-
Medical journals
- Career
Regulatory network of competitively interacting RNAs and effectiveness of rectal tumors radiotherapy
Authors: D. S. Kutilin; M. A. Gusareva; N. G. Kosheleva; O. I. Kit
Authors‘ workplace: National Medical Research Centre for Oncology, Russian Federation, Rostov-on-Don
Published in: Klin Onkol 2022; 35(4): 297-306
Category: Original Articles
doi: https://doi.org/10.48095/ccko2022297Overview
Background: Currently, rectal tumors radiotherapy effectiveness reaches an acceptable level only in a small number of patients (they have a complete clinical response), which is associated with the formation of malignant cells radioresistance. A comprehensive study that integrates various epigenetic parameters would explain a number of molecular mechanisms of rectal tumor cells radioresistance and identify new biomarkers. In the last decade, using high-throughput sequencing, the competitively interacting RNAs regulatory network (long non-coding RNAs, miRNAs and mRNAs) has been shown. Purpose: The aim of the study was to analyze the features of competitively interacting RNAs regulatory network functioning in patients with rectal cancer who are radioresistant and sensitive to radiotherapy. The study was performed on 500 patients with diagnosed rectal cancer. Radiotherapy was performed on a Novalis TX linear particle accelerator according to the standard protocol (single focal dose 2.4 Gy, total focal dose 54.0 Gy). Total RNA preparations were isolated from paired biopsy fragments of tumor and non-tumor tissues of the rectum (obtained by video-colonoscopy). The relative abundance of mRNA, miRNA and lncRNA transcripts was assessed by the RT-qPCR method. Using bioinformatic analysis, the probability of potential interactions between the investigated mRNA, miRNA and lncRNA was determined. It has been shown that the effectiveness of radiotherapy depends on the level of miRNA (miRNA-195-5p; miRNA-4257; miRNA-5187-5p; miRNA-149-5p; miRNA-138 -1-3p; miRNA-6798-5p; miRNA-6819-5p; miRNA-4728-5p; miRNA-1249-5p; miRNA-557; miRNA-1273h-5p; miRNA-6737-5p; miRNA-6808-5p; miRNA-3202; miRNA-5195-3p; miRNA-130b-3p) and lncRNA (XIST, HELLPAR, NEAT1, AC008124. 1, LINC01089, LINC01547, and VASH1-AS1) expression, which regulate the DNA repair system (H2AX, RBBP8) and apoptosis (BCL2). Conclusion: A comprehensive study of competitively interacting RNAs regulatory network and radiotherapy effectiveness of rectal tumors made it possible to establish the mechanisms of radioresistance formation and its biomarkers.
Keywords:
rectal cancer – miRNA – Gene expression – DNA repair – Long non-coding RNAs – apoptosis – radiotherapy effectiveness – competitively interacting RNA network
Introduction
In 2020, more than 339 000 deaths from rectal cancer (RC) were registered, which makes this nosology the leading one in terms of mortality among many oncological diseases [1]. Chemoradiotherapy is currently used to relieve symptoms and local control of RC. However, it is important to note that not every patient responds positively to radiotherapy (RT), which is associated with radioresistance/radiosensitivity of malignant cells. Despite the long-term improvement RT algorithms for RC, it is still difficult to predict the volume of clinical response [2]. Over the past 30 years, an impressive amount of research has been carried out to study the mechanisms of radioresistance and a significant list of its molecular markers has been proposed, none of which, however, has entered clinical practice. At present, more than ever before, it is relevant to conduct a comprehensive study that integrates various molecular parameters and allows both to explain a number of molecular mechanisms of radioresistance and to propose a list of new markers.
In the last decade, the complexity of the human genome has been identified using high-throughput RNA sequencing. The existence of a regulatory network of competing endogenous RNA (ceRNA), consisting of long non-coding RNA (lncRNA), microRNA (miRNA), and mRNA, has been shown [3].
MicroRNAs are short, non-coding RNAs that regulate gene expression by catalyzing the destruction of mRNA or by inhibiting the translation of mRNA into protein. MicroRNAs make a significant contribution to the initiation and development of various molecular events, including the initiation of oncogenesis, progression and metastasis of tumors, which makes microRNAs potential biomarkers for assessing the progression and prognosis of intestinal malignant tumors. Although miRNAs regulate the expression of genes encoding proteins, mainly through degradation or silencing of mRNA, there is growing evidence that miRNAs can interact with lncRNA, which, in turn, also regulates the expression of target genes [4]. Long noncoding RNAs (lncRNAs) are RNAs over 200 nucleotides in size that regulate the biological activity of cells in a variety of ways, including transcriptional regulation, posttranscriptional regulation, and translation regulation. lncRNAs control posttranscriptional regulation, being a component of the ceRNAs network and acting as a molecular “sponge” (sorbent) of microRNAs [5,6].
The involvement of the ceRNA regulatory network in the initiation and progression of intestinal tumors has been confirmed in a number of studies [7]. However, the role of the imbalance of the ceRNA regulatory network (lncRNA – miRNA – mRNA) in patients with rectal cancer and the formation of radioresistance remains to be elucidated [8].
The study of the regulatory network of microRNAs-long noncoding RNAs-mRNAs is of great importance both for elucidating the molecular mechanisms underlying carcinogenesis and for creating a panel of new biomarkers [4].
Therefore, the aim of the study was to analyze the features of competitively interacting RNAs (lncRNA, miRNA, mRNA) regulatory network functioning in radioresistant and radiation-sensitive patients with rectal cancer.
Materials and methods
Clinical characteristics of patients
The study included 500 patients (200 women, 300 men) with diagnosed RC (age 28–85 years, median age 62.2 ± 4) who underwent inpatient treatment in 2019–2021 at the National Medical Research Centre for Oncology. Each patient signed a voluntary informed consent to participate in the study. Permission for the study was obtained from the ethical committee of the National Medical Research Center for Oncology (protocol No. 14). The sample of histologically confirmed tumors consisted of adenocarcinomas, of which 21% of tumors were classified as local (T3N0M0) and 79% of cases were those with regional lymph nodes metastases (T3N1-2M0). Remote conformal RT in combination with fluoropyrimidine chemotherapy was performed on a Novalis TX linear particle accelerator five times a week (single focal dose of 2.0 Gy to a total focal dose of 54.0 Gy) [9]. To isolate RNA, we used paired fragments of biopsy of conditionally normal and tumor rectum tissues obtained by video-colonoscopy (VCS) before RT.
To evaluate the performed RT effectiveness, a combination of approaches was used: MRI, VCS and histological analysis. Histological analysis of the surgical material made it possible to classify the samples according to the Tumor Regression Grade system, the system was proposed by A.M. Mandard and collaborators [10]. It includes 5 tumor regression grades, depending on the presence of residual tumor cells and the fibrosis degree. When response evaluating, qualitative changes in tumor cells are also taken into account, such as vacuolization and/or eosinophilia of the cytoplasm, nuclear pycnosis, necrosis, as well as the degree of inflammatory infiltration, including the presence of giant cells. Efficiency assessment using the MRI and VCS approaches also had several gradations:
1. complete response (complete pathomorphosis, regression);
2. partial response (incomplete pathomorphosis, partial regression, stabilization);
3. lack of response (progression).
RNA extraction, evaluation of gene and micro-RNA expression
The tissue sample was homogenized by mechanical action method with the Trizol (QIAzol, Qiagen) addition. Total RNA isolation was performed using the RNeasy Plus Universal Kits (Qiagen) according to the manufacturer‘s instructions. To eliminate genomic DNA contamination, the isolated RNA was treated with DNase-1. The reaction for complementary DNA (cDNA) synthesis was carried out using the REVERTA-L kit (Interlabservice, Russia) according to the manufacturer‘s instructions.
Real-time qPCR (RT-qPCR) was used to assess the relative expression of 18 genes. Highly specific synthetic oligonucleotides (primers) were developed using the Primer-BLAST program and GenBank database (National Center for Biotechnology Information) (Suppl. tab. 1).
1. miRNAs identified using the TarPmiR algorithm*. 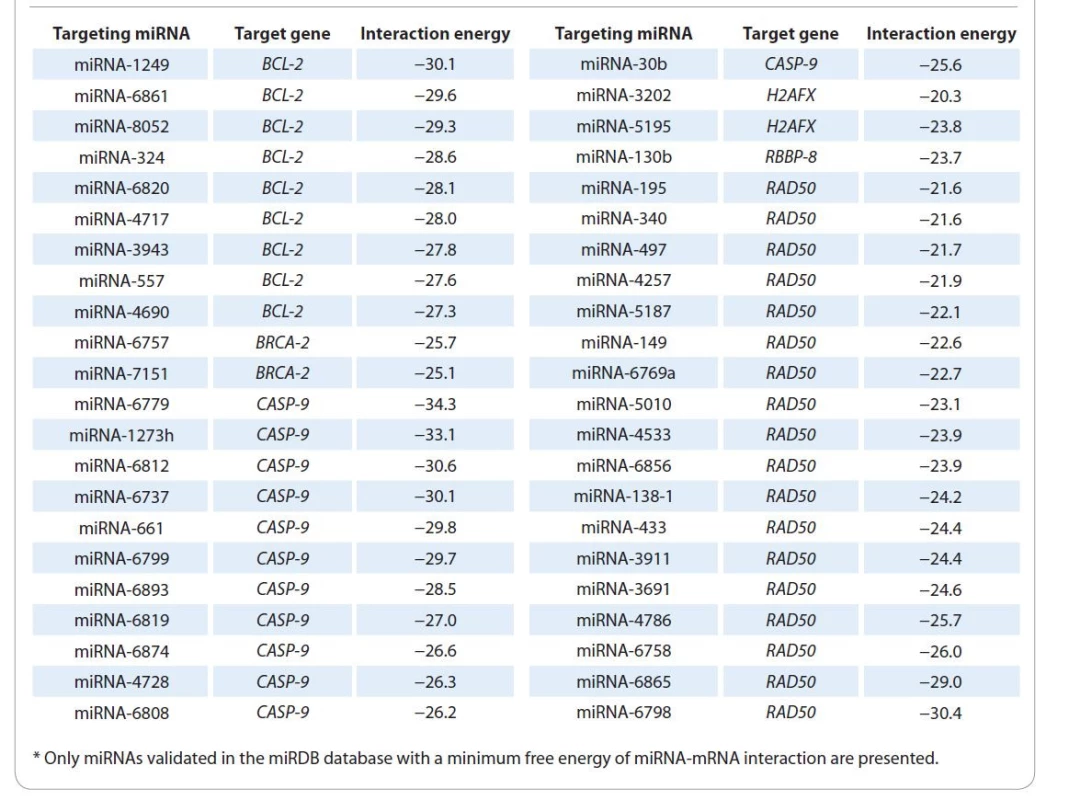
The amplification reaction was carried out in a mixture consisting of 1× PCR buffer, 0.2 mM deoxyribonucleoside triphosphates, 1.5 mM MgCl2, 0.6 mM synthetic oligonucleotides, 0.1 U/μl Taq polymerase and 15 ng complementary DNA. The mixture was incubated in a Bio-Rad CFX 96 Real-Time thermal cycler under the following temperature conditions: 94 °С for 240 seconds (sec) and then 39 cycles: 94 °С for 9 sec, 57 °С for 25 sec, and 72 °С for 45 sec. The relative expression (RE) of genetic loci was calculated using the formula RE = E-DDCt, taking into account the normalization for the reference genes (GAPDH, ACTB, B2M) and the mRNA level of corresponding target genes in normal tissue samples. The normalization scheme was as follows: 1 – calculation of the geometric mean C (t) of the reference genes; 2 – calculating the threshold cycle difference (DC (t)): C (t) target – C (t) reference; 3 – median DC (t) of each target gene for conditionally normal and tumor tissue; 4 – the DDC (t) calculation (the ratio DC (t) median of tumor tissue /DC (t) median of normal tissue); 5 – calculation of the final result E-DDC (t) [11].
The design of synthetic oligonucleotide sequences required for evaluating miRNA expression was performed using the I. Balcells algorithm. Primer-BLAST was used to design primers for long non-coding RNAs. Small non-coding RNA U6 was used as a reference to determine microRNA and lncRNA expression. To assess microRNA expression, the isolated total RNA was subjected to reverse transcription with specific RT-primers simultaneously with polyadenylation reaction [12]. The change in miRNA and lncRNA relative expression was also assessed by the RT-qPCR method.
Statistical and bioinformatic analysis
It was performed in the R language in the RStudio, version 8.10.173.987. The normality of distribution was assessed using the modified Shapiro-Wilk test (N > 50), the significance of intergroup differences was determined using the Mann-Whitney test (the Bonferroni correction), the relationships analysis of studied parameters was performed using the Spearman correlation coefficient (r).
The hierarchical clustering and Euclidean distance parameters were used for cluster analysis. The genetic loci grouping according to their biological functions was carried out using the functional module detection algorithm. The Q-value was calculated using the one-sided Fisher‘s exact test (the Benjamini-Hochberg correction) [13]. Bioinformatic search for miRNAs was carried out according to the reversed TarPmiR algorithm, which is based on the random forest machine learning algorithm (combines the method of random subspaces and the Breiman bagging method), which makes it possible to predict the binding site of mRNA and miRNA [14]. To assess the participation of differentially expressed miRNAs in key cellular signaling pathways, the over-representation analysis (ORA) was performed. Statistical significance in the ORA was calculated using Fisher‘s exact test [15].
Analysis of miRNA and lncRNA interaction was carried out as follows:
1. HITS-CLIP, PAR-CLIP and CLASH data were extracted from the Gene Expression Omnibus database and processed using FASTX-Toolkit v0.0.13, and further analyzed using PARalyzer v1.1.
2. All coordinates of the binding sites were converted to assemblies hg19, mm9/mm10 and ce6/ce10, respectively, using the UCSC LiftOver Tool. The genomic coordinates of the conserved miRNA target sites predicted by TargetScan, miRanda/mirSVR, PITA, Pictar and RNA22 were also assembled and transformed into hg19, mm9/mm10 and ce6/ce10 assemblies using LiftOver.
3. The coordinates obtained at step 2 were compared with the previously described CLIP clusters using BED - Tools [4].
Results
Radiotherapy efficacy and genetic loci expression
Data on the transcriptional activity of 18 genetic loci in rectum tumor tissue biopsy samples were used in cluster analysis (Hierarchical Clustering & Euclidean distance parameters), which made it possible to identify 2 clusters. In the first cluster, 85% of tumor tissue samples had increased expression of CASP9 gene and 70% decreased expression of BCL2, BRCA2, H2AX and RBBP8 genes. In the second cluster, 82% of patients have decreased expression of CASP9 and 79% have increased expression of BCL2, BRCA2, H2AX, RBBP8 and RAD50.
A comprehensive analysis of RT effectiveness results in 500 patients with RC gave the following results: in 150 RC patients, a complete response to RT was recorded; in 225 RC patients, a partial response to RT was recorded; and in 125 patients, there was no response to RT at all. At the same time, in tumor biopsy material from patients with a complete response to RT, the expression of H2AX and RBBP8 was decreased 3.3 and 2.5 times (P < 0.05), and CASP9 expression was increased 4.8 times (P < 0.005) relative to these parameters in normal tissue.
In tumor biopsy material in patients with partial/no response to RT, the expression of BCL2, BRCA2, H2AX, RBBP8 and RAD50 was statistically significantly (P < 0.05) increased by 4.2, 3.5, 3.2, 3.4 and 3.3 times accordingly, and CASP-9 expression was not statistically significantly different from its expression in normal tissue (Graph 1). That is, the transcriptional profile of BCL-2, BRCA2, H2AX, RAD50, CASP9 and RBBP8 is probably associated with the effectiveness of RT, which increases in RC-patients with CASP-9 overexpression and hypoexpression of H2AX and RBBP-8, and vice versa, the effectiveness of RT decreases with overexpression of BCL2, H2AX, RAD50, RBBP8 and BRCA2 genes.
Graph 1. Tumor tissue transcriptional profile in two groups of rectal cancer patients (with a complete response to radiotherapy – Group 1, and with partial response or no response to radiotherapy – Group 2). 
* statistically significant differences relative to conventionally normal rectal tissue (P < 0.05), ** statistically significant intergroup differences (P < 0.05) miRNA expression and RT efficiency
Using our modified bioinformatics algorithm TarPmiR, 1 927 microRNAs targeting BCL2, BRCA2, H2AX, RAD50, CASP9 and RBBP8 were identified. Of these miRNAs, only 109 were validated in the miRDB database (Graph 2), including 44 miRNAs that form strongest complexes with the corresponding target genes (minimum free energy of interaction in a miRNA-mRNA pair): 9 miRNAs for BCL2 gene, 2 miRNAs for BRCA2, 12 miRNAs for CASP9, 2 miRNAs for H2AX, 1 miRNA for RBBP8 gene and 18 mRNAs for RAD50 gene (Tab. 1).
Graph 2. miRNA transcriptional profile in rectal cancer patients with complete tumor regression or its absence. 
* statistically significant differences relative to normal tissue (P < 0.05), ** statistically significant intergroup differences (P < 0.05) For these 44 miRNAs, their expression in RC patients’ tissues was determined. Differential expression of a number of miRNAs, both intergroup and relative to normal tissue, was found. In the group of patients with insignificant tumor regression / lack of dynamics after RT (N = 350, low efficiency of RT), there was a statistically significant (P < 0.05) decrease in the expression of miRNA-1249-5p, miRNA-6820-3p, miRNA-4717-5p, miRNA -3943 and miRNA-557 2.9, 2.0, 3.3, 3.3 and 2.5 times, respectively, relative to normal tissue, and a decrease in the expression of miRNA-6820-3p and miRNA-557 1.8 and 2.3 times, respectively, relative to patients with complete tumor regression (N = 150, high efficiency of RT). A statistically significant (P < 0.05) 2.1 fold increase in miRNA-4690-5p expression relative to normal tissue was also found (Graph 2). miRNA-1249-5p, miRNA-4690-5, miRNA-6820-3p, miRNA-4717-5p, miRNA-3943 and miRNA-557 target the BCL2 gene.
miRNA-6757 expression in tumor tissue in patients with partial/no response to RT is statistically significant (P < 0.05); it was 5.0 times and 4.4 times lower than normal tissue and tumor tissue of patients with complete response to therapy, respectively. BRCA2 is the target gene of this miRNA, and its decrease should lead to an increase in this locus expression (Graph 2).
It was also found that the expression of miRNA-1273h, miR-6812-5p, miRNA-6737, miRNA-661, miRNA-4728, miRNA-30b and miRNA-6808 was statistically significantly (P < 0.05) decreased 2.5, 2.5, 3.3, 2.0, 5.0, 2.0 and 5.0 times, respectively, in tumor tissue of patients with a complete response to therapy relative to normal tissue. An increase in expression of miRNA-1273h 2.3 times (P < 0.05), miRNA-4728 4.0 times (P < 0.05), miRNA - -6819 4.0 times (P < 0.05), miRNA-6737 3.0 times (P < 0.05), miRNA-6874 3.3 times (P < 0.05) and miRNA-6808-5p 5.0 times (P < 0.005) was found in tumor tissue of RC patients with a partial response to therapy relative to tumor tissue in patients with a complete response to therapy. The expression of miRNA-6812 was reduced in tumor tissue of patients with complete and partial response to therapy 2.5 (P < 0.05) and 2.0 (P < 0.005) times, respectively, relative to normal rectal tissue.
In RC patients with a complete response to RT, statistically significant (P < 0.05) changes in the miRNA-5195 and miRNA-3202 expression were revealed. Thus, in tumor tissue, the expression of miRNA-3202 was 5.1 times higher than in normal tissue; and 4.3 times higher than in tumor tissue in RC patients with partial tumor response/no response to RT. Expression of miRNA-5195 was 1.8 times higher than in normal tissue, and 3.6 times higher than expression in tumor tissue in RC with partial/no response to RT (Graph 2). These miRNAs target mRNA of the histone protein H2AX.
In the course of the study, it was found that in tumor tissue of RC-patients with a complete response to therapy, miRNA-130b expression level was increased 6.0 times (P < 0.05) relative to normal tissue. At the same time, in RC patients with a partial response to therapy in tumor tissue, the expression level of this miRNA was decreased 5.0 times (P < 0.05).
Accordingly, in patients of the first group (complete response to therapy), the expression of miRNA-130b is 30.0 times higher (P < 0.0005) compared to expression in second group patients (partial response to RT), which may contribute to a decrease RBBP-8 gene transcriptional activity (regulates cell proliferation) in patients with a complete response to RT and increase its expression in RC-patients with a partial response to RT (Graph 2).
Out of 18 miRNAs targeting the RAD50 gene, only 8 have aberrant expression in rectal tumor tissue (miRNA-195-5p, miRNA-5187-5p, miRNA-149-5p, miRNA-138-1-3p, miRNA-433 - 3p, miRNA-3911, miRNA-6865-5p, miRNA-6798-5p). Thus, the expression of miRNA-195-5p relative to normal rectal tissue is 2.3 (P < 0.05) times higher in patients with complete tumor regression and 3.3 times (P < 0.05) lower in patients with incomplete tumor regression. At the same time, the differences in miRNA-195-5p expression between these groups are 7.7-fold (P < 0.05) (Graph 2). The expression of miRNA-5187-5p and miRNA-149-5p is increased 4.3 and 5.1 times (P < 0.05) in tumor tissue in patients with a complete response to therapy relative to normal tissue. The difference between 2 groups of patients in the expression of these microRNAs is 2.9 and 2.8 times (P < 0.05), respectively. There is also a statistically significant (P < 0.05) 2.7-fold difference in the expression of miRNA-4257 between these groups of patients. The expression of miRNA-138-1-3p, miRNA-433-3p, and miRNA-3911 is statistically significantly (P < 0.05) increased in patients with complete regression relative to expression in normal tissue by 2.5, 2.8 and 3.7 times, respectively. The expression of miRNA-6798-5p relative to normal rectal tissue is 3.5 (P < 0.05) times higher in patients with complete tumor regression and 5.0 times (P <0.05) lower in patients with incomplete tumor regression. At the same time, the differences in miRNA-6798-5p expression between these groups are 17.5 times (P < 0.005) (Graph 2). The expression of miRNA-6865-5p is reduced 5.0 times (P < 0.05) in tumor tissue in patients with incomplete tumor regression relative to normal rectal tissue.
The over-representation analysis (ORA) (Suppl. tab. 2) was performed for differentially expressed miRNAs in tumor tissues of two groups of RC patients. The ORA results show that miRNAs differentially expressed in two groups of patients, in addition to regulating the transcriptional activity of BCL2, CASP9, BRCA2, RAD50, H2AX and RBBP8 genes, are involved in the control of key tumor cells survival and proliferation signaling pathway (Suppl. tab. 2). Thus, rectal tumor tissue sensitivity, which determines the effectiveness of RT, is also associated with the differential expression of miRNA-195, miRNA-4257, miRNA-5187, miRNA-149, miRNA-138, miRNA-6798, miRNA-6820, miRNA-557, miRNA-6757, miRNA-1249, miRNA-1273h, miRNA-6737, miRNA-6819, miRNA-6874, miRNA-4728, miRNA-6808, miRNA-3202, miRNA-5195-3p and miRNA-130b.
2. List of lncRNAs interacting with several miRNAs differentially expressed in patients with different radiotherapy efficacy. 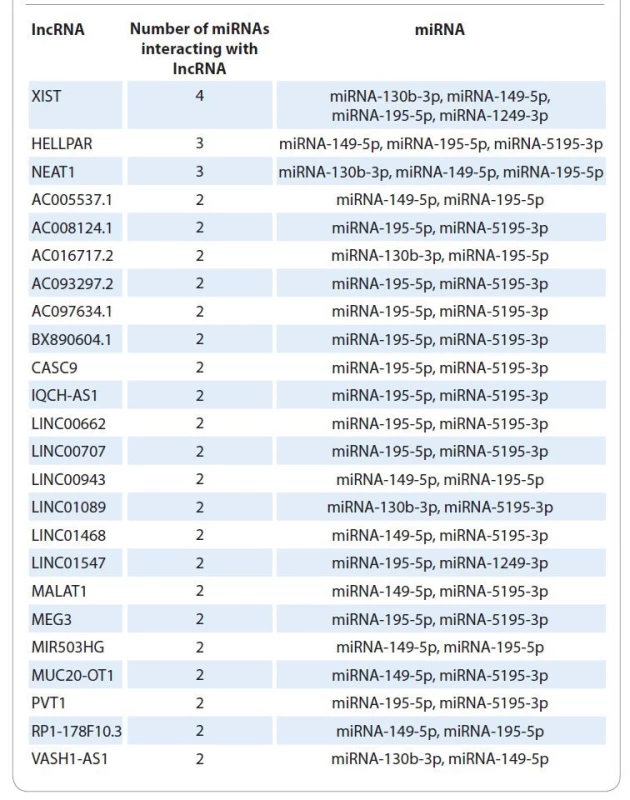
Mathematical prediction of potential interactions probability between miRNAs and long noncoding RNAs
The use of bioinformatic algorithms made it possible to identify for 5 (miR-130b, miR-149, miR-195, miR-1249, miR-5195) of 44 miRNAs interactions with 202 long noncoding RNAs (Suppl. tab. 3).
For miR-130b interactions with 21 lncRNAs were predicted, for miR-149 with 90 lncRNAs, for miR-1249 with 6 lncRNAs, and for miR-5195 with 33 lnc--RNAs. From this list of long noncoding RNAs 24 interacted with several miRNAs differentially expressed in patients with different RT efficacy (Tab. 2, Fig. 1).
Fig. 1. Euler-Venn diagram of overlapping interactions of 5 miRNAs and 202 lncRNA. 
The next stage of the work was the analysis of the representation of 24 lncRNA transcripts in biological samples of two RC patients’ groups.
Features of lncRNA expression and radiotherapy effectiveness
Analysis of long non-coding RNA transcripts relative abundance showed differences in this indicator for two groups of patients with different responses to RT.
Thus, in patients with a complete response to RT, a statistically significant (P < 0.05) decrease in the representation of lncRNA XIST, HELLPAR, NEAT1, AC008124.1 and AC016717.2 transcripts was found by 2.5, 3.3, 2.0, 2.5 and 5.0 times, respectively, relative to normal tissue and in patients with partial/no response to RT a statistically significant (P < 0.05) increase in the representation of lncRNA XIST, HELLPAR, NEAT1, AC008124.1, LINC01089, LINC01547 and VASH1-AS1 transcripts was found in 2.9, 3.9, 3.5, 3.7, 2.6, 4.7 and 1.9 times, respectively, relative to normal tissue.
At the same time, statistically significant differences (P < 0.05) in the representation levels of lncRNA XIST, HELLPAR, NEAT1, AC008124.1, AC016717.2, LINC01089, LINC01547 and VASH1-AS1 transcripts, i.e. 7.3, 13.0, 7.0, 9.3, 5.0, 3.1, 4.8 and 2.2 times, respectively, were found between two patients’ groups (Graph 3). Thus, the data obtained on the representation of long noncoding RNA transcripts will make it possible to model the network of molecular interactions underlying the radioresistance of tumor cells.
Graph 3. Relative representation (relative to normal tissue) of long noncoding RNAs transcripts in rectum tumor tissue of two groups of patients with complete or partial response/non-response to radiotherapy. 
* statistically significant differences relative to normal tissue (P < 0.05), ** statistically significant intergroup differences (P < 0.05). Discussion
The completed research stages allowed us to collect the necessary information on all components of competitively interacting endogenous RNA (ceRNA) regulatory network, including long non-coding RNA (lncRNA), miRNA and mRNA (gene expression).
A relationship was established between the transcriptional profiles of BCL2, BRCA2, H2AX, RAD50, CASP9 and RBBP8 genes and RT effectiveness. Thus, overexpression of BCL2, H2AX, RAD50, RBBP8 and BRCA2 genes is associated with low efficacy of RT, and overexpression of CASP-9 and, at the same time, hypoexpression of H2AX and RBBP8 was associated with high efficacy of RT. The use of modern bioinformatics algorithms made it possible to identify miRNAs targeting these genetic loci. These miRNAs expression in the tissues of cancer patients was evaluated. Thus, the target gene of miRNAs miRNA-1273h, miRNA-4728, miRNA-6819, miRNA-6737, miRNA-6874 and miRNA-6808-5p is CASP9; therefore, their high level of expression should contribute to a decrease in the level of mRNA of the initiator caspase-9, which is necessary to trigger the process of apoptosis. miRNAs targeting BCL2 gene in the tumor tissue of patients with low RT efficiency are predominantly hypoexpressive, both relative to normal tissue and relative to tumor tissue of patients with a complete response to RT. Accordingly, a decrease in the expression of these miRNAs (miRNA-1249-5p, miRNA-4690-5, miRNA-6820-3p, miRNA-4717-5p, miRNA-3943, miRNA-557) can lead to the overexpression of BCL-2 genetic locus, the product of which inhibits caspases and apoptosis [16]. Moreover, the overexpression of miRNA-3202/miRNA-5195, targeting mRNA histone protein H2AX, which triggers the process of changing chromatin conformation upon DNA damage under the influence of radiation [17], can promote H2AX hypoexpression, and vice versa.
miRNAs have many targets [18]; therefore, an over-representation analysis was carried out for differentially expressed miRNAs in tumor tissues of two groups of RC-patients, which made it possible to reveal their participation in a variety of cellular signaling pathways, including regulation of the cell cycle and apoptosis.
Comprehensive analysis of these data, combined with the data of mathematical prediction of interaction miRNA and lncRNA and their validation by PCR, allowed us to form a model of the regulatory network of competitively interacting RNAs under RT for rectal tumors (Fig. 2).
Fig.2. Features of competitively interacting RNAs regulatory network in patients with different responses to radiotherapy. 
According to this model, a decrease in the representation of lncRNA XIST, HELLPAR, NEAT1, AC008124.1 and AC016717.2 transcripts in patients with a complete response to RT leads to a decrease in the efficiency of interaction between lncRNA and targeting miRNAs (miRNA-130b-3p, miRNA-149-5p, miRNA-195-5p, miRNA-1249-3p, miRNA-5195-3p). Disruption of the lncRNA - -miRNA complex formation leads to an increase in the amount of free miRNA miRNA-130b-3p, miRNA-149-5p, miRNA - -195-5p, miRNA-5195-3p and their effective interaction with target genes (mRNA). Accordingly, the transcripts representation of miRNA target RBBP8 genes (miRNA-130b-3p, miRNA-149-5p, miRNA-195-5p) and H2AX (miR-5195-3p) decreases, and, consequently, the DNA repair efficiency decreases.
The opposite effect is observed in patients with partial response to RT. An increase in the representation of lncRNA XIST, HELLPAR, NEAT1, AC008124.1, LINC01089, LINC01547 and VASH1-AS1 transcripts in these patients leads to an increase in the efficiency of interaction between lncRNA and miRNA (miRNA-130b-3p, miRNA-149-5p, miRNA-195-5p, miRNA-1249-3p, miRNA-5195-3p). The formation of the lncRNA-miRNA complex leads to a decrease in the amount of free miRNAs miRNA-130b-3p, miRNA-149-5p, miRNA-195-5p and miRNA-5195-3p and a decrease in their interaction with target genes (mRNA).
Accordingly, the abundance of miRNA target genes transcripts RBBP8 (miRNA-130b-3p, miRNA-149-5p, miRNA - -195-5p) and H2AX (miRNA-5195-3p) increases, and, consequently, an increase in the efficiency of DNA repair and radioresistance formation (Fig. 2).
Conclusion
Thus, our comprehensive study of competitively interacting RNAs regulatory network features and rectal tumors RT effectiveness made it possible to establish both the mechanisms of radioresistance formation and its predictors. RT efficiency for RС depends on the miRNA expression level (miRNA-195-5p, miRNA-4257, miRNA-5187-5p, miRNA-149 - -5p, miRNA-138-1-3p, miRNA-6798-5p, miRNA-6819-5p, miRNA-4728-5p, miRNA-1249-5p, miRNA-557, miRNA - 1273h-5p, miRNA-6737-5p, miRNA-6808-5p, miRNA-3202, miRNA-5195-3p, miRNA-130b-3p) and lncRNA (XIST, HELLPAR, NEAT1, AC008124.1, LINC01089, LINC01547 and VASH1-AS1), providing effective regulation of the DNA repair system (H2AX and RBBP8) and apoptosis (BCL2).
Acknowledgement
The study was carried out within the state task framework „Search for rectal cancer radioresistance predictors and personalized neoadjuvant therapeutic approaches development“.
The authors declare they have no potential conflicts of interest concerning drugs, products, or services used in the study.
Autoři deklarují, že v souvislosti s předmětem studie nemají žádné komerční zájmy.
The Editorial Board declares that the manuscript met the ICMJE recommendation for biomedical papers.
Redakční rada potvrzuje, že rukopis práce splnil ICMJE kritéria pro publikace zasílané do biomedicínských časopisů.Denis Kutilin, PhD
National Medical Research
Oncology Center
14 line str., 63
Rostov-on-Don
Rostov region
344037, Russian Federation
e-mail: k.denees@yandex.ru
Submitted/Obdrženo: 30. 11. 2021
Accepted/Přijato: 18. 2. 2022
Sources
1. Sung H, Ferlay J, Siegel RL et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71 (3): 209–249. doi: 10.3322/caac.21 660.
2. Feeney G, Sehgal R, Sheehan M et al. Neoadjuvant radiotherapy for rectal cancer management. World J Gastroenterol 2019; 25 (33): 4850–4869. doi: 10.3748/wjg.v25.i33.4850.
3. Wang L, Cho KB, Li Y et al. Long noncoding RNA (lncRNA) -mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci 2019; 20 (22): 5758. doi: 10.3390/ijms20225758.
4. Novikova IA, Timoshkina NN, Kutilin DS. Differential microRNA expression in tumor and normal colon tissues. Yakut Med J 2020; 4 (72): 74–82.
5. Cao C, Zhang T, Zhang D et al. The long noncoding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene 2017; 36 (8): 1112–1122. doi: 10.1038/onc.2016.278.
6. Bailerley OA, Gareev IF, Pavlov VN et al. Exosomal long noncoding RNAs as biomarkers and therapeutic targets in cancer. Creative Surg Oncol 2019; 9 (4): 297–304. doi: 10.24060/2076-3093-2019-9-4-297-304.
7. Liu Q, Deng J, Wei X et al. Integrated analysis of competing endogenous RNA networks revealing five prognostic biomarkers associated with colorectal cancer. J Cell Biochem 2019; 120 (7): 11256. doi: 10.1002/jcb.28403.
8. Tang F, Lu Z, Wang J et al. Competitive endogenous RNA (ceRNA) regulation network of lncRNAs, miRNAs, and mRNAs in Wilms tumour. BMC Med Genomics 2019; 12 (1): 194. doi: 10.1186/s12920-019-0644-y.
9. Babaei M, Jansen L, Balavarca Y et al. Neoadjuvant therapy in rectal cancer patients with clinical stage II to III across European countries: variations and outcomes. Clin Colorectal Cancer 2018; 17 (1): e129–e142. doi: 10.1016/j.clcc.2017.09.002.
10. Mandard AM, Dalibard F, Mandard JC et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer 1994; 73 (11): 2680–2686. doi: 10.1002/1097-0142 (19940601) 73 : 11<2680:: aid-cncr2820731105>3.0.co; 2-c.
11. Kutilin DS. Regulation of gene expression of cancer/testis antigens in colorectal cancer patients. Mol Biol 2020; 54 (4): 580–595. doi: 10.31857/S0026898420040096.
12. Balcells I, Cirera S, Busk PK. Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol 2011; 11 (1): 70. doi: 10.1186/1472-6750-11-70.
13. Krishnan A, Zhang R, Yao V et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci 2016; 19 (11): 1454–1462. doi: 10.1038/nn.4353.
14. Ding J, Li X, Hu H. TarPmiR: a new approach for microRNA target site prediction. Bioinformatics 2016; 32 (18): 2768–2775. doi: 10.1093/bioinformatics/btw 318.
15. Backes C, Khaleeq QT, Meese E et al. miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res 2016; 44 (W1): W110–116. doi: 10.1093/nar/gkw 345.
16. Urbańska K, Orzechowski A. Unappreciated role of LDHA and LDHB to control apoptosis and autophagy in tumor cells. Int J Mol Sci 2019; 20 (9): 2085. doi: 10.3390/ijms20092085.
17. Scully R, Xie A. Double strand break repair functions of histone H2AX. Mutat Res 2013; 750 (1–2): 5–14. doi: 10.1016/j.mrfmmm.2013.07.007.
18. Abdelsattar ZM, Wong SL, Regenbogen SE et al. Colorectal cancer outcomes and treatment patterns in patients too young for average-risk screening. Cancer 2016; 122 (6): 929–934. doi: 10.1002/cncr.29716.
For Supplementary tables, see the online version of the article at www.linkos.cz
Labels
Paediatric clinical oncology Surgery Clinical oncology Haematology ENT (Otorhinolaryngology) General practitioner for adults Urology
Article was published inClinical Oncology

2022 Issue 4-
All articles in this issue
- Onkologická léčba 21. století a Kaplan-Meierovy křivky
- Rosai-Dorfman-Destombes disease – histiocytic disorder with infl ammatory manifestation
- Patients with neuregulin 1 (NRG1) rearranged cancer are suitable for the theranostic approach and targeted therapy
- Olanzapine in oncology palliative care
- Immunotherapy for cancer treatment
- How fatigue affects return to work in breast cancer patients
- Informace z České onkologické společnosti
- Slow increase of bilirubin concentration during administration of lenalidomide, bortezomib and dexamethasone for multiple myeloma (unmasking previously undiagnosed Gilbert syndrome) and disappearance of necrobiotic xanthogranuloma after complete remission of multiple myeloma
- Regulatory network of competitively interacting RNAs and effectiveness of rectal tumors radiotherapy
- Concurrent weekly cisplatin and simultaneous integrated boost intensity-modulated radiotherapy of locally advanced squamous cell carcinoma of the head and neck
- Microwave ablation of a solitary colorectal liver metastasis complicated by stomach perforation and gastrocutaneous fistula – a case report
- Clinical Oncology
- Journal archive
- Current issue
- Online only
- About the journal
Most read in this issue- Olanzapine in oncology palliative care
- Immunotherapy for cancer treatment
- Rosai-Dorfman-Destombes disease – histiocytic disorder with infl ammatory manifestation
- How fatigue affects return to work in breast cancer patients
Login#ADS_BOTTOM_SCRIPTS#Forgotten passwordEnter the email address that you registered with. We will send you instructions on how to set a new password.
- Career

