-
Medical journals
- Career
Ingavirin might be a promising agent to combat Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2)
Authors: Ivan Malík 1,2; Gustav Kovac 2; Tereza Padrtova 3; Lucia Hudecova 2
Authors‘ workplace: Department of Pharmaceutical Chemistry Faculty of Pharmacy, Comenius University in Bratislava 1; Institute of Chemistry, Clinical Biochemistry and Laboratory Medicine, Faculty of Medicine 2; Department of Chemical Drugs Faculty of Pharmacy, Masaryk University 3
Published in: Čes. slov. Farm., 2020; 69, 107-111
Category: Review Articles
Overview
The Severe Acute Respiratory Coronavirus 2 (SARS--CoV-2) and Coronavirus Disease-19 (COVID-19) pandemic, caused by the virus, have changed the world in just half a year. Lack of effective treatment, coupled with etiology of COVID-19, has resulted in more than 500,000 confirmed deaths at the time of writing, and the global economy is at an unseen unprecedented low level with unknown near - and long-term consequences. Ingavirin has been considered a non-toxic broad-spectrum antiviral with a complex mechanism of action. The molecule was originally designed for the prophylaxis and treatment of flu caused by both Influenza A and B viruses and for the treatment of viral causes of acute respiratory illness. The article hypothesized that the efficiency of given 1H-imidazol-4-yl heterocyclic scaffold-containing compound against SARS-CoV-2 might be connected with its ability to interfere with specific heterogeneous nuclear ribonucleoproteins (A1, for example). These specific cellular RNA-binding proteins showed affinity to Severe Acute Respiratory Coronavirus (SARS-CoV) nucleocapsid (N) protein, which shared high homology with the N protein of SARS-CoV-2 and the fact was expressed by a sequence identity of 90.52%. Impairing of the interactions between nuclear ribonucleoproteins and nucleocapsid (N) protein of SARS-CoV-2 might result in the inhibition of a viral replication cycle. Additional immunomodulating properties of ingavirin could be favorable for induction of adaptive immunity of host cells.
Keywords:
SARS-CoV-2 – COVID-19 – ingavirin – heterogeneous nuclear ribonucleoproteins – nucleocapsid (N) protein
Introduction
Coronavirus Disease-2019 (COVID-19) is an infectious illness caused by a novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), formerly known as novel Coronavirus (2019-nCoV) or Wuhan Coronavirus. The virus first originated in Wuhan, the capital of Hubei Province (China), spreading globally and affecting more than 200 countries till now. The disease has rapidly become a global health pandemic1).
Coronaviruses (CoVs) are a large family of pathogenic enveloped viruses with a positive-sense single-stranded RNA genome. CoVs belong to the Coronaviridae family of the Nidovirales order. The viruses have been classified into four genera that include α-, β-, γ-, and δ-CoVs. Among them, α - and β-CoVs infect mammals, γ-CoVs infect avian species, and δ-CoVs are able to infect both mammals and aves2).
The whole genome of a zoonotic SARS-CoV-2 (β-CoV, subfamily Coronavirinae, family Coronaviridae, order Nidovirales) is composed of approximately 30,000 nucleotides, which encodes many structural proteins (SPs) as well as non-structural proteins (NSPs).
The SPs include transmembrane spike (S), envelope (E), membrane (M) and highly immunogenic nucleocapsid (N) proteins, which are needed to produce a structurally complete viral particle. Their specific structural organizations and functions were described comprehensively in previous research3, 4).
Design and development of SARS-CoV-2 specific direct-acting antiviral drugs can be made possible by focusing on not to the SPs only but conserved enzymes (NSPs), such as main protease or 3C-like protease (Mpro or 3CLpro), papain-like protease (PLpro), helicase (non-structural protein 13; nsp13), non-structural protein 12 (nsp12) or RNA-dependent RNA polymerase (RdRp), are very promising viral targets as well5, 6).
The SARS-CoV-2 genome shared about 82% sequence identity with Severe Acute Respiratory Coronavirus (SARS-CoV) reported earlier and more than 92% sequence identity for SPs and essential enzymes3, 7).
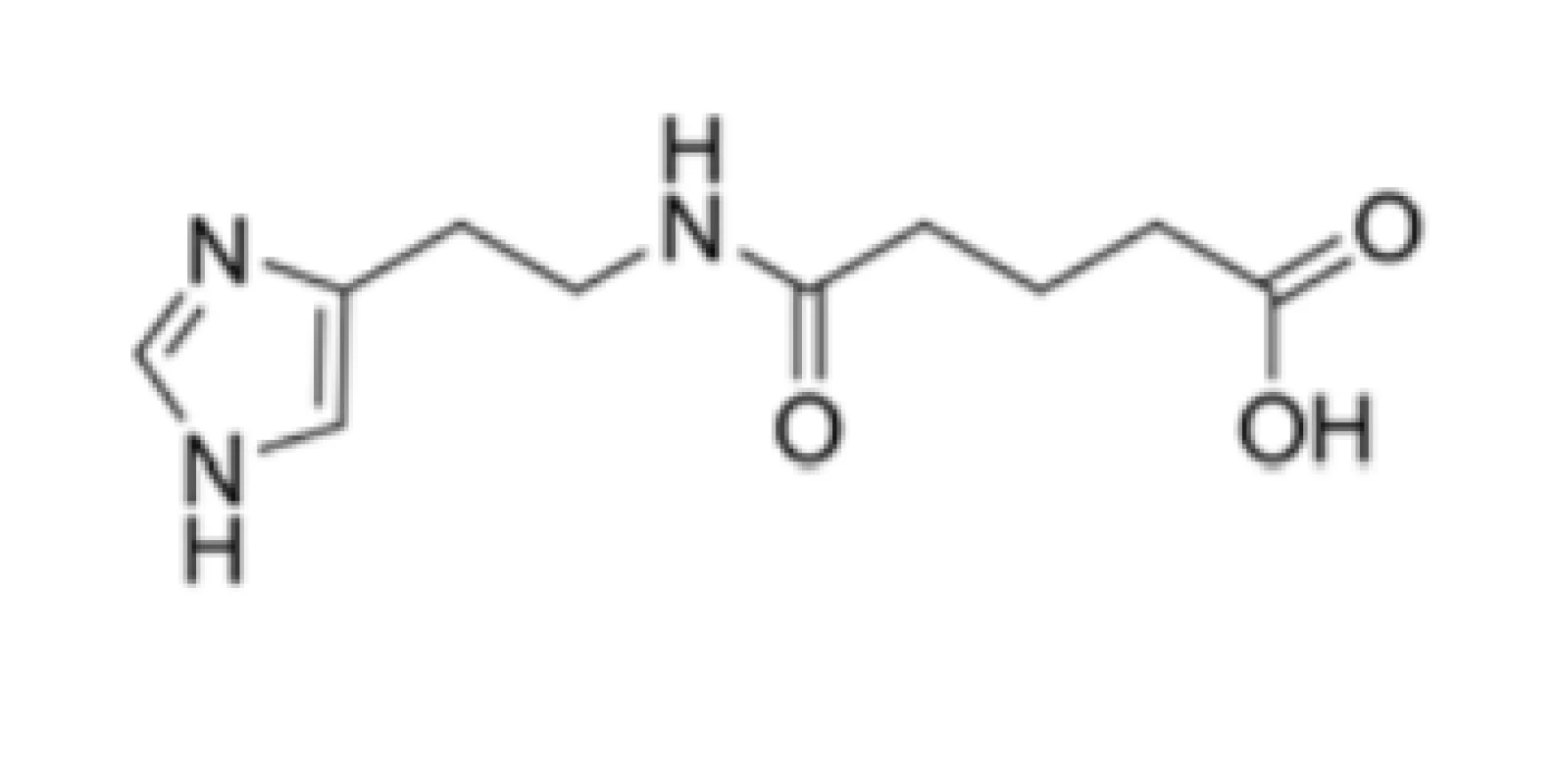
Many drugs (or their combinations) from various pharmacotherapeutic groups that could potentially target specific viral proteins (targets) or critical host cell processes have been evaluated in vitro against SARS-CoV-2 or as treatment interventions for COVID-19 in clinical trials8), however, minimal attention8–10) has been paid to ingavirin (Fig. 1) as a promising therapeutic modality.
Ingavirin acts as a powerful weapon against Influenza viruses and other viral causes of acute respiratory illness
Ingavirin (Fig. 1), a well-tolerated and safe 6-[2-(1H-imidazol-4-yl)ethylamino]-5-oxohexanoic acid (CAS Registry Number: 219694-63-0), was developed in the Russian Federation. The drug was recently added to a list of Influenza-limiting antivirals because of its direct interference with transportation of a newly synthesized viral nucleocapsid protein (nucleoprotein or N protein)11).
The molecule, also known as ingaviruin, was approved recently in the Russian Federation for the prophylaxis and treatment of flu caused by Influenza A (species Influenza A virus; genus Influenzavirus A; family Orthomyxoviridae) and B (Influenza B virus; Influenzavirus B; Orthomyxoviridae) virus as well as other acute respiratory viral infections (ARVI)12, 13). This compound was also efficient against both pandemic A/California/04/2009 and A/California/07/2009 strains and other strains of Influenza viruses, i.e., H3N2 or H5N114).
Ingavirin interacted with the N protein of Influenza A and B Virus, thus preventing oligomerization of the protein, a process essentially required for viral replication15). The compound was found to impair the biogenesis of concerned protein, lower efficiency of formation of mature conformationally compact N protein oligomers and retard the migration of newly synthesized N proteins from cytoplasm to a nucleus16–18).
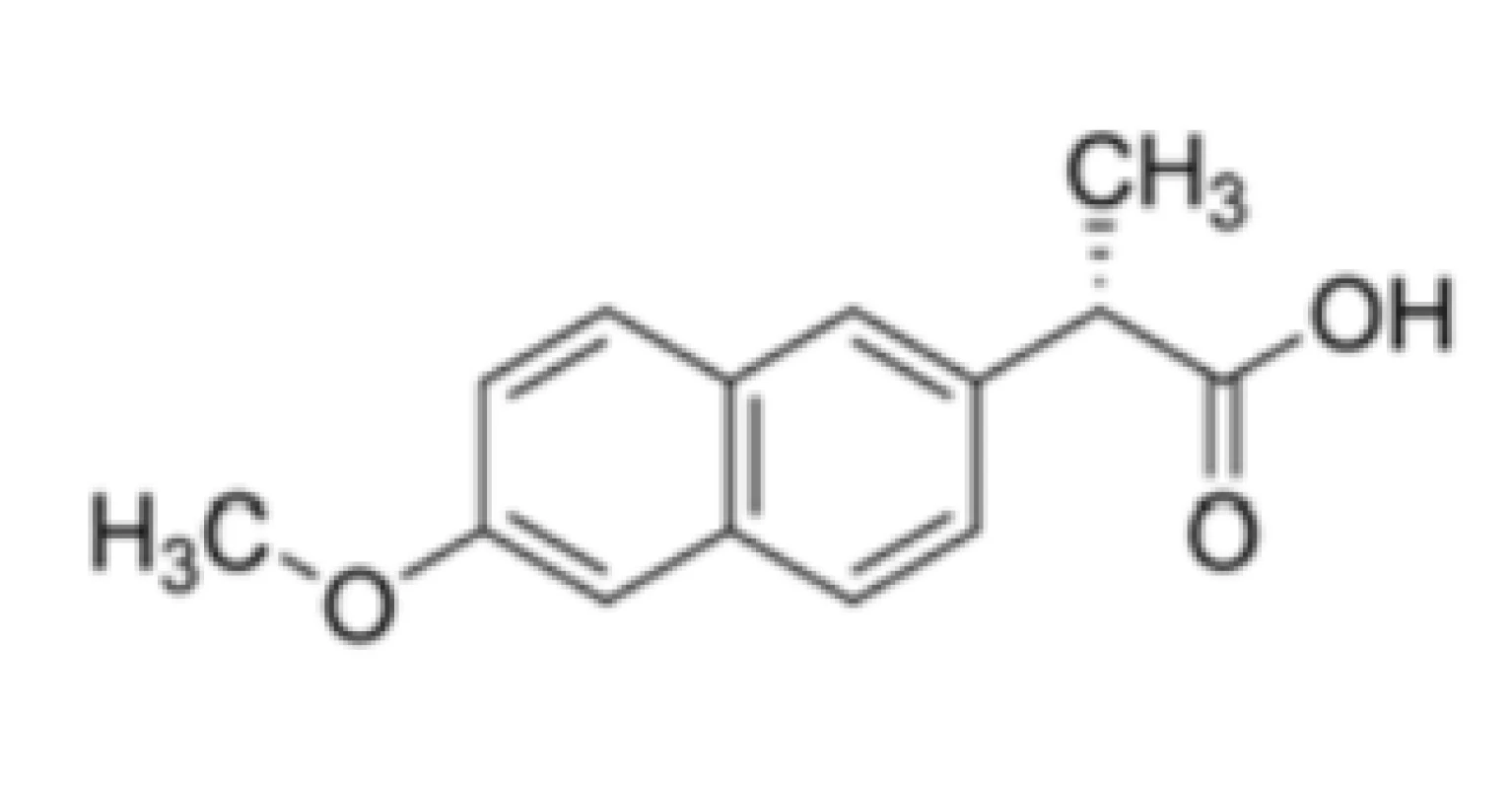
Besides, non-steroidal anti-inflammatory drug naproxen (Fig. 2) competed with RNA for binding to the N protein binding groove19) stabilizing protein monomers by altering the groove, in which an oligomerization loop interacted with RNA.
Ingavirin has been very efficient in vivo and in vitro against other RNA viruses, including Human Metapneumovirus (Metapneumovirus; Pneumovirinae; Paramyxoviridae)20), Human Respiratory Syncytial Virus (Paramyxovirus; Paramyxoviridae), Human Parainfluenza viruses (Paramyxovirus; Paramyxoviridae), the viruses, which can be found in Picornaviridae or Coronaviridae family as well as some DNA viruses (Adenoviridae)21).
Might be ingavirin effective against Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2)?
The N protein, an important antigen for SARS-CoV-2, is located within the virions in a complex with genomic RNA participating in RNA package and virus particle release. The nucleocapsid protein plays an essential role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication. It was observed that this highly charged basic protein was full of coils, highly disordered, formed a dimer by the interaction of its both C-terminal domains and could interact with non-specific nucleic acids with high affinity, i.e., the dimeric form might act as a fundamental functional unit in vivo. Very notable finding was that the SARS-CoV-2 N protein shared high homology with SARS-CoV N protein, which was defined by sequence identity of 90.52%22, 23).
The N protein of SARS-CoV has been regarded as one of the most crucial structural components of the virus being responsible for recognition of RNA stretch that served as a packaging signal leading to the formation of ribonucleoprotein. On the other hand, this protein shared 20–30% sequence identity (quite low homology) with N proteins of other CoVs24, 25).
Luo et al. discovered that the N protein of SARS-CoV showed high binding affinity to human heterogeneous nuclear ribonucleoproteins (hnRNPs), especially to the A1 type (hnRNP A1), which was related to pre-mRNA splicing in a nucleus and translation regulation in cytoplasm26).
Ingavirin interacted efficiently with the N protein of particular Influenza viruses and administration of concerned 1H-imidazol-4-yl cycle-containing small molecule in patients with asthma exacerbation associated with ARVI, including the one caused by SARS-CoV, increased effectiveness of the treatment. After being treated, fever periods were reduced, systemic manifestations were suppressed and, in addition, the number of bacterial complications was decreased by 15%11, 27).
Tsai et al. concluded that specific cellular RNA-binding proteins, including NS1-binding protein (NS1-BP) and specific heterogeneous nuclear ribonucleoproteins (hnRNPs), regulated Influenza A Virus RNA splicing28). The hnRNPs assisted in controlling the maturation of newly formed heterogeneous nuclear RNAs (hnRNAs/pre-mRNAs) into messenger RNAs (mRNAs), stabilized mRNA during their cellular transport and regulated their translation. Considering their functional diversity and complexity, hnRNPs acted as the key proteins in cellular nucleic acid modifications29).
Following the mentioned, the efficiency of ingavirin against SARS-CoV-2 might relate to its ability to interfere with hnRNP A1 and thus affecting the interactions between specific hnRNP A1 and viral N protein.
Additional beneficial immunomodulating properties of the drug were associated with the activation of a group of Toll-like and RIG-I-like receptor signalling pathways of innate and adaptive immunity and differentiation of hematopoietic cell precursors30). The receptors play a dominant role in first-line defence and in the induction of subsequent adaptive immunity31, 32).
Ingavirin, although not being interferon (INF) inducer itself, enhanced synthesis of both INF-α and INF-β receptors to INF and cell sensitivity to INF signaling33).
Conclusion
Ingavirin might be considered a promising anti-SARS-CoV-2 drug due to probable interactions with specific heterogeneous nuclear ribonucleoproteins of the virus. The interferences would result in the inhibition of viral replication. Moreover, the compound provided additional beneficial immunomodulating properties. On the other hand, the indisputable fact is that ingavirin was not originally designed or structurally optimized as the anti-SARS-CoV-2 agent for the treatment of COVID-19 but was repurposed. Continuous intensive systematic in silico investigation employing relevant computational techniques, in vitro and in vivo evaluations of specifically designed compounds, which affect selectively very key proteins (targets) of SARS-CoV-2 responsible for its attachment and replication within host cells, might open the window for efficient, highly specific and safe therapy of COVID-19.
Acknowledgements
Regarding the emerging situation with a COVID-19 pandemic, increase in number of people getting infected by SARS-CoV-2 daily, and absence of successful treatment, we decided to share our ideas with scientific community via this review. We would like to thank the Czech and Slovak Pharmacy Journal (Česká a slovenská farmacie) for the opportunity to publish the article.
Conflicts of interest: none.
Assoc. Prof. PharmDr. Ivan Malik, PhD.1,2 (∗) • G. Kovac2 • L. Hudecova2
1Department of Pharmaceutical Chemistry
Faculty of Pharmacy, Comenius University in Bratislava
Odbojarov 10, 832 32 Bratislava, Slovak Republic
2Institute of Chemistry, Clinical Biochemistry and Laboratory
Medicine, Faculty of Medicine
Slovak Medical University in Bratislava
Limbova 12, 833 03 Bratislava, Slovak Republic
e-mail: malik2@uniba.sk
T. Padrtova
Department of Chemical Drugs
Faculty of Pharmacy, Masaryk University
Palackého str. 1946/1, 612 42 Brno, Czech Republic
Sources
1. Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395, 497–506. doi:10.1016/S0140-6736(20)30183-5
2. Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020; 27, 325–328, doi:10.1016/j.chom.2020.02.001
3. Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., Chen W., Shi W., Tan, W. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020; 395, 565–574. doi:10.1016/S0140-6736(20)30251-8
4. Zumla A., Chan J. F., Azhar E. I., Hui D. S., Yuen K. Y. Coronaviruses – drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016; 15, 327–347. doi:10.1038/nrd.2015.37
5. McKee D. L., Sternberg A., Stange U., Laufer S., Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res. 2020; 157, art no. 104859 (9 pp.). doi:10.1016/j.phrs.2020.104859
6. Tiwari V., Beer J. C., Sankaranarayanan N. V., Swanson-Mungerson M., Desai, U. R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today 2020 [ahead of print]. doi:10.1016/j.drudis.2020.06.017
7. Gorbalenya A. E., Snijder E. J., Spaan W. J. M. Severe Acute Respiratory Syndrome Coronavirus phylogeny: Toward consensus. J. Virol. 2004; 78, 7863–7866. doi:10.1128/JVI.78.15.7863-7866.2004.
8. Subbarao K., Mahanty S. Respiratory virus infections: Understanding COVID-19. Immunity 2020; 52, 905–909. doi:10.1016/j.immuni.2020.05.004
9. https://clinicaltrials.gov/ (15. 6. 2020)
10. Fragkou P. C., Belhadi D., Peiffer-Smadja P., Moschopoulos C. D., Lescure F.-X., Janocha H., Karofylakis E., Yazdanpanah Y., Mentré F., Skevaki C., Laouénan C., Tsiodras S., on behalf of the ESCMID Study Group for Respiratory Viruses. Review of trials currently testing treatment and prevention of COVID-19. Clin. Microbiol. Infect. 2020 [article in press] (11 pp.). doi:10.1016/j.cmi.2020.05.019
11. Dzyublik A. Ya., Simonov S. S., Yachnik V. A. Clinical efficacy and safety of antiviral drug Ingavirin in patients with asthma exacerbations caused by an acute respiratory viral infection (ARVI). Pulmonologiya 2013; 43–50. doi:10.18093/0869-0189-2013-0-6-765-775
12. Chupakhin O. N., Charushin V. N., Rusinov V. L. Scientific foundations for the creation of antiviral and antibacterial preparations. Her. Russ. Acad. Sci. 2016; 86, 206–212. doi:10.1134/S1019331616030163
13. Loginova S. Y., Borisevich S. V., Maksimov V. A., Bondarev V. P., Nebolsin V. E. Therapeutic efficacy of Ingavirin, a new domestic formulation agaisnt Influenza A virus (H3N2). Antibiot. Khimioter. 2008; 53, 27–30.
14. Loginova S. Y., Borisevich S. V., Lykov M. V., Vedenina E. V., Borisevich G. V., Bondarev V. P., Nebolsin V. E., Chuchalin A. G. In vitro efficacy of Ingavirin against the Mexican pandemic subtype H1N1 of Influenza A virus, strains A/California/04/2009 and A/California/07/2009. Antibiot. Khimioter. 2009; 54, 15–17.
15. Zarubaev V. V., Garshinina A. V., Kalinina N. A., Shtro A. A., Belyaevskaya S. V., Slita A. V., Nebolsin V. E., Kiselev O. I. Activity of Ingavirin (6-[2-(1H-imidazol-4-yl)ethylamino]-5--oxohexanoic acid) against human respiratory viruses in in vivo experiments. Pharmaceuticals 2011; 4, 1518–1534. doi:10.3390/ph4121518
16. Loginova S. Ya., Borisevich S. V., Shkliaeva O. M., Maksimov V. A., Bondarev V. P., Nebolsin V. E. Prophylactic and therapeutic efficacies of Ingavirin, a novel Russian chemotherapeutic, with respect to Influenza pathogen A (H5N1). Antibiot. Khimioter. 2010; 55, 10–12.
17. Semenova N. P., Prokudina E. N., Livov D. K., Nebolsin V. E. Effect of the antiviral drug Ingaviruin on intracellular transformations and import into the nucleus of Influenza A virus nucleocapsid protein. Vopr. Virusol. 2010; 55, 17–20.
18. Zarubaev V. V., Nebolsin V., Garshinina A., Kalinina N., Shtro A., Kiselev O. Antiviral activity of Ingavirin (imidazolyl ethanamide pentandioic acid) against lethal influenza infection caused by pandemic strain A/California/07/09 (H1N1)v in white mice. Antiviral Res. 2010; 86, 50. doi:10.1016/j.antiviral.2010.02421
19. Monod A., Swale C., Tarus B., Tissot A., Delmas B., Ruigrok R. W., Crépin T., Slama-Schwok, A. Learning from structure-based drug design and new antivirals targeting the ribonucleoprotein complex for the treatment of influenza. Expert Opin. Drug Discov. 2015; 10, 345–371. doi:10.1517/17460441.2015.1019859
20. Isayeva E. I., Nebolsin V. E., Kozulina I. S., Morozova O. V. In vitro investigation of the antiviral activity of Ingavirin against Metapneumovirus. Vopr. Virusol. 2012; 57, 34–38.
21. Shuldyakov A. A., Lyapina E. P., Kuznetsov V. I., Erofeeva M. K., Pozdnyakova M. G., Maksakova V. L., Kotova O. S., Shelekhova S. E., Buzitskaya Zh. V., Amosova I. V., Gil A. Yu. Clinical and epidemiological efficacy of antiviral drug Ingavirin. Pulmonologiya 2012; 62–69. doi:10.18093/0869-0189-2012-0-4-62-69
22. Surjit M., Lal S. K. The SARS-CoV nucleocapsid protein: A protein with multifarious activities. Infect. Genet. Evol. 2008; 8, 397–405. doi:10.1016/j.meegid.2007.07.004
23. Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao Ch., Yang Y., Xie J., Ding Ch., Ma X., Weng J., Gao Y., He H., Jin T. Biochemical characterization of SARS--CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020; 527, 618–623. doi:10.1016/j.bbrc.2020.04.136
24. Marra M. A., Jones S. J. M., Astell C. R., Holt R. A., Brooks-Wilson A., Butterfield Y. S. N., Khattra J., Asano J. K., Barber S. A., Chan S. Y., Cloutier A., Coughlin S. M., Freeman D., Girn N., Griffith O. L., Leach S. R., Mayo M., McDonald H., Montgomery S. B., Pandoh P. K., Petrescu A. S., Robertson A. G., Schein J. E., Siddiqui A., Smailus D. E., Stott J. M., Yang G. S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T. F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones, S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G. A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R. C., Krajden M., Petric M., Skowronski D. M., Upton C., Roper R. L. The genome sequence of the SARS-associated coronavirus. Science 2003; 300, 1399–1404. doi:10.1126/science.1085953
25. Rota P. A., Oberste S. M., Monroe S. S., Nix A. W., Campagnoli R., Icenogle J. P., Peñaranda S., Bankamp B., Maher K., Chen M.-H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J. L., Chen Q., Wang D., Erdman D. D., Peret T. C. T., Burns C., Ksiazek T. K., Rollin P. E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Günther S., Osterhaus A. D. M. E., Drosten Ch., Pallansch M. A., Anderson L. J., Bellin W. J. Characterization of a novel coronavirus associated with Severe Acute Respiratory Syndrome. Science 2003; 300, 1394–1399. doi:10.1126/science.1085952
26. Luo H. B., Chen Q., Chen J., Chen K. X., Shen X., Jiang H. L. The nucleocapsid protein of SARS coronavirus (SARS_N) has a high binding affinity to the human cellular heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). FEBS Lett. 2005; 579, 2623–2628. doi:10.1016/j.febslet.2005.03.080
27. Solovyeva O. G. The experience of using the antiviral drug Ingavirin in the treatment of complicated forms of influenza and SARS. Pulmonologiya 2012; 62–66.
28. Tsai P.-L., Chiou N.-T., Kuss S., García-Sastre A., Lynch K. W., Fontoura B. M. A. Cellular RNA binding proteins NS1-BP and hnRNP K regulate Influenza A Virus RNA splicing. PLoS Pathog. 2013; 9, art. no. e1003460 (13 pp.). doi:10.1371/journal.ppat.1003460
29. Dreyfuss G., Matunis M. J., Piñol-Roma S., Burd C. G. hnRNP Proteins and the biogenesis of mRNA. Annu Rev. Biochem. 1993; 62, 289–321. doi:10.1146/annurev.bi.62.070193.001445
30. Sokolova T. M., Poloskov V. V., Shuvalov A. N., Burova O. S., Sokolova Z. A. Signaling TLR/RLR-mechanisms of immunomodulating action of Ingavirin and Thymogen preparations. Russian J. Biother. 2019; 18, 60–66. doi:10.17650/1726-9784-2019-18-1-60-66
31. Arpaia N., Barton G. M. Toll-like receptors: key players in antiviral immunity. Curr. Opin. Virol. 2011; 1, 447–454. doi:10.1016/j.coviro.2011.10.006
32. Loo Y.-M., Gale Jr. M. Immune signaling by RIG-I-like receptors. Immunity 2011; 34, 680–692. doi:10.1016/j.immuni.2011.05.003
33. Aschacher T., Krokhin A., Kuznetsova I., Langle J., Nebolsin V., Egorov A., Bergmann M. Effect of the preparation Ingavirin (imidazolyl ethanamide pentandioic acid) on the interferon status of cells under conditions of viral infection. Epidemiol. Infect. Dis. 2016; 21, 196–205.
Labels
Pharmacy Clinical pharmacology
Article was published inCzech and Slovak Pharmacy

2020 Issue 3-
All articles in this issue
- Medicinal products with controlled drug release for local therapy of inflammatory bowel diseases from perspective of pharmaceutical technology
- Metal complexes in medicine and pharmacy – the past and the present III
- Anti-inflammatory potential of composites of yeast glucan particles and geranylated flavonoid diplacone
- Ingavirin might be a promising agent to combat Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2)
- The study of hypoglycemic and hypolipidemic activity of Camelina sativa (L.) Crantz extracts in rats under conditions of high-fructose diet
- A pharmacodynamic study of a new gel containing an extract of Aloe vera and an extract of oak bark for potential treatment of periodontal diseases
- Czech and Slovak Pharmacy
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Medicinal products with controlled drug release for local therapy of inflammatory bowel diseases from perspective of pharmaceutical technology
- Ingavirin might be a promising agent to combat Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2)
- Metal complexes in medicine and pharmacy – the past and the present III
- A pharmacodynamic study of a new gel containing an extract of Aloe vera and an extract of oak bark for potential treatment of periodontal diseases
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career