-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Regulatory RNAs Involved in Bacterial Antibiotic Resistance
article has not abstract
Published in the journal: . PLoS Pathog 10(8): e32767. doi:10.1371/journal.ppat.1004299
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004299Summary
article has not abstract
What Are Small Regulatory RNAs?
An increasing number of RNAs have been recently shown to possess regulatory functions similar to those of proteins. In bacteria, these regulatory RNAs are usually noncoding and are short size (50–500 nts) transcripts that are often referred to as small RNAs (sRNAs) [1], [2]. These sRNAs are synthesized under specific environmental conditions and play a major role in the regulation of various cellular processes (Figure 1) [3]. Most of them act via an imperfect antisense base-pairing with their target mRNAs. Duplex formation usually results in inhibition or stimulation of mRNA target translation. In some cases, sRNAs can also bind proteins to influence their activities (e.g., 6S RNA). Compared to protein-dependent mechanisms, sRNAs require less energy, act faster and also allow a coordinated regulation of several targets. Owing to these characteristics, sRNAs allow efficient adaptation of bacteria to their ever-changing environment. Therefore, the possibility exists that some sRNAs may be involved in antibiotic resistance. In this report, we provide evidence that illustrates the growing number of sRNAs that influence bacterial resistance to antibiotics.
Fig. 1. Functional connections between antibiotics' modes of action and regulatory RNAs. 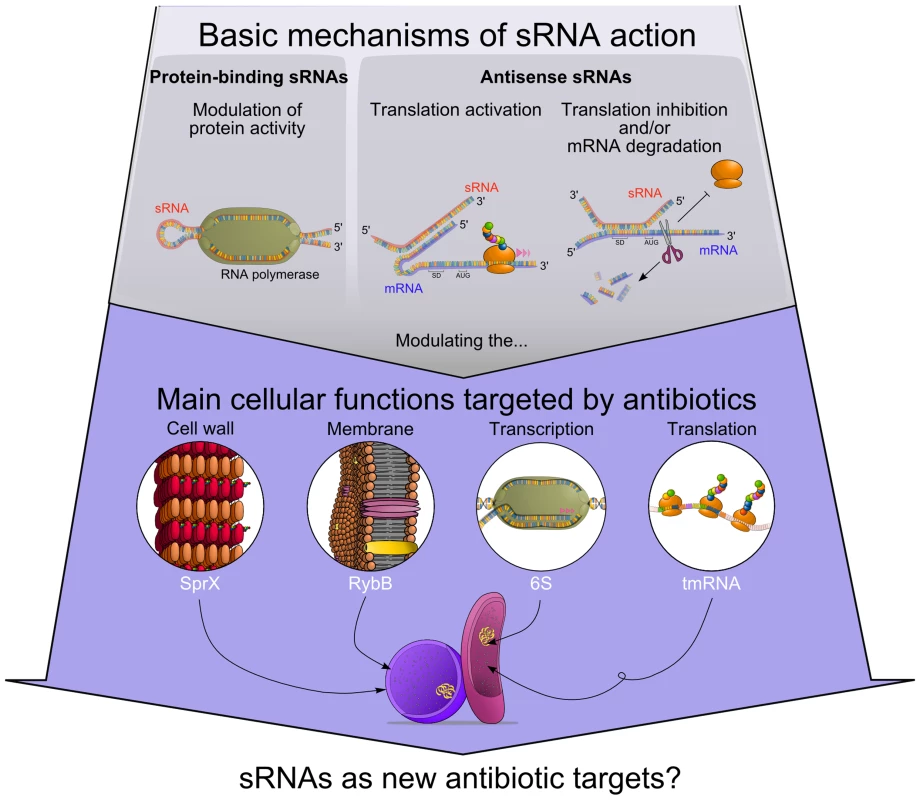
sRNAs modulate protein activity or influence target mRNAs fate. Some of those sRNA targets are part of cellular processes that can be affected by antibiotics. Accumulating evidence suggests that several sRNAs alter antibiotic resistance by modulating bacterial cell wall (SprX), cell membrane (RybB), transcription (6S RNA), and translation (tmRNA). Is There a Link between Antibiotic Exposure and sRNAs Expression in Bacteria?
In the last few years, several studies have shown a correlation between bacterial sRNA expression and antibiotic exposure. An initial study identified four sRNAs in Salmonella Typhimurium with an enhanced expression when the bacterium was challenged with antibiotics such as tigecycline or tetracycline [4], both of which inhibit bacterial protein synthesis. Remarkably, deletion of one of these sRNA genes that encodes SroA reduced Salmonella Typhimurium viability when exposed to tigecycline. Therefore, SroA sRNA activation in a strain exposed to tigecycline may influence bacterial resistance to that antibiotic. In an additional study using the Staphylococcus aureus strain as a model, the expression of some sRNAs correlated with exposure to four antibiotics (vancomycin, linezolid, ceftobiprole, and tigecycline) [5]. These recent data that suggest functional links between sRNA expression and the presence of antibiotics raise the important question whether sRNAs play a role in antibiotic resistance.
How Can sRNA Functions Lead to Altered Antibiotic Resistance?
Mechanisms of antibiotic resistance
Antibiotics can use miscellaneous mechanisms of action to target vital processes such as nucleic acid and protein synthesis, as well as envelope integrity. Strikingly, various bacterial sRNAs are involved in these functions (Figure 1) [3]. As a consequence, perturbation of sRNA activity may lead to alteration of cellular processes, and a potential outcome is modulation of bacterial antibiotic resistance.
sRNAs involved in RNA synthesis
Some antibiotics alter the transcriptional machinery, and sRNAs have been found to modulate this process. For instance, the 6S RNA functions through interaction with housekeeping forms of RNA polymerase holoenzyme in gram-negative and gram-positive bacteria. Even if 6S sRNA has not been shown to directly affect antibiotic resistance by regulating transcription, cells lacking 6S RNA are defective for persistence [6]. Since persistence is a process that makes bacteria highly tolerant to different antibiotics, data suggest a functional link between 6S RNA expression and antibiotic sensitivity.
sRNAs involved in protein synthesis
Protein synthesis involves translation of an mRNA. Many antibiotics interfere with protein synthesis, targeting various steps of the translation process [7]. Ribosomes stalled by protein synthesis inhibitors (e.g., chloramphenicol) can be rescued by a quality-control mechanism that involves the transfer-messenger RNA (tmRNA). However, cells lacking tmRNA are more sensitive to these inhibitors due to the defects in mRNA and protein quality monitoring [8]. In addition, mutations in tmRNA decrease persister cell survival, thus increasing susceptibility to a variety of antibiotics.
sRNAs involved in cell membrane integrity
Many antibiotics have to penetrate the bacterial cell envelope to be effective. For instance, the outer membrane of gram-negative bacteria functions as a selective barrier by combining a hydrophobic lipid bilayer with pore-forming proteins (porins). Therefore, any factor influencing bacterial membrane synthesis or permeability could lead to antibiotic resistance. There are two cell-penetrating pathways that can be used by antibiotics: the lipid-mediated pathway for macrolides (e.g., erythromycin) and other hydrophobic antibiotics, and the general diffusion porins for hydrophilic antibiotics (e.g., the family of β-lactams, gentamycin, and kanamycin) [9].
Lipopolysaccharides (LPS) are components of the outer membrane of gram-negative bacteria and provide a barrier against antibiotics using the lipid-mediated pathway [9]. The PhoP/PhoQ two-component system can activate many genes involved in LPS modifications and, by extent, can control resistance to antibiotics [10]. Interestingly, two sRNAs (MicA and GcvB) have been described as regulators of phoPQ mRNA in Escherichia coli [11]. Moreover, another sRNA, ArcZ, has been shown to directly regulate the expression of a phosphoethanolamine transferase, also involved in LPS modifications [12].
Alteration of outer membrane composition, particularly outer membrane proteins (Omp), represents one of the major mechanisms for antibiotic resistance. Thus, sRNAs such as MicF or MicC, which target the major porins OmpF and OmpC respectively, are likely to mediate antibiotic resistance. Moreover, at least six other sRNAs (InvR, MicA, OmrA/B, RseX, and RybB) have already been described as regulator of Omps [13], thus modulating outer membrane fluidity and permeability in gram-negative bacteria.
sRNAs involved in the regulation of membrane transporters
The active pumping of antibiotics out of the cells by efflux pump systems also contributes to antibiotic resistance [14]. As an example, the MtrCDE efflux pump and the inner membrane protein MtrF allow Neisseria gonorrhoea to resist various hydrophobic antimicrobials, including penicillin and erythromycin. Interestingly, NrrF sRNA reduces mtrF mRNA levels and therefore inhibits MtrF production [15].
One additional example is the overexpression of DsrA sRNA, which was found to induce multidrug resistance in E. coli [16]. In fact, through its positive control of the alternative sigma factor σS (or RpoS), DsrA triggers the expression of genes encoding the MdtEF multidrug efflux pump and thus affects antibiotic susceptibility. Since other sRNAs (ArcZ, RprA, and OxyS) are involved in rpoS regulation [17], these additional regulators can play a role in the control of the MdtEF efflux pump.
Similarly, the sRNA RyhB activates the translation of cirA mRNA that encodes a receptor and translocator of the antibiotic colicin Ia [18]–[20]. Under normal growth conditions, the Shine-Dalgarno (SD) sequence of cirA mRNA is not accessible for translation initiation. However, under iron starvation conditions, RyhB sRNA is expressed and pairs with cirA and that allows the accessibility to the SD sequence [18]. Consequently, the higher number of CirA transporter produced confers an increased sensitivity to colicin Ia. These results support the interpretation that RyhB modulates the sensitivity to colicin Ia through CirA.
sRNAs involved in cell wall turnover
Microbial cell wall is an essential structural component that is responsible for reproductive division and for maintaining cellular integrity. Furthermore, the cell wall limits entry of potentially harmful macromolecules into the cell. In a large scale analysis of S. aureus, it was observed that a sRNA was down-regulated when cells were challenged with vancomycin and ceftobiprole, two antibiotics that target the cell wall [5]. Remarkably, this down-regulated sRNA was antisense to the mecA gene encoding the penicillin-binding protein 2a (PBP2a) which induces a higher level of resistance to β-lactams. This observation suggests a putative role of this sRNA in antibiotic resistance. Recently, another S. aureus sRNA, SprX (also known as RsaOR), was shown to inhibit SpoVG protein synthesis, a protein encoded by the yabJ-spoVG operon involved in glycopeptide and oxacillin resistance [21].
Could sRNAs Be Used as Working Models to Design New Antibiotics?
RNAs represent an unexploited area of potential molecules for antibacterial design. The recent discovery of sRNAs as a general class of powerful regulators has revolutionized our understanding of gene regulation. sRNAs are involved in many cellular processes in response to stress or when cells are challenged with antibiotics. Unraveling the functions of sRNAs in virulence and host immunity will provide fundamental knowledge that can be used to develop next-generation antibiotics using sRNAs as original targets. However, many unanswered questions remain with respect to RNA biology, but we are convinced that sRNA-based therapeutic treatment of infectious diseases may become useful tools in the near future.
Zdroje
1. LalaounaD, Simoneau-RoyM, LafontaineD, MasseE (2013) Regulatory RNAs and target mRNA decay in prokaryotes. Biochim Biophys Acta 1829 : 742–747.
2. WatersLS, StorzG (2009) Regulatory RNAs in bacteria. Cell 136 : 615–628.
3. MichauxC, VerneuilN, HartkeA, GiardJC (2014) Physiological roles of small RNA molecules. Microbiology 160 : 1007–1019.
4. YuJ, SchneidersT (2012) Tigecycline challenge triggers sRNA production in Salmonella enterica serovar Typhimurium. BMC Microbiol 12 : 195.
5. HowdenBP, BeaumeM, HarrisonPF, HernandezD, SchrenzelJ, et al. (2013) Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure. Antimicrob Agents Chemother 57 : 3864–3874.
6. TrotochaudAE, WassarmanKM (2004) 6S RNA function enhances long-term cell survival. J Bacteriol 186 : 4978–4985.
7. WilsonDN (2014) Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 12 : 35–48.
8. LiJ, JiL, ShiW, XieJ, ZhangY (2013) Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J Antimicrob Chemother 68 : 2477–2481.
9. DelcourAH (2009) Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794 : 808–816.
10. GroismanEA (2001) The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183 : 1835–1842.
11. CoornaertA, ChiaruttiniC, SpringerM, GuillierM (2013) Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet 9 : 3.
12. MoonK, SixDA, LeeHJ, RaetzCR, GottesmanS (2013) Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol Microbiol 89 : 52–64.
13. VogelJ, PapenfortK (2006) Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 9 : 605–611.
14. LiXZ, NikaidoH (2009) Efflux-mediated drug resistance in bacteria: an update. Drugs 69 : 1555–1623.
15. JacksonLA, PanJC, DayMW, DyerDW (2013) Control of RNA Stability by NrrF, an Iron-Regulated Small RNA in Neisseria gonorrhoeae. J Bacteriol 195 : 5166–5173.
16. NishinoK, YamasakiS, Hayashi-NishinoM, YamaguchiA (2011) Effect of overexpression of small non-coding DsrA RNA on multidrug efflux in Escherichia coli. J Antimicrob Chemother 66 : 291–296.
17. MandinP, GottesmanS (2010) Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 29 : 3094–3107.
18. SalvailH, CaronMP, BelangerJ, MasseE (2013) Antagonistic functions between the RNA chaperone Hfq and an sRNA regulate sensitivity to the antibiotic colicin. EMBO J 32 : 2764–2778.
19. JakesKS, FinkelsteinA (2010) The colicin Ia receptor, Cir, is also the translocator for colicin Ia. Mol Microbiol 75 : 567–578.
20. HantkeK (1990) Dihydroxybenzoylserine–a siderophore for E. coli. FEMS Microbiol Lett 55 : 5–8.
21. EyraudA, TattevinP, ChabelskayaS, FeldenB (2014) A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res 42 : 4892–4905.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial PathogensČlánek A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during TransmissionČlánek The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil ChemotaxisČlánek Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Regulatory RNAs Involved in Bacterial Antibiotic Resistance
- From Dandruff to Deep-Sea Vents: -like Fungi Are Ecologically Hyper-diverse
- Pathogenicity and Epithelial Immunity
- Mother–Infant HIV Transmission: Do Maternal HIV-Specific Antibodies Protect the Infant?
- Hell's BELs: acterial 3 igases That Exploit the Eukaryotic Ubiquitin Machinery
- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Primary Seronegative but Molecularly Evident Hepadnaviral Infection Engages Liver and Induces Hepatocarcinoma in the Woodchuck Model of Hepatitis B
- TLR2 Signaling Decreases Transmission of by Limiting Bacterial Shedding in an Infant Mouse Influenza A Co-infection Model
- Production of an Attenuated Phenol-Soluble Modulin Variant Unique to the MRSA Clonal Complex 30 Increases Severity of Bloodstream Infection
- Inhibition of the TRAIL Death Receptor by CMV Reveals Its Importance in NK Cell-Mediated Antiviral Defense
- Early Mucosal Sensing of SIV Infection by Paneth Cells Induces IL-1β Production and Initiates Gut Epithelial Disruption
- Limited HIV Infection of Central Memory and Stem Cell Memory CD4+ T Cells Is Associated with Lack of Progression in Viremic Individuals
- Virus-Specific Regulatory T Cells Ameliorate Encephalitis by Repressing Effector T Cell Functions from Priming to Effector Stages
- A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during Transmission
- The HIV-1 Envelope Transmembrane Domain Binds TLR2 through a Distinct Dimerization Motif and Inhibits TLR2-Mediated Responses
- Infection with MERS-CoV Causes Lethal Pneumonia in the Common Marmoset
- VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: Identification of the Local Environmental Source as Arboreal
- Diverse Host-Seeking Behaviors of Skin-Penetrating Nematodes
- Capsid Protein VP4 of Human Rhinovirus Induces Membrane Permeability by the Formation of a Size-Selective Multimeric Pore
- The Murine Gammaherpesvirus Immediate-Early Rta Synergizes with IRF4, Targeting Expression of the Viral M1 Superantigen to Plasma Cells
- Characterization of an Insecticidal Toxin and Pathogenicity of against Insects
- The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil Chemotaxis
- Histone Deacetylase Inhibitors Impair the Elimination of HIV-Infected Cells by Cytotoxic T-Lymphocytes
- A Locus Encompassing the Epstein-Barr Virus Kinase Regulates Expression of Genes Encoding Viral Structural Proteins
- Distinct APC Subtypes Drive Spatially Segregated CD4 and CD8 T-Cell Effector Activity during Skin Infection with HSV-1
- Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
- Adoptive Transfer of EBV Specific CD8 T Cell Clones Can Transiently Control EBV Infection in Humanized Mice
- Schistosome Feeding and Regurgitation
- EVM005: An Ectromelia-Encoded Protein with Dual Roles in NF-κB Inhibition and Virulence
- Rabies Virus Hijacks and Accelerates the p75NTR Retrograde Axonal Transport Machinery
- Why HIV Virions Have Low Numbers of Envelope Spikes: Implications for Vaccine Development
- Identification of Anti-virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity
- HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization
- Cooperation between Epstein-Barr Virus Immune Evasion Proteins Spreads Protection from CD8 T Cell Recognition across All Three Phases of the Lytic Cycle
- Parasite Extracellular Vesicles: Mediators of Intercellular Communication
- RC1339/APRc from Is a Novel Aspartic Protease with Properties of Retropepsin-Like Enzymes
- Cyclic di-GMP-dependent Signaling Pathways in the Pathogenic Firmicute
- Non-random Escape Pathways from a Broadly Neutralizing Human Monoclonal Antibody Map to a Highly Conserved Region on the Hepatitis C Virus E2 Glycoprotein Encompassing Amino Acids 412–423
- Neutrophil Elastase Causes Tissue Damage That Decreases Host Tolerance to Lung Infection with Species
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation
- Why Sexually Transmitted Infections Tend to Cause Infertility: An Evolutionary Hypothesis
- Late Engagement of CD86 after Influenza Virus Clearance Promotes Recovery in a FoxP3 Regulatory T Cell Dependent Manner
- Determinants of Influenza Transmission in South East Asia: Insights from a Household Cohort Study in Vietnam
- A Novel Signal Transduction Pathway that Modulates Quorum Sensing and Bacterial Virulence in
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- A Cysteine Protease Inhibitor of Is Essential for Exo-erythrocytic Development
- EBNA3C Augments Pim-1 Mediated Phosphorylation and Degradation of p21 to Promote B-Cell Proliferation
- On the Front Line: Quantitative Virus Dynamics in Honeybee ( L.) Colonies along a New Expansion Front of the Parasite
- Assembly and Architecture of the EBV B Cell Entry Triggering Complex
- NLR-Associating Transcription Factor bHLH84 and Its Paralogs Function Redundantly in Plant Immunity
- The PDZ-Binding Motif of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Is a Determinant of Viral Pathogenesis
- Strain-Specific Properties and T Cells Regulate the Susceptibility to Papilloma Induction by Papillomavirus 1
- Human Cytomegalovirus pUL79 Is an Elongation Factor of RNA Polymerase II for Viral Gene Transcription
- The GAP Activity of Type III Effector YopE Triggers Killing of in Macrophages
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- Pathogenicity and Epithelial Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



