-
Články
- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
- Kongresy
- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Homeostatic Interplay between Bacterial Cell-Cell Signaling and Iron in Virulence
Pathogenic bacteria use interconnected multi-layered regulatory networks, such as quorum sensing (QS) networks to sense and respond to environmental cues and external and internal bacterial cell signals, and thereby adapt to and exploit target hosts. Despite the many advances that have been made in understanding QS regulation, little is known regarding how these inputs are integrated and processed in the context of multi-layered QS regulatory networks. Here we report the examination of the Pseudomonas aeruginosa QS 4-hydroxy-2-alkylquinolines (HAQs) MvfR regulatory network and determination of its interaction with the QS acyl-homoserine-lactone (AHL) RhlR network. The aim of this work was to elucidate paradigmatically the complex relationships between multi-layered regulatory QS circuitries, their signaling molecules, and the environmental cues to which they respond. Our findings revealed positive and negative homeostatic regulatory loops that fine-tune the MvfR regulon via a multi-layered dependent homeostatic regulation of the cell-cell signaling molecules PQS and HHQ, and interplay between these molecules and iron. We discovered that the MvfR regulon component PqsE is a key mediator in orchestrating this homeostatic regulation, and in establishing a connection to the QS rhlR system in cooperation with RhlR. Our results show that P. aeruginosa modulates the intensity of its virulence response, at least in part, through this multi-layered interplay. Our findings underscore the importance of the homeostatic interplay that balances competition within and between QS systems via cell-cell signaling molecules and environmental cues in the control of virulence gene expression. Elucidation of the fine-tuning of this complex relationship offers novel insights into the regulation of these systems and may inform strategies designed to limit infections caused by P. aeruginosa and related human pathogens.
Published in the journal: . PLoS Pathog 6(3): e32767. doi:10.1371/journal.ppat.1000810
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000810Summary
Pathogenic bacteria use interconnected multi-layered regulatory networks, such as quorum sensing (QS) networks to sense and respond to environmental cues and external and internal bacterial cell signals, and thereby adapt to and exploit target hosts. Despite the many advances that have been made in understanding QS regulation, little is known regarding how these inputs are integrated and processed in the context of multi-layered QS regulatory networks. Here we report the examination of the Pseudomonas aeruginosa QS 4-hydroxy-2-alkylquinolines (HAQs) MvfR regulatory network and determination of its interaction with the QS acyl-homoserine-lactone (AHL) RhlR network. The aim of this work was to elucidate paradigmatically the complex relationships between multi-layered regulatory QS circuitries, their signaling molecules, and the environmental cues to which they respond. Our findings revealed positive and negative homeostatic regulatory loops that fine-tune the MvfR regulon via a multi-layered dependent homeostatic regulation of the cell-cell signaling molecules PQS and HHQ, and interplay between these molecules and iron. We discovered that the MvfR regulon component PqsE is a key mediator in orchestrating this homeostatic regulation, and in establishing a connection to the QS rhlR system in cooperation with RhlR. Our results show that P. aeruginosa modulates the intensity of its virulence response, at least in part, through this multi-layered interplay. Our findings underscore the importance of the homeostatic interplay that balances competition within and between QS systems via cell-cell signaling molecules and environmental cues in the control of virulence gene expression. Elucidation of the fine-tuning of this complex relationship offers novel insights into the regulation of these systems and may inform strategies designed to limit infections caused by P. aeruginosa and related human pathogens.
Introduction
Microbes translate environmental cues to coordinate and modulate gene expression such that they can adapt to different niches and overcome hostile environments. Adaptation and coordination of gene expression is particularly important for pathogenic microorganisms that need to colonize dynamic host environments since their ability to sense and respond to host environmental cues is critical for their survival. In bacteria, modulation and coordination of gene expression are also influenced by population density via the regulated production of small molecules that serve as intricate signals impacting the expression of virulence factor genes. Many studies have addressed the role of quorum sensing (QS) communication networks in virulence where by diffusible intercellular auto-inducers factor and environmental signals bacterial cultures mediate pathogenicity by coordinating the expression of a large array of genes [1],[2]. Nevertheless, less is known regarding how environmental cues are translated in the context of QS signaling and how environmental cues and QS are integrated to promote the ability of a pathogen to survive and colonize particular niches within their host environments. The processing and integration of environmental inputs in QS becomes even more complex when a pathogen is able to occupy more than one niche.
Pseudomonas aeruginosa is a ubiquitous and an extremely versatile Gram-negative bacterium with an astounding ability to survive in many different environments and to infect multiple hosts ranging from amoebas to humans [3]. This pathogen has an extensively studied complex QS communication network that facilitates cross-talk between organisms and impacts many P. aeruginosa group-related behaviors including virulence [4],[5],[6],[7],[8],[and 9]. There are at least three known QS systems in P. aeruginosa: two are dependent on the acyl-homoserine-lactone (AHL) QS transcription factors LasR and RhlR [10] and a third is dependent on the 4-hydroxy-2-alkylquinolines (HAQs) LysR-type transcription factor MvfR [11],[12]. MvfR activation is mediated by the cell-cell signaling molecules 4-hydroxy-2-heptylquinoline (HHQ) and 3,4-dihydroxy-2-heptylquinoline (PQS), and leads to the positive regulation of many virulence-related factors, a large number of which are also controlled by the QS signal acyl-homoserine-lactone (AHL)-mediated RhlR and LasR circuitry.
The MvfR pathway is a critical virulence component essential for the full virulence of P. aeruginosa in multiple hosts [13],[14],[15] and is connected to LasR and RhlR by: (i) the dependence of mvfR expression at the early growth stages as a result of positive control by LasR [16], (ii) the conversion of HHQ into PQS controlled by PqsH [17],[18] whose expression is mediated by LasR [19],[20], and (iii) the negative effects of RhlR on the pqs operon [16],[21], which is responsible for the synthesis of all HAQs [11],[14],[19],[22],[23] including the MvfR ligands HHQ and PQS [12],[17],[21].
The QS regulons MvfR, LasR and RhlR respond not only to QS signal molecules but also to environmental signals [24], including host factors [25],[26],[27],[28] and other environmental cues such as phosphate [29], magnesium [30] and iron [31],[32],[33],[34],[35]. Iron acquisition is controlled by a large set of P. aeruginosa genes activated in response to iron starvation [36],[37],[38], including two siderophore complexes, pyoverdine and pyochelin [39],[40], and several ferric uptake regulators, among them are the general iron uptake regulator Fur, Fur-regulated pyoverdine siderophore-specific extracytoplasmic sigma factor PvdS, several ECF sigma factors, and the AraC regulator PchR, which regulates pyochelin uptake [40]. In low iron conditions, PvdS binds to iron-starvation (IS) boxes to induce the transcription of many genes involved in the iron starvation response [41]. The intricate relationship between QS and iron is exemplified by a series of findings demonstrating that iron starvation induced QS systems [26],[32],[34] and that the QS regulators MvfR [11], LasR/RhlR [42] and VqsR [31],[43],[44] were found to be responsible for the induction of many iron response genes. Moreover, MvfR contains an IS box in its promoter [36], and PQS production is positively-affected by two Fur-regulated small RNAs, Prrf 1 and 2 [45]. Adding to the complexity of how environmental cues such as iron levels affect QS and how iron is integrated into QS to modulate virulence gene expression is the ability of PQS to bind iron [46], to act as an iron trap molecule [47], and to form a toxic complex against the host [48].
MvfR activation by HHQ and PQS leads to the upregulation of the anthranilic acid (AA) - biosynthetic encoding genes phnAB, and pqsA-E operon [11],[12],[14] that have a conserved genomic organization in P. aeruginosa and in HAQs-producing Burkholderia species [49], to produce more HAQs leading to the upregulation of the MvfR-regulon in a positive feedback loop. Although the fifth gene of the pqs operon pqsE (PA14_51380), which encodes a predicted GloB, Zn-dependent hydrolase [50] and member of the metallo-beta-lactamase super family (Pfam PF00753), is not required for HAQ synthesis [12],[19], it is co-regulated together with the pqsA-D genes. We have shown that PqsE is essential for complete P. aeruginosa virulence in mice because it controls the expression of a number of MvfR regulon-dependent genes [11]. Although PqsE was previously implicated as the PQS response gene [19],[20], it was recently shown to act independently of MvfR and PQS [51]. Thus, the PqsE functions associated with the integration and translation of the QS cell-cell signals has yet to be resolved.
Here we examine the interplay between environmental cues and cell-cell signaling molecules and assess how they are integrated in the modulation of MvfR regulon gene expression. To elucidate the QS multi-layered regulation, we also examine the functional dependency of the MvfR regulon components, especially PqsE, and PQS and HHQ, on the Rhl regulon. The findings presented offer new insights into the highly complex P. aeruginosa virulence-associated regulatory loops that may aid in understanding and controlling its pathogenicity.
Results
Dissection of the QS MvfR regulon reveals a key component functioning independently of the cell-cell signaling molecules PQS and HHQ
To elucidate how multi-layered regulatory networks sense and respond to external and internal cell signals to modulate gene expression, we studied the role of MvfR pathway components in integrating and translating signals from PQS and HHQ in the activation of the MvfR regulon genes. To this end, we measured pyocyanin production as an index. This secreted P. aeruginosa phenazine was chosen since its production is dependent on the MvfR pathway components, including the cell-cell signaling molecules, PQS and HHQ, and their corresponding biosynthetic enzymes PqsA-D, their AA precursor, PqsE, and on its Phz biosynthetic operons (Figure 1A and [11]). Here we found that overexpression of PqsE under a constitutive promoter (pDN19pqsE) in pqsA− and mvfR− mutant cells not producing HAQs restored pyocyanin production (Figure 1A). In contrast, overexpression of mvfR under a constitutive promoter in a pqsE− background did not restore pyocyanin production (Figure 1A) even when HHQ, PQS, or PA14 cell-free supernatants were added (data not shown). These results highlight the crucial role of PqsE in the regulation of MvfR regulon-dependent factors and demonstrate that PqsE possesses activation properties that are independent of HAQ-mediated signals (Table S1). To assess PqsE mode of action on pyocyanin production, we co-cultured pqsE− cells constitutively expressing the phenazine biosynthetic operon phzA2-G2 with pqsE− cells harboring the phzM and phzS genes essential to pyocyanin synthesis [52] and assessed pyocyanin production. As shown in Figure 1B, approximately 60% of the pyocyanin production was restored, indicating that PqsE participated in pyocyanin production regulation rather than in its synthesis.
Fig. 1. PqsE, a key mediator of the MvfR regulon activation, functions independently of AA and its derivatives. 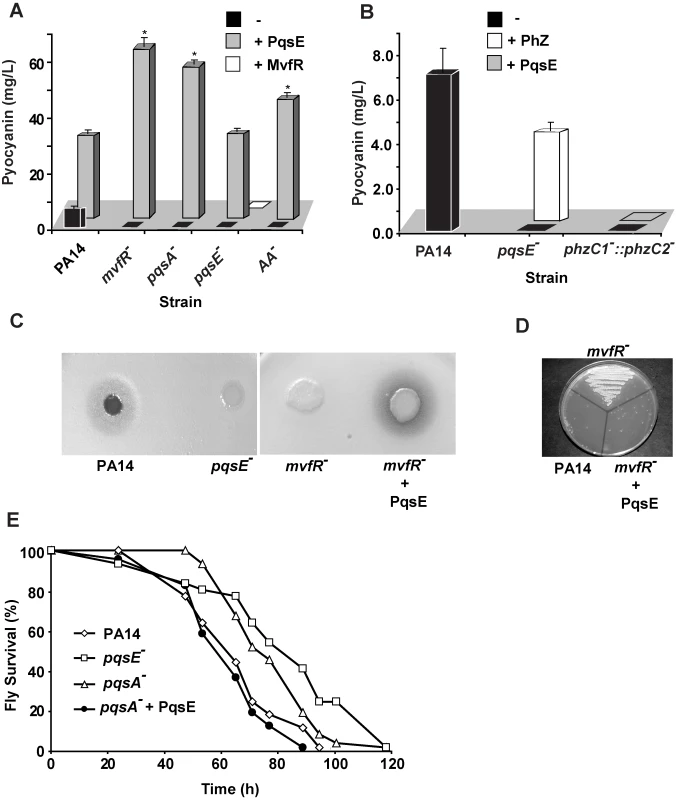
(A and B) Pyocyanin production was measured from PA14 and mutants with and without constitutive expression of PqsE or MvfR as a consequence of the presence of pDN19pqsE or pDN18mvfR plasmids, respectively. (A) AA− is a triple mutant with non-functional phnAB, trpE and kynBU that does not produce anthranilate. Production of pyocyanin (+ Phz) was achieved by co-culturing two sets of cells one constitutively expressing phzA2-G2, and the other phzM and phzS genes encoding the phenazines and pyocyanin biosynthetic genes respectively. Asterisks in A show strains harboring the plasmid pDN19pqsE that are significantly different (P value <0.01) from PA14 harboring that plasmid. (C–D) PqsE is essential for the virulence of P. aeruginosa against Cryptococcus neoformans independently of HAQs. PqsE was constitutively expressed in mvfR− mutant cells. An empty vector served as a control (−). (C) 1 µL of bacterial culture was spotted onto YPD top-agar where yeast cells were plated. Yeast killing zones were formed only around the PA14 and mutant cells expressing PqsE. (D) The death of yeast cells within the killing zone was demonstrated by assessing their viability on YPD plates. (E) PqsE causes fly mortality in absence of HAQs. Survival kinetics of Drosophila melanogaster was assessed using a fly feeding assay. The survival kinetics of pqsA− and pqsE− infected flies was significant different (P value <0.005) form that of PA14-infected flies. However, the kinetics of pqsA− + PqsE- infected flies did not differ significantly from that of the PA14-infected flies(P value = 0.27). Second, we tested whether the precursor of all HAQs, AA was required for PqsE function instead. To this end we used a triple mutant strain deficient in phnAB, trpE and kynBU (AA− mutant) unable to produce any AA since all three AA synthesis pathways were knocked out [53]. Expression of PqsE in this triple mutant also resulted in high levels of pyocyanin production (Figure 1A) corroborating with the above results and demonstrating that PqsE function did not require AA or any of its derivatives to promote production of the MvfR regulon-dependent factor pyocyanin.
Third, since PqsE controlled the regulation of one of the key MvfR-regulated factors, pyocyanin, we sought to define the impact of this factor in the regulation of all MvfR-dependent virulence genes. We carried out whole genome expression studies and compared the expression profiles of a pqsE− mutant to those of the PA14 parental strain, an mvfR− mutant and to those of PA14 and an mvfR− over-expressing pqsE strain (NCBI GEO, accession number #GSE17147). These results showed that PqsE profoundly affected the expression of 90% of the MvfR-regulated genes, including at least thirty-six known and predicted transcription factors (Tables S1B and S2). Of the PqsE-dependent genes, 241 were found to be negatively regulated and 384 positively regulated by PqsE (Table S1). At least 75 positively-regulated genes encoded for putative or known virulence factors (Table S1) [11],[42]. Importantly, included among the positively-regulated virulence transcriptional factors was the QS AHL regulator rhlR [38] and iron response genes, including the iron starvation sigma factor pvdS and genes involved in the synthesis of the siderophore complex pyochelin (Table S3A).
To confirm that PqsE overexpression also restores virulence functions apart from restoring their expression independently of the signaling molecules PQS and HHQ, we used two assays. The first is based on the observation that virulent P. aeruginosa strains; including PA14 kill yeast [54],[55],[56]; and the second is based on that P. aeruginosa can infect and kill Drosophila melanogaster [57],[58],[59], and that mvfR mutant cells exhibit attenuated virulence in flies [57]. As illustrated in Figure 1C–D, a zone of yeast growth inhibition was observed around PA14, but not around the mvfR−, or pqsE− mutants following plating of C. neoformans KN99α 5 mm from the bacterial colony on a YPD plate (Figure 1D). The killing zone was restored following PqsE overexpression in mvfR− backgrounds (Figure 1C–D). In agreement flies infected with pqsA− or pqsE− mutants cells exhibited significant delayed in mortality compared to that caused by the WT or the pqsA− cells expressing pqsE (Figure 1E) demonstrating again that PqsE is crucial for P. aeruginosa pathogenicity and independent of PQS and HHQ.
MvfR-dependent gene regulation relies on the functional cooperation between RhlR and PqsE
Comparison of the pqsE transcriptome (Table S1) to lasR/rhlR [42] revealed that almost half (46%) of the genes regulated by LasR/RhlR were also regulated by PqsE (Figure S3A) indicating a relationship between AHL - and MvfR-mediated QS regulons. This relationship is also extended to the negative effects that both components have on the transcription of the pqs operon ([16] and Table S1 and Figure 2A). A green fluorescent protein (GFP) reporter gene [32] fused to the pqs operon promoter (Figures 2B), quantitative PCR analysis (Figure S2D) and quantification of HHQ and PQS levels (Figure 2C) further validated the above finding. Moreover, in agreement, Figure 2D shows that HAQ synthesis down-regulation paralleled the accumulation of AA (HAQ precursor) followed by an increase in antABC gene expression that encodes enzymes for AA degradation (Table S1).
Fig. 2. The homeostatic regulation of the signaling molecules HHQ and PQS is orchestrated by PqsE. 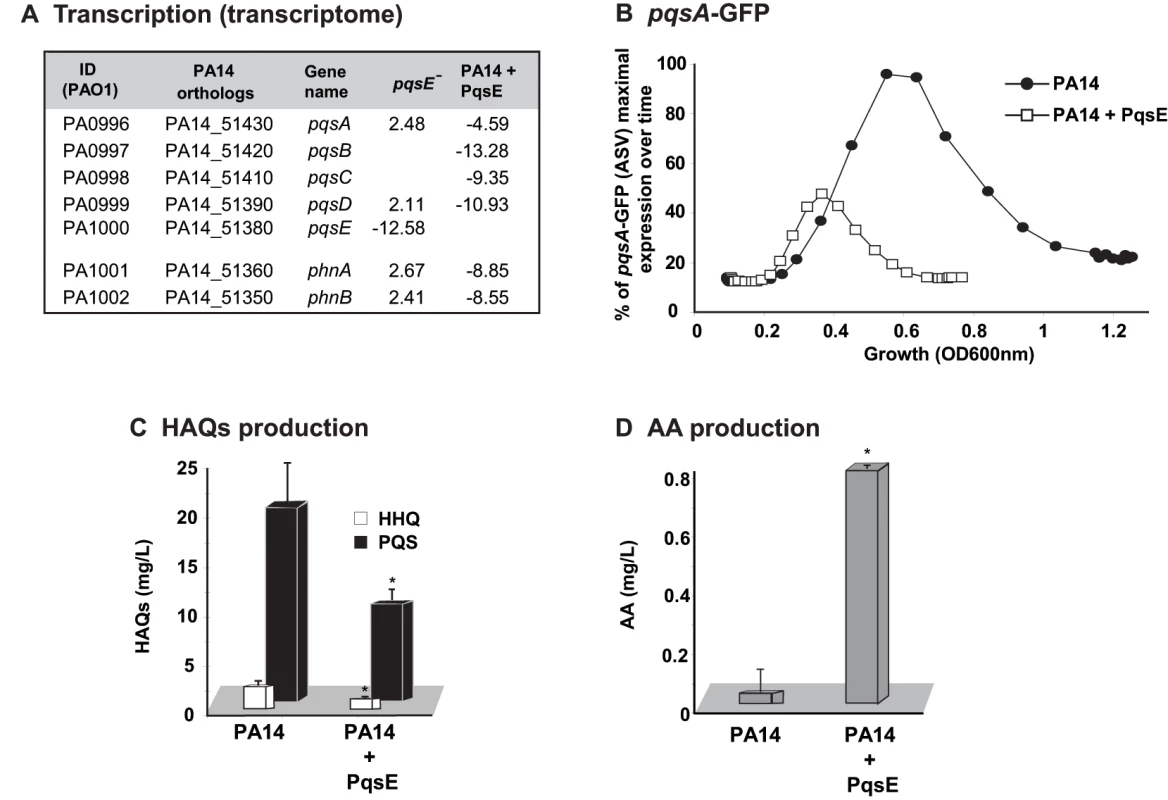
Effect of PqsE on pqs operon gene expression, and production of HAQs and AA. (A) Fold change in expression of phn and pqs operons in pqsE− mutant and PA14 constitutively expressing PqsE versus PA14. (B) GFP intensity derived from a pqsA-GFP(ASV) reporter fusion; (C) HAQs and (D) AA levels as assessed by LC-MS. t-tests (p = 0.001 for HHQ and p = 0.004 for PQS) showed that the difference between PA14 and PA14+PqsE is statistically significant. To determine whether there was indeed a functional relationship between the respective communication-systems components RhlR and PqsE in the regulation of the MvfR regulon signal production and whether they together affected signal integration, we proceeded to assess whether there was a RhlR-PqsE codependency in the negative regulation of HAQ biosynthesis. Figures 3A and S4B show that overexpression of PqsE in a rhlR− mutant did not result in a downregulation of the promoter-derived expression of the pqs operon in contrast to the overexpression of PqsE in the wild-type (WT) strain PA14 where expression of the pqs operon was downregulated (Figure 2 and Figure S2D). These results indicate that PqsE negative control of the activity of the MvfR regulon depends on RhlR.
Fig. 3. MvfR network regulation requires finely tuned cooperation between the MvfR component PqsE and the AHL QS regulator RhlR. 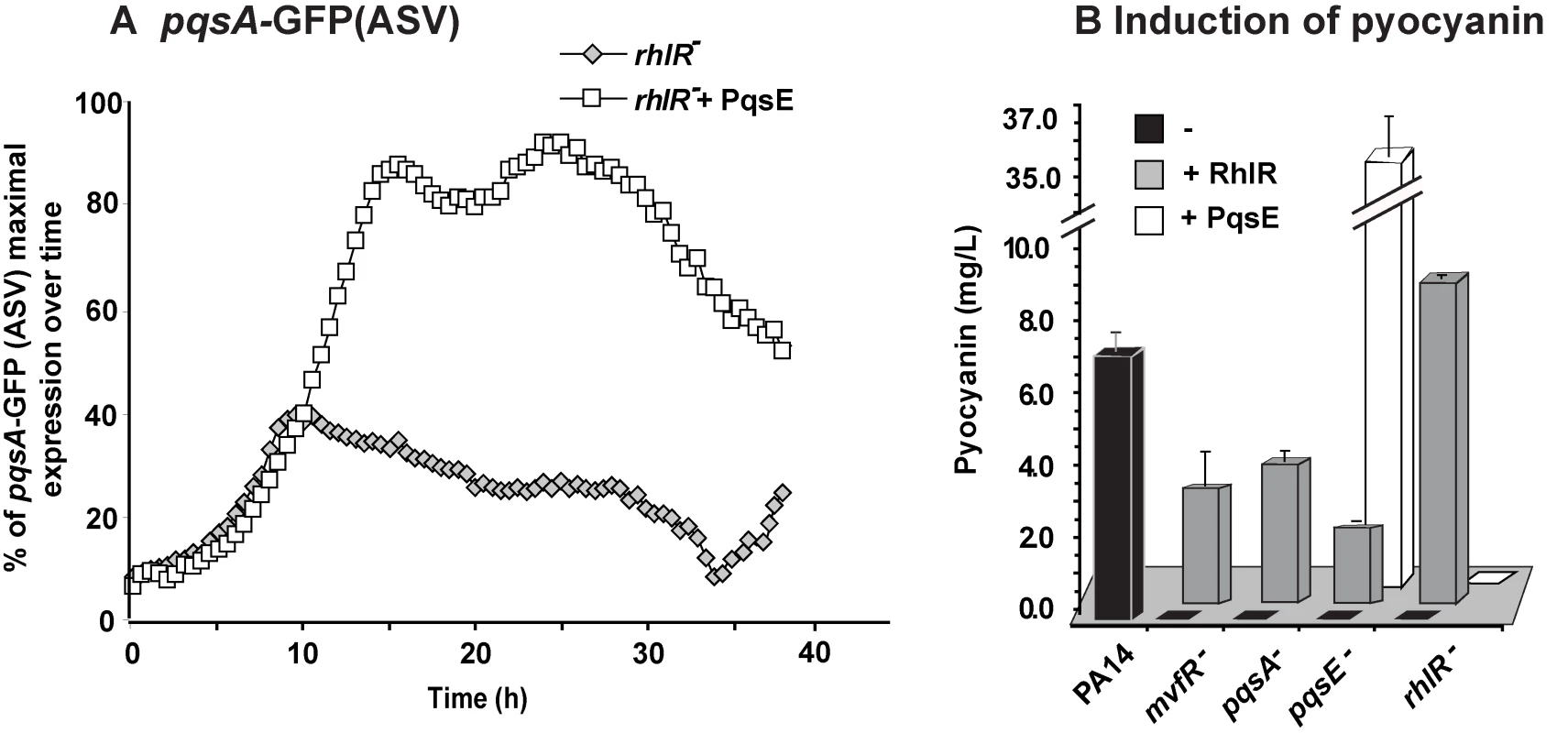
(A) The expression of pqsA was determined by measuring GFP emission. A pqsA-GFP (ASV) fusion in the rhlR mutant harboring pDN19pqsE was used to determine pqsA expression levels. (B) Pyocyanin levels were measured from various PA14 mutants harboring either pDN19pqsE or pUCP20rhlR plasmids. Empty vector served as control. Second, we examined whether there was an RhlR-PqsE codependency in signal integration by MvfR-regulon virulence genes downstream of PqsE. To this end, we assessed whether PqsE overproduction in rhlR− cells could restore pyocyanin production since it was completely abolished in both pqsE− [11],[19] and rhlR− [38] mutants. Figure 3B shows that PqsE did not restore pyocyanin production in rhlR− while RhlR expression partially (∼30%) restored pyocyanin production in pqsE− mutant cells. This finding suggests that PqsE also depends on RhlR in the positive regulation of pyocyanin production and that RhlR acts downstream of PqsE. Interestingly, Figure S5 shows that pyoverdine levels are higher in rhlR− than in PA14 but not in pqsE− mutant cells. Moreover, PqsE or RhlR overproduction in rhlR− or pqsE− mutant cells respectively did not fully downregulated pyoverdine production, while PqsE or RhlR overproduction in the corresponding mutant cells did (Figure S5). This finding suggests RhlR-PqsE codependency in the homeostatic regulation of pyoverdine.
Based on the above findings, it is likely that the PqsE-RhlR activities were not limited to controlling downstream genes associated only with pyocyanin or pyoverdine production if the high number of genes co-regulated by PqsE and the Las/Rhl system are considered (Figure S3A).
Signal integration studies reveal a homeostatic negative feedback regulation by HHQ and PQS on cell-cell signaling and PqsE-controlled genes, respectively
The pyocyanin levels produced by the non-HAQs producing mutants pqsA−, mvfR− and AA− [12],[19],[22],[53] overexpressing pqsE were higher than the levels produced by the HAQs-producing PA14 parental strain carrying the same plasmid (Figure 1A). This difference raised the question regarding whether the presence and/or levels of HAQs had dose-dependent negative effects on pyocyanin levels. To this end we assessed the effect of exogenously-added HAQs on pyocyanin levels by using 20 mg/L of PQS or HHQ, a concentration corresponding to the approximate maximal physiologic levels reached by PA14 or pqsH− strains respectively at stationary phase ([17] and Figure S4A). Figure 4A shows that the pyocyanin levels in either pqsA::pqsH− or mvfR− mutants overexpressing pqsE were significantly lower in the presence of either HHQ or PQS. Figure 4B shows that PQS concentrations (up to 1 mg/L) induced pyocyanin production in both pqsH− and pqsA−::pqsH− cells but concentrations >1 mg/L decreased pyocyanin production in a dose-dependent manner in all strains tested (Figure 4B) without significantly affecting cell growth (data not shown). This concentration-dependent decrease in pyocyanin levels was independent of PqsE function and phz operon regulation since it was also observed in pqsE− cells constitutively expressing phz genes (Figure 4C). The PQS-mediated down-regulation was not specific to PA14 cells as it was also observed in the PA01 P. aeruginosa strain (Figure 4C).
Fig. 4. Negative homeostatic feedback regulation on MvfR regulon products and activity is mediated via cell-cell signaling molecule concentration. 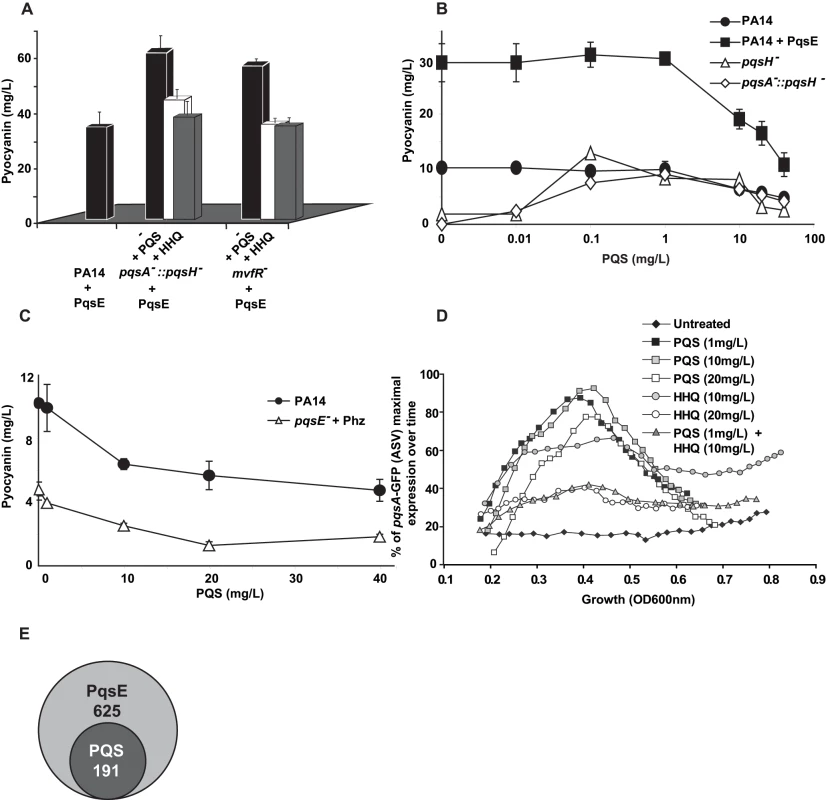
(A) Pyocyanin levels were assessed in PA14 and mutants cells harboring the plasmid pDN19pqsE with or without the addition of PQS or HHQ (20 mg/L). t-tests (p<0.05) showed that the difference between untreated and PQS/HHQ treated cells was statistically significant (B–C) Pyocyanin levels were determined following the addition of PQS over a broad-range of concentrations using a PQS non-producing strain (B) or using a narrow range of PQS concentrations in PQS-producing strains (C). PqsE was constitutively expressed (+PqsE). The empty vector was used as a control. phz genes were expressed following co-culture of pqsE− cells constitutively expressing phzA2-G2 with pqsE− cells constitutively expressing the phzM and phzS genes. The cells were grown in the presence of exogenously added PQS and pyocyanin production measured by measuring the OD600 nm. (D) The expression of pqsA was determined using a pqsA-GFP (ASV) fusion in a pqsA-::pqsH− double mutant in the presence of various concentrations of HHQ and PQS. (E) A Venn diagram showing the number of PqsE-regulated genes counterbalanced by PQS. The data was adapted from Table S1. To determine whether high physiological levels of PQS and/or HHQ negatively-impact pqs operon gene expression, we conducted experiments using pqsA−::pqsH− cells harboring the pqsA-GFP (ASV) reporter gene. Figure 4D shows that 20 mg/L HHQ negatively-impacted pqsA gene expression compared to 10 mg/L. PqsA gene expression was not affected by any of the PQS concentrations tested. Interestingly, a negative effect on pqsA gene expression, similar to that observed following treatment with 20 mg/L HHQ, was also observed when the two HAQs were added together in sub-inhibitory concentrations (1 mg/L PQS +10 mg/L of HHQ). This result is indicating that together HHQ and PQS have synergistic inhibitory effect and implying also that high activation of the pqs operon led to its down-regulation.
To further elucidate the role of PQS on PqsE-dependent gene regulation, we compared the transcriptional profiles of mvfR− mutant cells overexpressing PqsE in the absence or presence of 20 mg/L PQS (Table S1). High PQS concentrations negatively affected the expression of 191 of 625 (31%) PqsE-regulated genes (Figure 4E and Table S1). This effect was more apparent among the known and putative virulence factors where the expression of 64% of the PqsE-regulated genes, (including chitinase, halovibrin, cellulase, pyocins, lectin, and elastase genes) was significantly reduced by more than 2-fold upon PQS addition (Table S1). The addition of PQS further increased the expression of only 7 genes: fpvA, the major pyoverdine receptor; gatC, a Glu-tRNA amidotransferase subunit C; sucA, a 2-oxoglutarate dehydrogenase; bkdA1, a 2-oxoisovalerate dehydrogenase and of three hypothetical proteins; PA4642, PA1343 and PA2405 (Table S1). Interestingly, transcription of phz operon genes was not modified by the addition of PQS although pyocyanin production was affected (Figure 4A), suggesting that PQS may be acting post-transcriptionally in this case.
Homeostatic feedback modulation of the MvfR regulon is fine-tuned by an iron starvation response
As shown in Table S3A, PqsE positively-affected the expression of 43 iron starvation-related genes [36],[37] including the iron starvation sigma factor PvdS [41],[60], the pyochelin regulator PchR [61], vqsR [31],[62] and PA2384 [63]. Interestingly, PqsE negatively regulated only 6 iron related genes, bfrB and the siderophore pyoverdine associated genes pvdA pvdF, pvdJ, pvdN and pvdQ (Table S3A) reflected also in the pyoverdine levels (Figure S5). It is noteworthy that PqsE acted differentially on the siderophores, serving as a positive regulator of pyochelin and a negative regulator of pyoverdine (Figure S5). In addition, Table S3A reveal that HAQs are also involved in the control of iron-related genes by PqsE since constitutive expression of pqsE triggered this effect in the mvfR− background cells lacking HAQs but not in PA14 cells.
To examine how iron starvation is translated in the context of MvfR signaling, we first examined whether there is a relationship between iron starvation and the regulation of PQS and MvfR regulon genes. We compared pqsA transcription using a pqsA-GFP (ASV) reporter in PA14 cells grown in the absence (D-TSB medium) or presence of high iron levels. Figure 5A demonstrates that iron significantly reduced pqsA transcription. Subsequently, we examined the effect of iron directly on the induction of pqs operon transcription in presence only of PQS and not of other HAQs in pqsA−::pqsH− mutant cells. Using 1 mg/L PQS, an amount sufficient to fully induce pqs operon transcription and increasing concentrations of FeCl3 Figure 5B shows an iron concentration-dependent effect on pqsA gene expression.
Fig. 5. Homeostatic interplay between PQS and iron: Iron fine-tunes PQS activities. 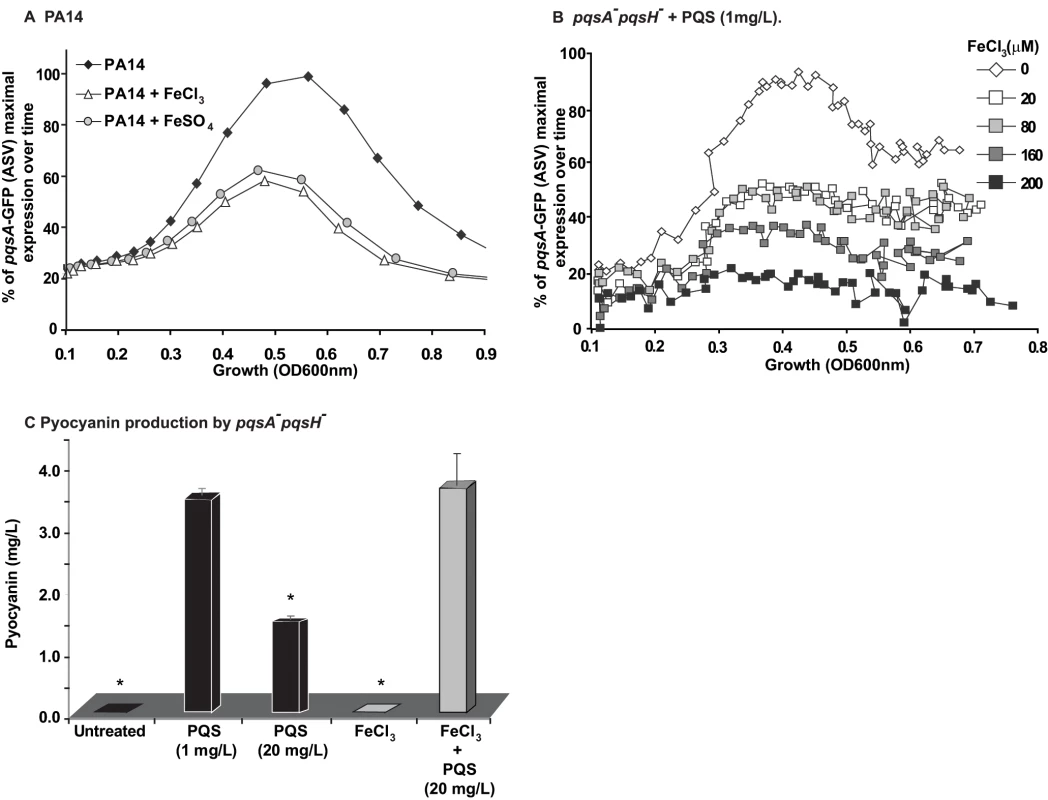
The effect of iron on MvfR induction was tested using the pqsA-GFP reporter in PA14 (A) and PA14 pqsA−::pqsH cells treated with PQS (1 mg/L) (B). The effect of iron on pyocyanin production was tested when PQS was supplied at 1 mg/L or 20 mg/L (C). The cells were grown in low iron medium D-TSB or in media supplemented with iron (FeCl3 or FeSO4, 200 µM). Asterisks show samples that are statistically significant different (P value<0.05) from the PQS 1 mg/L treated sample. We next examined if iron could also counterbalance the downstream effects of PQS on PqsE-dependent genes by assessing the effect of HAQs and iron on pyocyanin production. Figure 5C shows that the addition of iron abolished the reduction in pyocyanin production conferred by PQS (20 mg/L) and restored pyocyanin production to that observed in the presence of 1 mg/L PQS. A similar effect was observed in PA14 cells and pqsA−::pqsH− cells overexpressing PqsE (Figure S6A) where the addition of 20 mg/L PQS decreased pyocyanin levels which were restored in the presence of iron. Since iron alone did not affect pyocyanin production in the experimental conditions used, it suggested that pyocyanin production was affected due to direct effect of iron on PQS. No significant difference in growth was observed between PA14 cells grown in absence or presence of various concentrations of iron (up to 250 µM, Figure S6B). Collectively, these findings indicate that iron counterbalanced PQS-dependent regulation by ‘fine-tuning’ its activity, possibly by reducing PQS activity when it is in a complex with it.
Discussion
In this work, we delineated paradigmatically the complex relationships between bacterial multi-layered regulatory QS circuitries, their signaling molecules, and the environmental cues to which they respond.
The intracellular communication system of P. aeruginosa possesses complex signal transduction systems allowing this versatile pathogen to regulate and coordinate virulence functions in the context of multiple hosts, environments, and competition from other microorganisms [7],[64],[65],[66]. Here we showed that one of these complex signal transduction systems, MvfR, responds to both positive and negative feedback loops that are interconnected with the RhlR QS complex system and that these interactions fine tune the production and concentration of secreted output signals that in turn serve as inputs to preserve a homeostatic regulation. Moreover, our experiments demonstrated that via the finely tuned cooperation and homeostatic interplay between the MvfR circuitry components PqsE, and PQS and HHQ with RhlR and iron, this pathogen governs and balances the intensity of its virulence response.
Although HHQ and PQS principally serve as MvfR ligands [17],[18], our results show that once maximal in vitro physiological levels are reached, they negatively impact their own production and the downstream PqsE regulated genes. PqsE, HHQ and PQS are essential molecules in the negative feedback auto-regulatory loops that contribute to this homeostatic regulation. Although the HHQ concentrations shown here are not attained in vitro because HHQ is fully converted into PQS, this effect is most likely relevant in vivo where we have shown that HHQ levels are higher than those of PQS [17]. In addition, in lasR− mutants that accumulated during chronic infections HHQ levels are also higher than PQS since PqsH responsible for the conversion of HHQ to PQS is under the control of LasR [67]. Nevertheless, we show that HHQ and PQS have together synergistic effect as a negative auto-regulators that down-regulated pqs operon transcription, reducing their own production and that of the other HAQs. Thus, jointly with PqsE, PQS and HHQ most probably contributed to the down-regulation of the pqs and phn operons observed during the late growth phase of P. aeruginosa (Figure S1).
In addition to being activator and auto-down-regulator PQS acted also as a homeostatic agent at high physiological concentrations by down-regulating most PqsE-dependent, downstream genes. Consistently, maximum pyocyanin production occurred only at low PQS concentrations that were sufficient to maximally activate the pqs operon. The homeostatic effect of PQS downstream of the PqsE genes was clearly independent of MvfR, PqsE and of other HAQs given that its effects were still apparent in pqsA− and mvfR− backgrounds. Interestingly, this effect appeared also to be post-transcriptional since PQS did not significantly impact phz operon transcription but affected pyocyanin production even when the phz operon was constitutively expressed. The mechanism behind this effect remains to be discovered. One intriguing possibility may be that PQS exerts its effect via RsmA and/or on small RNAs like rsmZ or prrF.
Previous studies have suggested that while PqsE is the PQS response protein [19],[20], it does not influence PQS production [11],[12]. Here we show that PqsE is a crucial player in orchestrating the homeostatic regulation of the signaling molecules HHQ and PQS as well as establishing a connection to the QS RhlR system, underscoring it as a key mediator of MvfR regulon activation and cooperation with the AHL QS system. Our findings also provide initial answers as to why PqsE, although not involved in the synthesis of HAQs in vivo or in vitro [11],[19],[20]), is tightly regulated together with the other pqs operon genes. Although our findings are primarily based on trans-regulatory studies, the overexpression of PqsE demonstrated for the first time that PqsE can impact HAQs concentrations by down-regulating their production. In corroboration, are both the AA accumulation and the transcriptional induction of the antABC genes responsible for AA degradation [68],[69] and shown to be regulated by prrF1 and prrF2 [45]. Since pqsE is co-transcribed by MvfR together with pqsA-D genes, the reduced production of HAQs mediated by PqsE indicates that pqsE gene transcription itself is also downregulated in a negative feedback mechanism that finely balances the regulatory loop.
Although PQS and HHQ signal molecules are critical to MvfR-dependent gene expression, their addition has failed to rescue pqsE- mutant cells to activate expression of many MvfR-regulated genes or to produce of pyocyanin [11],[17],[19],[20]. Here we found that overexpression of PqsE induced pyocyanin production and transcription of an additional approximately 600 MvfR-regulated genes independently of MvfR, HAQs and AA, demonstrating the crucial role of PqsE in activating MvfR regulon genes independently of the HAQs. Ultimately, expression of PqsE in an mvfR− or pqsA− strain restored P. aeruginosa virulence as determined by growth inhibition of yeast and flies feeding assay, indicating that PqsE did not need HAQs to confer virulence in these systems. Corroboratory results were reported by Farrow et al. [51] who showed in a qualitative manner that expression of PqsE in an mvfR− mutant restored pyocyanin production. These results together indicate that, at least with regard to the genes listed in Table S1, PQS and HHQ only act as inducers of MvfR to express PqsE that once expressed induces the P. aeruginosa virulence response without HAQs or MvfR. Thus, PqsE cannot be designated as the “quinolone signal response protein”. Nevertheless, it is not yet known how PqsE, a protein that belongs to the metallo-beta-lactamase super family without any known DNA binding motifs, regulates the transcription of so many genes. Its predicted hydrolase activity suggests that it may cleave or participate in the synthesis of small molecules. Due to the location of the pqsE gene in the pqs operon, the immediate candidates likely targeted by PqsE are HAQs. However following extensive LC/MS analyses, we were unable to detect any molecule that accumulated or diminished in concentration in pqsE− cultures compared to WT cultures (data not shown). In addition we were unable to complement pyocyanin production in a pqsE− culture by exogenously adding HAQs, AHLs or whole PA14 supernatants ([11] and data not shown). Nonetheless, collectively, our results indicate that PqsE is involved in a negative feedback loop that affects the regulation and integration of HAQs-mediated cell-cell signaling molecules and that is functionally dependent on RhlR. The exact nature of the co-dependency between PqsE and RhlR remains unclear. The downregulation of rhlR expression by ∼2 fold in a pqsE mutant is not sufficient to explain the striking transcriptional and phenotypic effects mediated by PqsE. Since PqsE is not predicted to be a transcriptional factor [50] it is highly likely that it may exert its effect on RhlR post-transcriptionally, and this effect may be perhaps extended to other transcriptional factors.
The MvfR affected gene list has a substantial overlap [11] with the previously published list of Rhl/Las-controlled genes [42], and the expression of almost all MvfR-regulated genes controlled by PqsE. Both PqsE activities (i.e., fine-tuning HAQs production by down-regulating the pqs operon, induction of pyocyanin production and downregulation of pyoverdine production) were dependent on RhlR apparently acting downstream but in a tight collaboration with PqsE. Recently, Farrow and colleagues showed that the addition of AHL C4-HSL (a RhlR inducer) to PAO1 pqsE− isogenic mutants also restored pyocyanin production [51]. These findings, although we did not reproduce them in PA14 cells, are in agreement with our findings that PqsE and RhlR functions are linked. However, the exact relationship between PqsE and RhlR, that is when or how they cooperate, remains elusive since RhlR in some cases functions in the absence of PqsE; for example, the RhlR-dependent C4-HSL levels in a pqsE− mutant strain were identical to the parental strain (data not shown) as also was previously shown for the mvfR− mutant [11].
The relationship between iron, QS regulation, and P. aeruginosa virulence is multifaceted [31],[32],[34],[36],[45],[63] and extremely complex. Data presented in this report demonstrate that the MvfR regulon represents a striking paradigm of the interplay between environmental signals and bacterial secreted cell-cell signal molecules that participate in positive and negative homeostatic regulatory loops. QS MvfR components control the transcription of many iron related genes, while iron related regulators control the expression of QS genes (see Table S3B) in addition to iron related genes. The relationship between iron and QS regulation is further strengthened through the iron-related regulators VqsR [43] and the PA2384 product [63] that were found to control the expression of phnAB and pqsA-E operons. Furthermore, the iron starvation sigma factor PvdS was shown to positively control the expression of mvfR via its IS box [36], iron was shown to control the pqs operon during biofilm formation [32], and the two small Fur-regulated RNAs Prrf 1 and 2 positively-regulated PQS production [45]. Our results showing that iron levels affected HAQs activities both as inducers of MvfR and as fine-balancers provide corroboration for the view that the MvfR regulon is closely linked with iron regulation. The complexity of the interplay between the MvfR regulon and iron control is further increased by: a. the ability of PQS but not HHQ to trap iron [47], which likely reduces available iron within the cell and promotes iron starvation, thereby affecting PqsE-mediated control of bacterial iron response genes, including the siderophores pyochelin and pyoverdine; and b. iron, especially in high concentrations, induces oxidative stress that was shown to affect and being affected by PQS [70]. Thus, it is possible that some of the phenotypic effects of PQS and iron shown here could be attributed to oxidative stress. Thus, it would be of importance to further investigate the contribution of iron, as a nutrient, a signal molecule, and an oxidative stress inducer in QS and P. aeruginosa virulence.
The existence of a tight interconnection between iron concentrations, QS, and virulence in P. aeruginosa is likely due to iron conditions encountered in vivo [71],[72] serving as a signal indicating a hostile environment requiring expression of virulence or fitness-related genes. When host tissues become damaged as a consequence of virulence factor production, the resulting increase in iron concentrations should down-regulate virulence factor concentrations, thereby reducing bacterial virulence that may favor host survival and potentially chronic infection.
A complete understanding of the regulation of the multiple P. aeruginosa virulence networks, in particular the mechanisms of the homeostatic and down-regulation processes (Figure 6), will be essential for the development of drugs targeting QS inhibition [73],[74]. The findings presented in this study may aid in the design of anti-infective therapies tailored to interfere with virulence pathways and provide a paradigm for understanding the complex QS networks of other bacterial pathogens besides that of P. aeruginosa.
Fig. 6. Schematic of the positive and negative homeostatic interplay among the MvfR regulon components PqsE, and PQS and HHQ with RhlR and iron. 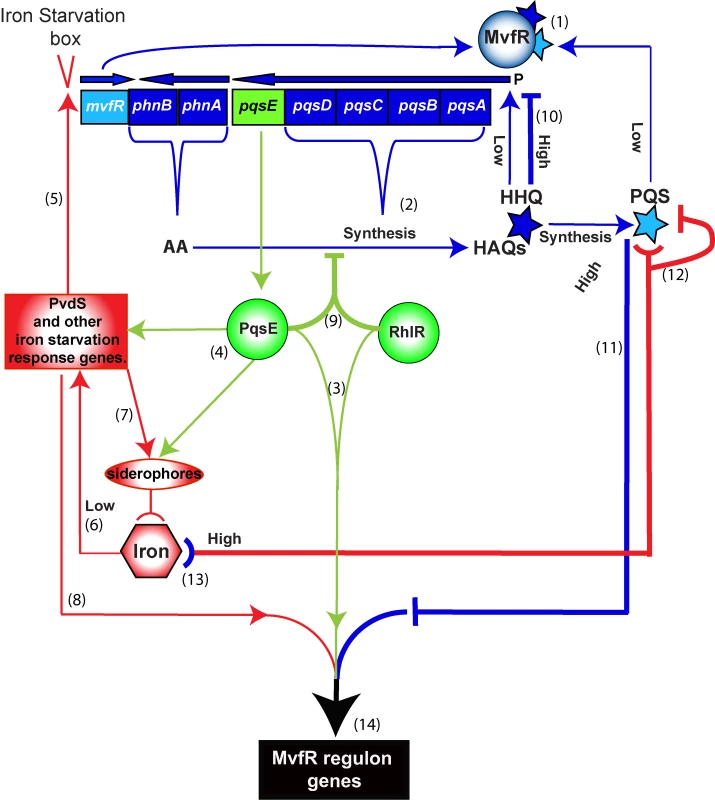
PqsE (green), HHQ and PQS (blue) and iron (red) play a dual role in up- or down-regulating the MvfR regulon. The outcome—that is the level of downstream gene expression translated into the bacterial virulence response—is the integrated sum of these interactions. Positive loops (thin lines): (1) MvfR is induced by HHQ and its derivative PQS to express phn and pqs operons, which are in turn (2) responsible for the synthesis of HAQs. PqsE is not required for HAQ synthesis and does not need AA or its derivatives for its “bottleneck” function, (3) controlling the expression of many virulence factors in cooperation with the AHL regulator RhlR. (4) PqsE also controls many iron starvation response genes, such as PvdS and siderophores. (5) PvdS in turn up-regulates the transcription of mvfR via an iron starvation box. (6) Low iron conditions also contribute to the induction of PvdS and other iron-related regulators to activate the iron response including (7) uptake of iron into the cell by siderophores as well as (8) induction of the virulence response. Negative loops (thick lines): (9) PqsE in cooperation with RhlR down-regulates the expression of the phn and pqs operons, thus reducing HAQ production. When a threshold concentration of HHQ is reached, (10) HHQ down-regulates the pqs operon. (11) PQS at high physiological levels in turn counterbalances the expression of PqsE-controlled genes, including many virulence factors. High levels of iron in presence of low levels of PQS, reduce P. aeruginosa virulence, at least in part, by (12) binding and inactivating PQS. In contrast, when PQS is at high physiological levels its inactivation by iron will increase virulence by reducing the negative PQS counterbalance and thus sustain the positive loops that include (13) iron starvation as a result of PQS trapping iron. (14) The integration of these processes enforces a fine-tuning of MvfR regulon gene expression levels, therefore determining the magnitude of virulence. Materials and Methods
Bacterial strains, growth conditions, and plasmids
Table S4 lists bacterial strains and plasmids used in this study. P. aeruginosa were routinely grown in Luria Bertani (LB) broth at 37°C for 18 h, and diluted to OD600 nm 0.05 and grown to the desired OD600 nm. For low iron media the bacteria were grown in D-TSB medium [36] that was treated with Chelex 100 beads (Bio-Rad, Hercules, CA) and for high iron FeCl3 or FeSO4 were added at concentrations of 200 µM. The E. coli JM109 strain was used for sub-cloning and plasmid propagation. The E. coli S17-1 strain was used for conjugation between E. coli and P. aeruginosa by the pEX18Ap-derivative allelic replacement method [75]. Antibiotics used included ampicillin (Amp) (100 µg/ml), carbenicillin (Crb) (300 µg/ml), gentamycin (Gnt) (15/60 µg/ml), kanamycin ((Kan), 50/200), tetracycline (Tet) (15/200 µg/ml) and chloramphenicol (Cam) (15/50 µg/ml) for E. coli and P. aeruginosa respectively.
DNA manipulations
The plasmid overexpressing PqsE was generated by PCR amplification of the pqsE gene from PA14 genomic DNA using primer pairs GX119 and GX120 (Table S4). The PCR product was digested with HindIII/XbaI and sub-cloned into the pDN19 plasmid vector under plac promoter to generate pDN19pqsE that constitutively expresses pqsE. Construct integrity was confirmed by DNA sequencing. Plasmids were introduced into E. coli or P. aeruginosa PA14 by electroporation. Non polar deletions were generated by pEX18AP allelic replacement using sucrose selection. Fragments with the size of about 1 kb flanking the desired genes were cloned into the pEX18Ap plasmid vector and introduced into E. coli by electroporation followed by conjugation to P. aeruginosa. Alternatively, the λ-Red recombinase method was used to generate chromosomal deletions or insertions [53].
Reporter genes
Two kinds of reporter genes were used: 1) translational and transcriptional fusions to lacZ where the β-galactosidase activity assay was performed in triplicate as described [76] and; 2) a pqsA-GFP (ASV) fusion consisting of a pqsA promoter upstream to a short-lived GFP that allows for the detection of pqs operon up or down regulation carried on the plasmid pAC37 [32]. Overnight cultures were diluted to an OD600 nm of 0.05 in black, clear bottom sterile 96-well assay plates (Corning Inc., Corning, NY). The plates were incubated for 50 h at 37°C in an Infinite F200 plate reader (Tecan Group Ltd, Männedorf, Switzerland). Every 30 min the plates were shaken for 2 min and read at 600 nm and fluorescence detected by excitation at 485 nm and emission at 535 nm. The results are expressed as an average of 3–6 observations that were normalized to a strain that did not carry the plasmid pAC37.
RNA isolation, generation and analysis of transcriptome data
Bacteria were respectively grown overnight at 37°C, diluted to an OD600 nm of 0.05 in 25 ml LB with the corresponding antibiotics at 37°C until the OD600 nm reached 3.0. The total RNA was isolated with the RNeasy Mini kit (QIAGEN Inc., Valencia, CA) and cDNA synthesis and labeling performed according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). The P. aeruginosa PAO1 GeneChip® Genome array (Affymetrix) was used for hybridization, staining, washing and scanning according to the manufacturer's instructions. Experiments were independently performed in triplicate. Affymetrix DAT files were processed using the Affymetrix Gene Chip Operating System (GCOS) to create .cel files. The raw intensity .cel files were normalized by robust multi-chip analysis (RMA) (Bioconductor release 1.7) with PM-only models. Array quality control metrics generated by the Affymetrix Microarray Suite 5.0 were used to assess hybridization quality. Normalized expression values were analyzed with SAM (Significance Analysis of Microarray) using the permuted unpaired two-class test. Genes whose transcript levels exhibited either a 2-fold or up or down regulation and had a q value <6% were further analyzed. The results of the GeneChip® arrays were imported to GeneSpring 7.3 (Agilent Technologies, Inc., Palo Alto, CA) and the expression signals of the GeneChip® arrays were normalized to the constant value of 1.0 and the ratio cut-off was set to 2-fold. Annotations were performed using the database http://pseudomonas.com/. The transcriptome results were (in part) validated by assessing β-galactosidase expression and RT-PCR of selected genes (Figure S2). The data are deposited in NCBI GEO with accession number #GSE17147.
Quantitative real-time RT - PCR
Cells from each triplicate experiment were harvested at an OD600 nm of 2, 3 and 4. Total RNA was subsequently isolated using the RiboPure-Bacteria RNA Isolation kit (Ambion, Austin, TX) according to the manufacturer's instructions. cDNAs were synthesized with random reverse primers using the Reverse Transcription RETROscript kit (Ambion) according to the manufacturer's instructions. Specific primers (Table S4) for the amplification of products of approximately 200 base pairs were designed using the Primer3 algorithm (http://frodo.wi.mit.edu/primer3/) and analyzed by In Silico simulation of PCR amplifications (http://insilico.ehu.es/) and by the Primer Analysis Software NetPrimer (Premier Biosoft International, http://www.premierbiosoft.com/netprimer/index.html) for the detection of expressed pqsA, pqsE and rpoD that served as the normalizer genes [77]. Quantitative RT-PCR was carried out using the Brilliant II SYBR Green QPCR Master Mix (Stratagene) with a RT Fluorescence Detection System MX3005P (Stratagene, La Jolla, CA) in a 25 µl final volume. The efficiency of each pair of primers was determined by a standard curve of 8 dilutions of 1∶4 of PA14 genomic DNA. The relative expression ratios were calculated and analyzed using MXPro analysis software, version 4.01 (Stratagene) using a mathematical model that included an efficiency correction. The fold induction of mRNA was determined from the threshold values that were first normalized for rpoD gene expression that served as a normalizer and then for the threshold value of the WT strain harboring the pDN19 plasmid at an OD600 nm of 2 that served as the calibrator. The data are expressed as the average of triplicate samples.
HAQs detection
The quantification of HAQs concentration in bacterial culture supernatants and in vivo from rectus adominus muscle of burned and infected mice was performed by LC/MS as described previously [17],[78]. The HAQs were separated on a C18 reverse-phase column connected to a mass spectrometer using a water/acetonitrile gradient [78]. Positive electrospray in the MRM mode with 2×10−3 mTorr argon and 30 V as the collision gas were employed to quantify HAQs using the ion transitions HHQ 244>159, HHQ-D4 248>163, HQNO 260>159, PQS 260>175, and PQS-D4 264>179. The pseudomolecular ions of each compound were monitored in full scan mode using the unsaturated PA14 HAQs response factors.
Pyocyanin production assay
Samples of 5 ml were spun down and the supernatants mixed with equal volumes of chloroform. The lower blue organic phase was collected and mixed with 5 ml of HCl (0.2 N). The upper reddish phase was collected and its OD52 onm was measured. The concentration of pyocyanin was determined by the formula: mg/L = OD52 onm×17.072 normalized to cell counts and the statistical significance was assessed using the Student's 2 tailed t-test assuming equal variance [79]. In order to assess the production of pyocyanin by expression of the phz genes we used a co-culture of cells harboring the pUCP-A2G2 and pUCP-MS plasmids [80]. All experiments were performed in triplicate.
Pyoverdine production detection
D-TSB medium was used to grow 200 µl of bacterial cells in 96 wells plate. Production of pyoverdine was assessed using a plate reader (Infinite F200, Tecan Group Ltd, Männedorf, Switzerland). Pyoverdine levels were determined every 30 minutes using excitation at 400 nm and emission at 460 nm and the values obtained were normalized to cell growth (OD600 nm). Pyoverdine concentrations were calculated using a calibration curve of fluorescence of a range of concentrations of pyoverdine (Sigma Aldrich, US).
Yeast killing assay
Yeast (Cryptococcus neoformans KN99 α, Candida albicans ATCC #90028 DAY185 strain or Saccharomyces cerevisiae YJM310 strain) were plated for 2 days on YPD agar (Difco) plates at 30°C. A colony was picked and grown for 18 h in liquid YPD media (Difco) at 30°C with shaking (200 rpm). The yeast was diluted 1∶100 in 4 ml soft YPD agar (0.6% agar) and poured onto an YPD plate that was dried for 30 min in a laminar flow hood. A 1 µl drop of an overnight culture of the desired bacterial strain was put on top of the yeast lawn and the plate incubated for 2–3 days at 30°C. A dead yeast zone was formed around a by PA14 bacterial colony bun not around mutants such e.g., mvfR −, pqsA − and pqsE −. The viability of yeast in these zones was tested by plating yeast from distance of 5 mm from the bacterial colonies on YPD plates.
Fly infection
Fly infection feeding assay was performed as previously described in [58],[59]. Briefly, 45 female Oregon-R flies per group, 5–7 days old, were fed with a mixture of 4 ml of LB bacterial culture at OD600 nm 3.0 with 1 ml of 20% sucrose. Thus, feeding mix contained a final concentration of 80% LB containing ∼2×109 bacterial cells per ml and 4% sucrose. An autoclaved cotton ball was placed at the bottom of each fly vial and was impregnated with 5 ml of the feeding mix. The 45 flies per treatment group were sub-divided in three fly vials (15 flies in each), sealed with a clean cotton ball, and incubated at 25°C. Fly survival was recorded twice a day until all flies succumbed to the infection. Statistical analysis of the survival curves was preformed using the log-rank test (Mantel-Haenszel) of the Kaplan-Meier estimate of survival using the software MedCalc (http://www.medcalc.be/). Two independent experiments gave similar results.
Supporting Information
Zdroje
1. FuquaC
ParsekMR
GreenbergEP
2001 Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35 439 468
2. JointI
Allan DownieJ
WilliamsP
2007 Bacterial conversations: talking, listening and eavesdropping. An introduction. Philos Trans R Soc Lond B Biol Sci 362 1115 1117
3. CornelisP
2008 Pseudomonas: Genomics and Molecular Biology;
CornelisP
244 Horizon Scientific Press
4. KirisitsMJ
ParsekMR
2006 Does Pseudomonas aeruginosa use intercellular signalling to build biofilm communities? Cell Microbiol 8 1841 1849
5. SchusterM
GreenbergEP
2006 A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296 73 81
6. VenturiV
2006 Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev 30 274 291
7. HeurlierK
DenervaudV
HaasD
2006 Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int J Med Microbiol 296 93 102
8. SmithRS
IglewskiBH
2003 P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol 6 56 60
9. DubernJF
DiggleSP
2008 Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol Biosyst 4 882 888
10. ShinerEK
RumbaughKP
WilliamsSC
2005 Inter-kingdom signaling: deciphering the language of acyl homoserine lactones. FEMS Microbiol Rev 29 935 947
11. DézielE
GopalanS
TampakakiAP
LépineF
PadfieldKE
2005 The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol Microbiol 55 998 1014
12. DézielE
LépineF
MilotS
HeJ
MindrinosMN
2004 Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101 1339 1344
13. RahmeLG
TanMW
LeL
WongSM
TompkinsRG
1997 Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci U S A 94 13245 13250
14. CaoH
KrishnanG
GoumnerovB
TsongalisJ
TompkinsR
2001 A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci U S A 98 14613 14618
15. RahmeLG
AusubelFM
CaoH
DrenkardE
GoumnerovBC
2000 Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci U S A 97 8815 8821
16. XiaoG
HeJ
RahmeLG
2006 Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology 152 1679 1686
17. XiaoG
DézielE
HeJ
LépineF
LesicB
2006 MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol 62 1689 1699
18. WadeDS
CalfeeMW
RochaER
LingEA
EngstromE
2005 Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol 187 4372 4380
19. GallagherLA
McKnightSL
KuznetsovaMS
PesciEC
ManoilC
2002 Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol 184 6472 6480
20. DiggleSP
WinzerK
ChhabraSR
WorrallKE
CamaraM
2003 The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol 50 29 43
21. McGrathS
WadeDS
PesciEC
2004 Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol Lett 230 27 34
22. LépineF
MilotS
DézielE
HeJ
RahmeLG
2004 Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom 15 862 869
23. LépineF
DekimpeV
LesicB
MilotS
LesimpleA
2007 PqsA is required for the biosynthesis of 2,4-dihydroxyquinoline (DHQ), a newly identified metabolite produced by Pseudomonas aeruginosa and Burkholderia thailandensis. Biol Chem 388 839 845
24. Soberon-ChavezG
Aguirre-RamirezM
OrdonezL
2005 Is Pseudomonas aeruginosa only “sensing quorum”? Crit Rev Microbiol 31 171 182
25. ZaborinaO
LépineF
XiaoG
ValuckaiteV
ChenY
2007 Dynorphin Activates Quorum Sensing Quinolone Signaling in Pseudomonas aeruginosa. PLoS Pathog 3 e35 doi:10.1371/journal.ppat.0030035
26. DuanK
SuretteMG
2007 Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol 189 4827 4836
27. WagnerVE
FrelingerJG
BarthRK
IglewskiBH
2006 Quorum sensing: dynamic response of Pseudomonas aeruginosa to external signals. Trends Microbiol 14 55 58
28. WuL
EstradaO
ZaborinaO
BainsM
ShenL
2005 Recognition of host immune activation by Pseudomonas aeruginosa. Science 309 774 777
29. JensenV
LonsD
ZaouiC
BredenbruchF
MeissnerA
2006 RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J Bacteriol 188 8601 8606
30. GuinaT
WuM
MillerSI
PurvineSO
YiEC
2003 Proteomic analysis of Pseudomonas aeruginosa grown under magnesium limitation. J Am Soc Mass Spectrom 14 742 751
31. CornelisP
AendekerkS
2004 A new regulator linking quorum sensing and iron uptake in Pseudomonas aeruginosa. Microbiology 150 752 756
32. YangL
BarkenKB
SkindersoeME
ChristensenAB
GivskovM
2007 Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153 1318 1328
33. BollingerN
HassettDJ
IglewskiBH
CostertonJW
McDermottTR
2001 Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J Bacteriol 183 1990 1996
34. KimEJ
WangW
DeckwerWD
ZengAP
2005 Expression of the quorum-sensing regulatory protein LasR is strongly affected by iron and oxygen concentrations in cultures of Pseudomonas aeruginosa irrespective of cell density. Microbiology 151 1127 1138
35. MasseE
ArguinM
2005 Ironing out the problem: new mechanisms of iron homeostasis. Trends Biochem Sci 30 462 468
36. OchsnerUA
WildermanPJ
VasilAI
VasilML
2002 GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol 45 1277 1287
37. PalmaM
WorgallS
QuadriLE
2003 Transcriptome analysis of the Pseudomonas aeruginosa response to iron. Arch Microbiol 180 374 379
38. OchsnerUA
KochAK
FiechterA
ReiserJ
1994 Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol 176 2044 2054
39. VasilML
2007 How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20 587 601
40. CornelisP
MatthijsS
Van OeffelenL
2009 Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22 15 22
41. ViscaP
LeoniL
WilsonMJ
LamontIL
2002 Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol Microbiol 45 1177 1190
42. SchusterM
LostrohCP
OgiT
GreenbergEP
2003 Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185 2066 2079
43. JuhasM
WiehlmannL
HuberB
JordanD
LauberJ
2004 Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology 150 831 841
44. JuhasM
WiehlmannL
SalunkheP
LauberJ
BuerJ
2005 GeneChip expression analysis of the VqsR regulon of Pseudomonas aeruginosa TB. FEMS Microbiol Lett 242 287 295
45. OglesbyAG
FarrowJM3rd
LeeJH
TomarasAP
GreenbergEP
2008 The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J Biol Chem 283 15558 15567
46. BredenbruchF
GeffersR
NimtzM
BuerJ
HausslerS
2006 The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ Microbiol 8 1318 1329
47. DiggleSP
MatthijsS
WrightVJ
FletcherMP
ChhabraSR
2007 The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol 14 87 96
48. ZaborinA
RomanowskiK
GerdesS
HolbrookC
LepineF
2009 Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci U S A 106 6327 6332
49. VialL
LépineF
MilotS
GroleauMC
DekimpeV
2008 Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J Bacteriol 190 5339 5352
50. YuS
JensenV
SeeligerJ
FeldmannI
WeberS
2009 Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein. Biochemistry 48 10298 10307
51. FarrowJM3rd
SundZM
EllisonML
WadeDS
ColemanJP
2008 PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol 190 7043 7051
52. MavrodiDV
BonsallRF
DelaneySM
SouleMJ
PhillipsG
2001 Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183 6454 6465
53. LesicB
RahmeLG
2008 Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol Biol 9 20
54. HoganDA
KolterR
2002 Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296 2229 2232
55. KaleliI
CevahirN
DemirM
YildirimU
SahinR
2007 Anticandidal activity of Pseudomonas aeruginosa strains isolated from clinical specimens. Mycoses 50 74 78
56. KerrJR
1994 Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J Clin Microbiol 32 525 527
57. LauGW
GoumnerovBC
WalendziewiczCL
HewitsonJ
XiaoW
2003 The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect Immun 71 4059 4066
58. ApidianakisY
PitsouliC
PerrimonN
RahmeL
2009 Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci U S A
59. ApidianakisY
RahmeLG
2009 Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat Protoc 4 1285 1294
60. TiburziF
ImperiF
ViscaP
2008 Intracellular levels and activity of PvdS, the major iron starvation sigma factor of Pseudomonas aeruginosa. Mol Microbiol 67 213 227
61. HeinrichsDE
PooleK
1996 PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J Bacteriol 178 2586 2592
62. JuhasM
EberlL
TummlerB
2005 Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol 7 459 471
63. ZhengP
SunJ
GeffersR
ZengAP
2007 Functional characterization of the gene PA2384 in large-scale gene regulation in response to iron starvation in Pseudomonas aeruginosa. J Biotechnol 132 342 352
64. GirardG
BloembergGV
2008 Central role of quorum sensing in regulating the production of pathogenicity factors in Pseudomonas aeruginosa. Future Microbiol 3 97 106
65. BjarnsholtT
GivskovM
2007 The role of quorum sensing in the pathogenicity of the cunning aggressor Pseudomonas aeruginosa. Anal Bioanal Chem 387 409 414
66. WinstanleyC
FothergillJL
2009 The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol Lett 290 1 9
67. D'ArgenioDA
WuM
HoffmanLR
KulasekaraHD
DézielE
2007 Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64 512 533
68. BundyBM
CampbellAL
NeidleEL
1998 Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobacter sp. strain ADP1. J Bacteriol 180 4466 4474
69. UrataM
MiyakoshiM
KaiS
MaedaK
HabeH
2004 Transcriptional regulation of the ant operon, encoding two-component anthranilate 1,2-dioxygenase, on the carbazole-degradative plasmid pCAR1 of Pseudomonas resinovorans strain CA10. J Bacteriol 186 6815 6823
70. HausslerS
BeckerT
2008 The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog 4 e1000166 10.1371/journal.ppat.1000166
71. HaasB
KrautJ
MarksJ
ZankerSC
CastignettiD
1991 Siderophore presence in sputa of cystic fibrosis patients. Infect Immun 59 3997 4000
72. RatledgeC
DoverLG
2000 Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54 881 941
73. LesicB
LépineF
DézielE
ZhangJ
ZhangQ
2007 Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog 3 e126 doi:10.1371/journal.ppat.0030126
74. RasmussenTB
GivskovM
2006 Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol 296 149 161
75. SchweizerHP
1992 Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol 6 1195 1204
76. MillerJH
1972 Experiments in molecular genetics. Cold Spring Harbor, N.Y. Cold Spring Harbor Laboratory 352 355
77. SavliH
KaradenizliA
KolayliF
GundesS
OzbekU
2003 Expression stability of six housekeeping genes: A proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microbiol 52 403 408
78. LépineF
DézielE
MilotS
RahmeLG
2003 A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim Biophys Acta 1622 36 41
79. EssarDW
EberlyL
HaderoA
CrawfordIP
1990 Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172 884 900
80. MavrodiDV
BlankenfeldtW
ThomashowLS
2006 Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 44 417 445
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 3- Stillova choroba: vzácné a závažné systémové onemocnění
- Perorální antivirotika jako vysoce efektivní nástroj prevence hospitalizací kvůli COVID-19 − otázky a odpovědi pro praxi
- Diagnostika virových hepatitid v kostce – zorientujte se (nejen) v sérologii
- Jak souvisí postcovidový syndrom s poškozením mozku?
- Familiární středomořská horečka
-
Všechny články tohoto čísla
- All Mold Is Not Alike: The Importance of Intraspecific Diversity in Necrotrophic Plant Pathogens
- Tsetse EP Protein Protects the Fly Midgut from Trypanosome Establishment
- Perforin and IL-2 Upregulation Define Qualitative Differences among Highly Functional Virus-Specific Human CD8 T Cells
- N-Acetylglucosamine Induces White to Opaque Switching, a Mating Prerequisite in
- Origin and Evolution of Sulfadoxine Resistant
- Rapid Evolution of Pandemic Noroviruses of the GII.4 Lineage
- Natural Strain Variation and Antibody Neutralization of Dengue Serotype 3 Viruses
- Fine-Tuning Translation Kinetics Selection as the Driving Force of Codon Usage Bias in the Hepatitis A Virus Capsid
- Structural Basis of Cell Wall Cleavage by a Staphylococcal Autolysin
- Direct Visualization by Cryo-EM of the Mycobacterial Capsular Layer: A Labile Structure Containing ESX-1-Secreted Proteins
- Lipopolysaccharide Is Synthesized via a Novel Pathway with an Evolutionary Connection to Protein -Glycosylation
- MicroRNA Antagonism of the Picornaviral Life Cycle: Alternative Mechanisms of Interference
- Limited Trafficking of a Neurotropic Virus Through Inefficient Retrograde Axonal Transport and the Type I Interferon Response
- Direct Restriction of Virus Release and Incorporation of the Interferon-Induced Protein BST-2 into HIV-1 Particles
- RNAIII Binds to Two Distant Regions of mRNA to Arrest Translation and Promote mRNA Degradation
- Direct TLR2 Signaling Is Critical for NK Cell Activation and Function in Response to Vaccinia Viral Infection
- The Essentials of Protein Import in the Degenerate Mitochondrion of
- Dynamic Imaging of Experimental Induced Hepatic Granulomas Detects Kupffer Cell-Restricted Antigen Presentation to Antigen-Specific CD8 T Cells
- An Accessory to the ‘Trinity’: SR-As Are Essential Pathogen Sensors of Extracellular dsRNA, Mediating Entry and Leading to Subsequent Type I IFN Responses
- Innate Killing of by Macrophages of the Splenic Marginal Zone Requires IRF-7
- Exoerythrocytic Parasites Secrete a Cysteine Protease Inhibitor Involved in Sporozoite Invasion and Capable of Blocking Cell Death of Host Hepatocytes
- Inhibition of Macrophage Migration Inhibitory Factor Ameliorates Ocular -Induced Keratitis
- Membrane Damage Elicits an Immunomodulatory Program in
- Fatal Transmissible Amyloid Encephalopathy: A New Type of Prion Disease Associated with Lack of Prion Protein Membrane Anchoring
- Nucleophosmin Phosphorylation by v-Cyclin-CDK6 Controls KSHV Latency
- A Combination of Independent Transcriptional Regulators Shapes Bacterial Virulence Gene Expression during Infection
- Inhibition of Host Vacuolar H-ATPase Activity by a Effector
- Human Cytomegalovirus Protein pUL117 Targets the Mini-Chromosome Maintenance Complex and Suppresses Cellular DNA Synthesis
- Dispersion as an Important Step in the Biofilm Developmental Cycle
- Kaposi's Sarcoma-Associated Herpesvirus ORF57 Protein Binds and Protects a Nuclear Noncoding RNA from Cellular RNA Decay Pathways
- Differential Regulation of Effector- and Central-Memory Responses to Infection by IL-12 Revealed by Tracking of Tgd057-Specific CD8+ T Cells
- The Human Polyoma JC Virus Agnoprotein Acts as a Viroporin
- Expansion, Maintenance, and Memory in NK and T Cells during Viral Infections: Responding to Pressures for Defense and Regulation
- T Cell-Dependence of Lassa Fever Pathogenesis
- HIV and Mature Dendritic Cells: Trojan Exosomes Riding the Trojan Horse?
- Endocytosis of the Anthrax Toxin Is Mediated by Clathrin, Actin and Unconventional Adaptors
- A Capsid-Encoded PPxY-Motif Facilitates Adenovirus Entry
- Homeostatic Interplay between Bacterial Cell-Cell Signaling and Iron in Virulence
- Serological Profiling of a Protein Microarray Reveals Permanent Host-Pathogen Interplay and Stage-Specific Responses during Candidemia
- YfiBNR Mediates Cyclic di-GMP Dependent Small Colony Variant Formation and Persistence in
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Kaposi's Sarcoma-Associated Herpesvirus ORF57 Protein Binds and Protects a Nuclear Noncoding RNA from Cellular RNA Decay Pathways
- Endocytosis of the Anthrax Toxin Is Mediated by Clathrin, Actin and Unconventional Adaptors
- Perforin and IL-2 Upregulation Define Qualitative Differences among Highly Functional Virus-Specific Human CD8 T Cells
- Inhibition of Macrophage Migration Inhibitory Factor Ameliorates Ocular -Induced Keratitis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Autoři: prof. MUDr. Vladimír Palička, CSc., Dr.h.c., doc. MUDr. Václav Vyskočil, Ph.D., MUDr. Petr Kasalický, CSc., MUDr. Jan Rosa, Ing. Pavel Havlík, Ing. Jan Adam, Hana Hejnová, DiS., Jana Křenková
Autoři: MUDr. Irena Krčmová, CSc.
Autoři: MDDr. Eleonóra Ivančová, PhD., MHA
Autoři: prof. MUDr. Eva Kubala Havrdová, DrSc.
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



