-
Medical journals
- Career
Polymorphisms Contribution to the Determination of Significant Risk of Specific Toxicities in Multiple Myeloma
Authors: M. Almaši 1; S. Ševčíková 2; H. Šváchová 2; B. Sáblíková 2; P. Májková 2; R. Hájek 1,2,3
Authors‘ workplace: Laboratory of Experimental Hematology and Cell Immunotherapy, Department of Clinical Hematology, University Hospital Brno, Czech Republic 1; Babak Myeloma Group, Department of Pathological Physiology, Faculty of Medicine, Masaryk University, Brno, Czech Republic 2; Department of Internal Medicine – Hematooncology, University Hospital Brno, Czech Republic 3
Published in: Klin Onkol 2011; 24(Supplementum 1): 39-42
Overview
The introduction of new drugs improved clinical response of patients with diagnosed multiple myeloma (MM); however, MM is still an incurable disease that leads to frequent relapses. Individual genetic variability can significantly affect therapeutic response, sensitivity and toxicity. Analysis of single nucleotide polymorphisms (SNPs) to study genetic changes is the genomic method that can obtain information for improving the effectiveness of treatment with minimum undesirable toxicity followed by individual treatments. The aim of this paper is to explain the possibility of detection and evaluation of polymorphisms associated with toxicity of treatment in patients with MM.
Key words:
single nucleotide polymorphisms (SNPs) – genotyping – real-time PCR – allelic discrimination – toxicity – multiple myeloma
This work was supported by research grants of The Ministry of Education, Youth and Sports: LC06027, MSM0021622434; research projects of IGA of The Ministry of Health: NS10207, NS10406, NT11154 and grant of GACR GAP304/10/1395.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
The Editorial Board declares that the manuscript met the ICMJE “uniform requirements” for biomedical papers.Introduction
Multiple myeloma (MM) does not involve only factors of the tumor cells themselves, but also host factors, as genetic variability of each individual can significantly affect the therapeutic response, as well as sensitivity and/or toxicity. The knowledge of pharmacogenetics can potentially aid in the discovery, development and ultimately individualization of anticancer drugs. Identification of genetic variations that predict drug response is the first step towards translation of pharmacogenetics into clinical practice [1].
Single nucleotide polymorphisms (SNPs) are inherited and do not change from generation to generation, creating genetic variability in populations, based on the substitution of a single nucleotide (A, G, C or T) in the DNA sequence. This substitution must occur in a population with a frequency greater than 1%, otherwise it is a single-point mutation [2]. Two of the three SNPs include substitution of cytosine (C) for thymine (T). The frequency of these polymorphisms, ranging from 2–40%, accounts for 90% of total genetic variation. In the human population, over 99% of genomic DNA is identical; yet, such a small variability in DNA is an important factor in diversity of each individual’s predisposition to disease, response to treatment, etc. Most SNPs are not responsible for any specific illness, but can be used as biological markers through their occurence close to the gene responsible for disorder pathogenesis.
In large genome-wide association studies (GWAS), thousands of polymorphisms are monitored simultaneously; those that occur more frequently in patients than in healthy voluntaries are identified as candidates and studied. After verifying the results of independent studies and finding the mechanism of action of genes, a proposed panel of candidate genes for investigation of individuals with increased risk of developing the disease is proposed. The International Myeloma Foundation (IMF) has created a comprehensive myeloma specific bank of DNA samples ‚Bank On A Cure’ – BOAC (http//:www.myeloma.org), to create a truly collaborative bank for research capable of analyzing complex genetic information from thousands of MM patients.
Association studies evaluate individual polymorphisms separately or in combination. A comparison of nucleotide sequences of the same chromosomes of two individuals found on an average of one SNP per 1 200 pairs of nucleotides. SNPs on a chromosome or a segment of a chromosome, which tend to be inherited together, form a haplotype [3]. Study of polymorphisms and haplotypes can be used to predict the risk of illness or specify the impact of genetic predisposition to increased risk of side effects of treatment.
Significant Specific Toxicity of Treatment in MM
Introduction of new immunomodulatory drugs (IMiDs), such as thalidomide and lenalidomide, improved clinical outcome in MM patients. However, the use of these IMiDs is associated with higher risk of VTE (venous thromboembolism) in 12% or 8% of cases, respectively [4]. Use of IMiDs in combination with dexamethasone or chemotherapy increases the risk of VTE up to 25% [5–7]. The US Food and Drug Administration (FDA) and European Medicines Evaluation Agency (EMEA) have published recommendations suggesting the use of thromboprophylaxis with IMiDs treatment [8,9]. Therefore, in MM patients, low-molecular weight heparines (LMWHs) are used as primary drug of thromboprophylaxis [10,11].
Despite this, VTE develops in a significant number of patients with MM. The main interest is to find an accurate method for prediction of VTE risk, allowing individualization of treatment and protection of MM patients from serious toxicity. Extensive observations published by Johnson et al indicate that polymorphisms of DNA damage response genes and cytokine-mediated apoptosis are of great importance in the development of VTE after thalidomide treatment [12]. In this study, the authors built a classification tree from 7 SNPs enabling prediction of individual risk of VTE in MM patients. Findings in our validation set (111 patients treated with thalidomide, in total 19 % of these patients developed VTE) did not indicate the possible use of SNPs in clinical practice for prediction of VTE risk [13]. In general, further studies are need-ed to verify if SNPs-based approach is eligible for prediction of VTE risk in MM patients.
Another example of severe toxicity of treatment is peripheral neuropathy (PN), occurring mainly after treatment with bortezomib and thalidomide [14,15]. Using a custom-built single nucleotide polymorphism (SNP) array, Johnson et al tested the association of thalidomide-related peripheral neuropathy (TrPN) with 3,404 SNPs. They report that an individual risk of developing peripheral neuropathy after thalidomide treatment can be mediated by polymorphisms in genes governing repair mechanisms and inflammation in the peripheral nervous system [16]. Results of Broyl et al suggest significant SNPs associated with early-onset bortezomib-induced peripheral neuropathy (BiPN) in apoptotic genes. SNPs in inflammatory genes and DNA repair genes were associated with late-onset BiPN [17]. On the other hand, in the study of Favis et al, genes associated with immune function (CTLA4, CTSS), reflexive coupling within Schwann cells (GJE1), drug binding (PSMB1) and neuron function (TCF4, DYNC1I1) are associated with BiPN [18] (Tab. 1).
1. SNPs in genes associated with peripheral neuropathy (PN). 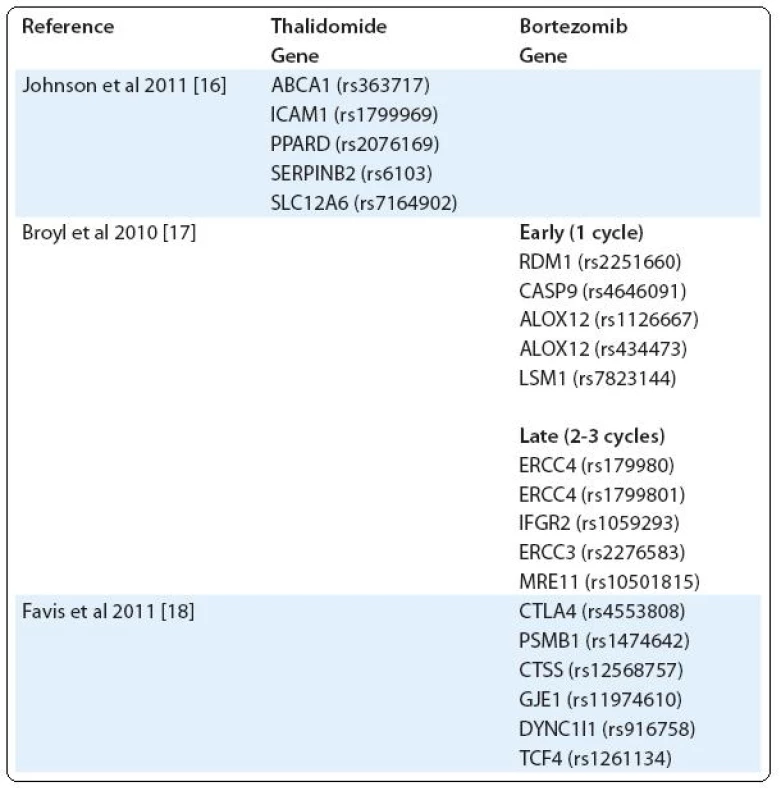
Methodology for Determination of Polymorphisms
For genotyping, different methodologies are used. At present, in large genome-wide association studies (GWAS), research focuses on the analysis of SNPs using chip technology. These chips allow simultaneous analysis of a large number of polymorphisms located at various loci in the genome. For validation of polymorphisms associated with toxicity, real-time PCR is currently used.
In our institute, analyses of gene polymorphisms using real-time PCR allelic discrimination is performed on Step-One Real-Time PCR instrument (Applied Biosystems) using standard TaqMan genotyping assays. Each TaqMan assay is composed of two probes for each allele. TaqMan probes are based on 5´–3´ exonuclease activity of Taq polymerase, which cleaves labeled TaqMan probe, when hybridized with complementary target sequences. Each probe has a different fluorophore for each allele. If the probe sequence is complementary to the DNA sequence, fluorescence signal is detected by the instrument. Different colors then identify individuals as wild-type homozygotes, mutant homozygotes or heterozygotes with two variants (alleles) of gene (Fig. 1).
Fig. 1. Results of SNP analysis of VEGFA (rs699947) gene: red and blue colour points identify homozygotes (allele 1 – AA or allele 2 – CC); green colour points identify heterozygotes for both alleles (AC) (Almasi, 2009). 
Genomic DNA is isolated from whole peripheral blood using MagNA Pure DNA Isolator (Roche). DNA concentration is measured on Nanodrop ND-1000 (NanoDrop Technologies, Inc.). Analyses of gene polymorphisms, using standard TaqMan genotyping assays, are carried out according to manufacturer’s instructions. In brief, probes, primers and Taq--Man universal PCR master mix are obtained from Applied Biosystems. Reaction solution of 10 μl contained 0,5 μl Taq-Man Genotyping assay mix (consisting of 20 × mix of unlabeled PCR primers and TaqMan MGB probe, FAM and VIC dye-labeled), 8 μl of PCR mixture reagent and 10 ng of genomic DNA. Reactions are run according to manufacturer’s instructions. The polymerase chain reaction consists of pre-PCR read 60°C for 30 sec, holding stage at 95°C for 10 min, 50 cycles of denaturation at 92°C for 15 sec, annealing at 60°C for 1 min 30 sec, and post-PCR read at 60°C for 30 sec.
Evaluation of Polymorphisms
Different studies have focused on finding a direct correlation between these polymorphisms and major toxicities in MM. All analyzed polymorphisms are first tested using χ2 for Hardy-Weinberg equilibrium (HWE), which examines the distribution of alleles in individuals in the population. Only when the polymorphism meets conditions of HWE, it may be subjected to further analysis. The most important statistical concept is odds ratio (OR). Homozygote most frequent allele is usually referred to as the reference group for calculation of the odds ratio, which reflects how many times specific toxicity has an increased or decreased risk of occurring in patients with different genotypes. If the value of OR > 1, this is a risk factor compared to the reference group. If OR < 1, it is a protective factor. When evaluating the interaction of more polymorphisms that are inherited together, haplotype analysis is used. One of the most popular programs is PHASE, estimating haplotypes from population genotypes.
Conclusion
Current data do not indicate potential application of polymorphisms for routine clinical application for major toxicities prediction in MM patients. Further studies of genetic polymorphisms associated with the occurrence of significant toxicity may contribute to more effective and rational prophylaxis for these complications of therapy in patients with MM.
prof. MUDr. Roman Hájek, CSc.
Babak Myeloma Group
Department of Pathological Physiology
Faculty of Medicine
Masaryk University
Kamenice 5
625 00 Brno
Czech Republic
e-mail: r.hajek@fnbrno.cz
Sources
1. Yong WP, Innocenti F, Ratain MJ. The role of pharmacogenetics in cancer therapeutics. Br J Clin Pharmacol 2006; 62(1): 35–46.
2. Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 2003; 33 (Suppl): 228–237.
3. Snustad DP, Siemens MJ. Genomika. In: Snustad DP, Siemens MJ (eds). Genetika. Brno: Masarykova univerzita 2009 : 461–503.
4. Bennett CL, Angelotta C, Yarnold PR et al. Thalidomide - and lenalidomide-associated thromboembolism among patients with cancer. JAMA 2006; 296(21): 2558–2560.
5. Cavo M, Zamagni E, Cellini C et al. Deep-vein thrombosis in patients with multiple myeloma receiving first-line thalidomide-dexamethasone therapy. Blood 2002; 100(6): 2272–2273.
6. Cavo M, Zamagni E, Tosi P et al. First-line therapy with thalidomide and dexamethasone in preparation for autologous stem cell transplantation for multiple myeloma. Haematologica 2004; 89(7): 826–831.
7. Rajkumar SV, Hayman S, Gertz MA et al. Combination therapy with thalidomide plus dexamethasone for newly diagnosed myeloma. J Clin Oncol 2002; 20(21): 4319–4323.
8. European Medicines Agency. Questions and answers on the positive opinion for Thalido mide Pharmion. 2008. Available from: http://www.emea.europa.eu/pdfs/human/opinion/Q&AThalidomide54352407en.pdf..
9. US Food and Drug Administration. Lenalidomide package insert. 2006. Available from: http://www.fda.gov/cder/foi/label/2006/021880s001.pdf.
10. Klein U, Kosely F, Hillengass J et al. Effective prophylaxis of thromboembolic complications with low molecular weight heparin in relapsed multiple myeloma patients treated with lenalidomide and dexamethasone. Ann Hematol 2009; 88(1): 67–71.
11. Palumbo A, Rajkumar SV, Dimopoulos MA et al. Prevention of thalidomide - and lenalidomide-associated thrombosis in myeloma. Leukemia 2008; 22(2): 414–423.
12. Johnson DC, Corthals S, Ramos C et al. Genetic associations with thalidomide mediated venous thrombotic events in myeloma identified using targeted genotyping. Blood 2008; 112(13): 4924–4934.
13. Almasi M, Sevcikova S, Slaby O et al. Association study of selected genetic polymorphisms and occurrence of venous thromboembolism in multiple myeloma patients treated with thalidomide. Clin Lymphoma Myeloma Leuk. [accepted for publication]
14. Chaudhry V, Cornblath DR, Polydefkis M et al. Characteristics of bortezomib - and thalidomide-induced peripheral neuropathy. J Peripher Nerv Syst 2008; 13(4): 275–282.
15. Delforge M, Bladé J, Dimopoulos MA et al. Treatment-related peripheral neuropathy in multiple myeloma: the challenge continues. Lancet Oncol 2010; 11(11): 1086–1095.
16. Johnson DC, Corthals SL, Walker BA et al. Genetic factors underlying the risk of thalidomide-related neuropathy in patients with multiple myeloma. J Clin Oncol 2011; 29(7): 797–804.
17. Broyl A, Corthals SL, Jongen JL et al. Mechanisms of peripheral neuropathy associated with bortezomib and vincristine in patients with newly diagnosed multiple myeloma: a prospective analysis of data from the HOVON-65/GMMG-HD4 trial. Lancet Oncol 2010; 11(11): 1057–1065.
18. Favis R, Sun Y, van de Velde H et al. Genetic variation associated with bortezomib-induced peripheral neuropathy. Pharmacogenet Genomics 2011; 21(3): 121–129.
Labels
Paediatric clinical oncology Surgery Clinical oncology
Article was published inClinical Oncology

2011 Issue Supplementum 1-
All articles in this issue
- Editorial (CZ)
- Radiotherapeutic methods
- Multiple Myeloma
- Monoclonal Gammopathy of Undeterminated Significance: Introduction and Current Clinical Issues
- Sample Processing and Methodological Pitfalls in Multiple Myeloma Research
- Flow Cytometry in Monoclonal Gammopathies
- Flow Cytometric Phenotyping and Analysis of T Regulatory Cells in Multiple Myeloma Patients
- Genomics in Multiple Myeloma Research
- Polymorphisms Contribution to the Determination of Significant Risk of Specific Toxicities in Multiple Myeloma
- Oligonucleotide-based Array CGH as a Diagnostic Tool in Multiple Myeloma Patients
- Visualization of Numerical Centrosomal Abnormalities by Immunofluorescent Staining
- Impact of Nestin Analysis in Multiple Myeloma
- Editorial (EN)
- List of authors and reviewers
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Multiple Myeloma
- Flow Cytometric Phenotyping and Analysis of T Regulatory Cells in Multiple Myeloma Patients
- Monoclonal Gammopathy of Undeterminated Significance: Introduction and Current Clinical Issues
- Flow Cytometry in Monoclonal Gammopathies
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

