-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
The Baculovirus Uses a Captured Host Phosphatase to Induce Enhanced Locomotory Activity in Host Caterpillars
The baculovirus is a classic example of a parasite that alters the behavior or physiology of its host so that progeny transmission is maximized. Baculoviruses do this by inducing enhanced locomotory activity (ELA) that causes the host caterpillars to climb to the upper foliage of plants. We previously reported that this behavior is not induced in silkworms that are infected with a mutant baculovirus lacking its protein tyrosine phosphatase (ptp) gene, a gene likely captured from an ancestral host. Here we show that the product of the ptp gene, PTP, associates with baculovirus ORF1629 as a virion structural protein, but surprisingly phosphatase activity associated with PTP was not required for the induction of ELA. Interestingly, the ptp knockout baculovirus showed significantly reduced infectivity of larval brain tissues. Collectively, we show that the modern baculovirus uses the host-derived phosphatase to establish adequate infection for ELA as a virion-associated structural protein rather than as an enzyme.
Published in the journal: . PLoS Pathog 8(4): e32767. doi:10.1371/journal.ppat.1002644
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002644Summary
The baculovirus is a classic example of a parasite that alters the behavior or physiology of its host so that progeny transmission is maximized. Baculoviruses do this by inducing enhanced locomotory activity (ELA) that causes the host caterpillars to climb to the upper foliage of plants. We previously reported that this behavior is not induced in silkworms that are infected with a mutant baculovirus lacking its protein tyrosine phosphatase (ptp) gene, a gene likely captured from an ancestral host. Here we show that the product of the ptp gene, PTP, associates with baculovirus ORF1629 as a virion structural protein, but surprisingly phosphatase activity associated with PTP was not required for the induction of ELA. Interestingly, the ptp knockout baculovirus showed significantly reduced infectivity of larval brain tissues. Collectively, we show that the modern baculovirus uses the host-derived phosphatase to establish adequate infection for ELA as a virion-associated structural protein rather than as an enzyme.
Introduction
Viruses and other parasites are known to usurp or alter the behavior of their hosts for their own benefit. This type of behavior modification by animal and even plant viruses is widely observed in arthropod hosts [1], [2]. One of the earliest documented examples of such behavior modification is Wipfelkrankheit or tree-top disease of caterpillars [3]. A hallmark of this disease is enhanced locomotory activity (ELA) that causes the diseased caterpillars to migrate to the upper foliage of the host plant where they die. We now know that the causative agent of Wipfelkrankheit is a large, double-stranded DNA virus in the family Baculoviridae. Baculoviruses form a large group of arthropod-specific pathogens that commonly attack lepidopteran insects [4]. The baculovirus genome is large, 80 to over 160 kbp, and generally encodes more than 100 potential genes of which more than 10% appear to be derived from an ancestral host [5]. Baculoviruses produce two types of progeny during their infection cycle: the budded virus (BV) and occlusion-derived virus (ODV). BVs are involved in spread of the virus within an infected host. ODVs on the other hand are occluded within an occlusion body (OB) that protects and transmits the ODV from insect-to-insect via oral infection [6], [7].
At a late stage of infection, baculovirus-infected lepidopteran larvae often display ELA [3], [8], [9] and climb to the top of the host plant where they die and liquefy after death. It is believed that this behavior results in the dispersal of progeny OBs over a larger surface area thus improving the chance of virus transmission to other hosts. We have previously identified a protein tyrosine phosphatase (ptp) gene of the baculovirus, Bombyx mori nucleopolyhedrovirus (BmNPV) that induces wandering-like ELA in the silkworm B. mori. This gene was identified by behavioral screening of silkworms against a library of gene knockout mutants of BmNPV. Interestingly, the BmNPV ptp gene appears to have been acquired by an ancestral BmNPV from an ancestral silkworm [8]. Unlike silkworms that are infected with wild-type BmNPV, silkworms that are infected with a ptp-deleted BmNPV do not show ELA. The protein encoded by ptp, PTP, shows dephosphorylation activity with protein and RNA as substrate [10]–[13], however, the role that PTP plays in the induction of ELA is still unknown. Recently, a knockout mutant of the baculovirus Lymantria dispar nucleopolyhedrovirus (LdMNPV) has also been shown to exhibit reduced ELA in comparison to wild-type LdMNPV in the European gypsy moth [9]. Specifically, gypsy moths infected with an ecdysteroid UDP-glucosyltransferase (egt) gene deletion mutant of LdMNPV show reduced climbing behavior. Here we surprisingly show that baculovirus PTP induces ELA as a structural protein, and not as an enzyme. Furthermore, we show that baculovirus infection of brain tissues appears to be important for the induction of ELA.
Results
BmNPV-induced ELA in larval B. mori requires PTP protein but not PTP-associated phosphatase activity
We previously reported that a ptp gene deletion mutant of BmNPV (BmPTPD) does not induce ELA in larval B. mori at a late stage of infection [8]. This suggested that baculovirus-induced ELA involves the dephosphorylation of an unknown protein or RNA target by baculovirus PTP. To test this hypothesis, we generated BmPTP-C119S (Figure 1A), a mutant virus that expressed a PTP that was nearly deficient in phosphatase activity (Supplementary Figure S1A). This mutagenesis was based on previous studies showing that mutation of cysteine 119 to serine (C119S) in the P-loop motif of the closely related PTP of Autographa californica NPV (AcMNPV) almost completely abolishes phosphatase activity [10], [12]. To our surprise BmPTP-C119S induced ELA in 5th instar B. mori in a manner similar to that induced by wild-type BmNPV (Figure 1B). This indicated that the phosphatase activity of PTP is not required for the induction of ELA.
Fig. 1. Effect of mutation of the BmNPV ptp gene on virus-induced ELA in 5th instar B. mori. 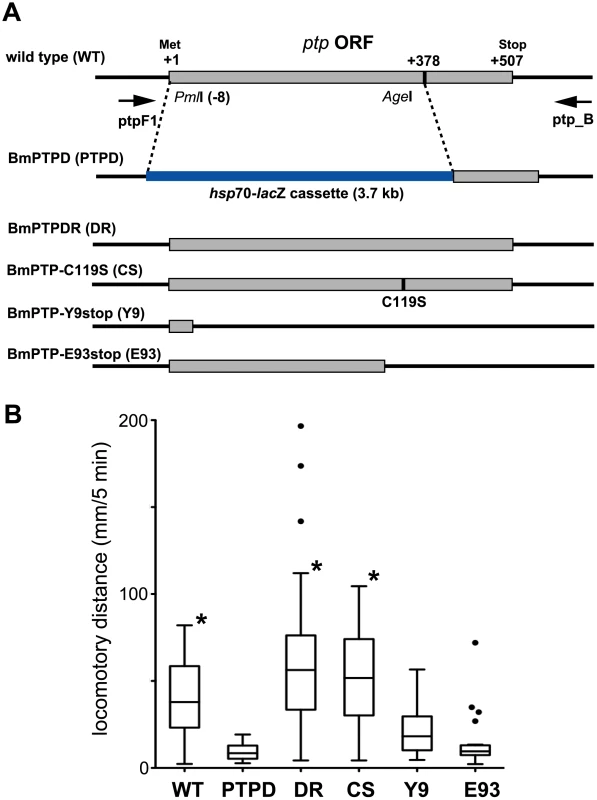
(A) Schematic representation of the ptp gene locus of wild-type (WT) and mutant BmNPVs. The locations of PCR primers (ptpF1 and ptp_B) used in the genotyping experiments are represented by the arrows. In BmPTPD (PTPD), nucleotides −8 to 377 of the ptp gene are replaced by a hsp70-lacZ gene cassette (3.7 kbp). BmPTPDR (DR) is a repair mutant in which the hsp70-lacZ gene cassette of BmPTPD was replaced with the original BmNPV sequence. BmPTP-C119S (CS) contains a point mutation within the ptp gene which results in an amino acid residue substitution (C119S) within the predicted P-loop motif that is required for phosphatase activity. BmPTP-Y9stop (Y9) and BmPTP-E93stop (E93) contain point mutations within the ptp gene which generate stop codons at Tyr-9 and Glu-93, respectively, but the RNA structures of the respective transcripts are likely to be unchanged. (B) Induction of ELA in 5th instar B. mori injected with wild-type or mutant BmNPVs. Distances traveled at 90 h p.i. are shown by box-and-whisker diagrams. The boxes represent the median and 25–75 percentile ranges of the distances traveled. The whiskers indicate the most extreme data points, which were no more than 1.5 times the interquartile range from the boxes. The dots indicate outliers predicted by Prism software. BmPTPD did not induce any ELA during the assay period. *p<0.05, Kruskal-Wallis analysis with Dunn's post test in comparison to the value obtained for BmPTPD. The abbreviations of the viruses are the same as in A. In order to determine whether the full-length PTP protein or RNAs transcribed from the ptp locus is required for the induction of ELA, we next generated BmPTP-Y9stop and BmPTP-E93stop (Figure 1A). These BmNPV mutants each carried a ptp gene with a point mutation in the coding region that generated a premature stop codon. These mutations likely had little effect on the structure of the expressed mRNAs, however, the expressed proteins were only 9 or 93 amino acid residues in length. In a manner similar to that observed with BmPTPD, BmPTP-Y9stop and BmPTP-E93stop were both unable to induce ELA in larval B. mori (Figure 1B). This indicated that the PTP protein itself is required for the induction of ELA but not mRNAs transcribed from the ptp gene.
PTP binds ORF 1629, a WASP-like capsid protein
Our mutagenesis experiments indicated that the phosphatase activity of PTP is not required for the induction of ELA, thus we next used a yeast two-hybrid (Y2H) screening system to identify proteins that likely interact with PTP. The Y2H screening identified 5 clones which potentially interact with PTP (Figure 2A). Four of the clones (#12h-3, -4, -11, and -16) contained BmNPV-derived sequences whereas one clone (#2d-2) contained a B. mori-derived sequence of unknown function. The protein encoded by clone #12h-3 showed the strongest interaction with PTP (Figure 2A). This clone contained a nearly full-length (1572 nts) open reading frame (ORF) corresponding to ORF1629 of BmNPV (Supplementary Figure S2A). ORF1629 encodes a WASP-like protein that localizes at one end of the nucleocapsid structure [14], [15]. The deduced protein encoded by the clone #12h-11 corresponded to the C-terminal 347 amino acid residues (64% of full-length) of ORF1629 and exhibited a moderately strong interaction with PTP (Supplementary Figure S2A). Further analysis in yeast revealed that (i) the C-terminal of ORF1629 is critical for interaction with PTP (Supplementary Figure S2A), and (ii) the N-terminal 90 amino acid residues of PTP do not interact with ORF1629. The inability of the N-terminal of PTP to interact with ORF1629 is consistent with our locomotion assay results showing that BmPTP-E93stop did not induce ELA (Figure 1B).
Fig. 2. PTP interacts with ORF1629 in BmNPV-infected cells. 
(A) A yeast two-hybrid screen was performed to identify interactions between BmNPV PTP and proteins in BmNPV-infected (12 h p.i.) BmN cells or in epidermal tissues from BmNPV-infected (2 d p.i.) 5th instar B. mori. This screening identified 5 PTP-interacting clones (12h-3, 12h-11, 12h-4, 12h-16, and 2d-2) by X-gal and 3-AT assays. Clones 12h-3, 12h-11, 12h-4, and 12h-16 were derived from BmNPV-infected BmN cells whereas clone 2d-2 was derived from BmNPV-infected larval B. mori. A legend showing the location of positive standards and PTP-interacting clones (streaked in triplicate) is shown to the right. (B) Interaction of PTP and ORF1629 in BmNPV-infected BmN cells. BmN cells were inoculated with BmNPV (WT) or BmPTPD-wt (D-wt) at an MOI of 5 or mock-infected (mock). BmPTPD-wt expresses FLAG-tagged PTP under an authentic ptp promoter. At 72 h p.i., the cells were harvested and immunoprecipitated with anti-FLAG antibody, and then subjected to western blot analysis (left panels) with anti-ORF1629 antibody or anti-FLAG antibody. The right “Input” panels show western blot analysis using whole cell extracts. In order to examine whether PTP interacts with ORF1629 in BmNPV-infected BmN cells, we next generated BmPTPD-wt. BmPTPD-wt is a derivative of BmPTPD that expresses FLAG-tagged PTP under an authentic ptp promoter immediately upstream of the polh gene (Supplementary Figure S3B–C). The authentic promoter of ptp was identified by 5′-RACE analyses (Supplementary Figure S3A). Immunoprecipitation experiments with anti-FLAG antibody and cell extracts from cells infected with BmPTPD-wt clearly showed that PTP interacts strongly with ORF1629 (Figure 2B), confirming the results of the Y2H experiments.
PTP is a BV structural component localized in the virion envelope
Because ORF1629 is a known structural protein, we speculated that PTP is also a structural protein that is associated with the BV envelope or capsid. Western blot analysis of BV-derived proteins that were fractionated into envelope and capsid components showed that PTP is primarily localized in the BV envelope (Figure 3A).
Fig. 3. PTP is an envelope-associated protein required for the production of normal virions. 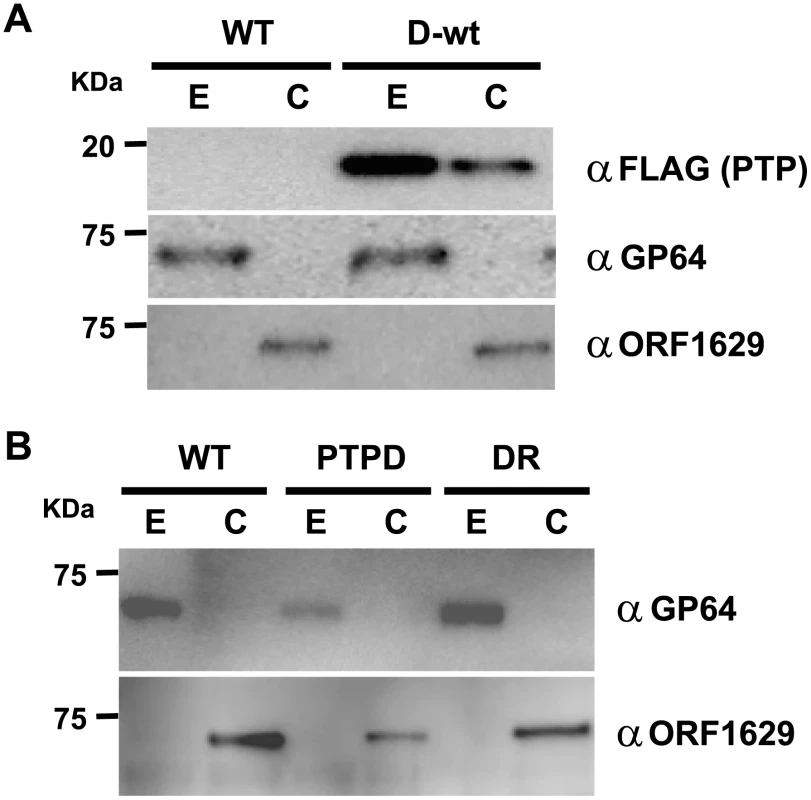
(A) Localization of PTP in the envelope and capsid fractions of budded virus. Western blot analysis of envelope (E) and capsid (C) fractions of budded virus (BV) of BmNPV or BmPTPD-wt were performed with anti-FLAG, anti-GP64 or anti-ORF1629 antibodies. (B) Localization of GP64 and ORF1629 in PTP-deficient BV. Western blot analysis of envelope (E) and capsid (C) fractions of BV of BmNPV, BmPTPD, and BmPTPDR were performed with anti-GP64 or anti-ORF1629 antibody. In order to examine whether the loss of PTP has any effects on the structural properties of the BV, we investigated the relative levels of ORF1629 and GP64 in BVs that were isolated from BmN cells infected with BmNPV, BmPTPD or BmPTPDR (a repair virus of BmPTPD) by western blot analysis. GP64 is a major envelope protein of BV that is essential for cell-to-cell infection [16]. The amounts of both GP64 and ORF1629 were reduced in the envelope and capsid, respectively, of BmPTPD BVs, in comparison to the corresponding BV fractions of BmNPV and BmPTPDR (Figure 3B). These findings indicated that disruption of ptp results in the formation of abnormal BVs with potentially reduced virus infectivity and/or replication.
Loss of PTP leads to reduced progeny production in BmN cells and silkworm larvae
The role of PTP in the productive infection of BmNPV was investigated in BmN cells and silkworm larvae at 3 and 4 days postinfection (d p.i.), respectively. These time points were chosen because the production of BV and OB of wild-type BmNPV peak at these times. In BmN cells, BmPTPD produced about 50% fewer BVs and OBs in comparison to wild-type BmNPV at 3 d p.i. (Supplementary Figure S4). The reduction in BmPTPD OB production in BmN cells was consistent with that found in Sf9 cells infected with a ptp-deleted AcMNPV [17], [18]. Similar reductions in BV and OB production were also observed in BmN cells infected with BmPTP-Y9stop - and BmPTP-E93stop (Supplementary Figure S4). In contrast, BmPTP-C119S and BmPTPDR produced wild-type levels of BV and OB in BmN cells (Supplementary Figure S4). These results indicated that the expression of full-length PTP is required for the production of wild-type levels of BV and OB in BmN cells.
We next examined if the dramatic drop in BV and OB production that was found in BmN cells also occurred in BmPTPD-infected silkworm larvae. The production of BV and OB in the hemolymph of BmPTPD-infected larvae was less than 50% of that found in wild-type BmNPV-infected larvae at 4 d p.i. (Figure 4A–B). In contrast, the production of BV and OB in larvae infected with BmPTP-C119S or BmPTPDR was similar to that found in BmNPV-infected larvae (Figure 4A–B). These in vivo findings were essentially identical to those found in vitro and suggested again that the PTP protein, but not its enzymatic activity, is essential for normal BV and ODV production. In addition, we did not observe significant differences in the median lethal dose (LD50) of BmNPV, BmPTPD, BmPTPDR, and BmPTP-C119S in 5th instar B. mori (Supplementary Table S1), suggesting that the absence of PTP does not alter the virulence of BmNPV.
Fig. 4. BmPTPD produces fewer progeny in 5th instar B. mori and shows a delay in late gene expression in BmN cells. 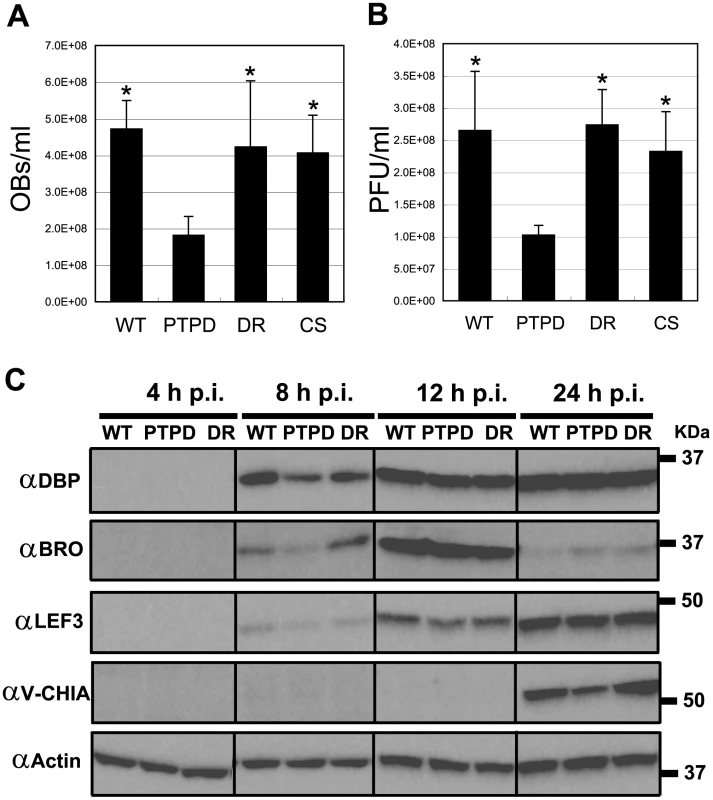
Production of OBs (A) and BVs (B) in the hemolymph of larvae infected with BmNPV, BmPTPD, BmPTPDR or BmPTP-C119S at 4 d p.i. Data shown are means ± standard deviation (SD) (N = 4). *p<0.05, one-way ANOVA with Tukey's post test in comparison to BmPTPD. (C) Western blot analysis of the expression of viral gene products in BmN cells infected with BmNPV, BmPTPD or BmPTPDR. The proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with antibodies that recognize BmNPV early-expressed (DBP, BRO, and LEF3) or late-expressed (V-CHIA) proteins or actin. Similar results were obtained in two independent experiments. Abbreviations: WT, BmNPV; PTPD, BmPTPD; DR, BmPTPDR; and CS, BmPTPD-C119S. In order to investigate why BmPTPD produced fewer progeny, the expression profiles of a series of known baculovirus early and late gene products were examined by western blot analysis. Western blot analysis clearly showed that the expression of both early (DBP, BRO, and LEF3) and late (V-CHIA) proteins was delayed in BmPTPD-infected BmN cells (Figure 4C). These results indicated that loss of PTP caused a delay in the infection cycle, a delay that presumably led to the production of fewer BVs and OBs.
In order to examine the effects of ptp deletion in larval B. mori in greater detail, we measured the expression of viral early/late (gp64) and very late (polh) class genes in 16 tissues that were isolated from BmNPV - or BmPTPD-infected larvae. qRT-PCR analyses showed that, with the exception of a few tissues (i.e., corpora allata, prothoracic glands, and hemocytes), the relative expression levels of gp64 and polh were much lower in BmPTPD-infected larvae than in BmNPV-infected larvae (Figure 5A). The expression of polh in the brain of BmPTPD-infected larvae at 4 d p.i. showed the most dramatic (67%) reduction (Figure 5B). These findings were consistent with the western blot analyses indicating that the replication cycle of BmPTPD was generally delayed but went further to show that the reduction in virus replication was most pronounced in the larval brain.
Fig. 5. Reduced expression of viral genes in tissues of larvae infected with a ptp-disrupted virus. 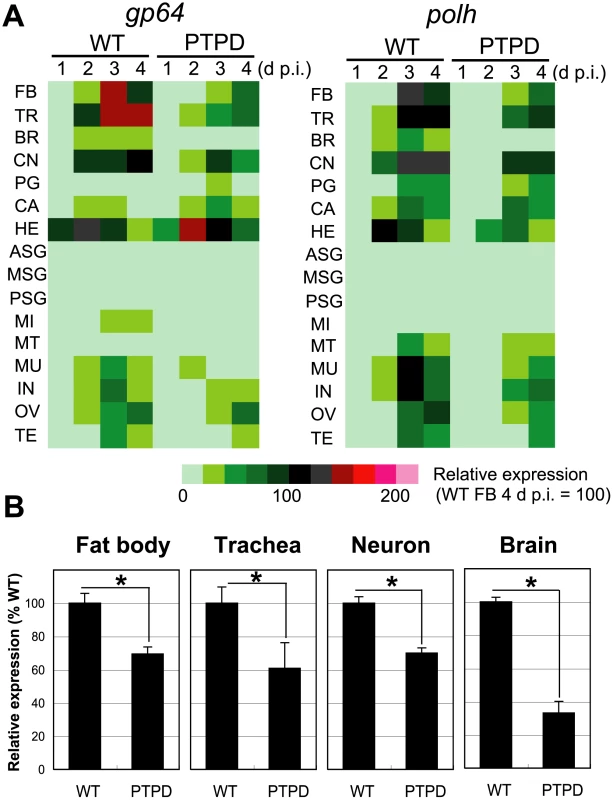
(A) Heatmaps of viral gene expression in 16 tissues of 5th instar B. mori infected with BmNPV (WT) or BmPTPD (PTPD). The tissues were dissected from virus-infected larvae at 1, 2, 3, and 4 d p.i., and the expression of the early/late and very late genes gp64 and polh, respectively, were quantified by qRT-PCR. Tissues from 5 to 30 larvae were mixed and used for the preparation of cDNAs. Abbreviations: FB, fat body; TR, trachea; BR, brain; CN, central nerve; PG, prothoracic gland; CA, corpora allata; HE, hemocyte; ASG, anterior silk gland; MSG, middle silk gland; PSG, posterior silk gland; MI, midgut; MT, Malpighian tubule; MU, muscle; IN, integument; OV, ovary; and TE, testis. (B) Expression of polh in fat body, trachea, central nerve, and brain. Tissues were dissected from four individual larvae at 4 d p.i. First strand cDNAs were generated from individual larvae and qRT-PCR was performed using primers that targeted the polh gene. Data shown are means ± SD (N = 4). *p<0.05, Student's t-test. Discussion
The manipulation of the behavior of caterpillars by baculoviruses has been known for over 100 years as Wipfelkrankheit. Recent developments in molecular biological and genomic tools have led to the identification of two baculovirus genes, ptp and egt, that are involved in altering host behavior [8], [9]. Interestingly, the baculovirus appears to have obtained both ptp and egt from an ancestral host. Hoover et al. [9] hypothesize that the egt gene product (EGT), a protein that is known to inactivate 20-hydroxyecdysone, controls the climbing behavior of NPV-infected gypsy moth larvae by hormonal regulation. On the other hand, the mechanistic action of how the ptp gene establishes behavioral control of host larvae remains elusive. In this study, we attempted to unravel this intriguing mystery by dissecting the functions of ptp/PTP in BmNPV-infected silkworms. We surprisingly found that the phosphatase activity of PTP appears not to be required for the behavioral control. In addition, we found that BmNPV induces ELA only when ptp mRNA is translated as a full-length protein, suggesting a non-enzymatic role for PTP. Our analyses revealed that PTP is a structural component of BV that is required for the production of mature BVs with full infectivity. We also found that loss of PTP dramatically reduces virus gene expression in several host tissues, especially in the brain.
Previous biochemical studies show that baculovirus PTP has the ability to remove phosphate groups from protein and RNA substrates [10]–[13]. In this study we confirmed that BmNPV PTP is also a functional phosphatase (Supplementary Figure S1A). Biologically, PTP and baculovirus LEF-4 have been predicted to play coordinated roles in 5′ cap formation of baculovirus late mRNAs [18]. A double-knockout mutant of ptp and lef-4 of AcMNPV, however, does not show defects in mRNA cap formation and replicates normally in cultured cells [18]. Thus, the overall biological significance of the phosphatase activity of baculovirus PTP is still unknown. Interestingly, the ptp gene is conserved in Group I NPVs (e.g., BmNPV) but not in Group II NPVs (e.g., LdMNPV). Group II NPVs, however, are also able to induce ELA even though they lack the ability to produce PTP. This conundrum can be explained by the presence of the egt gene, a gene that is found in both Group I and II NPVs [4]. PTP and EGT appear to induce different types of ELA. PTP is involved in wandering-like ELA that is dramatically enhanced by light and shows positive phototropism [8], whereas EGT is involved in the induction of vertical climbing behavior [9]. Thus, the baculovirus-induced “wandering” and “climbing” behaviors appear to be regulated by different viral genes but appear to work in concert in Group I NPVs to improve transmission of the virus. In addition, Group I and Group II NPVs have unique BV envelope structures (e.g., Group I NPVs use GP64 as an envelope fusion protein for host cell attachment whereas in Group II NPVs use the F protein [19]). These structural differences may lead to unique tissue tropism and modes of ELA induction by Group I and Group II NPVs. Additionally, there may be other baculovirus genes that are involved in induction of ELA but their roles in ELA may be difficult to identify if they are essential for other viral functions or if host-derived genes can partially substitute for their functions.
Modern baculoviruses have apparently captured a number of essential and non-essential ‘auxiliary’ genes from ancestral host insects by horizontal gene transfer [5]. The authentic biological function of these captured genes or their products may be maintained, modified or lost in modern baculoviruses so that they confer selective advantages. The viral fibroblast growth factor (vfgf) gene is a clear example of a captured ancestral host gene whose authentic function has been maintained during evolution [20], [21]. The protein encoded by vfgf, vFGF, transmits its signaling via a host FGF receptor that, when activated, causes the migration of hemocytes to virus-infected tissues. vFGF is thus able to usurp the host's signaling pathway in order to recruit hemocytes which, following infection, can disseminate the virus to other tissues and increase systemic infection. BmNPV ptp is another example of a captured ancestral host gene [8]. In the case of BmNPV ptp, however, the biological importance of the PTP protein appears to have changed over time from a protein with enzymatic significance to one that has structural significance for establishing infection in larval tissues that are critical for the induction of ELA. In this study, we show that PTP binds strongly to ORF1629, a baculovirus structural protein that is phosphorylated during the infection cycle [22]. We hypothesize that in modern baculoviruses, the ability of PTP to bind ORF1629 or some other target became more important because of the role it plays in increasing virus transmission. In contrast, the ability of PTP to dephosphorylate a potential protein or RNA substrate appears not to be as important or perhaps taken over by a host phosphatase or only required when the virus has to replicate under unusual conditions. Alternatively, the PTP protein may have a dual function as a structural protein in the induction of ELA and as a phosphatase enzyme perhaps during earlier stages of infection, in specific tissues, or different host developmental stages. The ptp gene is thus the first example of a host-derived gene whose product is utilized by the modern baculovirus in a completely different manner from how it was likely utilized in the ancestral host.
Wandering is a normal ELA behavior that occurs towards the end of the last larval instar that causes caterpillars to search for an appropriate location to undergo metamorphosis [23]. Wandering behavior is regulated by a combination of internal (e.g., larval size, hormones) and external (e.g., photoperiod) processes. In larval Manduca sexta the brain exerts a net inhibitory influence that prevents wandering behavior during the caterpillar feeding stage [24], [25]. At the hormonal level, wandering is induced by exposure to the 20-hydroxyecdysone which causes the brain to become excitatory during the wandering stage. We hypothesize that baculovirus infection of caterpillar brain also leads to an excitatory state leading to the induction of the wandering-like ELA that we observe in BmNPV-infected silkworms. Electrophysiological studies of the locomotory patterns in the brain and subesophageal ganglion from baculovirus-infected larvae will allow us to understand what occurs in the central nervous system during virus-induced ELA. Our current hypothesis suggests that the baculovirus plays a direct role in the induction of ELA by infecting the brain. However, other more subtle factors such as baculovirus-induced changes in host energy metabolism, signal transduction, sensitivity to light or gravity, etc. may also play roles in the induction of the various types of ELA.
In conclusion, we show here that PTP functions to induce wandering-like ELA in baculovirus-infected caterpillars as a structural protein and likely not as an enzyme. Notably, we found that virus propagation was markedly reduced in brain tissues when ptp was deleted from the BmNPV genome. These results tell an amazing story of how the modern baculovirus has evolved to use a captured host gene in a different way from how it was likely used by the ancestral host. Collectively, we conclude that PTP augments baculovirus infection of the brain and possibly other tissues that play critical roles in the induction of ELA.
Materials and Methods
Insects, cell lines, and viruses
Larval B. mori were reared as described previously [26]. BmN (BmN-4) cells were cultured at 27°C in TC-100 medium supplemented with 10% fetal bovine serum [26]. The T3 strain of BmNPV was used as the wild-type virus. The construction of BmPTPD (a ptp deletion mutant) and BmPTPDR (a repair virus of BmPTPD), have been reported previously [8] (see Figure 1A). The titers of BmNPV and mutant BmNPVs were determined by plaque assay on BmN cells [26].
Generation of BmPTP-C119S, BmPTP-Y9stop, BmPTP-E93stop, and BmPTPD-wt
BmNPV genomic DNA containing ptp and its flanking regions were cloned into pcDNA3.1(-) and used as a template to generate mutations in the ptp gene. Mutagenesis was performed by overlapping PCR [26] and confirmed by DNA sequencing. The resultant plasmids were transfected with Bsu36I-digested BmPTPD DNA (a Bsu36I restriction endonuclease site is uniquely found within the lacZ gene cassette of BmPTPD) into BmN cells using Lipofectin reagent (Invitrogen). Five days after transfection, the medium was collected and stored at 4°C until use. Three recombinant BmNPVs expressing PTP-C119S (BmPTP-C119S), PTP-Y9stop (BmPTP-Y9stop), and PTP-E93stop (BmPTP-E93stop) (Figure 1A) were isolated by the identification of plaques that did not express β-galactosidase [26]. The presence of the mutated ptp genes in these constructs was confirmed by polymerase chain reaction (PCR) using primers ptpF1 and ptp_B (Supplementary Table S2).
BmPTPD-wt, a repair mutant of BmPTPD that expresses a FLAG-tagged PTP under an authentic ptp gene promoter (inserted immediately upstream of the polh gene) was generated by a two step process. Firstly, the FLAG-tagged ptp gene driven by the authentic ptp gene promoter (identified by 5′-RACE) was amplified by PCR using BmNPV DNAs and primers ptpEPS1 and ptpEPS3 (Supplementary Table S2). The amplicon was inserted into the transfer vector pBmEPS1 [27], and the recombinant transfer plasmid was transfected with Bsu36I-digested BmNPV-abb [27] genomic DNA into BmN cells using Cellfectin reagent (Invitrogen) [28]. A recombinant BmNPV (T3-wt) expressing the FLAG-tagged PTP was plaque-purified by the identification of plaques that were OB-positive. In the second step, the authentic ptp gene of T3-wt was disrupted by transfection of T3-wt genomic DNA with a plasmid carrying a lacZ gene cassette flanked by ptp gene sequences [8] into BmN cells using Cellfectin reagent (Invitrogen). BmPTPD-wt (Supplementary Figure S3), a recombinant BmNPV expressing FLAG-tagged PTP under the authentic ptp gene promoter (but not expressing authentic PTP) was identified by the formation of plaques expressing β-galactosidase [29] and by PCR using the primer sets BmEPS_F1/BmEPS_R1 and ptpF2/ptpG2 (Supplementary Table S2). Expression of FLAG-tagged PTP by BmPTPD-wt was confirmed by western blot analysis with anti-FLAG antibody (Sigma).
Locomotion assay
Locomotion assays were performed as reported previously with minor modifications [30]. Briefly, 5th instar B. mori (24 larvae per treatment) were starved for several hours, injected with 50 µl of a viral suspension containing 1×105 PFU, and returned to the artificial diet at 27°C. Infected larvae were photographed at 3 h intervals from 84 to 132 h postinfection (h p.i.). At each 3 h interval, the 24 infected larvae (separated into 4 groups of 6 larvae) were placed in the center of a piece of paper marked with concentric circles (the radius of each circle was 5 mm greater than the previous circle, with a maximum radius of 100 mm). Photographs were taken with a digital camera at 1 min intervals until 5 min after release. The coordinates of each larva, at the midpoint of the third and fourth abdominal segments, was determined at each time point after release using ImageJ software (Rasband WS (2006) ImageJ. Bethesda, Maryland: U. S. National Institutes of Health, rsb.info.nih.gov/ij/). The distance moved during each 1 min-long interval was determined and summed up to derive total locomotory distance in 5 min. The locomotory distance of dead larvae was designated as zero.
Yeast two-hybrid screening
Yeast two-hybrid (Y2H) screening was performed using the PROQUEST two-hybrid system (Gibco BRL) as described previously [31]. The Y2H screening used a cDNA library that was generated from BmNPV-infected BmN cells as described previously [31], as well as a cDNA library that was constructed using mRNAs purified from epidermal tissues from BmNPV-infected larvae (2 d p.i.).
Western blotting and immunoprecipitation
BmN cells were infected with BmNPV, BmPTPD, or BmPTPDR at an MOI of 5 and harvested at 48 h p.i. Biochemical fractionation of the BmN cells was performed as described previously [32]. Procedures for the isolation of BVs and fractionation of BV components were reported previously [33]. SDS-PAGE and western blotting were performed using anti-FLAG antibody, anti-GP64 antibody (Santa Cruz Biotechnology), anti-ORF1629 antibody [14] (a gift from George F. Rohrmann), anti-LEF3 antibody [34], [35] (a gift from Eric B. Carstens), anti-BRO antibody [36], anti-DBP antibody [37], and anti-actin antibody (Santa Cruz Biotechnology) as described previously [20]. Immunoprecipitation experiments were performed as described previously [20].
OB and BV production in larval B. mori
Fifth instar B. mori (4 larvae per virus) were inoculated with virus and reared as described above. OBs that were released into the hemolymph, at 96 h p.i., were quantified from individual larva using a hemocytometer as described previously [26]. Hemolymph BV titer was determined by plaque assay on BmN cells [26].
Quantitative reverse transcription-PCR (qRT-PCR)
Fifth instar B. mori were inoculated and reared as described above. Total RNA was prepared using Trizol reagent (Invitrogen) from 16 tissues (brain, corpora allata, central nerve, prothoracic gland, fat body, trachea, hemocyte, testis, ovary, anterior silk gland, middle silk gland, posterior silk gland, midgut, Malpighian tubule, integument, and muscle) that were dissected from BmNPV - or BmPTPD-infected, 5th instar B. mori (5 to 30 larvae/tissue) at 1, 2, 3, and 4 d p.i. For the experiments shown in Figure 5B, total RNA was prepared using Trizol reagent from four tissues (brain, central nerve, fat body, and trachea) that were dissected from four individual 5th instar larvae. First-strand cDNAs were synthesized from 0.2 µg of total RNA, and qRT-PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems) using primers that were previously described [29]. PCR was performed using the StepOne real-time PCR system (Applied Biosystems) [29].
Statistical analysis
Statistical analysis was performed using Prism 5 software (Graphpad). One-way analyses of variance (ANOVA) was performed with post hoc Tukey's test comparing each of the treatment group means with the mean of the control group. Locomotion assay data were subjected to Kruskal-Wallis analysis with post hoc Dunn's test. Student's t-test was used to compare values obtained in the qRT-PCR experiments (Figure 5B).
Supporting Information
Zdroje
1. HurdH 2003 Manipulation of medically important insect vectors by their parasites. Annu Rev Entomol 48 141 161
2. StaffordCAWalkerGPUllmanDE 2011 Infection with a plant virus modifies vector feeding behavior. Proc Natl Acad Sci U S A 108 9350 9355
3. GoulsonD 1997 Wipfelkrankheit: manipulation of host behavior by a baculovirus. Oecologia 109 219 228
4. RohrmannGF 2011 Baculovirus Molecular Biology. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/books/NBK49500/
5. KatsumaSKawaokaSMitaKShimadaT 2008 Genome-wide survey for baculoviral host homologs using the Bombyx genome sequence. Insect Biochem Mol Biol 38 1080 1086
6. GranadosRRLawlerKA 1981 In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108 297 308
7. KeddieBAAponteGWVolkmanLE 1989 The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science 243 1728 1730
8. KamitaSGNagasakaKChuaJWShimadaTMitaK 2005 A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc Natl Acad Sci U S A 100 14121 14126
9. HooverKGroveMGardnerMHughesDPMcNeilJ 2011 A gene for an extended phenotype. Science 333 1401
10. TakagiTTaylorGSKusakabeTCharbonneauHBuratowskiS 1998 A protein tyrosine phosphatase-like protein from baculovirus has RNA 5′-triphosphatase and diphosphatase activities. Proc Natl Acad Sci U S A 95 9808 9812
11. GrossCHShumanS 1998 Characterization of a baculovirus-encoded RNA 5′-triphosphatase. J Virol 72 7057 7063
12. ShengZCharbonneauH 1993 The baculovirus Autographa californica encodes a protein tyrosine phosphatase. J Biol Chem 268 4728 4733
13. KimDWeaverRF 1993 Transcription mapping and functional analysis of the protein tyrosine/serine phosphatase (PTPase) gene of the Autographa californica nuclear polyhedrosis virus. Virology 195 587 595
14. RussellRLFunkCJRohrmannGF 1997 Association of a baculovirus-encoded protein with the capsid basal region. Virology 227 142 152
15. GoleyEDOhkawaTMancusoJWoodruffJBD'AlessioJA 2006 Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314 464 467
16. MonsmaSAOomensAGBlissardGW 1996 The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J Virol 70 4607 4616
17. LiYMillerLK 1995 Properties of a baculovirus mutant defective in the protein phosphatase gene. J Virol 69 4533 4537
18. LiYGuarinoLA 2008 Roles of LEF-4 and PTP/BVP RNA triphosphatases in processing of baculovirus late mRNAs. J Virol 82 5573 5583
19. PearsonMNRohrmannGF 2002 Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J Virol 76 5301 5304
20. KatsumaSDaimonTMitaKShimadaT 2006 Lepidopteran ortholog of Drosophila Breathless is a receptor for the baculoviral fibroblast growth factor. J Virol 80 5474 5481
21. KatsumaSHorieSShimadaT 2008 The fibroblast growth factor homolog of Bombyx mori nucleopolyhedrovirus enhances systemic virus propagation in B. mori larvae. Virus Res 137 80 85
22. VialardJERichardsonCD 1993 The 1,629-nucleotide open reading frame located downstream of the Autographa californica nuclear polyhedrosis virus polyhedrin gene encodes a nucleocapsid-associated phosphoprotein. J Virol 67 5859 5866
23. DominickOSTrumanJW 1984 The physiology of wandering behaviour in Manduca sexta. I. Temporal organization and the influence of the internal and external environments. J Exp Biol 110 35 51
24. DominickOSTrumanJW 1986 The physiology of wandering behaviour in Manduca sexta. IV. Hormonal induction of wandering behaviour from the isolated nervous system. J Exp Biol 121 133 151
25. DominickOSTrumanJW 1986 The physiology of wandering behaviour in Manduca sexta. III. Organization of wandering behaviour in the larval nervous system. J Exp Biol 121 115 132
26. NakanishiTGotoCKobayashiMKangWKSuzukiT 2010 Comparative studies of a lepidopteran baculovirus-specific protein FP25K: Development of a novel BmNPV-based vector with a modified fp25K gene. J Virol 84 5191 5200
27. KangWTristemMMaedaSCrookNEO'ReillyDR 1998 Identification and characterization of the Cydia pomonella granulovirus cathepsin and chitinase genes. J Gen Virol 79 2283 2292
28. KatsumaSShimadaTKobayashiM 2004 Characterization of the baculovirus Bombyx mori nucleopolyhedrovirus gene homologous to the mammalian FGF gene family. Virus Genes 29 211 217
29. KatsumaSShimadaT 2009 Bombyx mori nucleopolyhedrovirus ORF34 is required for efficient transcription of late and very late genes. Virology 392 230 237
30. KokushoRZhangC-XShimadaTKatsumaS 2011 Comparative analysis of budded virus infectivity of Bombyx mandarina and B. mori nucleopolyhedroviruses. Virus Genes 43 313 317
31. KangWKImaiNSuzukiMIwanagaMMatsumotoS 2003 Interaction of Bombyx mori nucleopolyhedrovirus BRO-A and host cell protein laminin. Arch Virol 148 99 113
32. JarvisDLBohlmeyerDAGarciaAJr 1991 Requirements for nuclear localization and supramolecular assembly of a baculovirus polyhedrin protein. Virology 185 795 810
33. GuarinoLASmithGDongW 1995 Ubiquitin is attached to membranes of baculovirus particles by a novel type of phospholipid anchor. Cell 80 301 309
34. ItoESahriDKnippersRCarstensEB 2004 Baculovirus proteins IE-1, LEF-3, and P143 interact with DNA in vivo: a formaldehyde cross-linking study. Virology 329 337 347
35. ChenTSahriDCarstensEB 2004 Characterization of the interaction between P143 and LEF-3 from two different baculovirus species: Choristoneura fumiferana nucleopolyhedrovirus LEF-3 can complement Autographa californica nucleopolyhedrovirus LEF-3 in supporting DNA replication. J Virol 78 329 339
36. KangWSuzukiMZemskovEOkanoKMaedaS 1999 Characterization of baculovirus repeated open reading frames (bro) in Bombyx mori nucleopolyhedrovirus. J Virol 73 10339 10345
37. OkanoKMikhailovVSMaedaS 1999 Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J Virol 73 110 119
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Fungal Biofilms
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 4- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Infekční komplikace virových respiračních infekcí – sekundární bakteriální a aspergilové pneumonie
-
Všechny články tohoto čísla
- Systematic Review of Mucosal Immunity Induced by Oral and Inactivated Poliovirus Vaccines against Virus Shedding following Oral Poliovirus Challenge
- The Arbuscular Mycorrhizal Symbiosis: Origin and Evolution of a Beneficial Plant Infection
- Modelling the Evolutionary Dynamics of Viruses within Their Hosts: A Case Study Using High-Throughput Sequencing
- Structural Basis of Cytotoxicity Mediated by the Type III Secretion Toxin ExoU from
- Fungal Biofilms
- The Problem of Auto-Correlation in Parasitology
- T Regulatory Cells Control Susceptibility to Invasive Pneumococcal Pneumonia in Mice
- Small-Molecule Inhibitors of Dengue-Virus Entry
- The Baculovirus Uses a Captured Host Phosphatase to Induce Enhanced Locomotory Activity in Host Caterpillars
- Matrix Metalloprotease 9 Mediates Neutrophil Migration into the Airways in Response to Influenza Virus-Induced Toll-Like Receptor Signaling
- Necrotrophism Is a Quorum-Sensing-Regulated Lifestyle in
- Modeling of the N-Glycosylated Transferrin Receptor Suggests How Transferrin Binding Can Occur within the Surface Coat of
- The Role of Cofactors in Prion Propagation and Infectivity
- The Role of Mast Cells in the Defence against Pathogens
- The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens
- Systematic Review of Mucosal Immunity Induced by Oral and Inactivated Poliovirus Vaccines against Virus Shedding following Oral Poliovirus Challenge
- The Arbuscular Mycorrhizal Symbiosis: Origin and Evolution of a Beneficial Plant Infection
- Modelling the Evolutionary Dynamics of Viruses within Their Hosts: A Case Study Using High-Throughput Sequencing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



