-
Medical journals
- Career
PGC‐1 isoforms and their target genes are expressed differently in human skeletal muscle following resistance and endurance exercise
Authors: Mika Silvennoinen *,1; Juha P. Ahtiainen 1; Juha J. Hulmi 1; Satu Pekkala 2; Ritva S. Taipale 1; Bradley C. Nindl 3; Tanja Laine 1; Keijo Häkkinen 1; Harri Selänne 4,5; Heikki Kyröläinen 1; Heikki Kainulainen 1
Authors‘ workplace: Department of Biology of Physical Activity, University of Jyväskylä, Jyväskylä, Finland 1; Department of Health Sciences, University of Jyväskylä, Jyväskylä, Finland 2; The Military Performance Division, The Unites States Army Research Institute of Environmental Medicine, Natick, Massachusetts 3; Jyväskylä Central Hospital, Jyväskylä, Finland 4; LIKES Research Center for Sport and Health Sciences, Jyväskylä, Finland 5
Published in: Physiological Reports, 3, 2015, č. 10, s. 1-12
Category: Original Research
doi: https://doi.org/10.14814/phy2.12563© 2015 The Authors. Physiological Reports published by Wiley Periodicals, Inc. on behalf of the American Physiological Society and The Physiological Society.
This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited.Overview
The primary aim of the present study was to investigate the acute gene expression responses of PGC‐1 isoforms and PGC‐1α target genes related to mitochondrial biogenesis (cytochrome C), angiogenesis (VEGF‐A), and muscle hypertrophy (myostatin), after a resistance or endurance exercise bout. In addition, the study aimed to elucidate whether the expression changes of studied transcripts were linked to phosphorylation of AMPK and MAPK p38. Nineteen physically active men were divided into resistance exercise (RE, n = 11) and endurance exercise (EE, n = 8) groups. RE group performed leg press exercise (10 × 10 RM, 50 min) and EE walked on a treadmill (~80% HRmax, 50 min). Muscle biopsies were obtained from the vastus lateralis muscle before, 30 min, and 180 min after exercise. EE and RE significantly increased the gene expression of alternative promoter originated PGC‐1α exon 1b ‐ and 1bxs'‐derived isoforms, whereas the proximal promoter originated exon 1a‐derived transcripts were less inducible and were upregulated only after EE. Truncated PGC‐1α transcripts were upregulated both after EE and RE. Neither RE nor EE affected the expression of PGC‐1β. EE upregulated the expression of cytochrome C and VEGF‐A, whereas RE upregulated VEGF‐A and downregulated myostatin. Both EE and RE increased the levels of p‐AMPK and p‐MAPK p38, but these changes were not linked to the gene expression responses ofPGC‐1 isoforms. The present study comprehensively assayed PGC‐1 transcripts in human skeletal muscle and showed exercise mode‐specific responses thus improving the understanding of early signaling events in exercise‐induced muscle adaptations.
Keywords:
PGC1-1b, PGC-1a, physical activity, splice variant.Introduction
Skeletal muscle comprises about 40–50% of body mass in humans (lean) and plays significant roles in locomotion, heat production, and whole‐body metabolism. Skeletal muscle has an outstanding capability to adapt to a variety of external stimuli that explains, in part, the marked differences observed in physical performance (e.g., endurance and strength) and health profiles between individuals (Hawley et al. 2014). The question remains, Which specific mechanisms are involved in the response to different types of physical activities? Because the effects are so diverse, mechanisms comprise multiple signaling cascades that form a complex network with each other (Egan et al. 2010). Yet, there have been attempts to identify single signaling cascades or molecules that could work as a master regulator for controlling exercise‐specific adaptations (Atherton et al.2005; Baar and Esser 1999). In recent years, the transcriptional coactivator peroxisome proliferator‐activated receptor‐γ coactivator (PGC)‐1α has been under a thorough investigation. PGC‐1α has been identified as a regulator of mitochondrial biogenesis, angiogenesis, antioxidant defense, and inflammatory proteins (Olesen et al. 2010).
A single bout of prolonged endurance exercise transiently increases PGC‐1α mRNA content in human and rat skeletal muscle (Pilegaard et al. 2003; Gidlund et al. 2015; Baar et al. 2002; Chinsomboon et al. 2009). Furthermore, other types of physical activity (e.g., sprint and resistance exercise) can also result in an increase in the gene expression of PGC‐1α (Gibala 2009; Ydfors et al. 2013). There are several intracellular signaling pathways that may contribute to eliciting the exercise‐induced PGC‐1α gene expression response including calcium signaling, AMPK and MAPK signaling, ROS‐mediated regulation, and β‐adrenergic signaling (Olesen et al. 2010).
Recently the existence of several different splice variants of PGC‐1α has been found in skeletal muscle. The splice variants differ from their starting exon (exon 1a, exon 1b, and exon 1b'/1c) and via alternative 3′ splicing, which produce either full‐length PGC‐1α protein (PGC‐1α) or shorter N‐truncated protein (NT‐PGC‐1α). Exon 1a‐derived PGC‐1α mRNAs are transcribed from the canonical proximal promoter, while exon 1b ‐ and 1b'‐derived mRNAs are transcribed from an alternative promoter ~14‐kb upstream from the canonical one (Ruas et al. 2012; Zhang et al. 2009). Lately, the roles of different splice variants of PGC‐1α in the initial signaling events of exercise‐induced skeletal muscle adaptations have been under heavy investigation. Ruas et al. (2012) showed that the expression of PGC‐1α4, exon 1b originated N‐truncated PGC‐1α transcript, results in robust skeletal muscle hypertrophy in vitro and in vivo. The study suggested that PGC‐1α4 is preferentially induced in mouse and human muscle during resistance exercise, and this would lead to muscle hypertrophy via induced expression of IGF1 and repressed expression of myostatin. However, the studies of Lundberg et al. (2014) and Ydfors et al. (2013) have questioned the preferential induction of PGC‐1α4 expression after resistance exercise by showing that also endurance exercise has effects on the response of this transcript in human skeletal muscle. In fact, the effects of endurance exercise were shown already in the study of Ruas et al. (2012). Interestingly, the study of Thom et al. (2014) showed that hypoxia specifically induces truncated forms of PGC‐1α (NT‐PGC‐1α and PGC‐1α4), which induces VEGF expression and angiogenesis, while having only a little effect on mitochondrial genes. Previous studies have also shown that both resistance and endurance exercise are able to induce expression of alternative and proximal promoter originated PGC‐1α transcripts, the alternative promoter originated transcripts being much more inducible compared to proximal promoter originated transcripts (Lundberg et al. 2014; Ydfors et al. 2013).
Exact primer design is an essential part of studies measuring mRNA levels of different splice variants. Since the most of the previous studies (Lundberg et al. 2014; Norrbom et al. 2011; Ydfors et al. 2013) investigating exercise‐induced acute gene expression responses of different PGC‐1αsplice variants in human skeletal muscle did not report exact primer sequences, it was impossible to evaluate the specificity of the used primers. To increase knowledge of early signaling events that drive the exercise mode‐specific adaptations, and to confirm previous findings with well‐defined primers, the present study aimed to investigate the acute gene expression responses of differentPGC‐1 isoforms after a single bout of high‐load resistance exercise (leg press protocol) or moderate intensity endurance exercise (uphill walking on treadmill) in human skeletal muscle. In addition, we aimed to determine how these two different exercise protocols affect the expression of a selection of known PGC‐1α target genes related to mitochondrial biogenesis, angiogenesis, and muscle hypertrophy. Furthermore, we wanted to elucidate whether the expression changes of studied transcripts were linked to phosphorylation changes of known PGC‐1α regulators AMPK and p38 MAPK.
Materials and Methods
Ethical approval
The study was conducted according to the Declaration of Helsinki, and ethical approval was granted by the ethics committees of the University of Jyväskylä and the Central Finland Health Care District, Jyväskylä, Finland. Each subject was carefully informed of all potential risks and discomforts and, thereafter, signed an informed consent document.
Study subjects
Two different experimental setups were included in the study: resistance and endurance exercise. The studies were linked to the larger military research project. A total of 22 healthy male reservists, who were physically active but not endurance or strength athletes, were recruited for the present investigation. Two subjects withdrew from the endurance exercise group and one subject was excluded from this group because of inadequate muscle samples. Therefore, the final study groups included 11 subjects in resistance exercise group (RE) and eight in endurance exercise group (EE). The general characteristics of the groups, which have been partly published previously (Ahtiainen et al. 2015), were as follows (mean ± SD): RE (n = 11, 26.0 ± 4.6 years, 182 ± 8 cm, 78.6 ± 11.7 kg, MVC 339.7 ± 99.7 kg, CMJ 34.0 ± 3.8 cm, 1RM 207.0 ± 26.6 kg, VO2max 59.9 ± 5.3 mL·kg−1·min−1) and EE (n = 8, 27.0 ± 3.6 years, 181 ± 9 cm, 72.5 ± 11.5 kg, MVC 221.8 ± 33.2 kg, CMJ 30.9 ± 3.6 cm, 1RM 167.3 ± 33.6 kg, VO2max 65.3 ± 5.2 mL·kg−1·min−1).
Exercise protocols
The protocols and equipment used with both exercise modes have been described in detail elsewhere (Ahtiainen et al. 2015). In brief, RE was a single high‐load hypertrophic type of resistance exercise consisting 10 sets of 10 repetition maximum (10 × 10 RM) using a bilateral leg press device (David 210, David Health Solutions Ltd., Helsinki, Finland). The starting load was 70% of 1RM. Thereafter, the loads were adjusted so that each subject would be able to perform a maximum of 10 repetitions for each set. The recovery between the sets was 2 min, except a 10 min rest that was applied between the fifth and sixth set. EE consisted of 50 min of strenuous walking on a treadmill. To increase exercise load additional weight (16.5 kg) were carried in backpack (OJK‐1, Telineyhtymä, Kotka, Finland). At minutes 0 : 00–5 : 00 and 40 : 00–45 : 00, the speed of the treadmill was 4.5 km·h−1 and the slope 4.0 degrees. At minutes 5 : 00–10 : 00 and 45 : 00–50 : 00, the speed was 7.0 km·h−1 and the slope 4.0 degrees. During the minutes 10 : 00–40 : 00, the walking speed was individually controlled and adjusted in 5‐min intervals by heart rate (HR) and blood lactate measurements. The criteria for walking speed included (1) blood lactate concentration approximately 4.0 mmol·L−1; and (2) HR between 75% and 85% of the individual HR maximum (HRmax) that was determined by a maximal endurance capacity test before the study.
The subjects were familiarized with the upcoming exercises and testing protocols 4–5 days before the study. To control effects of nutrition and hydration status, the subjects fasted for 12 h before the first (PRE) biopsy. Immediately after the biopsy, the subjects ate an energy bar (170 kcal, protein 7 g, carbohydrate 21 g, and fat 5.5 g) and drank 0.5 L of water. Two hours after the exercise loading, the subjects were allowed to eat again an energy bar and drink water (ad libitum). The energy bars and water were given to avoid lack of energy and disturbance of fluid balance during the measurement day lasting 8.5 h. Moreover, the amount of given energy was selected so that we could minimize the known effects of caloric restriction, fasting, and feeding on exercise responses (Ranhotra 2010; Canto et al. 2010). The subjects were asked to refrain from vigorous physical activity for 2 days before the study.
Muscle strength and explosive power
To assess basal strength characteristics and muscular fatigue produced by exercise, the following measurements were performed before and after exercise: maximal voluntary bilateral isometric force (MVC) of leg extensor muscles (electromechanical dynamometer, Department of Biology of Physical Activity, University of Jyväskylä, Jyväskylä, Finland) (Ahtiainen et al. 2015), maximal dynamic bilateral 1RM leg press (David 210, David Health Solutions Ltd, Helsinki, Finland), and countermovement jump (CMJ).
Blood lactate
In both EE and RE, fingertip blood lactate samples were collected before and immediately after exercises into capillary tubes, which were placed in a 1‐mL hemolyzing solution and analyzed automatically (EKF diagnostic, Biosen, Barleben, Germany). To adjust speeds in EE, blood lactate was monitored (Lactate Pro LT‐1710 analyser 35, Arkray Inc., Kyoto, Japan) during the exercise in 5‐min intervals.
Muscle biopsy procedure
The first (PRE) muscle biopsy was obtained from VL (right leg) 3 h before exercise. Biopsies were taken with a 5‐mm Bergström biopsy needle together with suction, midway between the patella and greater trochanter. Muscle depth was kept constant in different biopsies within subject by markings on the needle. The muscle sample was cleaned of any visible connective and adipose tissue as well as blood and frozen immediately in liquid nitrogen (−180°C) and stored at −80°C. The postexercise biopsy samples were taken from VL (left leg) after 30 and 180 min of recovery. The recovery protocol was identical in both exercise groups. The subjects were physically passive during the recovery.
Western blot analysis
Muscle biopsy specimens were hand‐homogenized in ice‐cold buffer (20 mmol·L−1 HEPES [pH 7.4], 1 mmol·L−1 EDTA, 5 mmol·L−1 EGTA, 10 mmol·L−1 MgCl2, 100 mmol·L−1 b‐glycerophosphate, 1 mmol·L−1 Na3PO4, 2 mmol·L−1 DTT, 1% Triton X‐100, 0.2% sodium deoxycholate, 30 μg·mL−1leupeptin, 30 μg·mL−1 aprotinin, 60 μg·mL−1 PMSF, and 1% phosphatase inhibitor cocktail; P 2850; Sigma, St. Louis, MO) at a dilution of 15 μL·mg−1 of wet weight muscle. Homogenates were rotated for 30 min at 4°C, centrifuged at 10,000 g for 10 min at 4°C to remove cell debris, and stored at −80°C. Total protein content was determined using the bicinchoninic acid protein assay (Pierce Biotechnology, Rockford, IL).
Aliquots of muscle lysate, containing 30 μg of total protein, were solubilized in Laemmli sample buffer and heated at 95°C for 10 min to denature proteins, and were then separated by SDS‐PAGE for 90 min at 200 V using 4–20% gradient gels on Criterion electrophoresis cell (Bio‐Rad Laboratories, Hercules, CA). All samples from each subject were run on the same 18‐sample gel. Proteins were transferred to PVDF membranes at 350 mA constant current for 3 h on ice at 4°C. Membranes were blocked in TBS with 0.1% Tween 20 (TBS‐T) containing 5% nonfat dry milk for 1 h and then incubated overnight at 4°C with rabbit polyclonal primary antibodies. Antibodies recognizing phosphorylated p38MAPKThr180/Tyr182 and AMPKαThr172 were purchased from Cell Signaling Technology (Danvers, MA) and these primary antibodies were diluted 1 : 2000 in TBS‐T containing 2.5% nonfat dry milk. For measuring protein levels of nontruncated full‐length splice variants of PGC‐1α, the antibody (1 : 3000, Calbiochem, Merck KGaA, Darmstadt, Germany) against C‐terminus of protein (amino acids 777–797) was used. Membranes were then washed (5 × 5 min) in TBS‐T, incubated with secondary antibody (horseradish peroxidase‐conjugated anti‐rabbit IgG; Cell Signaling Technology) diluted 1 : 25,000 in TBS‐T with 2.5% milk for 1 h followed by washing in TBS‐T (5 × 5 min). Proteins were visualized by ECL according to the manufacturer's protocol (SuperSignal west femto maximum sensitivity substrate, Pierce Biotechnology) and quantified (band intensity × volume) using a ChemiDoc XRS in combination with Quantity One software (version 4.6.3. Bio‐Rad Laboratories).
The uniformity of protein loading was confirmed by staining the membrane with Ponceau S. Our earlier studies and preliminary experiments confirmed a proportional linear relation between the protein loaded and the strongest band in Ponceau S at ∼42 kDa in quantification between 5 and 60 μg of total protein loaded (Hulmi et al. 2012). A less well proportional and linear relationship was found between GAPDH, α‐actin, and staining of myosin heavy chain left in the gel after blotting. Therefore, all of the results were normalized to the corresponding Ponceau S staining value at ∼42 kDa. In addition, to reduce gel‐to‐gel variation, the results of all samples within the gel were normalized to the mean value of all PRE samples within the gel. Quantification of p‐p38 MAPKThr180/Tyr182 was based on the average of two visible bands at 42 and 44 kDa. Phosphorylated AMPKαThr172 was quantified using the visible band at 62 kDa and nontruncated PGC‐1α using the band at 100 kDa.
RNA extraction and cDNA synthesis
Total RNA was isolated from the muscle biopsy with Trizol reagent (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Muscle samples were homogenized with a FastPrep FP120 (Thermo Fisher Scientific, Waltham, MA) tissue homogenizer by using Lysing Matrix D FP120 (Thermo Fisher). The quality of RNA was confirmed by spectrophotometry (NanoDrop; Thermo Fisher Scientific) and agarose gel electrophoresis. Reverse transcription of mRNA was performed from total RNA (5 μg) by using anchored oligo(dT)20 primers (Oligomer, Helsinki, Finland) and a SuperScript III Reverse Transcriptase kit (Life Technologies) according to the manufacturer's instructions. The cDNA samples were stored in −20°C.
Real‐time quantitative PCR
Cytochrome C (CYC), glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), and vascular endothelial growth factor A (VEGF‐A) mRNAs were quantified using TaqMan primers and hydrolysis probes (Assay IDs: CYC: Hs01588974_g1; GAPDH: Hs03929097_g1; VEGF‐A: Hs00900055_m1), TaqMan Gene Expression Master Mix (Life Technologies), and ABI 7300 Real‐Time quantitative PCR System (Life Technologies). The PCR cycle parameters used were: +50°C for 2 min, +95°C for 10 min, 40 cycles at +95°C for 15 sec, and +60°C for 1 min. The other studied transcripts (PGC‐1α exon 1a‐, 1b ‐ and 1b' ‐derived mRNAs, total NT‐PGC‐1α, total PGC‐1α, PGC‐1β, and myostatin) were quantified using iQ SYBR Supermix (Bio‐Rad Laboratories) and CFX96 Real‐Time PCR Detection System (Bio‐Rad Laboratories). The PCR cycle parameters used with this system were as follows: +95°C for 10 min, 40 cycles at +95°C for 10 sec, at gene‐specific annealing temperature (61°C, except 56 for total NT‐PGC‐1α) for 30 sec and at +72°C for 30 sec, followed by 5 sec at +65°C.
The primer sequences recognizing total NT‐PGC‐1α and PGC‐1α exon 1a‐derived transcripts were copied from Ruas et al. (2012) and myostatin from Kim et al. (2007). Other primers were designed with Primer3 (web version 4.0.0) software (Koressaar and Remm 2007; Untergasser et al. 2012). The structure of 5′ region of the human PGC‐1α gene is illustrated in Figure 1A. In addition, the detailed description of exon structure and primer design of measured PGC‐1α transcripts is presented in Figure 1B. The sequences of primer sets used with the SYBR green method are listed in Table 1. Following pilot RT‐qPCR runs performed with the SYBR green method, the specificity of each primer set was monitored by the melting curves and by agarose gel (3%) electrophoresis (Fig. 2). In addition, optimal gene‐specific annealing temperatures were determined. The amplification efficiencies for each gene were between 95% and 105%.
Figure 1. (A) The schematic structure of the 5′ region of the human PGC‐1α gene (Miura et al. 2008). White boxes indicate untranslated exon regions and gray boxes translated coding regions. Straight lines between boxes indicate introns. When mRNA is formed from the gene, two completely distinct first exons of PGC‐1α (exon 1a and 1b) can be spliced to the common exon 2. Furthermore, exon 1b can be spliced to the common exon 2 in two different ways producing either PGC‐1α exon 1b‐ or 1b'‐derived transcripts. In addition to differences in the starting exon, the splice variants of PGC‐1α differ via alternative 3′ splicing. The exon subsequent to exon 6 may be either exon 7a (ex7a) or exon 7b (ex7b). Ex7a is the exon insert, which contains an in‐frame stop codon resulting in the shorter N‐truncated proteins (NT‐PGC‐1α). Ex7b is present in nontruncated full‐length PGC‐1α proteins, which are translated using all 13 exons of PGC‐1α. The proximal promoter drives the transcription of PGC‐1α exon 1a‐derived transcripts (truncated and nontruncated) and the alternative promoter the transcription of PGC‐1α exon 1b and 1b'‐derived transcripts (truncated and nontruncated). (B) Detailed descriptions of primer pairs and their possible cDNA targets are presented. Primer pairs for detecting PGC‐1α exon 1a‐, 1b‐, 1b'‐derived transcripts are specific for different first exons, but do not separate truncated and nontruncated PGC‐1α splice variants. Primer pair for total NT‐PGC‐1α is specific for truncated PGC‐1α splice variants, but do not separate differences in first exon. The total PGC‐1α primer pair was designed to detect all truncated and nontruncated splice variants of PGC‐1α. The possible exon structures of mRNA targets for each primer pair are illustrated with gray boxes. The structure is presented only from the first exon to Ex7a or Ex7b because the mRNA structure is common in following exons (Ex8–Ex13). Binding sites of target specific forward (F) and reverse (R) primers are pointed out by arrows drawn next to the cDNA sequence in question. The sequences of all primers are also listed in Table 1. The sequences are amplicon sequences plus five nucleotides in the 3′ and 5′ end. The sequences of every other exon are bolded to separate subsequent exons from each other. The sequences were constructed by using human PGC‐1α cDNA sequence (Ensembl: ENST00000264867), human PGC‐1α gene sequence with its upstream regions (NCBI, Gene ID: 10891), and published cDNA structures of murine PGC‐1α splice variants (Wen et al. 2014; Ruas et al. 2012). 
1. RT-qPCR primers used with the SYBR green method 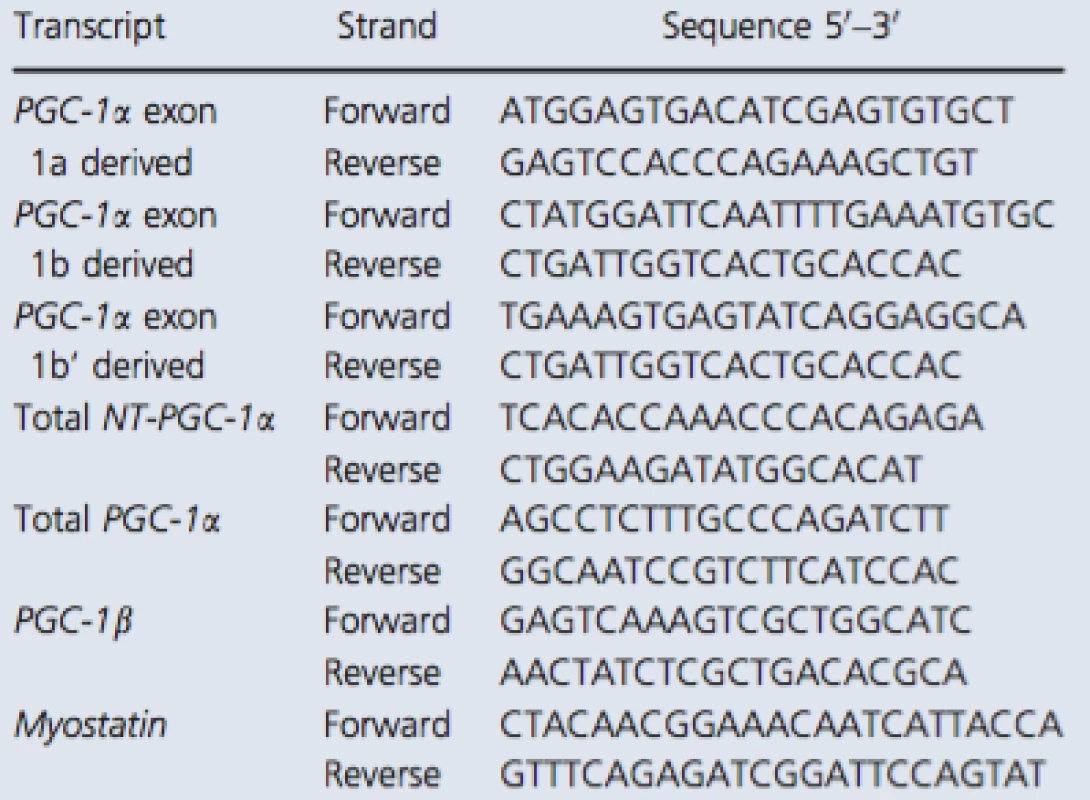
Figure 2. RT‐qPCR products on agarose gel. The bands correspond to the calculated amplicon sizes: PGC‐1α exon 1a‐ (127 bp, lane 2), 1b‐ (153 bp, lane 3), 1b' (188 bp, lane 4)‐derived transcripts, total NT‐PGC‐1α (172 bp, lane 5), total PGC‐1α (241 bp, lane 6), PGC‐1β (104 bp, lane 7), and myostatin (243 bp, lane 8). 50‐bp DNA ladder at lanes 1 and 9. 
Each sample was analyzed in duplicate, and a nontemplate control was included in each run. Relative gene expression levels of all target transcripts were normalized using RNA:cDNA hybrid concentrations which were measured using a Quant‐iT™ PicoGreen® assay (Life Technologies) according to the manufacturer's recommendations. This method of normalization has been validated especially for human exercise studies and muscle biopsy samples (Lundby et al. 2005). The stability of RNA:cDNA hybrid concentrations was compared to stability of GAPDH, one of the most common reference genes used in exercise studies. The average differences between time points in both exercise modes were similar in GAPDH and RNA:cDNA hybrid concentrations. However, the variation within time points in both exercise modes were lower in RNA:cDNA hybrid concentrations compared to GAPDH levels. The RNA:cDNA hybrid concentrations are presented in (Fig. 3G). The normalized relative gene expression results (R) of each sample were expressed in relation to PRE average and calculated using following formula:
Figure 3. The expression profiles of PGC‐1α exon 1a‐derived transcripts (A), PGC‐1α exon 1b‐derived transcripts (B), PGC‐1α exon 1b'‐derived transcripts (C), total NT‐PGC‐1α (D), total PGC‐1α (E), and PGC1β (F) at 30 min (30′) and 180 min (180′) after resistance (RE, n = 11) and endurance (EE, n = 8) exercises. The gene expression data of each sample are normalized to corresponding relative concentration value of RNA:cDNA hybrids (G). The gene expression changes are presented in relation to PRE average. Black lines represent median responses to exercise and gray lines represent individual responses. Error bars represent interquartile range. *Wilcoxon matched pairs signed‐rank test with Holm–Bonferroni correction P < 0.05 versus PRE, **P < 0.01 versus PRE. 
Statistical analyses
Conventional statistical methods were used to obtain means, standard deviations (SD), medians, interquartile ranges (IQR), and percentiles. The Shapiro–Wilk test was used to test the normality of the variables and the Levene's test was used to analyze the homogeneity of variances. Due to the small sample size and random violations in the normal distribution assumption and the homogeneity of the variance assumption, a nonparametric Friedman's two‐way ANOVA by ranks test was used to determine differences between time points (PRE, 30′ and 180′) within groups (RE and EE). Wilcoxon matched‐pair signed‐rank test with Holm–Bonferroni correction was used for the post hoc analysis. A Spearman's rank correlation test was used to test if the changes in p38 MAPKThr180/Tyr182 or AMPKαThr172 phosphorylation were associated with exercise‐induced gene expression changes. The significance level was set at P < 0.05. All statistical analyses were carried out using IBM SPSS statistics 20 software (IBM Corporation, Armonk, NY).
Results
The effects of exercise bouts
RE led to a decrease of 45 ± 16% (P < 0.01) in MVC and 17 ± 24% (NS, P = 0.07) in CMJ from pre ‐ to postexercise. Blood lactate increased to 11.1 ± 3.0 mmol·L−1 (P < 0.01) immediately after RE. During the loading in EE, averaged treadmill speed was 6.2 ± 0.4 km·h−1 and the slope 4.1 ± 0.8 degrees, the blood lactate was 4.2 ± 1.0 mmol·L−1 and HR was 83 ± 8% of the HRmax. Immediately after EE, MVC was 9 ± 14% (NS, P = 0.06) and CMJ 7 ± 6% (P < 0.05) lower than before the exercise, and blood lactate was 2.4 ± 1.5 mmol·L−1 (P < 0.01). These results (except CMJ) have been previously published (Ahtiainen et al. 2015).
Gene expression responses of PGC‐1 transcript variants after RE and EE
The average basal state (PRE) Cq values (all subjects) for studied PGC‐1 transcripts were as follows: PGC‐1α exon 1a‐derived transcripts 25.6 ± 1.2, PGC‐1α exon 1b‐derived transcripts 31.3 ± 3.1, PGC‐1α exon 1b'‐derived transcripts 33.8 ± 1.7, total NT‐PGC‐1α 29.7 ± 1.4, totalPGC1α 24.1 ± 1.2, and PGC1β 30.0 ± 1.6. The expression of PGC‐1α exon 1a‐derived transcripts increased by 1.7‐fold (P < 0.05, Fig. 3A) 30 min after EE. After RE no significant response was detected. The expression of PGC‐1α exon 1b‐derived transcripts was significantly increased in response to both RE and EE (Fig. 3B), while the peak changes were detected 180 min after exercise. At this time point, the average gene expression change was 170‐fold (P < 0.05) after EE and 997‐fold (P < 0.01) after RE. The expression of PGC‐1α exon 1b'‐derived transcripts were low in the present basal conditions. In the RE group, the transcripts were detected in six PRE samples from 11 and in EE group in three PRE samples from eight. However, after RE and EE the expression of PGC‐1α exon 1b'‐derived transcripts was substantially increased (Fig. 3C). When PRE values below the detection limit (Cq value before of which amplification was repeatable and specific) were replaced with the detection limit values (Cq 34.9), the average change was 67‐fold at 180 min after RE (P < 0.01) and ninefold at 180 min after EE (P < 0.05). The expression of truncated PGC‐1α transcripts (total NT‐PGC‐1α) (Fig. 3D) was increased at 180 min after RE (fivefold, P < 0.01) and 30 min after EE (1.7‐fold, P < 0.01). The expression of total PGC1α (all truncated and nontruncated splice variants together) (Fig. 3E) was increased at 180 min after RE (fourfold, P < 0.01) and 30 min after EE (1.8‐fold, P < 0.05). PGC1β had no significant gene expression response to either of the exercises (Fig. 3F).
Gene expression responses of PGC‐1α‐regulated genes after RE and EE
The gene expression of mitochondrial marker cytochrome c was increased at 30 min after EE (1.7‐fold, P < 0.05, Fig. 4A), but there were no responses to RE. However, the gene expression of angiogenesis regulator VEGF‐A was increased both after EE (threefold, P < 0.05) and RE (twofold,P < 0.05). EE‐induced response was significant already 30 min after exercise, but RE‐induced response did not appear until 180 min after exercise (Fig. 4B). RE decreased (2.5‐fold, P < 0.05) the gene expression of myostatin, the known inhibitor of muscle growth, without response to EE (Fig. 4C).
Figure 4. Gene expression changes of known PGC‐1α target genes cytochrome C [(A), CYC], VEGF‐A (B), and myostatin (C) at 30 min (30′) and 180 min (180′) after resistance (RE, n = 11) and endurance (EE, n = 8) exercises. The gene expression changes are presented in relation to PRE average. Black lines represent median responses to exercise and gray lines individual responses. Error bars represent interquartile range. *Wilcoxon matched pairs signed‐rank test with Holm–Bonferroni correction P < 0.05 versus PRE, **P < 0.01 versus PRE. ![Figure 4.
Gene expression changes of known PGC‐1α target genes cytochrome C [(A), CYC], VEGF‐A (B), and myostatin (C) at 30 min (30′) and 180 min (180′) after resistance (RE, n = 11) and endurance (EE, n = 8) exercises. The gene expression changes are presented in relation to PRE average. Black lines represent median responses to exercise and gray lines individual responses. Error bars represent interquartile range. *Wilcoxon matched pairs signed‐rank test with Holm–Bonferroni correction P < 0.05 versus PRE, **P < 0.01 versus PRE.](https://pl-master.mdcdn.cz/media/image/075f8d4fe6d44e4c808027fa8951ed59.jpg?version=1711889551)
The level of PGC‐1α proteins, p‐p38 MAPKThr180/Tyr182 and p‐AMPKαThr172, and their associations with studied gene expression changes
The protein level of full‐length nontruncated PGC‐1α was increased at 180 min after RE (2.3‐fold,P < 0.05, Fig. 5A) without changes after EE (n = 7). The results of phosphorylated p38 MAPKThr180/Tyr182 and AMPKαThr172 have been previously published from pre to post 30 min (Ahtiainen et al. 2015). The present study adds to that study a new time point (180 min) and because of this, the data were reanalyzed using different statistics, and the gel‐to‐gel variation was normalized differently. The level of the active phosphorylated form of p38 MAPKThr180/Tyr182 was increased by ninefold (P < 0.05, Fig. 5B) at 30 min after RE, but not after 180 min or EE. The level of p‐AMPKαThr172 was increased by fourfold at 30 min after RE (Fig. 5C), but not anymore after 180 min. There was also a trend (P = 0.07) for increased levels of p‐AMPKαThr172 at 30 min after EE.
Figure 4. Gene expression changes of known PGC‐1α target genes cytochrome C [(A), CYC], VEGF‐A (B), and myostatin (C) at 30 min (30′) and 180 min (180′) after resistance (RE, n = 11) and endurance (EE, n = 8) exercises. The gene expression changes are presented in relation to PRE average. Black lines represent median responses to exercise and gray lines individual responses. Error bars represent interquartile range. *Wilcoxon matched pairs signed‐rank test with Holm–Bonferroni correction P < 0.05 versus PRE, **P < 0.01 versus PRE. ![Figure 4.
Gene expression changes of known PGC‐1α target genes cytochrome C [(A), CYC], VEGF‐A (B), and myostatin (C) at 30 min (30′) and 180 min (180′) after resistance (RE, n = 11) and endurance (EE, n = 8) exercises. The gene expression changes are presented in relation to PRE average. Black lines represent median responses to exercise and gray lines individual responses. Error bars represent interquartile range. *Wilcoxon matched pairs signed‐rank test with Holm–Bonferroni correction P < 0.05 versus PRE, **P < 0.01 versus PRE.](https://pl-master.mdcdn.cz/media/image/075f8d4fe6d44e4c808027fa8951ed59.jpg?version=1711889551)
There were no strong associations between the exercise‐induced changes in p‐p38 MAPKThr180/Tyr182 or p‐AMPKαThr172 levels and the gene expression responses of PGC‐1 isoforms,PGC‐1β, and myostatin in either of the exercise groups. However, the level change of p‐AMPKαThr172 (ratio 30′/PRE) associated with the corresponding changes in VEGF‐A (rs = 0.821,P = 0.023) and CYC (rs = 0.75, P = 0.052) at 30 min after EE. Similar associations were also found after EE when the both time points after exercise were pooled (ratio 30′/PRE and ratio 180′/PRE) and analyzed together (VEGF‐A: rs = 0.608, P = 0.036; CYC: rs = 0.601, P = 0.039).
Discussion
The main findings of this study were that (1) both EE and RE induced significant increase in the gene expression of alternative promoter originated PGC‐1α exon 1b ‐ and 1b'‐derived isoforms, whereas the proximal promoter originated PGC‐1α exon 1a‐derived transcripts were less inducible and were upregulated only after EE; (2) truncated PGC‐1α transcripts were upregulated markedly after RE and slightly after EE; (3) based on the marker gene expression changes, EE induced responses typical for angiogenesis and mitochondrial biogenesis, while RE induced responses typical for angiogenesis and muscle hypertrophy; (4) both EE and RE increased the levels of phosphorylated p38 MAPK and AMPKα, but these changes were not linked to the gene expression responses of PGC‐1 isoforms.
The primary aim of the present study was to investigate the acute mRNA responses of differentPGC‐1 isoforms after high‐load resistance exercise and moderate intensity endurance exercise. The RE and EE protocols used in the present study differed significantly from protocols used in other human studies with a similar aim. The RE protocol was designed to represent common hypertrophic RE for leg muscles. It consisted of significantly more sets (10 vs. 4) and had shorter recovery periods (2 vs. 5 min) between sets compared to study of Ydfors et al. (2013) who also used leg press loading in their study. Other studies (Lundberg et al. 2014; Norrbom et al. 2011) have used knee extension ergometers in the RE loading. The present EE protocol consisted of strenuous walking on treadmill. Even though being common recreational exercise mode, the effects of walking exercises on expression of PGC‐1 isoforms have not been studied earlier. All previous studies (Popov et al. 2014; Ydfors et al. 2013; Gidlund et al. 2015) investigating PGC‐1 splice variant responses have used a bicycle loading for EE. The tests of maximal strength (MVC), explosive strength (CMJ), and lactate levels indicated that RE was anaerobic exercise that predictably induced significant fatigue. As intended, the intensity of EE was close to the onset of blood lactate accumulation (4.0 mmol·L−1) and EE induced lesser muscular fatigue in leg extensors compared to RE.
The current results together with previous studies (Lundberg et al. 2014; Ydfors et al. 2013; Norrbom et al. 2011; Gidlund et al. 2015) showed clearly that the expression of alternative promoter driven PGC‐1α transcripts (exon 1b and 1b'‐derived) are strongly induced after different types of RE and EE, whereas transcripts originating from proximal promoter (PGC‐1α exon 1a‐derived) are much less inducible. In the present study, proximal promoter driven transcripts were induced only after EE and the magnitude of the response was slight compared to strong responses seen in alternative promoter‐driven transcripts after EE (Fig. 4A–C). Our study is the first that reports the RE‐induced responses of PGC‐1α exon 1b'‐derived transcripts in human skeletal muscle. Popov et al. (2014) have reported earlier the responses of these transcripts after EE, but the reverse primer used in the detection of these transcripts contained two mismatch nucleotides, which may have impaired the specificity and function of this primer pair. The same primer pair with mismatch nucleotides was used earlier also in the study of Ruas et al. (2012) for detection of PGC‐1α3. The rodent studies have shown that alternative promoter originated PGC‐1α isoforms, which were also induced in the present study, promote angiogenesis (Chinsomboon et al. 2009), mitochondrial biogenesis, and improve fatty acid oxidation capacity in skeletal muscles (Miura et al. 2008). Despite different exercise modes and protocols, both RE and EE increased the expression of truncated PGC‐1α transcripts (total NT‐PGC‐1α) similarly to earlier studies (Gidlund et al. 2015; Popov et al. 2014; Ydfors et al. 2013). Interestingly, it has been shown recently that hypoxia specifically induces truncated forms of PGC‐1α (NT‐PGC‐1α and PGC‐1α4), which induces VEGFexpression and angiogenesis, while having only a little effect on mitochondrial genes (Thom et al.2014). In this study, neither RE nor EE modulated the expression of PGC‐1β thus supporting the assumption that PGC‐1β is not transcriptionally regulated after exercise (Meirhaeghe et al. 2003).
According the conventional dogma, chronic EE (low‐intensity to high‐volume loading) favors skeletal muscle adaptations (e.g., mitochondrial biogenesis, angiogenesis and improved β‐oxidation) enhancing oxidative capacity with modest effect on muscle size (Holloszy and Booth 1976). Conversely, RE (high–intensity to low‐volume loading) increases protein accretion promoting muscle hypertrophy and force (Hawley et al. 2014). The present increased gene expression of mitochondrial marker cytochrome c by EE but not RE suggests that mitochondrial adaptation was induced preferentially by EE. In contrast, angiogenic response seemed to be stimulated by both EE and RE because both exercise types increased the mRNA expression of angiogenesis regulatorVEGF‐A. As expected, RE decreased the gene expression of myostatin, the known target of PGC‐1α4 (Ruas et al. 2012) and an inhibitor of muscle growth, but the EE did not have any effect. The downregulation of myostatin may be a part of hypertrophic response induced by RE (Hulmi et al.2007). Yet, the importance of the role of myostatin in hypertrophic response to RE in humans is still unclear and needs further experimental evidence.
To check whether the activation changes of known signaling molecules regulating PGC‐1αexpression would explain the expression changes of specific isoforms of PGC‐1, the levels of p‐p38 MAPKThr180/Tyr182 and p‐AMPKαThr172 were measured. The levels of p‐p38 MAPK were increased after RE and levels of p‐AMPKα after both RE and EE. However, the phosphorylation changes of these signaling molecules were not associated with the gene expression responses ofPGC‐1 isoforms. This result is not fully consistent with the study of Norrbom et al. (2011), which indicated that AMPK is a major regulator of PGC‐1α transcripts from the proximal promoter, but the expression of transcripts from an alternative promoter are regulated by β‐adrenergic signaling in combination with AMPK. Even if AMPK activation did not explain PGC‐1 isoform responses, we found that VEGF‐A and CYC responses 30 min after EE were associated with a corresponding change in the level of p‐AMPKα suggesting that AMPK activation may partly explain VEGF‐A andCYC responses seen after EE.
There were some limitations in our study. Because two separate exercise groups were used instead of a crossover design, the experimental setup did not allow direct comparison of gene expression responses between RE and EE. This study included walking endurance exercise, and it should be noted that another type of endurance exercise than continuous strenuous walking might induce different kind of cellular signaling responses in the front thigh muscles. It is also acknowledged that the time point: immediately after exercise could have been more optimal for detecting peak AMPK and p38 MAPK phosphorylation responses. However, the phosphorylation of these signaling proteins may not drop dramatically during the first 30 min after exercise (Bartlett et al. 2012; Sriwijitkamol et al. 2007). The present and previous studies (e.g., Bartlett et al. 2012; Lundberg et al. 2014; Popov et al. 2014; Gidlund et al. 2015) have shown that the studied transcripts seem to peak during the first 3 h after exercise. Thus, the selected time points can be considered justified and valid for the gene expression response detection of the studied genes. Our setup measured only acute exercise responses without physiological outcome variables of long‐term training effects, which could be connected to measured acute exercise responses.
In conclusion, this study comprehensively assayed PGC‐1 transcripts in human skeletal muscle and showed specific response profiles after the present RE and EE. The study established that both alternative promoter originated PGC‐1α exon 1b ‐ and 1b'‐derived transcripts are strongly induced after EE and RE, whereas the proximal promoter originated PGC‐1α exon 1a‐derived transcripts are less inducible and were upregulated only after EE. Truncated PGC‐1α transcripts were upregulated both after EE and RE, thus there was no clear exercise‐type specificity observed in the responses of these transcripts. The expression changes of marker genes suggested that EE induced responses typical for angiogenesis and mitochondrial biogenesis, while RE induced responses typical for angiogenesis and muscle hypertrophy. Our results improve the understanding of exercise‐type specific early signaling events and support the idea that gene expression responses of PGC‐1α isoforms may have an important role in exercise‐induced muscle adaptations. However, future studies are still required to verify the roles of different PGC‐1α isoforms in the mechanisms of muscle adaptation.
Conflict of Interests
None declared.
Acknowledgements
The authors thank Ms. Sheila Gagnon, M.Sc., Opri Jokelainen, M.Sc., Suvi Pulkkinen, M.Sc., Sira Karvinen, M.Sc., Mira Tuovinen, Mr. Risto Puurtinen, and Mrs. Aila Ollikainen for their work in data collection and analyses.
Funding Information
This work was financially supported by a grant from the Scientific Advisory Board for Defence, the David Sports Ltd., and the Finnish Cultural Foundation Central Fund (Kainulainen). Support from the National Doctoral Programme of Musculoskeletal Disorders and Biomaterials (TBDP) is also acknowledged.
- Manuscript Received: September 2, 2015.
- Manuscript Accepted: September 6, 2015.
Correspondence
Mika Silvennoinen, Department of Biology of Physical Activity, University of Jyväskylä, P. O. Box 35, FIN‐40014 University of Jyväskylä, Jyväskylä, Finland.
Tel: +358 40 733 8864
Fax: +358 14 260 2071
E‐mail: mika.m.silvennoinen@jyu.fi
Sources
1. Ahtiainen, J. P., S. Walker, M. Silvennoinen, H. Kyröläinen, B. C. Nindl, K. Häkkinen, et al. 2015. Exercise type and volume alter signaling pathways regulating skeletal muscle glucose uptake and protein synthesis. Eur. J. Appl. Physiol. 115 : 1835–1845.
2. Atherton, P. J., J. Babraj, K. Smith, J. Singh, M. J. Rennie, and H. Wackerhage. 2005. Selective activation of AMPK-PGC - 1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J. 19 : 786–788.
3. Baar, K., and K. Esser. 1999. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am. J. Physiol. 276:C120–C127.
4. Baar, K., A. R. Wende, T. E. Jones, M. Marison, L. A. Nolte, M. Chen, et al. 2002. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 16 : 1879–1886.
5. Bartlett, J.D., C. Hwa Joo, T. S. Jeong, J. Louhelainen, A. J. Cochran, M. J. Gibala, et al. 2012. Matched work high - intensity interval and continuous running induce similar increases in PGC-1alpha mRNA, AMPK, p38, and p53 phosphorylation in human skeletal muscle. J. Appl. Physiol. (1985) 112 : 1135–1143.
6. Canto, C., L. Q. Jiang, A. S. Deshmukh, C. Mataki, A. Coste, M. Lagouge, et al. 2010. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 11 : 213–219.
7. Chinsomboon, J., J. Ruas, R. K. Gupta, R. Thom, J. Shoag, G. C. Rowe, et al. 2009. The transcriptional coactivator PGC-1alpha mediates exercise-induced angiogenesis in skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 106 : 21401 – 21406.
8. Egan, B., B. P. Carson, P. M. Garcia-Roves, A. V. Chibalin, F. M. Sarsfield, N. Barron, et al. 2010. Exercise intensity - dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J. Physiol. 588 : 1779–1790.
9. Gibala, M. 2009. Molecular responses to high-intensity interval exercise. Appl. Physiol. Nutr. Metab. 34 : 428–432.
10. Gidlund, E. K., M. Ydfors, S. Appel, H. Rundqvist, C. J. Sundberg, and J. M. Norrbom. 2015. Rapidly elevated levels of PGC-1alpha-b protein in human skeletal muscle after exercise: exploring regulatory factors in a randomized controlled trial. J. Appl. Physiol. (1985) 119 : 374–384.
11. Hawley, J. A., M. Hargreaves, M. J. Joyner, and J. R. Zierath. 2014. Integrative biology of exercise. Cell 159 : 738–749.
12. Holloszy, J. O., and F. W. Booth. 1976. Biochemical adaptations to endurance exercise in muscle. Annu. Rev. Physiol. 38 : 273–291.
13. Hulmi, J. J., J. P. Ahtiainen, T. Kaasalainen, E. Pöllänen, K. Häkkinen, M. Alen, et al. 2007. Post exercise myostatin and activin IIb mRNA levels: effects of strength training. Med. Sci. Sports Exerc. 39 : 289–297.
14. Hulmi, J. J., S. Walker, J. P. Ahtiainen, K. Nyman, W. J. Kraemer, and K. Häkkinen. 2012. Molecular signaling in muscle is affected by the specificity of resistance exercise protocol. Scand. J. Med. Sci. Sports 22 : 240–248.
15. Kim, J. S., J. K. Petrella, J. M. Cross, and M. M. Bamman. 2007. Load-mediated downregulation of myostatin mRNA is not sufficient to promote myofiber hypertrophy in humans: a cluster analysis. J. Appl. Physiol. (1985) 103 : 1488–1495.
16. Koressaar, T., and M. Remm. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23 : 1289–1291.
17. Lundberg, T. R., R. Fernandez-Gonzalo, J. Norrbom, H. Fischer, P. A. Tesch, and T. Gustafsson. 2014. Truncated splice variant PGC-1alpha4 is not associated with exercise - induced human muscle hypertrophy. Acta Physiol. (Oxf) 212 : 142–151.
18. Lundby, C., N. Nordsborg, K. Kusuhara, K. M. Kristensen, P. D. Neufer, and H. Pilegaard. 2005. Gene expression in human skeletal muscle: alternative normalization method and effect of repeated biopsies. Eur. J. Appl. Physiol. 95 : 351–360.
19. Meirhaeghe, A., V. Crowley, C. Lenaghan, C. Lelliott, K. Green, A. Stewart, et al. 2003. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator - activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochem. J. 373 : 155–165.
20. Miura, S., Y. Kai, Y. Kamei, and O. Ezaki. 2008. Isoform-specific increases in murine skeletal muscle peroxisome proliferator - activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to beta2-adrenergic receptor activation and exercise. Endocrinology 149 : 4527–4533.
21. Norrbom, J., E. K. Sallstedt, H. Fischer, C. J. Sundberg, H. Rundqvist, and T. Gustafsson. 2011. Alternative splice variant PGC-1alpha-b is strongly induced by exercise in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 301:E1092–E1098.
22. Olesen, J., K. Kiilerich, and H. Pilegaard. 2010. PGC-1alpha - mediated adaptations in skeletal muscle. Pflugers Arch. 460 : 153–162.
23. Pilegaard, H., B. Saltin, and P. D. Neufer. 2003. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J. Physiol. 546 : 851–858.
24. Popov, D. V., A. V. Bachinin, E. A. Lysenko, T. F. Miller, and O. L. Vinogradova. 2014. Exercise-induced expression of peroxisome proliferator-activated receptor gamma coactivator-1alpha isoforms in skeletal muscle of endurance - trained males. J. Physiol. Sci. 64 : 317–323.
25. Ranhotra, H. S. 2010. Long-term caloric restriction up - regulates PPAR gamma co-activator 1 alpha (PGC-1alpha) expression in mice. Indian J. Biochem. Biophys. 47 : 272–277.
26. Ruas, J. L., J. P. White, R. R. Rao, S. Kleiner, K. T. Brannan, B. C. Harrison, et al. , et al. 2012. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell 151 : 1319–1331.
27. Sriwijitkamol, A., D. K. Coletta, E. Wajcberg, G. B. Balbontin, S. M. Reyna, J. Barrientes, et al. 2007. Effect of acute exercise on AMPK signaling in skeletal muscle of subjects with type 2 diabetes: a time-course and dose-response study. Diabetes 56 : 836–848.
28. Thom, R., G. C. Rowe, C. Jang, A. Safdar, and Z. Arany. 2014. Hypoxic induction of vascular endothelial growth factor (VEGF) and angiogenesis in muscle by truncated peroxisome proliferator-activated receptor gamma coactivator (PGC)-1alpha. J. Biol. Chem. 289 : 8810–8817.
29. Untergasser, A., I. Cutcutache, T. Koressaar, J. Ye, B. C. Faircloth, M. Remm, et al. 2012. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115.
30. Wen, X., J. Wu, J. S. Chang, P. Zhang, J. Wang, Y. Zhang, et al. 2014. Effect of exercise intensity on isoform-specific expressions of NT-PGC-1 alpha mRNA in mouse skeletal muscle. Biomed. Res. Int. 2014 : 402175.
31. Ydfors, M., H. Fischer, H. Mascher, E. Blomstrand, J. Norrbom, and T. Gustafsson. 2013. The truncated splice variants, NT-PGC-1alpha and PGC-1alpha4, increase with both endurance and resistance exercise in human skeletal muscle. Physiol. Rep. 1:e00140.
32. Zhang, Y., P. Huypens, A. W. Adamson, J. S. Chang, T. M. Henagan, A. Boudreau, et al. 2009. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1alpha. J. Biol. Chem. 284 : 32813–32826.
Article was published inPhysiological Reports

2015 Issue 10-
All articles in this issue
- S100B and NSE serum concentrations after simulated diving in rats
- The non-antibiotic macrolide EM900 inhibits rhinovirus infection and cytokine production in human airway epithelial cells
- Noninvasive quantification of alveolar morphometry in elderly never- and ex-smokers
- Subtle modulation of ongoing calcium dynamics in astrocytic microdomains by sensory inputs
- Caffeine and contraction synergistically stimulate 5′‐AMP‐activated protein kinase and insulin‐independent glucose transport in rat skeletal muscle
- PGC‐1 isoforms and their target genes are expressed differently in human skeletal muscle following resistance and endurance exercise
- Physiological Reports
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- S100B and NSE serum concentrations after simulated diving in rats
- Subtle modulation of ongoing calcium dynamics in astrocytic microdomains by sensory inputs
- The non-antibiotic macrolide EM900 inhibits rhinovirus infection and cytokine production in human airway epithelial cells
- Noninvasive quantification of alveolar morphometry in elderly never- and ex-smokers
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career


