 mutants observed in cancer patients. Mol Biosyst 2008; 4 (6): 599–605. doi: 10.1039/b802481b.
37. Machackova E, Damborsky J, Valik D et al. Novel germline BRCA1 and BRCA2 mutations in breast and breast/ovarian cancer families from the Czech Republic. Hum Mutat 2001; 18 (6): 545. doi: 10.1002/humu. 1232.
38. Tavtigian SV, Deffenbaugh AM, Yin L et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 2006; 43 (4): 295–305. doi: 10.1136/jmg.2005.033878.
39. Shimelis H, Mesman RLS, Von Nicolai C et al. BRCA2 hypomorphic missense variants confer moderate risks of breast cancer. Cancer Res 2017; 77 (11): 2789–2799. doi: 10.1158/0008-5472.CAN-16-2568.
40. Thomassen M, Blanco A, Montagna M et al. Characterization of BRCA1 and BRCA2 splicing variants: a collaborative report by ENIGMA consortium members. Breast Cancer Res Treat; 132 (3): 1009–1023. doi: 10.1007/s10549-011-1674-0.
41. Chen CF, Li S, Chen Y et al. The nuclear localization sequences of the BRCA1 protein interact with the importin-alpha subunit of the nuclear transport signal receptor. J Biol Chem 1996; 271 (51): 32863–32868. doi: 10.1074/jbc.271.51.32863.
42. Spain BH, Larson CJ, Shihabuddin LS et al. Truncated BRCA2 is cytoplasmic: implications for cancer-linked mutations. Proc Natl Acad Sci USA 1999; 96 (24): 13920–13925. doi: 10.1073/pnas.96.24.13920.
43. Tram E, Savas S, Ozcelik H. Missense variants of uncertain significance (VUS) altering the phosphorylation patterns of BRCA1 and BRCA2. PLoS 2013; 8 (5): e62468. doi: 10.1371/journal.pone.0062468.
44. Svojgr K, Sumerauer D, Puchmajerova A et al. Fanconi anemia with biallelic FANCD1/BRCA2 mutations – case report of a family with three affected children. Eur J Med Genet 2016; 59 (3): 152–157. doi: 10.1016/j.ejmg.2015.11.013.
45. Reid S, Renwick A, Seal S et al. Biallelic BRCA2 mutations are associated with multiple malignancies in childhood including familial Wilms tumour. J Med Genet 2005; 42 (2): 147–51. doi: 10.1136/jmg.2004.022673.
46. Offit K, Levran O, Mullaney B et al. Shared genetic susceptibility to breast cancer, brain tumors, and Fanconi anemia. J Natl Cancer Inst 2003; 95 (20): 1548–1551. doi: 10.1093/jnci/djg072.
47. Fackenthal JD, Yoshimatsu T, Zhang B et al. Naturally occurring BRCA2 alternative mRNA splicing events in clinically relevant samples. J Med Genet 2016; 53 (8): 548–558. doi: 10.1136/jmedgenet-2015-103570.
48. Thirthagiri E, Klarmann KD, Shukla AK et al. BRCA2 minor transcript lacking exons 4-7 supports viability in mice and may account for survival of humans with a pathogenic biallelic mutation. Hum Mol Genet 2016; 25 (10): 1934–1945. doi: 10.1093/hmg/ddw066.
49. Acedo A, Hernández-Moro C, Curiel-García A et al. Functional classification of BRCA2 DNA variants by splicing assays in a large minigene with 9 exons. Hum Mutat 2015; 36 (2): 210–221. doi: 10.1002/humu.22725.
50. Fraile-Bethencourt E, Díez-Gómez B, Velásquez-Zapata V et al. Functional classification of DNA variants by hybrid minigenes: identification of 30 spliceogenic variants of BRCA2 exons 17 and 18. PLoS Genet 2017; 13 (3): e1006691. doi: 10.1371/journal.pgen.1006691.
51. Fraile-Bethencourt E, Valenzuela-Palomo A, Díez-Gómez B et al. Identification of eight spliceogenic variants in BRCA2 exon 16 by minigene assays. Front Genet 2018; 9 : 188. doi: 10.3389/fgene.2018.00188.
52. Colombo M, De Vecchi G, Caleca L et al. Comparative in vitro and in silico analyses of variants in splicing regions of BRCA1 and BRCA2 genes and characterization of novel pathogenic mutations. PLoS One 2013; 8 (2): e57173. doi: 10.1371/journal.pone.0057173.
53. Colombo M, Blok MJ, Whiley P et al. Comprehensive annotation of splice junctions supports pervasive alternative splicing at the BRCA1 locus: a report from the ENIGMA consortium. Hum Mol Genet 2014; 23 (14): 3666–3680. doi: 10.1093/hmg/ddu075.
54. Wong-Brown M, McPhillips M, Gleeson M et al. When is a mutation not a mutation: the case of the c.594-2A>C splice variant in a woman harbouring another BRCA1 mutation in trans. Hered Cancer Clin Pract 2016; 14 : 6. doi: 10.1186/s13053-015-0045-y.
55. de la Hoya M, Soukarieh O, López-Perolio I et al. Combined genetic and splicing analysis of BRCA1 c.[594-2A>C; 641A>G] highlights the relevance of naturally occurring in-frame transcripts for developing disease gene variant classification algorithms. Hum Mol Genet 2016; 25 (11): 2256–2268. doi: 10.1093/hmg/ddw094.
56. Sawyer SL, Tian L, Kähkönen M et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov 2015; 5 (2): 135–142. doi: 10.1158/2159-8290.CD-14-1156.
57. Steffensen AY, Dandanel M, Jønson L et al. Functional characterization of BRCA1 gene variants by mini-gene splicing assay. Eur J Hum Genet 2014; 22 (12): 1362–1368. doi: 10.1038/ejhg.2014.40.
58.Whiley PJ, Guidugli L, Walker LC et al. Splicing and multifactorial analysis of intronic BRCA1 and BRCA2 sequence variants identifies clinically significant splicing aberrations up to 12 nucleotides from the intron/exon boundary. Hum Mutat 2011; 32 (6): 678–687. doi: 10.1002/humu.21495.
59. Easton DF, Deffenbaugh AM, Pruss D et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet 2007; 81 (5): 873–883. doi: 10.1086/521032.
60. Walker LC, Whiley PJ, Couch FJ et al. Detection of splicing aberrations caused by BRCA1 and BRCA2 sequence variants encoding missense substitutions: implications for prediction of pathogenicity. Hum Mutat 2010; 31 (6): E1484–E1505. doi: 10.1002/humu.21267.
61. Rebbeck TR, Friebel TM, Friedman E et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat 2018; 39 (5): 593–620. doi: 10.1002/humu.23406.
62. Menéndez M, Castellsagué J, Mirete M et al. Assessing the RNA effect of 26 DNA variants in the BRCA1 and BRCA2 genes. Breat Cancer Res Treat 2012; 132 (3): 979–992. doi: 10.1007/s10549-011-1661-5.
63. Sweet K, Senter L, Pilarski R et al. Characterization of BRCA1 ring finger variants of uncertain significance. Breast Cancer Res Treat 2010; 119 (3): 737–743. doi: 10.1007/s10549-009-0438-6.
64. Thouvenot P, Ben Yamin B, Fourrière L et al. Functional assessment of genetic variants with outcomes adapted to clinical decision-making. PLoS Genet 2016; 12 (6): e1006096. doi: 10.1371/journal.pgen.1006096.
65. Lee MS, Green R, Marsillac SM et al. Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res 2010; 70 (12): 4880–4890. doi: 10.1158/0008-5472. CAN-09-4563.
66.Vallon-Christersson J, Cayanan C, Haraldsson K et al. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet 2001; 10 (4): 353–360. doi: 10.1093/hmg/10.4.353.
67. Houdayer C, Caux-Moncoutier V, Krieger S et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat 2012; 33 (8): 1228–1238. doi: 10.1002/humu.22101.
68. Tavtigian SV, Byrnes GB, Goldgar DE et al. Classification of rare missense substitutions, using risk surfaces, with genetic-and molecular-epidemiology applications. Hum Mutat 2008; 29 (11): 1342–1354. doi: 10.1002/humu.20896.
69. Preisler-Adams S, Schönbuchner I, Fiebig B et al. Gross rearrangements in BRCA1 but not BRCA2 play a notable role in predisposition to breast and ovarian cancer in high-risk families of German origin. Cancer Genet Cytogenet 2006; 168 (1): 44–49. doi: 10.1016/j.cancergencyto.2005.07.005.
70. Judkins T, Hendrickson BC, Deffenbaugh AM et al. Application of embryonic lethal or other obvious phenotypes to characterize the clinical significance of genetic variants found in trans with known deleterious mutations. Cancer Res 2005; 65 (21): 10096–10103. doi: 10.1158/0008-5472.CAN-05-1241.
71. Liu J, Pan Y, Ma B et al. “Similarity trap” in protein-protein interactions could be carcinogenic: simulations of p53 core domain complexed with 53BP1 and BRCA1 BRCT domains. Structure 2006; 14 (12): 1811–1821. doi: 10.1016/j.str.2006.10.009.
72. Théry JC, Krieger S, Gaildrat P et al. Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. Eur J Hum Genet 2011; 19 (10): 1052–1058. doi: 10.1038/ejhg.2011.100.
73. Bonnet C, Krieger S, Vezain M et al. Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet 2008; 45 (7): 438–446. doi: 10.1136/jmg.2007.056895.
74. Bonatti F, Pepe C, Tancredi M et al. RNA-based analysis of BRCA1 and BRCA2 gene alterations. Cancer Genet Cytogenet 2006; 170 (2): 93–101. doi: 10.1016/j.cancergencyto.2006.05.005.
75. Heramb C, Wangenstee T, Grindedal EM et al. BRCA1 and BRCA2 mutation spectrum – an update on mutation distribution in a large cancer genetics clinic in Norway. Hered Cancer Clin Pract 2018; 16 : 3. doi: 10.1186/s13053-017-0085-6.
76. Farrugia DJ, Agarwal MK, Pankratz VS, at al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res 2008; 68 (9): 3523–3531. doi: 10.1158/0008-5472.CAN-07-1587.
77. Biswas K, Das R, Alter BP et al. A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood 2011; 118 (9): 2430–2442. doi: 10.1182/blood-2010-12-324541.
78. Pölsler L, Fiegl H, Wimmer K et al. High prevalence of BRCA1 stop mutation c.4183C>T in the Tyrolean population: implications for genetic testing. Eur J Hum Genet 2016; 24 (2): 258–262. doi: 10.1038/ejhg.2015.108.
79. Karchin R, Agarwal M, Andrej Sali A et al. Classifying variants of undetermined significance in BRCA2 with protein likelihood ratios. Cancer Inform 2008; 6 : 203–216.
80. Guidugli L, Carreira A, Caputo SM et al. Functional assays for analysis of variants of uncertain significance in BRCA2. Hum Mutat 2014; 35 (2): 151–164. doi: 10.1002/humu.22478.
81. Mesman RL, Calléja FM, Hendriks G et al. The functional impact of variants of uncertain significance in BRCA2. Genet Med 2019; 21 (2): 293–302. doi: 10.1038/s41436-018-0052-2.
82. Biswas K, Das R, Eggington JM et al. Functional evaluation of BRCA2 variants mapping to the PALB2-binding and C-terminal DNA-binding domains using a mouse ES cell-based assay. Hum Mol Genet 2012; 21 (18): 3993–4006. doi: 10.1093/hmg/dds222.
83. Muller D, Rouleau E, Schultz I et al. An entire exon 3 germ-line rearrangement in the BRCA2 gene: pathogenic relevance of exon 3 deletion in breast cancer predisposition. BMC Med Genet 2011; 12 : 121. doi: 10.1186/1471-2350-12-121.
84. Colombo M, Lòpez-Perolio I, Meeks HD et al. The BRCA2 c.687T > A variant is not pathogenic: a model for clinical calibration of spliceogenicity. Hum Mutat 2018; 39 (5): 729–741. doi: 10.1002/humu.23411.
85. Joosse SA, Brandwijk KI, Devilee P et al. Prediction of BRCA2-association in hereditary breast carcinomas using array-CGH. Breast Cancer Res Treat 2012; 132 (2): 379–389. doi: 10.1007/s10549-010-1016-7.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2019 Issue Supplementum2-
All articles in this issue
- Recommendations for Preventive Care for Women with Rare Genetic Cause of Breast and Ovarian Cancer
- Risks of Solid Tumors in Heterozygous Carriers of Recessive Syndromes
- Karcinom prsu u nosiček mutací v genu BRCA1/2 – léčíme ho jinak? Zaměřeno na systémovou terapii u mutací v genu BRCA1/2
- Effectiveness of Neoadjuvant Therapy with Platinum-Based Agents for Patients with BRCA1 and BRCA2 Germline Mutations – A Retrospective Analysis of Breast Cancer Patients Treated at MMCI Brno
- Germline CHEK2 Gene Mutations in Hereditary Breast Cancer Predisposition – Mutation Types and their Biological and Clinical Relevance
- Contribution of Massive Parallel Sequencing to Diagnosis of Hereditary Ovarian Cancer in the Czech Republic
- Genetic Causes of Rare Pediatric Ovarian Tumors
- Gastrointestinal Polyposes and Lynch Syndrome – a Pathologist’s Perspective
- An Update on Inherited Colon Cancer and Gastrointestinal Polyposis
- BAP1 Syndrome – Predisposition to Malignant Mesothelioma, Skin and Uveal Melanoma, Renal and Other Cancers
- DICER1 Syndrome
- GAPPS – Gastric Adenocarcinoma and Proximal Polyposis of the Stomach Syndrome in 8 Families Tested at Masaryk Memorial Cancer Institute – Prevention and Prophylactic Gastrectomies
- Twenty Years of BRCA1 and BRCA2 Molecular Analysis at MMCI – Current Developments for the Classification of Variants
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Germline CHEK2 Gene Mutations in Hereditary Breast Cancer Predisposition – Mutation Types and their Biological and Clinical Relevance
- Risks of Solid Tumors in Heterozygous Carriers of Recessive Syndromes
- Recommendations for Preventive Care for Women with Rare Genetic Cause of Breast and Ovarian Cancer
- GAPPS – Gastric Adenocarcinoma and Proximal Polyposis of the Stomach Syndrome in 8 Families Tested at Masaryk Memorial Cancer Institute – Prevention and Prophylactic Gastrectomies
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

