-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
A Novel Signal Transduction Pathway that Modulates Quorum Sensing and Bacterial Virulence in
The rhl quorum-sensing (QS) system allows P. aeruginosa to regulate diverse metabolic adaptations and virulence. However, how rhl QS system is regulated remains largely unknown. Here, we report that two-component sensor BfmS controls rhl QS system by repressing its cognate response regulator BfmR, which directly suppresses the expression of rhl QS regulator RhlR gene and reduces the production of QS signal molecule N-butanoyl-L-homoserine lactone (C4-HSL). We find that BfmS is critical to the ability of P. aeruginosa to modulate the expression of virulence-associated traits and adapt to the host. Intriguingly, although wild-type BfmS is a repressor of BfmR, naturally occurring missense mutation (L181P, L181P/E376Q, or R393H) can convert its function from a repressor to an activator of BfmR, leading to BfmR activation, which in turn reduces the level of rhl QS signal C4-HSL. These results, therefore, provide important and novel insight into the regulation and evolution of P. aeruginosa virulence.
Published in the journal: . PLoS Pathog 10(8): e32767. doi:10.1371/journal.ppat.1004340
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004340Summary
The rhl quorum-sensing (QS) system allows P. aeruginosa to regulate diverse metabolic adaptations and virulence. However, how rhl QS system is regulated remains largely unknown. Here, we report that two-component sensor BfmS controls rhl QS system by repressing its cognate response regulator BfmR, which directly suppresses the expression of rhl QS regulator RhlR gene and reduces the production of QS signal molecule N-butanoyl-L-homoserine lactone (C4-HSL). We find that BfmS is critical to the ability of P. aeruginosa to modulate the expression of virulence-associated traits and adapt to the host. Intriguingly, although wild-type BfmS is a repressor of BfmR, naturally occurring missense mutation (L181P, L181P/E376Q, or R393H) can convert its function from a repressor to an activator of BfmR, leading to BfmR activation, which in turn reduces the level of rhl QS signal C4-HSL. These results, therefore, provide important and novel insight into the regulation and evolution of P. aeruginosa virulence.
Introduction
Pseudomonas aeruginosa is an important opportunistic pathogen that accounts for 10% of all hospital-acquired infections [1], [2]. Most notably, P. aeruginosa is the leading cause of chronic pulmonary infections and mortality in cystic fibrosis (CF) patients [3]. The success of P. aeruginosa relies on the production and precise coordination of numerous virulence-associated factors such as lipopolysaccharide, flagella, type IV pili, exopolysaccharide alginate, toxins, proteases, lipases, pyocyanin, and rhamnolipids, which are primarily controlled by regulatory systems such as the quorum-sensing (QS) system and the two-component system (TCS) [4]–[12].
P. aeruginosa has two well-characterized acyl-homoserine lactone (acyl-HSL) - based QS systems, las (LasR-LasI) and rhl (RhlR-RhlI) [4]–[6], [8]–[10]. In addition, a third Pseudomonas quinolone signal (PQS) acts as a link between the las and rhl QS systems, although PQS is not involved in sensing cell density [4]–[6], [8]–[10]. The synthase of LasI catalyzes the synthesis of N-(3-oxododecanoyl) homoserine lactone (3O-C12-HSL), whereas RhlI catalyzes the synthesis of N-butyryl-homoserine lactone (C4-HSL), which induces their respective cognate transcriptional regulators LasR and RhlR, responsible for the activation of numerous QS-controlled genes [4]–[6], [8]–[10]. The transcriptional regulator LasR is generally considered to sit at the top of the QS hierarchy in P. aeruginosa. LasR/3O-C12-HSL activates the transcription of rhlR, and RhlR/C4-HSL activates the transcription of rhlI and various virulence-associated genes [4]–[6], [8]–[10]. However, RhlR is able to control the expression of LasR-specific factors independent of LasR [13]. 2-(2-hydroxylphenyl)-thiazole-4-carbaldehyde (IQS) could activate the rhl system in a LasR-independent manner [14]. Thus, the regulation of the rhl QS system is much more complicated than previously thought. So far, LasR and Vfr are the two transcriptional regulators known to regulate the expression of rhlR directly, other than RhlR itself [4]–[6], [8]–[10], [15].
Pathogenic bacteria, including P. aeruginosa, probes its surrounding environment constantly and makes appropriate decisions during infection [7], [11], [12], [16]–[18]. An important molecular device to achieve sampling of environmental signals is the two-component system (TCS) [12], [16]. Classically, two-component systems are composed of an inner membrane-bound sensor, which is able to detect an environmental stimulus, and a response regulator, which is phosphorylated by the sensor kinase and which, in turn, modulates the expression of genes necessary for the appropriate physiological response [16]. Approximately 130 genes encoding for TCS components have been identified in the genome of P. aeruginosa [1], [7], [11]. This provides P. aeruginosa with a sophisticated capability to regulate diverse metabolic adaptations, virulence and antibiotic resistance processes [7]. In fact, a large number of TCSs or TCS components, such as GacSA, PhoPQ, SadARS, RetS, and LadS have been described as having a key role during the infection process [7], [8], [11]. However, the direct links between TCS and QS remain poorly understood [6]–[8], [11], [19]–[22].
The observation that the expression of BfmRS TCS was dramatically up-regulated in the lungs of cystic fibrosis patients compared to in vitro growth intrigued us [23]. We sought to determine the roles of BfmRS in virulence regulation in P. aeruginosa. In this study, we showed that BfmRS TCS directly controls rhl QS system and modulates the ability of P. aeruginosa to adapt to the host. We demonstrate that BfmRS TCS may play important roles in the regulation and evolution of bacterial virulence during long-term bacterial adaptation to lungs afflicted with cystic fibrosis.
Results
BfmS positively controls rhl QS in P. aeruginosa
In P. aeruginosa, BfmS (PA4102) is a putative two-component sensor kinase with uncharacterized functions although its cognate response regulator BfmR (PA4101) has been reported to play an important role in biofilm maturation [24], [25]. To probe the biological roles of BfmS, we generated a bfmS null mutant strain (ΔbfmS) as described in the Materials and Methods section and in our previous studies [26]. Interestingly, ΔbfmS strain was defective in green pigment and rhamnolipids (Figure 1A), which can be complemented by introducing the native copy of bfmS (Table S1 in Text S1) into the ΔbfmS strain (Figure 1A). Quantitative analysis of pyocyanin and rhamnolipids indicates that the deletion of bfmS results in a 3.5-fold decrease of pyocyanin production and a 5-fold decrease in rhamnolipid production respectively (Figure S1A and S1B). Given that rhamnolipids promote the swarming motility of P. aeruginosa [27], we next examined the swarming motility of a wild-type MPAO1 strain, a ΔbfmS strain and its complementary strain (ΔbfmS/p-bfmS). As shown in Figure S1C, deletion of bfmS abolished swarming whereas both wild-type MPAO1 strain and the complementary strain swarmed on the surface of plates at 36 h.
Fig. 1. Effect of bfmS deletion on the production of virulence-associated factors and the promoter activity of bfmR. 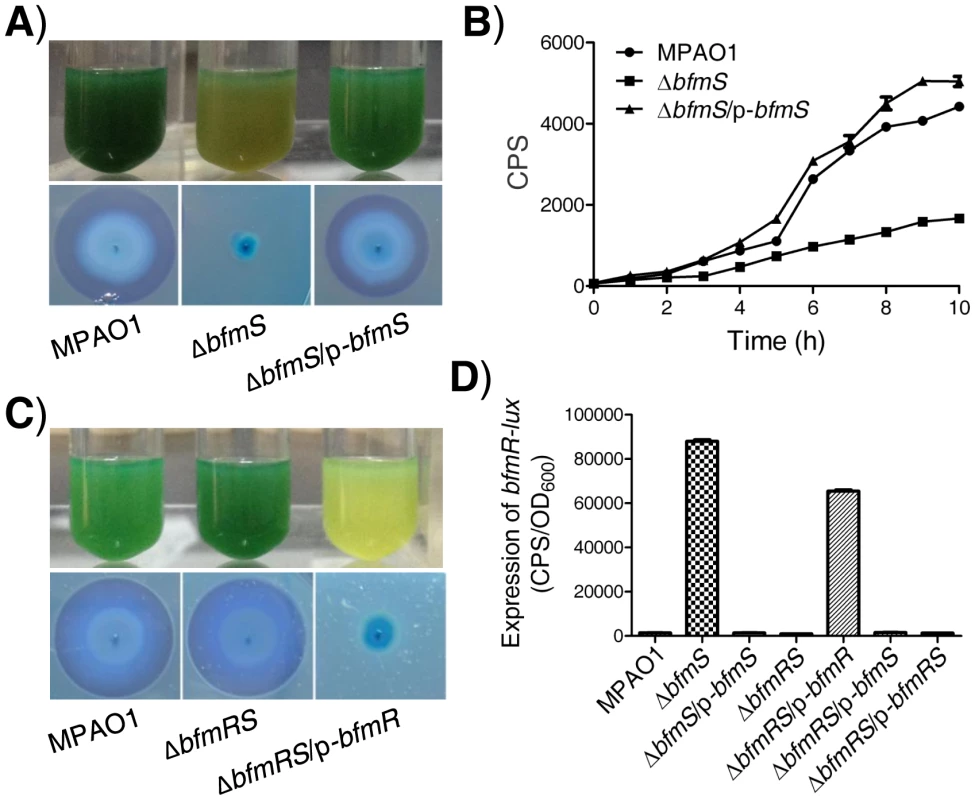
In all panels, MPAO1, ΔbfmS, and ΔbfmRS harbor plasmid PAK1900, respectively. A) Upper panel, P. aeruginosa MPAO1 and its derivatives were grown in Pyocyanin production broth (PPB) medium at 37°C for 36 h with shaking (250 rpm); the presence of the blue-green pigment indicates pyocyanin production. Lower panel, bacterial strains were inoculated onto a cetyltrimethylammonium bromide (CTAB) plate and incubated at 37°C for 24 h and then for 72 h at room temperature; the presence of a blue halo surrounding the colonies indicates the production of rhamnolipids. B) Relative amount of C4-HSL measured by the pDO100 (pKD-rhlA) system. MPAO1 and its derivatives were grown in M8-glutamate minimal medium supplemented with 0.2% glucose at 37°C for 24 h with shaking (250 rpm). Supernatants were subsequently prepared and measured for their relative C4-HSL contents. Plasmid pKD-rhlA carries the C4-HSL-responsive rhlA promoter fused to luxCDABE, so CPS (counts per second) values become an indirect measure of supernatant C4-HSL. C) Upper panel, MPAO1 and its derivatives were grown in PPB medium at 37°C for 24 h with shaking (250 rpm). Lower panel, MPAO1 and its derivatives were grown on CTAB plate and incubated at 37°C for 24 h and then for 72 h at room temperature. D) Expression of bfmR-lux in MPAO1 and its derivatives. Bacteria were grown in M8-glutamate minimal medium supplemented with 0.2% glucose at 37°C for 24 h with shaking (250 rpm) and then the bfmR-lux activity was measured. All experiments were independently repeated at least three times and the data shown represent comparable results. Values represent means ± standard error of the mean (SEM). The rhlAB operon is required for rhamnolipid synthesis [4]–[6], [9], [10]. We therefore constructed an rhlA promoter-lux fusion (rhlA-lux, Table S1 in Text S1) and measured its activity in a wild-type MPAO1 strain, a bfmS deletion strain (ΔbfmS), and its complementary strain (ΔbfmS/p-bfmS). The expression of rhlA-lux fusion in ΔbfmS was significantly lower than those of other strains when bacteria were grown in an M8-glutamate minimal medium supplemented with 0.2% glucose (Figure S1D). This result suggests that the decreased expression of rhlAB in ΔbfmS strain is likely responsible for the reduction in rhamnolipid production. Since the expression of rhlAB is positively controlled by the rhl quorum-sensing system in P. aeruginosa [4]–[6], [9], [10], we next sought to measure the RhlI-dependent autoinducer C4-HSL content in the wild-type MPAO1 strain, the ΔbfmS strain, and the complementary strain (ΔbfmS/p-bfmS). We used the pDO100 (pKD-rhlA) system [28] (Table S1 in Text S1) that carries a lux reporter fused with a rhlA promoter. As a result, supernatants prepared from either the wild-type MPAO1 strain or the complementary strain (ΔbfmS/p-bfmS), but not the ΔbfmS strain, markedly promoted the luminescence values and thereby C4-HSL levels (Figure 1B). We also observed that deletion of bfmS results in decreased rhlI promoter activity (Figure S1E). Based on these results, we conclude that BfmS positively controls rhl QS system in P. aeruginosa.
Transcriptional profiling of the bfmS deletion mutant strain
To further study the roles of bfmS, we used microarray analysis in order to compare the transcriptome of the ΔbfmS strain with that of the wild-type MPAO1 strain. As a result, we identified 131 genes with increased transcript levels (≥2-fold) (Table S2 in Text S1) and 71 genes with decreased transcript levels (≤2-fold) (Table S3 in Text S1) in the ΔbfmS strain versus wild-type MPAO1 strain. These 202 genes represent ∼3.6% of the total number of annotated genes in the P. aeruginosa PAO1 genome. Of those 202 genes, 42% encode hypothetical proteins of unknown functions (Tables S2 and S3 in Text S1). Grouping these genes according to their annotated function shows that they belong to several functional categories, primarily transport of small molecules, carbon compound catabolism, translation, and adaptation (Tables S2 and S3 in Text S1).
Among the 131 genes whose expression is up-regulated in the ΔbfmS strain, 7 genes were up-regulated more than 10-fold. Interestingly, these 7 genes, including PA4100, bfmR, PA4103, PA4104, PA4105, PA4106, and PA4107, are located at or near the bfmRS (PA4101-PA4102) loci (Figure S2A, Table S2 in Text S1). These microarray-based expression data are consistent with the operon predictions for P. aeruginosa, which suggested that PA4103 and PA4104 are organized into PA4103 operon (PA4103-PA4104) while PA4105, PA4106 and PA4107 are organized into PA4107 operon (PA4107-PA4106-PA4105) (www.pseudomonas.com). Among these genes, PA4100 encodes a dehydrogenase of unknown function, and bfmR encodes a two-component response regulator that acts as a biofilm maturation regulator, whereas PA4103, PA4104, PA4105, PA4106, and PA4107 encode hypothetical proteins. Although PA4103 contains a ferric reductase like transmembrane component (pfam01794) and PA4107 contains a calcium binding motif (cd00051), their biological functions are unknown. Further characterization of the functions of these genes may provide insight into the roles of bfmS in P. aeruginosa.
There are 11 genes whose expressions are down-regulated more than 10-fold in ΔbfmS strain (Figure S2, Table S3 in Text S1). Among them, 5 genes (rhlA, rhlB, antA, antB, and antC) are already known to be controlled by the rhl QS system [29], [30]. In addition, we observed that deletion of bfmS decreases the transcription of rhlI by approximately 66% (Table S3 in Text S1). We also found a moderate, 20% decrease in rhlR transcription in the ΔbfmS mutant compared with the parent, which is consistent with the results of an rhlR-lux reporter gene analysis (Figure S3A). These results further suggest that bfmS positively controls the rhl QS. To further confirm the differentially expressed genes identified by the microarray analysis, 12 genes representing a range of microarray signal intensity and expression profiles were subjected to real-time (RT) PCR analyses. There was a high degree of consistency among data generated by these two methods (Table S4 in Text S1), which assures the reliability of microarray analysis in determining transcriptional changes.
BfmS negatively controls BfmR, which is positively auto-regulated
Since deletion of bfmS led to the over-expression of bfmR (>90-fold) (Figure S2A, Table S2 in Text S1), we next sought to determine if the elevated bfmR contributes to the phenotypes observed in the ΔbfmS strain. We generated a bfmRS double mutant strain (ΔbfmRS) (Table S1 in Text S1) and performed phenotypic analysis. Interestingly, the ΔbfmRS strain and wild-type MPAO1 strain display similar phenotypes when bacteria are grown in Pyocyanin production broth (PPB) or on a cetyltrimethylammonium bromide (CTAB plate) (Figure 1C). The introduction of a wild-type bfmR gene (p-bfmR, Table S1 in Text S1) into ΔbfmRS strain restored its phenotypes similar to ΔbfmS strain (Figure 1C). These results suggest that the effect of bfmS deletion on either the pigment production or rhamnolipids production in P. aeruginosa is likely mediated through the over-expression of bfmR. Moreover, qRT-PCR analysis indicates that the expressions of at least 12 genes were significantly affected by the deletion of bfmS, whereas their altered expression levels caused by bfmS deletion were suppressed by additional deletion of bfmR. Thus, bfmR may mediate most, if not all, the output of bfmS.
We next tested if BfmS could affect the expression of BfmR. We constructed a bfmR promoter-lux fusion (bfmR-lux, Table S1 in Text S1) and then measured its activity in a wild-type MPAO1 strain, a bfmS deletion strain (ΔbfmS), its complementary strain (ΔbfmS/p-bfmS), a bfmRS double deletion strain (ΔbfmRS), and a ΔbfmRS strain harboring p-bfmR, p-bfmS or p-bfmRS (Table S1 in Text S1). As shown in Figure 1D, the activity of bfmR-lux in ΔbfmS strain was about 60-fold higher than that of the wild-type MPAO1 strain. Complementation with p-bfmS in the ΔbfmS strain restored the activity of bfmR-lux to levels similar to the wild-type strain (Figure 1D). In addition, the activity of bfmR-lux in the ΔbfmRS strain was similar to that observed in the wild-type MPAO1 strain; however, the introduction of p-bfmR, but not p-bfmS or p-bfmRS, to the ΔbfmRS strain dramatically increased the activity of bfmR-lux (>46-fold) (Figure 1D). Hence, BfmR can activate its own gene promoter in the absence of BfmS.
We next evaluated if the absence of BfmS causes an accumulation of BfmR in P. aeruginosa. To this end, we constructed an integration vector mini-ctx-BfmR-Flag (Table S1 in Text S1) and the resulting clone was mobilized into the wild-type MPAO1 and ΔbfmRS strain, yielding an MPAO1::BfmR-Flag strain and ΔbfmRS::BfmR-Flag strain, respectively. The ΔbfmRS::BfmR-Flag strain displayed a pigment-deficient phenotype as observed for either the ΔbfmS strain or the ΔbfmRS/p-bfmR strain (Figure S4A). The cell lysates of MPAO1::BfmR-Flag strain, ΔbfmRS::BfmR-Flag strain and its complementary strain (ΔbfmRS::BfmR-Flag/p-bfmS) were subjected to Western blot analysis using anti-FLAG antibodies. As shown in Figure S4B, a large amount of BfmR-Flag in the ΔbfmRS::BfmR-Flag strain was detected. In contrast, no detectable signal was obtained for the BfmR-Flag generated from the MPAO1::BfmR-Flag strain or the complementary strain (ΔbfmRS::BfmR-Flag/p-bfmS) (Figure S4B). Therefore, the absence of BfmS leads to an accumulation of its cognate response regulator BfmR.
BfmR directly binds to the promoters of its own, PA4107, PA4103, and rhlR
Since BfmR activates its own gene promoter, we next aimed to test if BfmR could bind its own promoter. We performed electrophoretic mobility shift assay (EMSA) using 6His-BfmR protein and DNA fragments containing bfmR, rhlA, and rhlC promoter regions, respectively. We found that 6His-BfmR could shift the bfmR promoter DNA, although it failed to shift the rhlA or rhlC promoter DNA (Figure 2A). We further determined the specific DNA sequence that BfmR can recognize in the bfmR promoter region by using a dye-primer-based DNase I footprint assay. We uncovered three BfmR-protected regions in the bfmR promoter DNA (Figure 2B). Interestingly, all three BfmR-protected regions contain a consensus sequence GATACAnnGC (where n is any nucleotides, Figure 2C).
Fig. 2. Direct binding of BfmR to its own promoter. 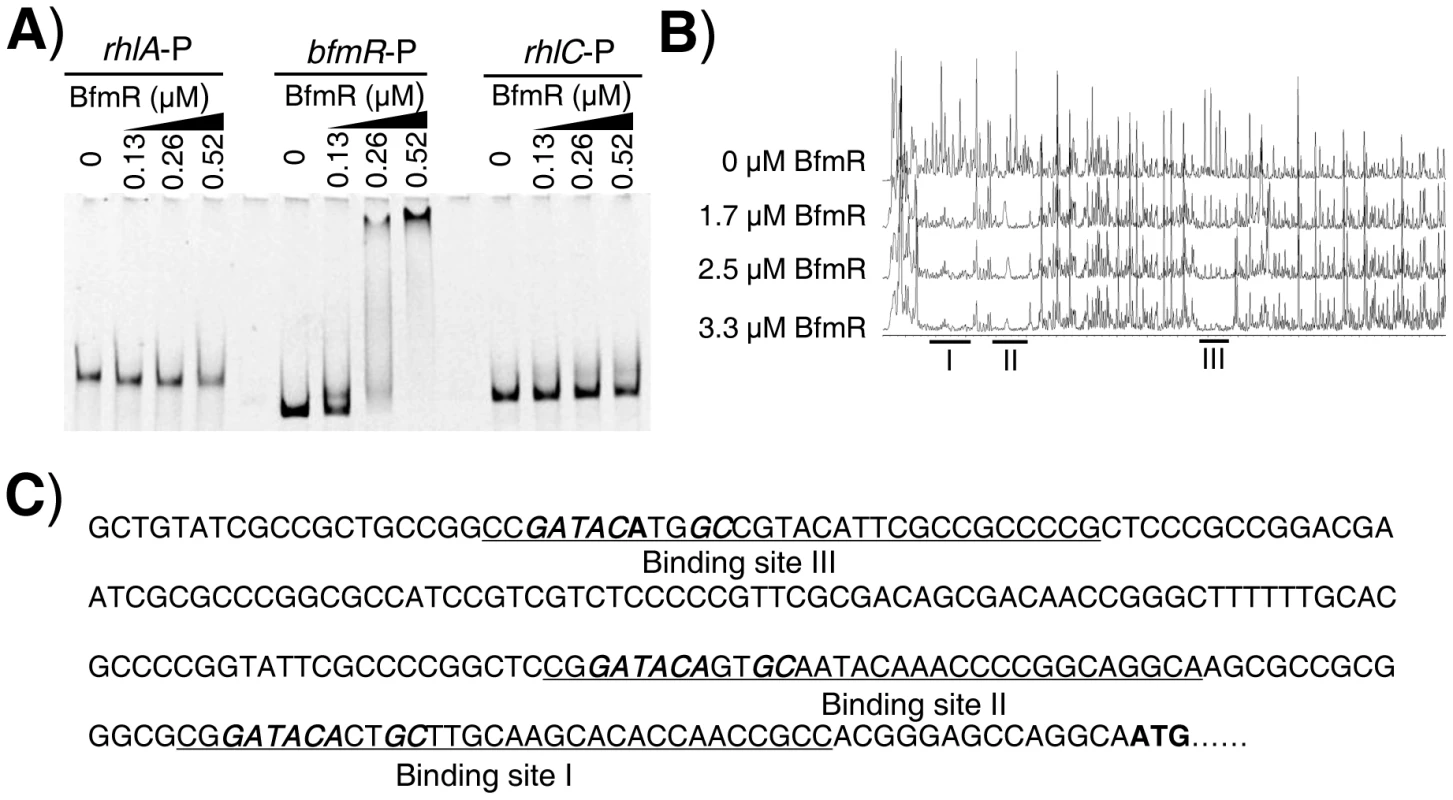
A) EMSA showing that 6His-BfmR binds to its own promoter but not to the promoter of either rhlA or rhlC. B) Dye primer-based DNase I footprint assays show the protection pattern of the bfmR promoter after digestion with DNase I following incubation in the absence or presence of different amounts of 6His-BfmR, as indicated. Three BfmR-protected regions in the bfmR promoter DNA were demonstrated. C) bfmR promoter sequence with a summary of the DNase I footprint assay results. The BfmR-protected regions are underlined, and the putative BfmR box is in bold and in italics. Start codon of bfmR is in bold. Using RAST (http://rsat.ulb.ac.be/rsat/), we found that 41 promoters (−1 bp to −400 bp of the coding region) of P. aeruginosa PAO1, including the PA4017 promoter, contain a putative BfmR-binding motif (GATACAnnGC) (Table S5 in Text S1). As expected, BfmR could shift the PA4107 promoter DNA (Figure S5A) in our EMSA analysis, although it failed to shift the rhlI promoter DNA (Figure S5A). Interestingly, we also observed that BfmR is able to bind to PA4103 promoter DNA (Figure S5B) that lacks a canonical BfmR-binding motif (GATACAnnGC) (Table S5 in Text S1). Using a dye-primer-based DNase I footprint assay, we uncovered that the BfmR-protected region of PA4103 promoter DNA contains a non-canonical BfmR-binding motif (GATACAnnAC, the mismatch is underlined) (Figure S5C), which is subsequently determined to be required for the BfmS-mediated regulation of PA4103 promoter activity (Figure S5D). Thus, it is likely that BfmR directly control the expression of the PA4107 and PA4103 operon.
As aforementioned, BfmR negatively controls the rhl QS system of P. aeruginosa, which does not need to bind promoter of rhlI (Figure S5A and S5B). These observations prompted us to determine if BfmR binds to the promoter of rhlR. EMSA analysis indicated that 6His-BfmR bound to the rhlR promoter DNA but not to the promoter region of rhlC that serves as a negative control (Figure 3A). Dye-primer-based DNase I footprint assay indicated that there were three BfmR-protected regions in the promoter of the rhlR (Figure 3B). Interestingly, protected region I (−220 to −193 from the start codon of rhlR) harbored a putative BfmR-binding motif (GATACTnnGC) with one mismatch (underlined) (Figure 3B), oriented in the opposite direction of the transcription of rhlR. Protected region II extends from nucleotide −151 to nucleotide −171 while protected region III extends from nucleotide −69 to nucleotide −85, relative to the start codon of rhlR, respectively (Figure 3B).
Fig. 3. BfmR regulates the expression of rhlR in a direct manner. 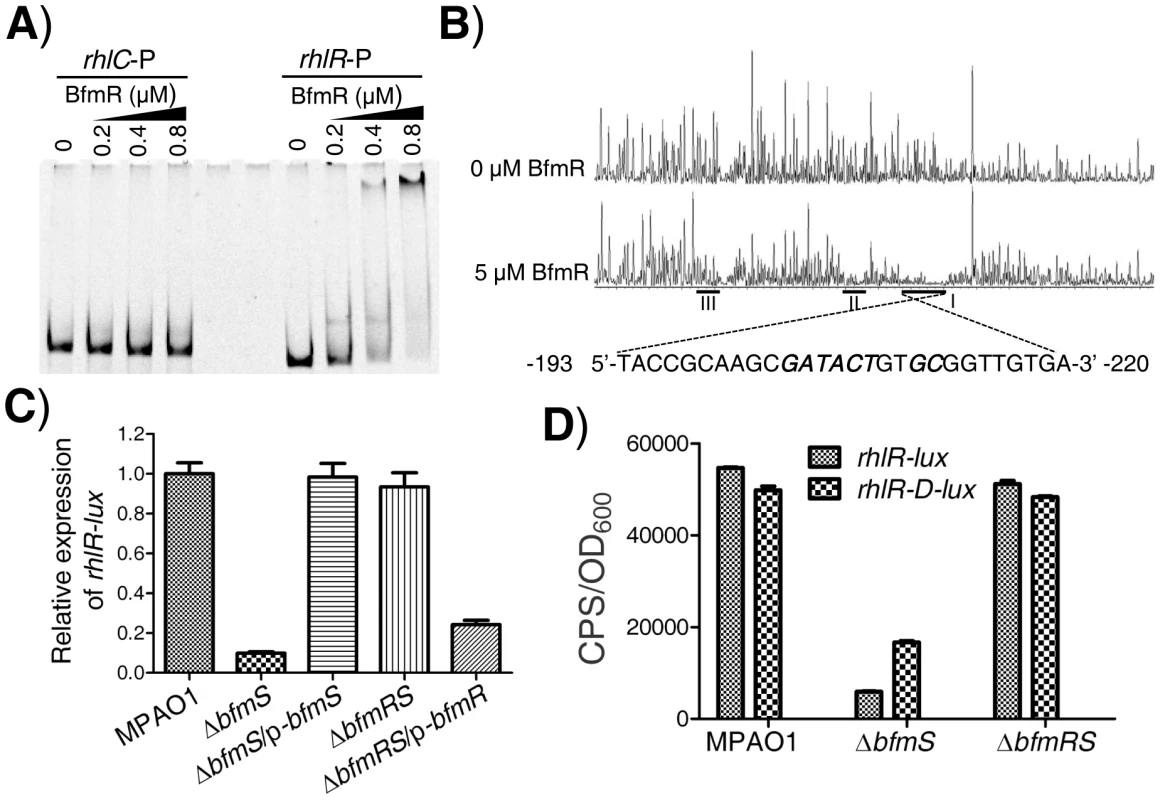
A) EMSA shows that 6His-BfmR directly binds to the promoter DNA of rhlR but not to that of rhlC. B) Electropherograms show the protection pattern of the rhlR promoter after digestion with DNase I following incubation in the absence or the presence of 6His-BfmR. There are three BfmR-protected regions (I, II, and III) in the promoter of the rhlR. BfmR-protected regions I harbors a putative BfmR-binding motif, which was highlighted in bold and in italics, as indicated. C) The relative expression of rhlR-lux in the wild-type MPAO1 (harboring PAK1900), the ΔbfmS strain (harboring PAK1900), the ΔbfmS/p-bfmS strain, the ΔbfmRS strain (harboring PAK1900), and the ΔbfmRS/p-bfmR strain. The relative gene expression in the wild-type MPAO1 was set to 1, and the other values were adjusted accordingly. D) The expression of rhlR-lux and rhlR-D-lux in the wild-type MPAO1, the ΔbfmS strain and the ΔbfmRS strain, as indicated. Bacteria were grown in low phosphate (0.32 mM) M8-glutamate minimal medium supplemented with 2% glucose at 37°C for 48 h. Values represent means ± SEM. The assays were independently repeated at least three times and the data shown represent comparable results. There are 44 bfmS-regulated genes harbor a consensus sequence (GATACAnnGC with or without one mismatch) in their promoter region (−1 bp to −400 bp of the coding region) (Tables S2 and S3 in Text S1). Additionally, there are 984 promoters (−1 bp to −400 bp of the coding region) harbor the consensus sequence (GATACAnnGC without or with one mismatch) in the PAO1 genome. These observations suggest that BfmR may serves as a global regulator affecting expression of a large number of genes.
BfmR represses the expression of rhlR in the absence of BfmS
We next elucidated if BfmR regulates the expression of rhlR. To do this, we measured rhlR promoter-lux fusion activity in the wild-type MPAO1 strain, the ΔbfmS strain, the complementary strain (ΔbfmS/p-bfmS), the ΔbfmRS strain, and the ΔbfmRS strain harboring p-bfmR (ΔbfmRS/p-bfmR). The low-phosphate (0.32 mM Pi) M8-glutamate minimal medium supplemented with 0.2% glucose, used as phosphate limitation, served to stimulate the expression of rhlR [21]. As shown in Figure 3C and S3B, the activity of rhlR-lux in the ΔbfmS strain was more than 8-fold lower than that observed in the wild-type MPAO1 strain. Complementation with p-bfmS in the ΔbfmS strain restored the activity of rhlR-lux similar to that of the wild-type strain (Figure 3C). Moreover, ΔbfmRS strain exhibited rhlR-lux activity similar to that observed in the wild-type MPAO1 strain, while the introduction of p-bfmR into the ΔbfmRS strain caused a 3.8-fold decrease in rhlR-lux activity (Figure 3C). Thus, it is likely that bfmS activates the expression of rhlR by repressing BfmR, which acts to negatively regulate rhlR expression and rhl QS. This notion was further substantiated by the observations that under low-phosphate growth conditions ΔbfmS also exhibits decreased rhlI (Figure S3C) and rhlA (Figure S3D) promoter activity, and lowered C4-HSL content (Figure S3E) as compared to wild-type MPAO1.
To determine if the putative BfmR-binding motif (GATACTnnGC) (Figure 3B) is involved in the BfmR-mediated inhibition of rhlR-lux activity, we deleted the first five residues (GATACT) in the consensus sequence (yielding rhlR-D-lux, Table S1 in Text S1), and examined the ability of the mutant sequence to permit the inhibition of the reporter gene in ΔbfmS strain. As shown in Figure 3D, the rhlR-lux activity in ΔbfmS strain was approximately 8-fold lower than that observed in the wild-type MPAO1 strain or in the ΔbfmRS strain. However, the rhlR-D-lux activity was about 2-fold lower in the ΔbfmS strain compared to the wild-type MPAO1 strain or the ΔbfmRS strain (Figure 3D). Thus, the five residues (GATACT) are required for the full inhibition of rhlR-lux activity in ΔbfmS strain, demonstrating the importance of these conserved binding-site residues. However, besides GATACT sequence elements, additional regulatory sequence elements within the promoter region of rhlR are most likely involved in BfmR-mediated inhibition of rhlR-lux activity, given that the rhlR-D-lux activity in ΔbfmS strain is still decreased, although to a much lesser extent than that of rhlR-lux (Figure 3D).
The Pta-AckA pathway modulates the activation of BfmR
Like many other response regulators [31], BfmR can be phosphorylated by acetyl phosphate (Figure S6A) and hence activated in vitro (Figure S6B). We further observed that the predicted phosphorylation site, aspartate residue D55, is required for the activation of BfmR in vitro and in vivo (Figure S7). As acetyl∼P is an intermediate in the acetate kinase (AckA)-phosphate acetyltransferase (Pta) pathway [31], we hypothesized that the AckA-Pta pathway may be involved in the activation of BfmR. Thus, we constructed a mutant strain (ΔbfmSΔackA-pta, Table S1 in Text S1) with deletion of both the bfmS gene and ackA-pta operon and measured the activity of bfmR-lux as well as the C4-HSL content in this strain and the ΔbfmS strain, respectively. As shown in Figure 4, the expression of bfmR-lux was lower (Figure 4A) while the C4-HSL content was higher (Figure 4B) in the ΔbfmSΔackA-pta strain than that of the ΔbfmS strain, respectively. The introduction of wild-type ackA-pta operon (p-ackA-pta, Table S1 in Text S1) into the ΔbfmSΔackA-pta strain was able to restore either the activity of bfmR-lux (Figure 4A) or the C4-HSL content to the level of the ΔbfmS strain (Figure 4B). Therefore, in the ΔbfmS strain, acetyl phosphate or the component that is dependent on the Acka-Pta pathway is required for the full activity of BfmR.
Fig. 4. Effect of the Pta-AckA pathway and carbon sources availability on the activation of BfmR in the absence of BfmS. 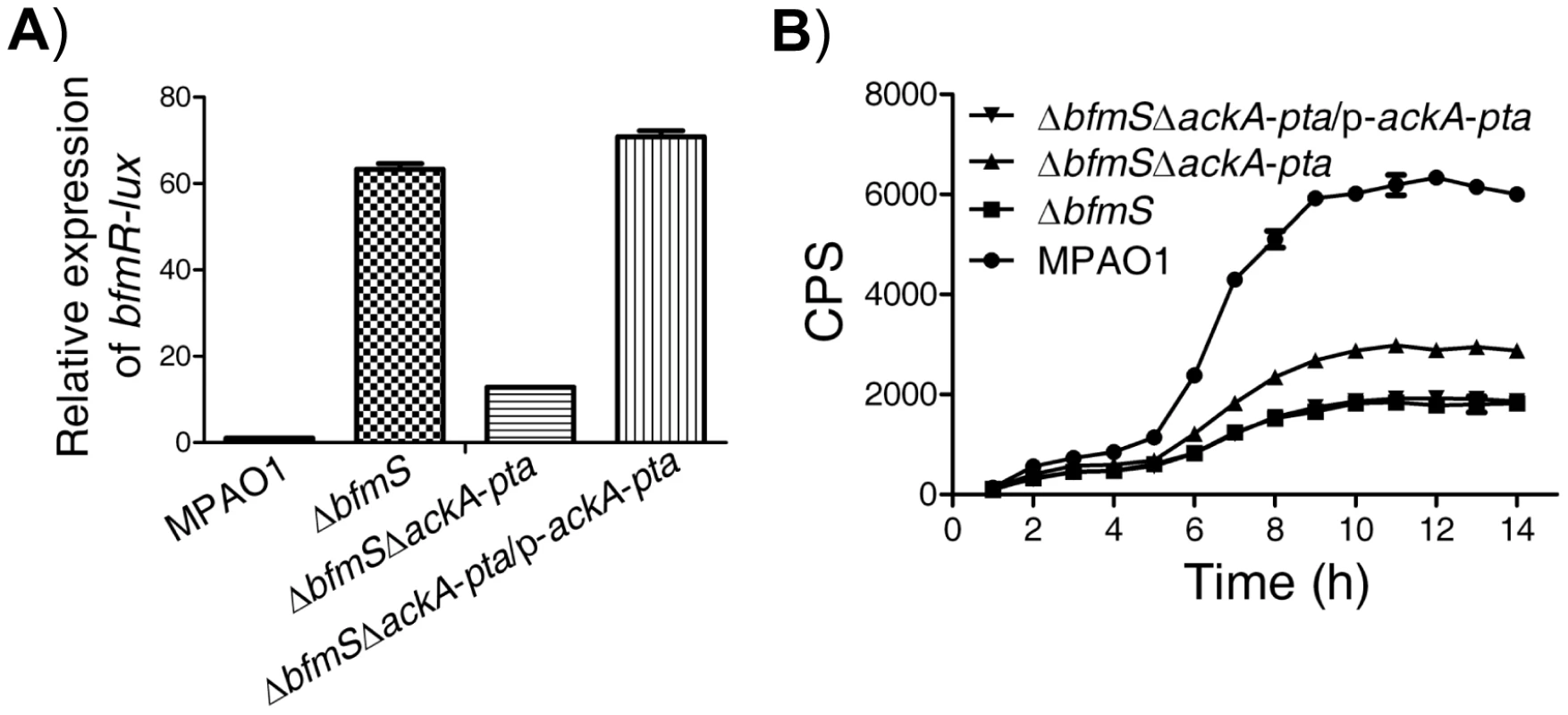
In all panels, MPAO1, ΔbfmS, and ΔbfmSΔackA-pta harbor plasmid PAK1900, respectively. A) The relative expression of bfmR-lux in the wild-type MPAO1, the ΔbfmS strain, the ΔbfmSΔackA-pta strain, and its complementary strain (ΔbfmSΔackA-pta/p-ackA-pta) when bacteria were grown in M8-glutamate minimal medium supplemented with 0.2% glucose at 37°C for 24 h with shaking (250 rpm). Values are relative to MPAO1 (set to 1). B) Relative amount of C4-HSL measured by the pDO100 (pKD-rhlA) system. MPAO1 and its derivatives were grown in M8-glutamate minimal medium supplemented with 0.2% glucose at 37°C for 24 h with shaking (250 rpm). Supernatants were subsequently prepared and measured for the relative C4-HSL contents. Values represent means ± SEM. The assays were independently repeated at least three times with similar results obtained, and the graphs show a set of representative data. BfmS regulates bacterial virulence and the ability of P. aeruginosa to adapt to the host environment
As BfmS modulates the rhl QS system that contributes significantly to the virulence of P. aeruginosa [4]–[6], [9], [10], we infected romaine lettuce leaves with P. aeruginosa to see if BfmS controls bacterial virulence. The pathogenicity assay revealed a significant difference in the manifestation of infection symptoms caused by the ΔbfmS strain compared to wild-type MPAO1 strain. Relative to wild-type MPAO1, the ΔbfmS strain failed to cause severe necrotic lesions of the leaves, which can be complemented by introducing the wild-type bfmS gene into the ΔbfmS strain (Figure 5A). In addition, the ΔbfmRS strain exhibited a virulence phenotype similar to that of a wild-type MPAO1 strain, while the introduction of p-bfmR into the ΔbfmRS strain led to a low virulence phenotype (Figure S8). Moreover, the constitutive expression of rhlR in ΔbfmS strain could restore either the virulence of ΔbfmS strain to the level of the wild-type MPAO1 strain (Figure 5B), suggesting that the decreased expression of rhlR is likely responsible for the attenuated virulence of ΔbfmS strain in the lettuce leaf model of P. aeruginosa infection.
Fig. 5. Effect of bfmS deletion on bacterial virulence and the ability of P. aeruginosa to adapt to the host. 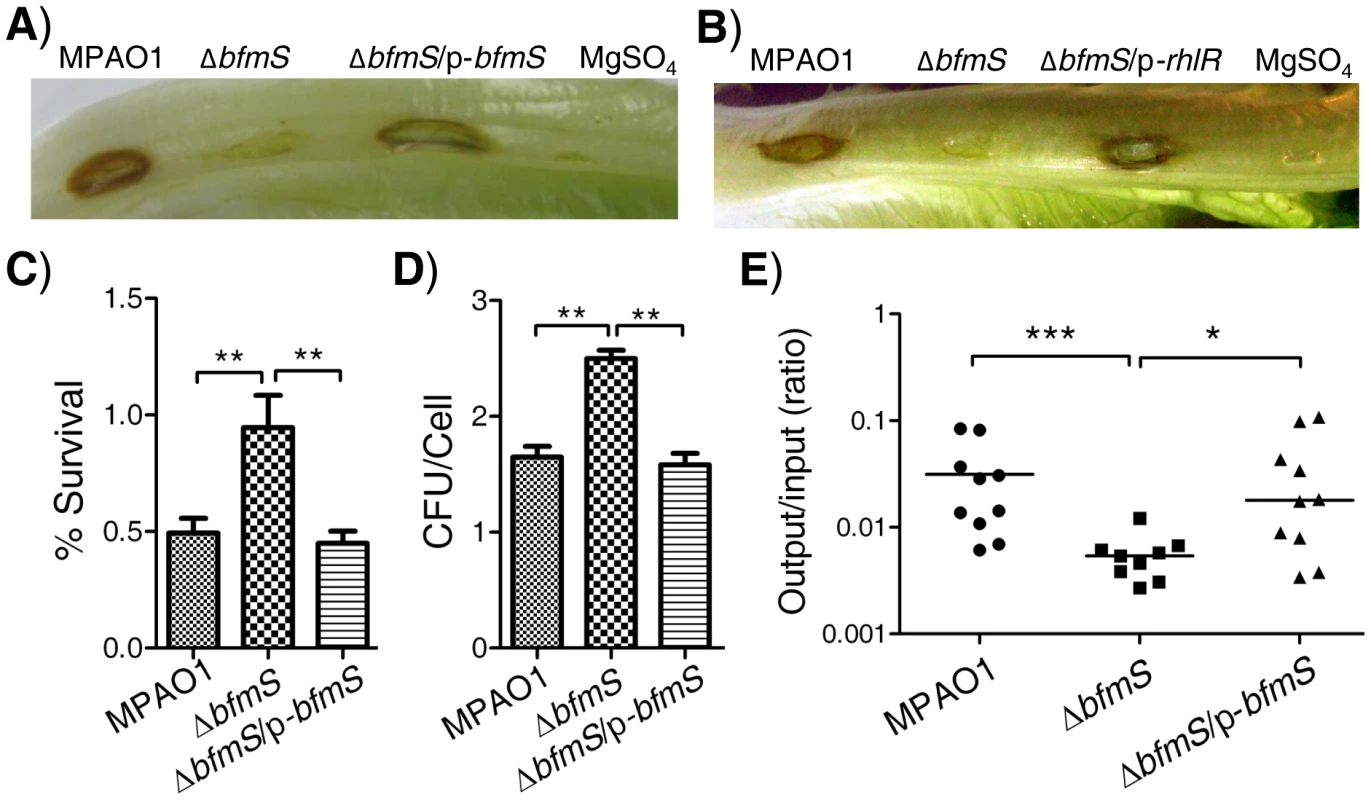
In all panels, MPAO1 and ΔbfmS harbor plasmid PAK1900, respectively. A) Photographs show lettuce midribs after three days of infection with 1×107 cfu of P. aeruginosa. Wild-type MPAO1 strain or ΔbfmS/p-bfmS strain shows necrosis and tissue maceration of infection. The ΔbfmS strain shows weak signs of infection. B) Constitutive expression of rhlR in ΔbfmS strain could restore the virulence to the level of the wild-type MPAO1 strain. In A) and B), lettuce leaves were inoculated with 10 mM MgSO4 as a control. C) Effect of P. aeruginosa inoculum on the survival rate of murine lung epithelial cell line 12 (MLE-12). D) Internalization of P. aeruginosa into the murine lung epithelial cell line 12 (MLE-12). In C) and D), values represent means ± SEM. E) Recovery of P. aeruginosa derivatives in a mouse model of acute pneumonia. Results are expressed as the ratio of cfu recovered per lung (output) to cfu present in the initial inoculum (input) and represent results from n = 9–10 mice per strain; the line shows the geometric mean for each group. CFU, Colony-Forming Unit. The Mann–Whitney test was used to calculate p-values (two-tailed). * p<0.05, ** p<0.01, *** p<0.001. Results are representative of two independent experiments. Since cytotoxicity and invasion of P. aeruginosa are useful traits for this pathogen [32], we further characterized BfmS to check if it regulates the cytotoxicity or the invasion of P. aeruginosa in a murine lung epithelial cell line (MLE-12), a widely used in vitro model for studying host-pathogen interactions [33]–[35]. Using an MTT assay, we found that about 50% MLE-12 cells were killed when challenged with wild-type MPAO1 strain; however, only 5% of MLE-12 cells were killed after inoculation with the ΔbfmS strain (Figure 5C). Using a colony forming unit (CFU) assay, we showed that deletion of bfmS significantly increases (p<0.01) the internalization of P. aeruginosa by approximately 50% (Figure 5D). Further, the invasive and cytotoxic phenotypes of ΔbfmS strain could be completely restored to the wild-type levels by the introduction of p-bfmS (Figure 5C and 5D). Thus, deletion of bfmS causes a loss of cytotoxic capacity while it enhances the invasion of P. aeruginosa MPAO1 to MLE-12 cells.
To further determine the virulence regulated by BfmS, a mouse model of acute pneumonia was used as described in our previous study [26]. C57BL/6J mice were intranasally infected with approximately 5×106 CFU of wild-type MPAO1, bfmS null mutant ΔbfmS, and its complementary strain ΔbfmS/p-bfmS. Figure 5E shows the CFU of bacteria recovered from the lungs compared to the initial inoculum at 18 h post infection, with a geometric mean indicated for each group. In this assay, wild-type MPAO1 was recovered in numbers approximately at 3.13% of the inoculum dose from lungs with a result 5.6-fold higher than that (0.56%) of the ΔbfmS strain. Further, bacteria of complementary strain (ΔbfmS/p-bfmS) were recovered from lungs with 3.43%, similar to that of the wild-type MPAO1 strain (Figure 5E). These results indicate that deletion of bfmS decreases P. aeruginosa survival in the mouse lungs in this model and thus reduced virulence.
Specific mutations in bfmS result in the activation of BfmR
The P. aeruginosa DK2 lineage is highly successful and has been isolated from ∼40 cystic fibrosis (CF) patients since the start of the sampling program in 1973 [36]. We noted that the DK2 lineage-specific mutations in BfmS are point mutations that cause two amino acid substitutions, proline replaces leucine 181 (L181P), and glutamine replaces glutamic acid (E376Q). Among them, L181P was fixed in the DK2 lineage before the year 1979, while E376Q was subsequently fixed in the DK2 lineage after 1991 [36]. We next investigated the regulatory effect associated with these amino acid substitutions observed in the BfmS. We created p-bfmSL181P, p-bfmSE376Q, and p-bfmSL181P/E376Q plasmids (Table S1 in Text S1) and introduced them to the ΔbfmS strain, and tested their effects on bfmR-lux activity. Interestingly, L181P and E376Q substitutions in BfmS caused a 16-fold and a 2-fold increase in the activity of bfmR-lux, respectively (Figure 6A). More significantly, the combined substitution (L181P/E376Q) led to a 27-fold increase in the activity of bfmR-lux (Figure 6A). Accordingly, L181P or L181P/E376Q substitutions in BfmS decreased the RhlI-dependent autoinducer C4-HSL content (Figure 6B). These data strongly suggest that mutations in specific residues of BfmS result in activation of BfmR.
Fig. 6. Effect of DK2 lineage-specific amino acid substitutions in BfmS on the activation of BfmR. 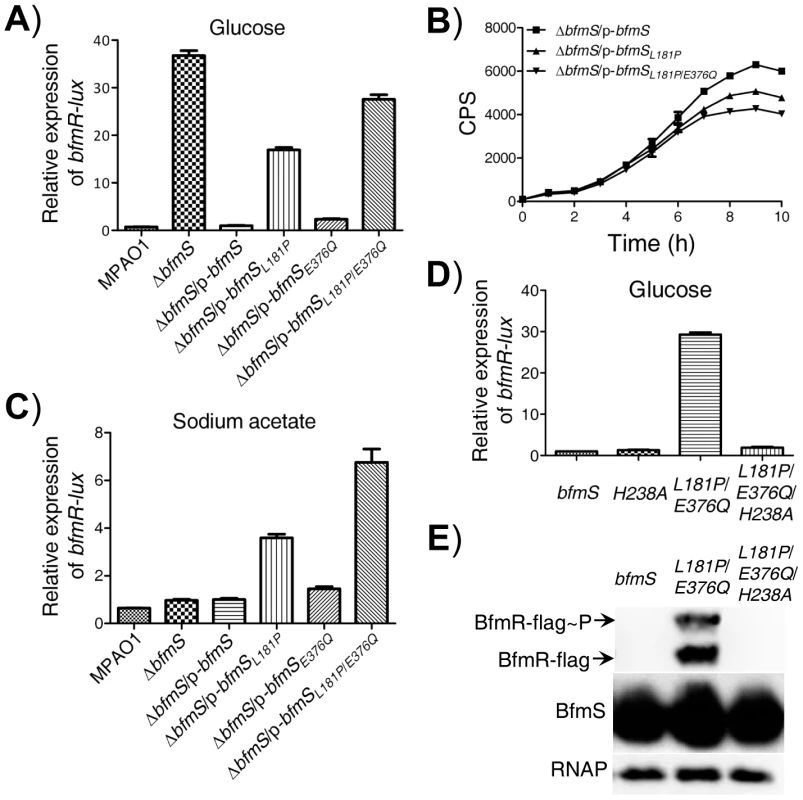
In all panels, MPAO1 and ΔbfmS harbor plasmid PAK1900, respectively. A) The relative expression of bfmR-lux in the wild-type MPAO1 and its derivatives when bacteria were grown in M8-glutamate minimal medium supplemented with 0.2% glucose at 37°C for 36 h with shaking (250 rpm), as indicated. Values are relative to MPAO1 (set to 1). B) Relative amount of C4-HSL measured by the pDO100 (pKD-rhlA) system. MPAO1 and its derivatives were grown in M8-glutamate minimal medium supplemented with 0.2% glucose at 37°C for 36 h with shaking (250 rpm). Supernatants were subsequently prepared and measured for the relative C4-HSL contents. C) The relative expression of bfmR-lux in the wild-type MPAO1 and its derivatives when bacteria were grown in M8-glutamate minimal medium supplemented with 0.082% sodium acetate at 37°C for 36 h with shaking (250 rpm). Values are relative to MPAO1 (set to 1). D) The bfmR-lux activity in ΔbfmS/p-bfmS strain (bfmS), ΔbfmS/p-bfmSH238A strain (H238A), ΔbfmS/p-bfmSL181P/E376Q (L181P/E376Q), and ΔbfmS/p-bfmSL181P/E376Q/H238A strain (L181P/E376Q/H238A) when bacteria were grown in M8-glutamate minimal medium supplemented with 0.2% glucose at 37°C for 36 h with shaking (250 rpm). Values are relative to ΔbfmS/p-bfmS strain (set to 1). In A) to D), results are representative of three independent experiments and values represent means ± SEM. E) Phos-tag analysis shows that amino acid substitution L181P/E376Q, but not the L181P/E376Q/H238A, causes overproduction of phosphorylated and unphosphorylated BfmR. Western blot analysis of BfmS showing that missense mutations in bfmS do not affect the protein levels of BfmS, and immunoblots for RNAP (RNA polymerase) served as loading control. Cell lysates of ΔbfmRS::BfmR-Flag/p-bfmS strain (bfmS), ΔbfmRS::BfmR-Flag/p-bfmSL181P/E376Q (L181P/E376Q), and ΔbfmRS::BfmR-Flag/p-bfmSL181P/E376Q/H238A strain (L181P/E376Q/H238A) were used as described in Materials and Methods section. Specific missense mutations convert BfmS function from a repressor to an activator of BfmR
The DK2 lineage-specific mutations (L181P, L181P/E376Q) may abolish the negative regulatory effects of BfmS on BfmR or alternatively, it may transform BfmS into a positive regulator of BfmR and therefore, activate BfmR (Figure 6A and 6B). To discriminate between these two possibilities, we tested the effect of these amino acid substitutions in BfmS on the activity of bfmR-lux when bacteria were grown in M8-glutamate minimal medium supplemented with sodium acetate. BfmS did not function as a negative regulator of BfmR when bacteria were grown in this media, given that the ΔbfmS strain exhibited similar bfmR-lux activity as the MPAO1 strain (Figure 6C). However, the ΔbfmS/p-bfmSL181P strain and ΔbfmS/p-bfmSL181P/E376Q strain displayed bfmR-lux activity 2.5-fold and 6-fold higher than that the activity observed in the reference strain (ΔbfmS/p-bfmS), respectively (Figure 6C). These results suggest that the L181P or L181P/E376Q amino acid substitution render BfmS as a positive regulator of BfmR. Further, the introduction of p-bfmSL181P or p-bfmSL181P/E376Q to the bfmRS double deletion mutant strain (ΔbfmRS) failed to increase the activity of bfmR-lux (Figure S9A), suggesting that the effect of L181P or L181P/E376Q substitutions in BfmS on the bfmR-lux activity is mediated through the activation of BfmR.
BfmS is a member of the HisKA subfamily of bacterial histidine kinases and it is predicted that the conserved H238 residue is required for its kinase activity [37]. We next investigated the regulatory role associated with this residue by changing His to Ala. We created p-bfmSH238A and p-bfmSL181P/E376Q/H238A plasmids (Table S1 in Text S1) and introduced them to the ΔbfmS strain, and examined their effects on the bfmR-lux activity. As shown in Figure 6D, H238A substitution in BfmS has no significant effect on the activity of bfmR-lux when bacteria were grown in M8-glutamate minimal medium supplemented with 0.2% glucose. However, H238A substitutions in the BfmSL181P/E376Q abolished its ability to induce the bfmR-lux activity (Figure 6D), which suggests that the H238 residue or the kinase activity is required for BfmSL181P/E376Q to activate BfmR. This hypothesis is further supported by the fact that amino acid substitution L181P/E376Q, but not L181P/E376Q/H238A, causes overproduction of phosphorylated and unphosphorylated BfmR (Figure 6E).
Furthermore, we noted that the RP73-specific mutation in bfmS causes the substitution of arginine by histidine at the codon 393 (R393H) [38]. The P. aeruginosa strain RP73 was isolated after 16.9 years of chronic lung infection in a CF patient [38]. Like the L181P substitution or the L181P/E376Q substitution, the R393H substitution in BfmS resulted in increased bfmR-lux activity (Figure S9B and S9C) and decreased level of C4-HSL (Figure S9D), suggesting that BfmSR393H activates BfmR. As the regulatory effects of these BfmS variants (BfmSL181P, BfmSL181P/E376Q, and BfmSR393H) on BfmR have been changed from negative to positive, we therefore term them “reverse function” mutants. Although we failed to obtain either the soluble full-length BfmS protein or the cytoplasmic region of BfmS that prevented us from the assays of the phosphatase/kinase activities of BfmS in vitro, our genetic analyses clearly indicate that specific missense mutations (L181P, L181P/E376Q, or R393H) can convert BfmS from a repressor to an activator of BfmR.
Discussion
In this study, we uncovered a novel signal transduction pathway, BfmS/BfmR/RhlR, for the regulation of the rhl QS system in P. aeruginosa. We demonstrated that BfmS has profound effects on the expression of virulence-associated traits and the ability of P. aeruginosa to adapt to the host. In addition, we found that deletion of bfmS leads to a dramatic increase in biofilm formation and that BfmR mediates this effect (Figure S10). This appearance is consistent with previous observations that BfmR is a biofilm maturation regulator [24], [25]. Intriguingly, BfmS is able to switch its function from a repressor to an activator of BfmR in cystic fibrosis (CF) isolates such as DK2 strains and RP73 strain. A proposed model for signal transduction by BfmRS TCS is shown in Figure 7.
Fig. 7. Model of the regulatory networks involving BfmRS in P. aeruginosa. 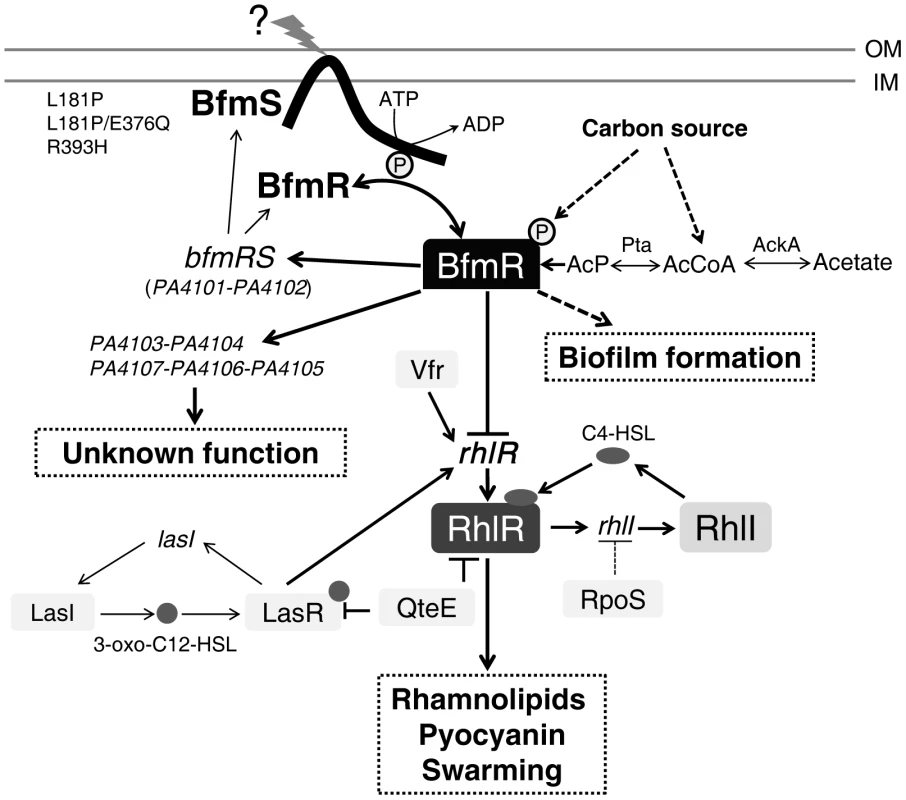
The lines show the interaction between the players: arrow, activation; hammerheads, repression; solid line, a direct influence or direct connection; dotted line, a putative or indirect connection. The question mark (“?”) denotes a yet-unidentified factor (factors) required for triggering the kinase or phosphatase activity of BfmS. QteE reduces LasR protein stability [80]. QteE also blocks RhlR accumulation, and this effect is independent of QteE's action on LasR [80]. Inactivation of RpoS causes elevated levels of rhlI (but not rhlR) transcription [81]. See text for details. OM, outer membrane. IM, inner membrane. AcP, acetyl phosphate. The prototypical two-component regulatory system is composed of a sensor kinase and a response regulator. In general, the sensor kinase senses an environment change and communicates it via phosphorylation to its cognate response regulator, and hence activates the response regulator's function. We demonstrated that BfmS functions as a negative regulator of BfmR (Figure 1D and Figure S4), whose activation requires phosphorylation on D55 (Figure S7). Therefore, BfmS might act as a phosphatase as opposed to a kinase for BfmR under our experimental conditions. In fact, many two-component sensors are bifunctional, catalyzing both the phosphorylation and dephosphorylation of their cognate response regulator [37], [39]–[41]. For some sensor kinases, the phosphatase activity may be the critical function in vivo [42]–[45]. Interestingly, the BfmS homologue in Pseudomonas syringae, RhpS, has also been shown to be a negative regulator of its cognate regulator RhpR in our previous studies [44], [45].
In the absence of BfmS, the Acka-Pta pathway can modulate the activity of BfmR (Figure 4). These observations suggest that the activation of BfmR is shaped by BfmS as well as by the nutritional status of P. aeruginosa. However, the expression of bfmR-lux in the ΔbfmSΔackA-pta strain was still about 12-fold higher than that in the wild-type MPAO1 strain (Figure 4A), indicating that acetyl phosphate or the component that is dependent on the Acka-Pta pathway is likely not the sole trigger of BfmR activation. This notion was further substantiated by the observation that the deletion of ackA-pta operon only partially alleviates the inhibition of QS signal C4-HSL production caused by the absence of bfmS (Figure 4B).
The rhlR gene encodes the transcriptional regulator RhlR, which has a central role in the quorum-sensing response, and is therefore very important for P. aeruginosa to co-ordinate its virulence in order to establish a successful infection [4], [6], [8]–[10], [46], [47]. We found that BfmR binds to and represses the rhlR promoter (Figure 3). A DNase I footprint analysis demonstrated that the BfmR-protected region (binding site I) of the rhlR promoter has a putative BfmR-binding motif (GATACTnnGC) that is crucial to the BfmR-mediated inhibition of rhlR-lux activity (Figure 3D), thereby reinforcing the likelihood that BfmR directly regulates rhlR. However, the detailed effect of BfmR on the rhlR gene expression awaits further study, since the rhlR promoter harbors multiple transcription start sites [15], regulatory sequences [15], and at least three BfmR-protected regions (Figure 3B). To our knowledge, this is the first evidence of a two-component regulator regulating rhlR in a direct manner. However, this finding is in contrast to a previous report suggesting that BfmR functions independently of QS signaling [24]. The exact cause of this discrepancy remains unknown. In the previous report [24], Petrova et al drew the conclusion based on the fact that deletion of bfmR has no significant effect on the transcript abundance of rhlA and lasB. Consistent with this, we observed that deletion of bfmRS has no significant effect on rhl-dependent phenotypes (Figure 1A and 1B) and the expression of rhlA (Table S4 in Text S1). However, deletion of bfmS alone leads to the activation of BfmR (Figure 1D and S4B), which in turn directly binds to the promoter and decreases the expression of the rhlR (Figure 3), causing the inhibition of the rhl QS system (Figure 1 and S3). Additionally, the decreased expression of rhlR in ΔbfmS strain (Figure 3C and 3D) may contribute to the attenuated virulence in lettuce leaves (Figure 5A) and the reduced production of QS signal C4-HSL, pyocyanin and rhamnolipids (Figure 1 and S1), since the constitutive expression of rhlR in ΔbfmS strain could restore these phenotypes to wild-type levels or higher (Figure 5B and S11). Moreover, the expression of rhlI in the ΔbfmS strain was significantly lower (>2-fold) than that of the wild-type strain (Tables S3 and S4 in Text S1). Thus, the bfmS deletion, which results in activation of BfmR, affects all aspects of the rhl QS system.
Deletion of bfmS impacts the expression of 202 genes that comprise 3.6% of the P. aeruginosa genome (Tables S2 and S3 in Text S1). These observations suggest that BfmS acts as a global regulator in P. aeruginosa. Besides regulating rhl quorum sensing, BfmS also regulates the expression of a large number of genes such as PA4103, PA4104, PA4105, PA4106 and PA4107, whose transcripts are likely independent of the quorum-sensing regulated [48]. Thus, it is not surprising that BfmS has a profound effect on the expression of virulence-associated traits and the ability of P. aeruginosa to adapt to the host (Figure 5). Interestingly, BfmS has a positive impact on acute virulence phenotypes (Figure 5A and 5E), but a negative effect on biofilm formation (Figure S10) that acts as a major virulence-associated trait contributing to chronic infections [49]. This formation suggests that BfmS may play an important role in mediating the switch between the acute and chronic infection lifestyles of P. aeruginosa.
P. aeruginosa can cause serious acute and chronic infections in humans and it underwent numerous genetic adaptations during evolution in the CF airways, resulting in remodeling of the regulatory networks to match the fluctuations in the environment of CF lung [36], [50]–[52]. bfmS in P. aeruginosa CF isolates are often found to undergo missense mutations. For instance, L181P (point mutations causing the substitution of leucine by proline at codon 181) or L181P/E376Q in at least 10 DK2 strains [36], R393H in RP73 strain [38], A21P/T120K/L164F in either LESB58 or LES431strain, L164F in c7447m strain, A4T in CIG1 strain, T120K/L163V/L164F in C3719 strain, D295N in CF5 strain, and P6S/L164F in CF614 strain (http://www.ncbi.nlm.nih.gov/). We found that specific missense mutations in bfmS gene (L181P, L181P/E376Q, and R393H) result in elevated BfmR activity (Figure 6 and Figure S9), which contributes to biofilm formation (Figure S10) [24], [25] as well as to the inhibition of the rhl QS system (Figures 1, 3, and S3). It is well known that biofilm formation enables P. aeruginosa to cause persistent infections [49] while the loss of quorum sensing is one of the dominating changes that occur during the adaptive process of the P. aeruginosa in the CF lung [36], [50]. Thus, the naturally occurring missense mutations in BfmS may provide a selective advantage to either DK2 strains or RP73 strain during the course of chronic infection in CF lungs. However, it should be noted that the P. aeruginosa community in the CF lung is very dynamic [50], [51], [53] and only a fraction of the isolates will most probably possess these mutations. Therefore, the role of these missense mutations in the chronic lung infection awaits further investigation.
Intriguingly, although BfmS functions as a negative regulator of BfmR (Figure 1D and S4B), the naturally occurring missense mutations in bfmS gene (L181P, L181P/E376Q, and R393H) can produce BfmS variants that no longer repress, but instead activate BfmR (Figure 6 and Figure S9). These “reverse function” mutants of BfmS may exhibit an elevated ratio of kinase to phosphatase activity on BfmR, given that the activation of BfmR requires the phosphorylation on D55 (Figure S7). In agreement with this notion, we found that H238, the conserved histidine residues predicted to be involved in the autophosphorylation of BfmS, is required for BfmSL181P/E376Q to activate BfmR (Figure 6D and 6E). The occurrence of BfmS “reverse function” mutants is not strain dependent, as evidenced by the fact that either DK2 lineage-specific mutations or RP73-specific mutation in bfmS could reverse its function against BfmR. The L181P mutation was located in the HAMP domain of BfmS, while the E376Q and the R393H mutation were located in ATP-binding domain. Currently, it is not clear how these missense mutations change the function of BfmS. Although further studies are needed to elucidate this elegant mechanism, our genetic analyses clearly indicated that naturally occurring missense mutations in P. aeruginosa gene could result in reverse of function, rather than simply loss (weakened) or gain (strengthened) of function.
Noticeably, bfmRS operon and BfmR-activated transcripts such as PA4103-4104 and PA4107-4106-4105 (Figure S2, Tables S2 and S4 in Text S1), were dramatically up-regulated in the lungs of cystic fibrosis patients compared to in vitro planktonic bacteria, indicating that BfmRS system is likely activated during chronic infection in CF lungs [23]. These genes also exhibit much higher gene expression levels in some P. aeruginosa CF isolates such as DK2-lineage strains (late stage infection isolates) [54] and E601 strain [55] compared to wild-type laboratory strain PAO1. These observations and results from the current study suggest that BfmRS TCS may sense and respond to environmental stress in CF lungs. We envision that further studies aimed at the characterization of the stimuli that BfmRS and/or its variants detect within the host could be of great importance to a full understanding of the mechanisms that make P. aeruginosa a successful pathogen and to the development of novel strategies to limit its infections.
Materials and Methods
Ethics statement
Animal experiments were performed in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China (11-14-1988). All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Public Health Clinical Center (Permit Number: 2013P201). The laboratory animal usage license number is SYXK-HU-2010-0098, certificated by Shanghai Committee of Science and Technology.
Bacterial strains, plasmids, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1 in Text S1. Unless noted otherwise, P. aeruginosa MPAO1 [56] and its derivatives were grown in Luria-Bertani (LB) broth, Pyocyanin production broth [57] (PPB: 20 g peptone, 1.4 g MgCl2, 10 g K2SO4, 20 ml glycerol per liter; pH 7.0), or M8-glutamate minimal medium [27] (6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 0.24 g MgSO4, 0.5 g glutamate per liter; pH 7.4) supplemented with 0.2% glucose, as indicated. E. coli cultures were grown in Luria-Bertani (LB) broth. All cultures were incubated at 37°C with shaking (250 rpm). For plasmid maintenance in P. aeruginosa, the medium was supplemented with 100 µg/ml carbenicillin or 100 µg/ml kanamycin when required. For plasmid maintenance in E. coli, the medium was supplemented with 100 µg/ml carbenicillin, 50 µg/ml kanamycin, 300 µg/ml trimethoprim, or 10 µg/ml gentamicin, as appropriate. For marker selection in P. aeruginosa, either 30 µg/ml gentamicin or 10 µg/ml tetracycline were used when required.
Construction of P. aeruginosa ΔbfmS, ΔbfmRS and ΔbfmSΔackA-pta mutants
For gene replacement, a SacB-based strategy [58] was employed as described in our previous study [26]. To construct the bfmS null mutant (ΔbfmS), polymerase chain reactions (PCRs) were performed in order to amplify sequences upstream (1,574 bp) and downstream (1,562 bp) of the intended deletion. The upstream fragment was amplified from MPAO1 genomic DNA using primers BfmSupF (with EcoRI site) and BfmSupR (with XbaI site), while the downstream fragment was amplified with primers, BfmSdownF (with XbaI site) and BfmSdownR (with HindIII site). The two PCR products were digested and then cloned into the EcoRI/HindIII-digested gene replacement vector pEX18Ap, yielding pEX18Ap::bfmSUD. A 1.8 kb gentamicin resistance cassette was cut from pPS858 with XbaI and then cloned into pEX18Ap::bfmSUD, yielding pEX18Ap::bfmSUGD. The resultant plasmid, pEX18Ap::bfmSUGD, was electroporated into MPAO1 with selection for gentamicin resistance. Colonies were screened for gentamicin sensitivity and loss of sucrose (5%) sensitivity, which typically indicates a double-cross-over event and thus marks the occurrence of gene replacement. The ΔbfmS strain was further confirmed by PCR.
A similar strategy was used to construct the ΔbfmRS strain as described above. Briefly, the upstream fragment (1,832 bp) of the intended deletion was amplified with primers BfmRupF (with EcoRI site) and BfmRupR (with XbaI site). The downstream fragment (1,562 bp) was amplified with primers, BfmSdownF (with XbaI site) and BfmSdownR (with HindIII site). A 1.8 kb gentamicin resistance cassette was cut from pPS858 with XbaI and then cloned into pEX18Ap::bfmRSUD, yielding pEX18Ap::bfmRSUGD.
Again, a similar strategy was used to construct the ΔbfmSΔackA-pta strain. Primers Acka-up-F (with KpnI site) and Acka-up-R (with BamHI site) amplified the upstream fragment (2,245 bp) of the intended deletion of ackA-pta operon in ΔbfmS. Primers Pta-domn-F (with BamHI site) and Pta-domn-R (with HindIII site) amplified the downstream fragment (1,709 bp). A 2.3 kb tetracycline resistance cassette was amplified from the integration vector mini-CTX-lacZ with primers, Mini-TC-F (with BamHI site) and Mini-TC-F (with BamHI site). The resultant plasmid, pEX18Ap::acka-ptaUTD, was electroporated into ΔbfmS strain with selection for tetracycline resistance. Colonies were screened for tetracycline sensitivity and loss of sucrose (5%) sensitivity, which typically indicate a double-cross-over event and thus mark the occurrence of gene replacement. PCR further confirmed the deletion of pta-acka loci.
Construction of chromosomal-borne BfmR-Flag strains
Primers bfmRflag-F (with HindIII site) and bfmRflag-R (with BamHI site) (Table S6 in Text S1) were used to perform PCR of the BfmR gene that was meant to fuse with the Flag-tag. A 1,586-bp PCR product covering the region from 848 bp upstream and the BfmR gene (not including the stop codon) was generated. The HindIII - and BamHI-digested PCR product was cloned into the HindIII and BamHI sites of the mini-CTX-lacZ [59] to generate mini-ctx-BfmR-Flag. The resulting plasmid was conjugated into P. aeruginosa MPAO1 and ΔbfmRS strains and the construct was integrated into the attB site as described previously though a diparental mating using E. coli S17 λ-pir as the donor, yielding a MPAO1::BfmR-Flag strain and a ΔbfmRS::BfmR-Flag strain, respectively (Table S1 in Text S1). In these mutant strains, parts of the mini-CTX-lacZ vector containing the tetracycline resistance cassette were deleted using a flippase (FLP) recombinase encoded on the pFLP2 plasmid.
Plasmid construction for the constitutive expression of bfmR, bfmS, bfmRS, rhlR, and acka-pta operon
To construct the plasmid for constitutive expression of bfmR, a 806 bp PCR product covering 15 bp of the bfmR upstream region, the bfmR gene, and 50 bp downstream of bfmR was amplified using primers BfmR(comp)Fwr (with HindIII site) and BfmR(comp)Rev (with BamHI site). The product was digested with HindIII and BamHI and ligated into PAK1900 [60] in the same orientation as plac to generate p-bfmR.
To construct the plasmid for the constitutive expression of bfmS, a 1,385 bp PCR product covering 30 bp of the bfmS upstream region, the bfmS gene, and 50 bp downstream of bfmS was amplified using primers BfmS(comp)Fwr (with HindIII site) and BfmS(comp)Rev (with BamHI site), and then cloned into PAK1900, yielding p-bfmS.
To construct the plasmid for the constitutive expression of bfmRS, a 2,107 bp PCR product covering 15 bp of the bfmR upstream region, the bfmRS operon, and 50 bp downstream of bfmS was amplified using primers BfmR(comp)Fwr (with HindIII site) and BfmS(comp)Rev (with BamHI site) and then cloned into PAK1900, yielding p-bfmRS.
To construct the plasmid for the constitutive expression of rhlR, a 770 bp DNA fragment covering 44 bp of the rhlR upstream region and the rhlR was amplified using primers RhlR-OE-F (with HindIII site) and RhlR-OE-R (with HindIII site) and then cloned into PAK1900. The construct with rhlR in the same orientation as plac was confirmed by DNA sequencing, yielding p-rhlR.
To construct the plasmid for constitutive expression of acka-pta operon, a 3,563 bp DNA fragment covering 136 bp of acka upstream region, the acka-pta operon, and a 65 bp downstream of pta was amplified using primers Acka-comp-F (with HindIII site) and Pta-comp-R (with BamHI site) and then cloned into PAK1900, yielding plasmid p-ackA-pta.
The five mutations, p-bfmRD55A, p-bfmSL181P, p-bfmSE376Q, and p-bfmSL181P/E376Q, and p-bfmSR393H were obtained using the QuikChange II site-directed mutagenesis kit (Stratagene). For generating p-bfmRD55A, the primer pair BfmR(D55A)-F/BfmR(D55A)-F was used. For generating p-bfmSL181P, primer pair PA4102L181P-F/PA4102L181P-R was used. For generating p-bfmSE376Q, the primer pair PA4102E376Q-F/PA4102E376Q-R was used. For generating p-bfmSL181P/E376Q, primer pairs PA4102L181P-F/PA4102L181P-R and PA4102E376Q-F/PA4102E376Q-R were used. For generating p-bfmSR393H, the primer pairs R393H-F/R393H-R was used.
All constructs were sequenced to ensure that no unwanted mutations resulted.
Construction, expression, and purification of 6His-BfmR, 6His-BfmRD55A, and BfmS34–154
Full-length of bfmR was cloned into pET28a with a thrombin-cleavable N-terminal His-tag. Primers bfmR-F (with NdeI site) and bfmR-R (XhoI) were used to amplify the bfmR gene from P. aeruginosa MPAO1 chromosomal DNA. The amplified fragments were ligated into similarly cut pET28a (Novagen) in order to produce the plasmids pET28a-6His-BfmR. pET28a-6His-BfmRD55A was obtained by using the primer pair BfmR(D55A)-F/BfmR(D55A)-R and a QuikChange II site-directed mutagenesis kit (Stratagene). The protein was expressed in the E. coli strain BL21 star (DE3) and purifications were performed as described in our previous studies [26], [61], [62]. Briefly, bacteria were grown at 37°C overnight in 10 ml of LB medium (containing 50 µg/ml kanamycin) with shaking (250 rpm). The next day, the cultures were transferred into 1 L of LB medium (containing 50 µg/ml kanamycin) incubated at 37°C with shaking (250 rpm) until the OD600 reached 0.6, and then IPTG (isopropyl-1-thio-β-d-galactopyranoside) was added to a final concentration of 1.0 mM. After 4 h incubation at 30°C with shaking (250 rpm), the cells were harvested by centrifugation and stored at −80°C. The cells were lysed at 4°C by sonication in lysis buffer [10 mM Tris (pH 7.4), 300 mM NaCl, 1 mM PMSF, and 2 mM DTT]. Clarified cell lysate was loaded onto a HisTrap HP column (Amersham Biosciences), washed with Ni-NTA washing buffer, and eluted with Ni-NTA elution buffer. The fractions containing 6His-BfmR or 6His-BfmRD55A were concentrated and loaded onto a Superdex-200 gel filtration column with a running condition of 10 mM Tris (pH 7.4), 300 mM NaCl, and 2 mM DTT. The purified protein was >90% pure as estimated by a 12% (wt/vol) SDS/PAGE gel.
The DNA sequence of the extracellular sensory domain of BfmS consisting of 121 residues (Gln34-Trp154) was amplified from MPAO1 genomic DNA with the primers PA4102-EX-F (with NcoI) and PA4102-EX-R (with BamHI) by PCR and was subsequently cloned into pET28b using NcoI and BamHI as the restriction enzymes. Following confirmation by DNA sequencing, the recombinant plasmid (pET28b-bfmS34–154) was transformed into E. coli strain BL21 star (DE3). The extracellular sensory domain of BfmS (designated BfmS34–154) was expressed and purified as described above with some modifications. Briefly, bacteria were grown at 37°C overnight in 10 ml of LB medium (containing 50 µg/ml kanamycin) with shaking (250 rpm). The next day, the cultures were transferred into 1 L of LB medium (containing 50 µg/ml kanamycin) incubated at 37°C with shaking (200 rpm) until the OD600 reached 0.6, and then IPTG was added to a final concentration of 1.0 mM. After 16 h incubation at 16°C with shaking (200 rpm), the cells were harvested by centrifugation and stored at −80°C. The cells were lysed at 4°C by sonication in lysis buffer [50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10% glycerol 1 mM PMSF, and 2 mM DTT]. Clarified cell lysate was loaded onto a HisTrap HP column (Amersham Biosciences), and eluted with Ni-NTA elution buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 20 mM imidazole 10% glycerol, and 1 mM DTT,). The fractions containing BfmS34–154 were concentrated and loaded onto a Superdex-75 gel filtration column with a running condition of 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10% glycerol, 1 mM DTT. The purified protein was >90% pure as estimated by a 12% (wt/vol) SDS/PAGE gel.
Monitoring gene expression by lux-based reporters
The plasmid pMS402 [63] carrying a promoterless luxCDABE reporter gene cluster was used to construct promoter-luxCDABE reporter fusions of the bfmR as described previously [28], [64]. For bfmR-lux, the bfmR promoter region (−463 to +18 of the start codon) was amplified by PCR using the primers PMS402-bfmR-F (with XhoI site) and PMS402-bfmR-R (with BamHI site). For rhlA-lux, the rhlA promoter region (−526 to −20 of the start codon) was amplified by PCR using the primers pms402-rhlA-F (with XhoI site) and pms402-rhlA-R (with BamHI site). For rhlR-lux, the rhlR promoter region (−450 to +19 of the start codon) was amplified by PCR using the primers pms402-rhlR-1F (with BamHI site) and pms402-rhlR-R (with BamHI site). The promoter oriented in the same direction as luxCDABE was selected for further analysis. To generate rhlR promoter mutant rhlR-D (deletion of GATACT, which is the central part of the putative BfmR-binding site on the reverse DNA strand), the DNA fragment was amplified using primers pms402-rhlR-1F/pms402-rhlR-R and subsequently cloned into pGEM-T vector. rhlR-D (rhlR promoter lacking putative BfmR-binding site) was obtained using a QuikChange II site-directed mutagenesis kit (Stratagene) and primer pair pms402-rhlR(D1)F/pms402-rhlR(D1)R. For 4103-lux, the PA4103 promoter region (−659 to +19 of the start codon) was amplified by PCR using primers pms402-p4103-F (with XhoI site) and pms402-p4103-R (with BamHI site). To generate PA4103 promoter mutant 4103-M (GATACA was mutated to ATATAT), primer pair p4103-mutation-F/p4103-mutation-R was used as described above. The promoter regions were cloned into the XhoI-BamHI site or BamHI site (for rhlR-lux) upstream of the lux genes on pMS402 and the cloned promoter sequences were confirmed by DNA sequencing. The constructs were transformed into MPAO1 or its derivatives by electroporation. Use of these lux-based reporters, gene expression under different conditions was measured as counts per second (cps) of light production with a 2104 EnVision Multilabel Plate Readers or Synergy 2 (Biotek). Relative light units were calculated by normalizing CPS to OD600.
Quantitative real-time PCR
The bacterial growth and the extraction of total RNAs were performed as described above. The total DNase-treated RNA (5 µg) was reversely transcribed to synthesize cDNA using the PrimeScript RT reagent Kit (Takara) with random primers according to the manufacturer's protocol. The resulting cDNA were diluted by 1∶2, 1∶4, and 1∶8 respectively. Triplicate quantitative assays were performed on 1 µl of each cDNA dilution with the THUNDERBIRD SYBR qPCR Mix and 300 nM primers using an Applied Biosystems 7500 Fast Real-Time PCR System. Dissociation curve analysis was performed in order to verify product homogeneity. The gene-specific primers used for Quantitative real-time PCR for PA4100, PA4103, PA4107, PA4108, ntrB, oprH, phoB, hmgA, rhlA, antA, nasA, and rhlI are listed in Table S6 in Text S1. The amplicon of 16S rRNA was used as an internal control in order to normalize all data. Relative expression levels of interest genes were calculated by the relative quantification method (ΔΔCT) as previously described [65], [66].
Sample preparation for in vivo detection of BfmR phosphorylation and Phos-tag gel electrophoresis
P. aeruginosa was grown at 37°C for 24 h on M8-glutamate minimal agar plate (M8-glutamate minimal medium supplemented with 0.2% glucose, and solidified with 2% agar). To prepare cell lysates for the Phos-tag gel assay, bacteria cells were scraped from the plate and immediately resuspended in 60 µl of lysis buffer [50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 1 mM MgCl2, 0.1% Triton X-100, 15 µg/ml DNaseI, 0.5 mM PMSF, 1 mM DTT) with 0.1% (vol/vol) Lysonase. Sufficient lysis was achieved by repeated pipetting up and down for 10 s followed by addition of 20 µl of 4×SDS loading buffer. Resulting cell lysates (10 µl) were immediately loaded onto a Phos-tag gel for electrophoresis.
BfmR-flag and BfmR-flag∼P were separated on 10% acrylamide gels containing 25 µM acrylamide-Phos-tag ligand (Wako Pure Chemical) and 50 µM MnCl2 as previously described [67]. Electrophoresis was performed at 30 mA at 4°C for 80 min in Tris-Glycine-SDS running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS, pH 8.4). After electrophoresis, the Phos-tag gel was washed 10 min at RT with Transfer Buffer [20%(v/v) methanol, 50 mM Tris, 40 mM glycine] supplied with 1 mM EDTA to remove Zn2+ from the gel, then the gel was incubated at room temperature with gentle shaking for another 10 min in Transfer Buffer twice to remove EDTA.
Western blot analysis
Samples resolved on gels were transferred to PVDF (Bio-Rad) membranes through semi-dry transfer assembly (Bio-Rad) for 30 min at room temperature. BfmR-Flag proteins were detected by Western blot analysis using a mouse anti-Flag monoclonal antibody (Cat#: AGM12165, Aogma) followed by a secondary, sheep anti-mouse IgG antibody conjugated to horseradish peroxidase (HRP) (Code#: NA931, GE Healthcare). For detection of ClpP protein, anti-ClpP polyclonal antibody and anti-rabbit IgG antibody conjugated to horseradish peroxidase (HRP) (Code#: NA934, GE Healthcare) were used. Anti-ClpP polyclonal antibody, which cross-reacts with the ClpP of Pseudomonas aeruginosa, was prepared by immunizing a rabbit with a Staphylococcus aureus full-length ClpP protein (NWMN_0736). For detection of BfmS protein, anti-BfmS polyclonal antibody (prepared by immunizing a rabbit with a BfmS34–154 protein, Shanghai Immune Biotech CO., Ltd) and anti-rabbit IgG antibody conjugated to horseradish peroxidase (HRP) (Code#: NA934, GE Healthcare) were used. For detection of RNAP protein, anti-RNAP (Neoclone, #WP003) antibody and anti-mouse IgG antibody conjugated to horseradish peroxidase (HRP) (Code#: NA931, GE Healthcare). Immunoblots for ClpP and RNAP served as loading control. The membrane is exposed to X-ray film (Kodak) or the chemiluminescent is detected by a Imaging Quant LAS-4000 (GE), according to the manufacturer's recommendation.
Measurement of pyocyanin production
Pyocyanin was extracted from culture supernatants and measured using previously reported methods [68]. Briefly, P. aeruginosa was grown in Pyocyanin production broth [57] (PPB: 20 g peptone, 1.4 g MgCl2, 10 g K2SO4, 20 ml glycerol per liter; pH 7.0) for 36 h at 37°C with shaking (250 rpm). The culture was subsequently centrifuged and filtered (pore size, 0.22 µm). 1.5 ml of chloroform was added to 2.5 ml of culture supernatant. After extraction, the chloroform layer was transferred to a fresh tube and mixed with 1 ml of 0.2 N HCl. After centrifugation, the top layer (0.2 N HCl) was removed and its absorption measured at 520 nm. Concentrations, expressed as micrograms of pyocyanin produced per ml of culture supernatant, were determined by multiplying the optical density at 520 nm (OD520) by 17.072.
Measurement of rhamnolipid production
Rhamnolipids production was estimated by inoculating strains on M8-based agar plates supplemented with 0.2% glucose(m/v), 2 mM MgSO4, 0.0005% (m/v) methylene blue, and 0.02% (m/v) cetyltrimethylammonium bromide, as described previously [68], [69]. The orcinol assay was used to directly assess the amount of rhamnolipids in the sample as previously described [28]. After a culture of 48 h in LB medium at 37°C with shaking (250 rpm), 1 ml of the culture supernatant was extracted twice with 2 ml of diethyl ether. The pooled ether fractions were evaporated to dryness and the remainder was dissolved in 100 µl of distilled water and mixed with 100 µl of 1.6% orcinol, and 800 µl of 60% sulfuric acid. After heating for 30 min at 80°C in the dark, the samples were cooled for 3 h at room temperature in the dark. Absorbance at 421 nm (A421) was measured. Rhamnolipid concentrations were calculated by comparing A421 values with those obtained for rhamnose standards between 0 and 1000 µg/ml, assuming that 1 µg of rhamnose corresponds to 2.5 µg of rhamnolipids.
Swarming motility assays
The motility assay was carried out as described previously [27], [68]. Swarming medium was based on M8-glutamate minimal medium [27] (6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 0.24 g MgSO4, 0.5 g glutamate per liter; pH 7.4), supplemented with MgSO4 (2 mM), glucose (0.2%), and Casamino acid (0.5%), and solidified with 0.5% agar. Bacteria were inoculated with a toothpick onto swarm agar plates. Swarm agar plates were incubated for 24 hours at 37°C and then incubated for more time at room temperature.
In vitro phosphorylation assays
Phosphorylation of 6His-BfmR was detected by utilizing the Pro-Q Diamond phosphorylation gel stain as described by the manufacturer (Invitrogen). Purified 6His-BfmR and 6His-BfmRD55A were incubated with buffer (10 mM Tris pH 8.0; 1 mM DTT; 5 mM MgCl2; 10 mM KCl; 50 mM acetyl phosphate) at 37°C for 30 min. The acetyl phosphate-treated samples of 6His-BfmR and 6His-BfmRD55A were resolved on a 12% SDS polyacrylamide gel, and then the gel was immersed in fixing solution (10% acetic acid, 50% methanol) for 30 min and subsequently washed three times with deionized water each for 10 min. The gel was stained with Pro-Q Diamond phosphoprotein gel stain for 60 min, followed by washing with deionized water for 30 min. The entire procedure was conducted at room temperature. Fluorescent output was recorded using an Tanon-5200 multi.
Electrophoretic mobility shift assay (EMSA)
The electrophoretic mobility shift experiments were performed as described in our previous studies with some modifications [26], [61], [62]. Briefly, 20 µl of the DNA probe mixture (30 to 50 ng) and various amounts of purified proteins in binding buffer (10 mM Tris-Cl, pH 8.0; 1 mM DTT; 10% glycerol; 5 mM MgCl2; 10 mM KCl) were incubated for 30 min at 37°C. When indicated, 50 mM acetyl phosphate was added to the solution. Native polyacrylamide gel (6%) was run in 0.5× TBE buffer at 85 V at 4°C. The gel was stained with GelRed nucleic acid staining solution (Biotium) for 10 min, and then the DNA bands were visualized by gel exposure to 260-nm UV light.
DNA probes were PCR-amplified from P. aeruginosa MPAO1 genomic DNA using the primers listed in Table S6 in Text S1. The probes for bfmR promoter, a 481 bp DNA fragment covering the promoter region of bfmR (from −463 to +18 of the start codon) was amplified using primers bfmR-F(EMSA) and bfmR-R(EMSA). For rhlR promoter, a 470 bp DNA fragment covering the promoter region of rhlR (from −450 to +20 of the start codon) was amplified using primers rhlR-F(EMSA) and rhlR-F(EMSA). For rhlI promoter, a 446 bp DNA fragment covering the promoter region of rhlI (from −444 to +2 of the start codon) was amplified using primers rhlI-F(EMSA) and rhlI-R(EMSA). For rhlA promoter, a 572 bp DNA fragment covering the promoter region of rhlA (from −591 to −19 of the start codon) was amplified using primers rhlA-F(EMSA) and rhlA-R(EMSA). For rhlC promoter, a 540 bp DNA fragment covering the promoter region of rhlC (from −549 to −9 of the start codon) was amplified using primers rhlC-F(EMSA) and rhlC-F(EMSA). For PA4103 promoter, a ca. 0.7 kb DNA fragment (4103-P) containing the promoter region of PA4103 (from −659 to +19 of the start codon) was amplified from plasmid 4103-lux DNA using primers pZE.05 and pZE.06. For PA4107 promoter, a 360 bp DNA fragment (4107-P) covering the promoter region of PA4107 (from −490 to −131 of the start codon) was amplified from P. aeruginosa MPAO1 genomic DNA using primers PA4108-F and PA4108-R. All PCR products were purified by using a QIAquick gel purification kit (QIAGEN).
Dye primer-based DNase I footprinting assay
The published DNase I footprint protocol was modified [70] in this study in the same way as described in our previous study [61]. Briefly, PCR was used to generate DNA fragments using the primer sets as detailed in Table S6 in Text S1. For amplification of bfmR promoter, primers bfmR-F(EMSA) and 6FAM-bfmR-R were used. For amplification of the rhlR promoter, primers rhlR-F(EMSA) and 6FAM-rhlR-R were used. For amplification of the PA4103 promoter, p4103-F (EMSA) and p4103-R-FAM were used. All PCR products were purified by with QIAquick gel purification kit (QIAGEN). 50 nM 6-carboxyfluorescein (6-FAM)-labeled promoter DNA and various amounts of 6His-BfmR (as indicated) in 50 µl of binding buffer (10 mM Tris-Cl, pH 8.0; 1 mM DTT; 10% glycerol; 5 mM MgCl2; 10 mM KCl; 50 mM acetyl phosphate) were incubated at room temperature for 10 min. 0.01 unit of DNase I was added to the reaction mixture and incubated for 5 more min. The digestion was terminated by adding 90 µl of quenching solution (200 mM NaCl, 30 mM EDTA, 1% SDS), and then the mixture was extracted with 200 µl of phenol-chloroform-isoamyl alcohol (25∶24∶1). The digested DNA fragments were isolated by ethanol precipitation. 5.0 µl of digested DNA was mixed with 4.9 µl of HiDi formamide and 0.1 µl of GeneScan-500 LIZ size standards (Applied Biosystems). A 3730XL DNA analyzer detected the sample, and the result was analyzed with GeneMapper software.
Analyses of gene expressions with oligonucleotide microarray
Overnight P. aeruginosa cultures were washed and diluted 100-fold in M8-glutamate minimal medium (6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 0.24 g MgSO4, 0.5 g Glutamate per liter; pH 7.4) supplemented with glucose (2 g/L). The bacteria were subsequently grown at 37°C for 48 h (OD600≈1.0) with shaking (250 rpm). Total RNA was immediately stabilized with RNAprotect Bacteria Reagent (Qiagen, Valencia, CA) and then extracted using a Qiagen RNeasy kit following the manufacturer's instructions. The total DNase-treated RNA samples were then analyzed by CapitalBio Corp for Chip (Affymetrix) assay. Briefly, samples were labeled according to the manufacturer (Affymetrix, Santa Clara, CA) and then hybridized to the Affymetrix GeneChip P. aeruginosa genome array (catalog number AFF-900339) for 16 h at 50°C though the use of the GeneChip hybridization oven at 60 rpm. Washing, staining, and scanning were performed using the Affymetrix GeneChip system. The data were normalized using Robust Multi-array Average (RMA) [71]. Gene expression analysis was performed using three independent mRNA samples for each strain. Microarray data were analyzed using SAM (Significance Analysis of Microarrays) software [72]. Criterion such as cutoff limitation for fold change ≥2 or ≤0.5 and q-value ≤5% was used to select differential expression genes. All data were submitted to the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-1983.
Bioassay of C4-HSL activity
The autoinducer of the rhl system, C4-HSL, was measured using an rhlA promoter-based P. aeruginosa strain, pDO100 (pKD-rhlA) [28]. This detection system was developed by fusing the C4-HSL-responsive rhlA promoter upstream of luxCDABE and introducing the construct into pDO100, a rhlI mutant strain [28]. Procedures were modified from the protocol described previously [28]. Briefly, the reporter strain pDO100 (pKD-rhlA) was grown in LB medium plus 100 µg/ml kanamycin overnight at 37°C with shaking (250 rpm) and diluted to an OD600 of 0.05 in fresh LB plus kanamycin. 90 µl was subsequently added to the wells of a 96-well microtitre plate. A 10 ml portion of the samples or medium control was added to the wells. The luminescence value was measured in a 2104 EnVision Multilabel Plate Readers or Synergy 2 (Biotek), and calculated from the luminescence value minus that of the medium control. The data are presented as CPS and are not normalized to OD600 of pDO100 (pKD-rhlA). In this assay, the growth curves of pDO100 (pKD-rhlA) are identical.
Infection of MLE-12 cells
Different strains of bacteria were grown overnight in Luria-Bertani (LB) broth at 37°C with shaking. Then, the bacteria were subject to pelleting by centrifugation at 5,000 g and resuspended in 10 ml of fresh LB broth and allowed to grow until the mid-logarithmic phase. OD600 nm was measured, and the density was adjusted to 0.25 OD (0.1 OD = 1×108 cells/ml). Mammalian cells were washed once with PBS after overnight culture in full medium, and changed to a serum-free and antibiotic-free medium immediately before infection. Cells were infected by various strains at a multiplicity of infection (m.o.i) of 10∶1 bacteria-to-cell ratio at indicated time points for 2 h. The cells were washed three times with PBS to remove surface bacteria and incubated with 100 µg/ml polymyxin B for 1 h. The cells were lyzed to evaluate the internalized bacteria using CFU assay on agar dishes as described in our previous studies [73], [74].
3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium Bromide (MTT) assay
The killing of MLE-12 cells by bacterial infection was performed by continuing incubation for 24 h and cell survival measured using MTT assay [75]. MLE-12 cells were cultured in 96-well plates as above. After incubation for 24 h, MTT dye was added to the cells in each well with at a final concentration of 1 µg/ml. Then, the cells were incubated at 37°C until the color developed. The yellow color may change to brown upon reduction by enzymes. The reaction was terminated by adding 100 µl of stop solution (10% DMSO, 10% SDS in 50 mM HEPES buffer). The plate was left at room temperature overnight. The next day, the 560-nm absorbance was read using a plate reader in order to quantify the dye conversion [76]. Background correction was done with controls containing only the dye.
Lettuce leaf model of infection
A lettuce leaf virulence assay was performed as described previously [77]–[79]. Briefly, P. aeruginosa strains were grown aerobically overnight at 37°C with shaking (250 rpm) in PPB broth or PPB broth containing carbenicillin (100 µg/ml) when appropriate, washed, resuspended, and diluted in sterile MgSO4 to a bacterial density of 1×109 CFU/ml. Lettuce leaves were prepared by washing with sterile distilled H2O and 0.1% bleach. Samples (10 µl) were then inoculated into the midribs of Romaine lettuce leaves. Containers containing Whatman paper moistened with 10 mM MgSO4 and inoculated leaves were kept in a growth chamber at 37°C for five days. Symptoms were monitored daily. As a control, lettuce leaves were inoculated with 10 mM MgSO4.
Mouse model of acute pneumonia
All P. aeruginosa strains were grown at 37°C overnight in PPB medium with shaking (250 rpm), diluted 100-fold in fresh PPB medium, and incubated at 37°C for 2.5–3.0 h until the cultures reached OD600 0.8. Bacteria were collected by centrifugation, washed, and suspended in PBS buffer. Viable P. aeruginosa were enumerated by colony formation on Pseudomonas isolation agar (PIA) (Difco) plates in order to quantify the infectious dose. Mouse infections were carried out as described previously [26], using 8-week-old female C57BL/6J mice obtained from Shanghai SLAC Laboratory Animal Co. Ltd. and housed under specified pathogen-free conditions. Mice were anaesthetized with pentobarbital sodium (intraperitoneal injection, 80 mg/kg) and intranasally infected with c. 5×106 cfu of each bacterial isolate. After that, animals were sacrificed 18 h post infection. Lungs were aseptically removed and homogenized in PBS plus 0.1% Triton X-100 in order to obtain single-cell suspensions. Serial dilutions of each organ were plated on Pseudomonas isolation agar (PIA) (Difco) plates. Bacterial burden per organ was calculated and is expressed as a ratio of the inoculum delivered per animal. Statistical analysis was performed using Prism software (GraphPad).
Statistical analysis
Two-tailed Student's t tests or Mann–Whitney test was used to calculate p-values (two-tailed) using Prism software (GraphPad), as indicated. * p<0.05, ** p<0.01, *** p<0.001.
Supporting Information
Zdroje
1. StoverCK, PhamXQ, ErwinAL, MizoguchiSD, WarrenerP, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406 : 959–964.
2. National Nosocomial Infections Surveillance S (2004) National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control 32 : 470–485.
3. LyczakJB, CannonCL, PierGB (2002) Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15 : 194–222.
4. SmithRS, IglewskiBH (2003) P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol 6 : 56–60.
5. WilliamsP, CamaraM (2009) Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12 : 182–191.
6. JimenezPN, KochG, ThompsonJA, XavierKB, CoolRH, et al. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76 : 46–65.
7. GooderhamWJ, HancockRE (2009) Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33 : 279–294.
8. BalasubramanianD, SchneperL, KumariH, MatheeK (2013) A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41 : 1–20.
9. RutherfordST, BasslerBL (2012) Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2 doi: 10.1101/cshperspect.a012427
10. SchusterM, GreenbergEP (2006) A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296 : 73–81.
11. RodrigueA, QuentinY, LazdunskiA, MejeanV, FoglinoM (2000) Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol 8 : 498–504.
12. RouxA, PayneSM, GilmoreMS (2009) Microbial telesensing: probing the environment for friends, foes, and food. Cell Host Microbe 6 : 115–124.
13. DekimpeV, DezielE (2009) Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155 : 712–723.
14. LeeJ, WuJ, DengY, WangJ, WangC, et al. (2013) A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol 9 : 339–343.
15. Croda-GarciaG, Grosso-BecerraV, Gonzalez-ValdezA, Servin-GonzalezL, Soberon-ChavezG (2011) Transcriptional regulation of Pseudomonas aeruginosa rhlR: role of the CRP orthologue Vfr (virulence factor regulator) and quorum-sensing regulators LasR and RhlR. Microbiology 157 : 2545–2555.
16. StockAM, RobinsonVL, GoudreauPN (2000) Two-component signal transduction. Annu Rev Biochem 69 : 183–215.
17. ShilohMU, ManzanilloP, CoxJS (2008) Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3 : 323–330.
18. TorresVJ, StauffDL, PishchanyG, BezbradicaJS, GordyLE, et al. (2007) A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1 : 109–119.
19. DongYH, ZhangXF, AnSW, XuJL, ZhangLH (2008) A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun Integr Biol 1 : 88–96.
20. ReimmannC, BeyelerM, LatifiA, WintelerH, FoglinoM, et al. (1997) The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol 24 : 309–319.
21. JensenV, LonsD, ZaouiC, BredenbruchF, MeissnerA, et al. (2006) RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J Bacteriol 188 : 8601–8606.
22. DieppoisG, DucretV, CailleO, PerronK (2012) The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS One 7: e38148.
23. SonMS, MatthewsWJJr, KangY, NguyenDT, HoangTT (2007) In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75 : 5313–5324.
24. PetrovaOE, SchurrJR, SchurrMJ, SauerK (2011) The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol 81 : 767–783.
25. PetrovaOE, SauerK (2009) A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5: e1000668.
26. LanL, MurrayTS, KazmierczakBI, HeC (2010) Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol Microbiol 75 : 76–91.
27. KohlerT, CurtyLK, BarjaF, van DeldenC, PechereJC (2000) Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol 182 : 5990–5996.
28. LiangH, DuanJ, SibleyCD, SuretteMG, DuanK (2011) Identification of mutants with altered phenazine production in Pseudomonas aeruginosa. J Med Microbiol 60 : 22–34.
29. SchusterM, LostrohCP, OgiT, GreenbergEP (2003) Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185 : 2066–2079.
30. OglesbyAG, FarrowJM3rd, LeeJH, TomarasAP, GreenbergEP, et al. (2008) The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem 283 : 15558–15567.
31. WolfeAJ (2005) The acetate switch. Microbiol Mol Biol Rev 69 : 12–50.
32. FleiszigSM, ZaidiTS, PrestonMJ, GroutM, EvansDJ, et al. (1996) Relationship between cytotoxicity and corneal epithelial cell invasion by clinical isolates of Pseudomonas aeruginosa. Infect Immun 64 : 2288–2294.
33. YuanK, HuangC, FoxJ, GaidM, WeaverA, et al. (2011) Elevated inflammatory response in caveolin-1-deficient mice with Pseudomonas aeruginosa infection is mediated by STAT3 protein and nuclear factor kappaB (NF-kappaB). J Biol Chem 286 : 21814–21825.
34. GuoQ, ShenN, YuanK, LiJ, WuH, et al. (2012) Caveolin-1 plays a critical role in host immunity against Klebsiella pneumoniae by regulating STAT5 and Akt activity. Eur J Immunol 42 : 1500–1511.
35. LiX, ZhouX, YeY, LiY, LiJ, et al. (2013) Lyn regulates inflammatory responses in Klebsiella pneumoniae infection via the p38/NF-kappaB pathway. Eur J Immunol doi: 10.1002/eji.201343972
36. YangL, JelsbakL, MarvigRL, DamkiaerS, WorkmanCT, et al. (2011) Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A 108 : 7481–7486.
37. WillettJW, KirbyJR (2012) Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet 8: e1003084.
38. JeukensJ, BoyleB, BianconiI, Kukavica-IbruljI, TummlerB, et al. (2013) Complete Genome Sequence of Persistent Cystic Fibrosis Isolate Pseudomonas aeruginosa Strain RP73. Genome Announc 1: e00568–13.
39. AminM, PorterSL, SoyerOS (2013) Split histidine kinases enable ultrasensitivity and bistability in two-component signaling networks. PLoS Comput Biol 9: e1002949.
40. HuynhTN, StewartV (2011) Negative control in two-component signal transduction by transmitter phosphatase activity. Mol Microbiol 82 : 275–286.
41. KenneyLJ (2010) How important is the phosphatase activity of sensor kinases? Curr Opin Microbiol 13 : 168–176.
42. RaivioTL, SilhavyTJ (1997) Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol 179 : 7724–7733.
43. KostakiotiM, HadjifrangiskouM, PinknerJS, HultgrenSJ (2009) QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol Microbiol 73 : 1020–1031.
44. DengX, LanL, XiaoY, KennellyM, ZhouJM, et al. (2010) Pseudomonas syringae two-component response regulator RhpR regulates promoters carrying an inverted repeat element. Mol Plant Microbe Interact 23 : 927–939.
45. XiaoY, LanL, YinC, DengX, BakerD, et al. (2007) Two-component sensor RhpS promotes induction of Pseudomonas syringae type III secretion system by repressing negative regulator RhpR. Mol Plant Microbe Interact 20 : 223–234.
46. LimmerS, HallerS, DrenkardE, LeeJ, YuS, et al. (2011) Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc Natl Acad Sci U S A 108 : 17378–17383.
47. RahmeLG, StevensEJ, WolfortSF, ShaoJ, TompkinsRG, et al. (1995) Common virulence factors for bacterial pathogenicity in plants and animals. Science 268 : 1899–1902.
48. WagnerVE, BushnellD, PassadorL, BrooksAI, IglewskiBH (2003) Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185 : 2080–2095.
49. CostertonJW, StewartPS, GreenbergEP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284 : 1318–1322.
50. SmithEE, BuckleyDG, WuZ, SaenphimmachakC, HoffmanLR, et al. (2006) Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103 : 8487–8492.
51. OliverA, CantonR, CampoP, BaqueroF, BlazquezJ (2000) High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288 : 1251–1254.
52. DamkiaerS, YangL, MolinS, JelsbakL (2013) Evolutionary remodeling of global regulatory networks during long-term bacterial adaptation to human hosts. Proc Natl Acad Sci U S A 110 : 7766–7771.
53. YangL, JelsbakL, MolinS (2011) Microbial ecology and adaptation in cystic fibrosis airways. Environ Microbiol 13 : 1682–1689.
54. YangL, RauMH, YangL, HoibyN, MolinS, et al. (2011) Bacterial adaptation during chronic infection revealed by independent component analysis of transcriptomic data. BMC Microbiol 11 : 184.
55. TralauT, VuilleumierS, ThibaultC, CampbellBJ, HartCA, et al. (2007) Transcriptomic analysis of the sulfate starvation response of Pseudomonas aeruginosa. J Bacteriol 189 : 6743–6750.
56. JacobsMA, AlwoodA, ThaipisuttikulI, SpencerD, HaugenE, et al. (2003) Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100 : 14339–14344.
57. BrintJM, OhmanDE (1995) Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol 177 : 7155–7163.
58. SchweizerHP, HoangTT (1995) An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158 : 15–22.
59. BecherA, SchweizerHP (2000) Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29 : 948–950, 952.
60. JansonsI, TouchieG, SharpR, AlmquistK, FarinhaMA, et al. (1994) Deletion and transposon mutagenesis and sequence analysis of the pRO1600 OriR region found in the broad-host-range plasmids of the pQF series. Plasmid 31 : 265–274.
61. LiuX, SunX, WuY, XieC, ZhangW, et al. (2013) Oxidation-sensing regulator AbfR regulates oxidative stress responses, bacterial aggregation, and biofilm formation in Staphylococcus epidermidis. J Biol Chem 288 : 3739–3752.
62. SunF, DingY, JiQ, LiangZ, DengX, et al. (2012) Protein cysteine phosphorylation of SarA/MgrA family transcriptional regulators mediates bacterial virulence and antibiotic resistance. Proc Natl Acad Sci U S A 109 : 15461–15466.
63. DuanK, DammelC, SteinJ, RabinH, SuretteMG (2003) Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol 50 : 1477–1491.
64. DuanK, SuretteMG (2007) Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol 189 : 4827–4836.
65. LanL, ChenW, LaiY, SuoJ, KongZ, et al. (2004) Monitoring of gene expression profiles and isolation of candidate genes involved in pollination and fertilization in rice (Oryza sativa L.) with a 10K cDNA microarray. Plant Mol Biol 54 : 471–487.
66. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 : 402–408.
67. BarbieriCM, StockAM (2008) Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem 376 : 73–82.
68. LiangH, DengX, JiQ, SunF, ShenT, et al. (2012) The Pseudomonas aeruginosa global regulator VqsR directly inhibits QscR to control quorum-sensing and virulence gene expression. J Bacteriol 194 : 3098–3108.
69. KohlerT, van DeldenC, CurtyLK, HamzehpourMM, PechereJC (2001) Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol 183 : 5213–5222.
70. ZianniM, TessanneK, MerighiM, LagunaR, TabitaFR (2006) Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17 : 103–113.
71. IrizarryRA, HobbsB, CollinF, Beazer-BarclayYD, AntonellisKJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4 : 249–264.
72. TusherVG, TibshiraniR, ChuG (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98 : 5116–5121.
73. YuanK, HuangC, FoxJ, LaturnusD, CarlsonE, et al. (2012) Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J Cell Sci 125 : 507–515.
74. KannanS, AudetA, KnittelJ, MullegamaS, GaoGF, et al. (2006) Src kinase Lyn is crucial for Pseudomonas aeruginosa internalization into lung cells. Eur J Immunol 36 : 1739–1752.
75. WuM, HuangH, ZhangW, KannanS, WeaverA, et al. (2011) Host DNA repair proteins in response to Pseudomonas aeruginosa in lung epithelial cells and in mice. Infect Immun 79 : 75–87.
76. WuM, BrownWL, StockleyPG (1995) Cell-specific delivery of bacteriophage-encapsidated ricin A chain. Bioconjug Chem 6 : 587–595.
77. GoldovaJ, UlrychA, HercikK, BrannyP (2011) A eukaryotic-type signalling system of Pseudomonas aeruginosa contributes to oxidative stress resistance, intracellular survival and virulence. BMC Genomics 12 : 437.
78. FiliatraultMJ, PicardoKF, NgaiH, PassadorL, IglewskiBH (2006) Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect Immun 74 : 4237–4245.
79. RahmeLG, TanMW, LeL, WongSM, TompkinsRG, et al. (1997) Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci U S A 94 : 13245–13250.
80. SiehnelR, TraxlerB, AnDD, ParsekMR, SchaeferAL, et al. (2010) A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 107 : 7916–7921.
81. WhiteleyM, ParsekMR, GreenbergEP (2000) Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol 182 : 4356–4360.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial PathogensČlánek A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during TransmissionČlánek The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil ChemotaxisČlánek Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 8- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- Regulatory RNAs Involved in Bacterial Antibiotic Resistance
- From Dandruff to Deep-Sea Vents: -like Fungi Are Ecologically Hyper-diverse
- Pathogenicity and Epithelial Immunity
- Mother–Infant HIV Transmission: Do Maternal HIV-Specific Antibodies Protect the Infant?
- Hell's BELs: acterial 3 igases That Exploit the Eukaryotic Ubiquitin Machinery
- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Primary Seronegative but Molecularly Evident Hepadnaviral Infection Engages Liver and Induces Hepatocarcinoma in the Woodchuck Model of Hepatitis B
- TLR2 Signaling Decreases Transmission of by Limiting Bacterial Shedding in an Infant Mouse Influenza A Co-infection Model
- Production of an Attenuated Phenol-Soluble Modulin Variant Unique to the MRSA Clonal Complex 30 Increases Severity of Bloodstream Infection
- Inhibition of the TRAIL Death Receptor by CMV Reveals Its Importance in NK Cell-Mediated Antiviral Defense
- Early Mucosal Sensing of SIV Infection by Paneth Cells Induces IL-1β Production and Initiates Gut Epithelial Disruption
- Limited HIV Infection of Central Memory and Stem Cell Memory CD4+ T Cells Is Associated with Lack of Progression in Viremic Individuals
- Virus-Specific Regulatory T Cells Ameliorate Encephalitis by Repressing Effector T Cell Functions from Priming to Effector Stages
- A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during Transmission
- The HIV-1 Envelope Transmembrane Domain Binds TLR2 through a Distinct Dimerization Motif and Inhibits TLR2-Mediated Responses
- Infection with MERS-CoV Causes Lethal Pneumonia in the Common Marmoset
- VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: Identification of the Local Environmental Source as Arboreal
- Diverse Host-Seeking Behaviors of Skin-Penetrating Nematodes
- Capsid Protein VP4 of Human Rhinovirus Induces Membrane Permeability by the Formation of a Size-Selective Multimeric Pore
- The Murine Gammaherpesvirus Immediate-Early Rta Synergizes with IRF4, Targeting Expression of the Viral M1 Superantigen to Plasma Cells
- Characterization of an Insecticidal Toxin and Pathogenicity of against Insects
- The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil Chemotaxis
- Histone Deacetylase Inhibitors Impair the Elimination of HIV-Infected Cells by Cytotoxic T-Lymphocytes
- A Locus Encompassing the Epstein-Barr Virus Kinase Regulates Expression of Genes Encoding Viral Structural Proteins
- Distinct APC Subtypes Drive Spatially Segregated CD4 and CD8 T-Cell Effector Activity during Skin Infection with HSV-1
- Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
- Adoptive Transfer of EBV Specific CD8 T Cell Clones Can Transiently Control EBV Infection in Humanized Mice
- Schistosome Feeding and Regurgitation
- EVM005: An Ectromelia-Encoded Protein with Dual Roles in NF-κB Inhibition and Virulence
- Rabies Virus Hijacks and Accelerates the p75NTR Retrograde Axonal Transport Machinery
- Why HIV Virions Have Low Numbers of Envelope Spikes: Implications for Vaccine Development
- Identification of Anti-virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity
- HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization
- Cooperation between Epstein-Barr Virus Immune Evasion Proteins Spreads Protection from CD8 T Cell Recognition across All Three Phases of the Lytic Cycle
- Parasite Extracellular Vesicles: Mediators of Intercellular Communication
- RC1339/APRc from Is a Novel Aspartic Protease with Properties of Retropepsin-Like Enzymes
- Cyclic di-GMP-dependent Signaling Pathways in the Pathogenic Firmicute
- Non-random Escape Pathways from a Broadly Neutralizing Human Monoclonal Antibody Map to a Highly Conserved Region on the Hepatitis C Virus E2 Glycoprotein Encompassing Amino Acids 412–423
- Neutrophil Elastase Causes Tissue Damage That Decreases Host Tolerance to Lung Infection with Species
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation
- Why Sexually Transmitted Infections Tend to Cause Infertility: An Evolutionary Hypothesis
- Late Engagement of CD86 after Influenza Virus Clearance Promotes Recovery in a FoxP3 Regulatory T Cell Dependent Manner
- Determinants of Influenza Transmission in South East Asia: Insights from a Household Cohort Study in Vietnam
- A Novel Signal Transduction Pathway that Modulates Quorum Sensing and Bacterial Virulence in
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- A Cysteine Protease Inhibitor of Is Essential for Exo-erythrocytic Development
- EBNA3C Augments Pim-1 Mediated Phosphorylation and Degradation of p21 to Promote B-Cell Proliferation
- On the Front Line: Quantitative Virus Dynamics in Honeybee ( L.) Colonies along a New Expansion Front of the Parasite
- Assembly and Architecture of the EBV B Cell Entry Triggering Complex
- NLR-Associating Transcription Factor bHLH84 and Its Paralogs Function Redundantly in Plant Immunity
- The PDZ-Binding Motif of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Is a Determinant of Viral Pathogenesis
- Strain-Specific Properties and T Cells Regulate the Susceptibility to Papilloma Induction by Papillomavirus 1
- Human Cytomegalovirus pUL79 Is an Elongation Factor of RNA Polymerase II for Viral Gene Transcription
- The GAP Activity of Type III Effector YopE Triggers Killing of in Macrophages
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- Pathogenicity and Epithelial Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



